Abstract
Although apoptosis is mechanistically well understood, a comprehensive understanding of how cells modulate their susceptibility towards apoptosis in a developing tissue is lacking. Here, we reveal striking dynamics in the apoptotic susceptibilities of different cell types in the Drosophila retina over a period of only 24 hours. Mitotic cells are extremely susceptible to apoptotic signals, while post-mitotic cells have developed several strategies to promote survival. For example, photoreceptor neurons accumulate the inhibitor of apoptosis, Diap1. In unspecified cells, Cullin-3-mediated degradation keeps Diap1 levels low. These cells depend on EGFR signaling for survival. As development proceeds, developmentally older photoreceptors degrade Diap1 resulting in increased apoptosis susceptibility. Finally, R8 photoreceptors have very efficient survival mechanisms independently of EGFR or Diap1. These examples illustrate how complex cellular susceptibility towards apoptosis is regulated in a developing organ. Similar complexities may regulate apoptosis susceptibilities in mammalian development and tumor cells may take advantage of it.
INTRODUCTION
Apoptosis is a major form of programmed cell death. Its biochemical mechanisms are evolutionarily conserved (reviewed in (Fuchs and Steller, 2011; Xu et al., 2009)). Essential for apoptosis are caspases, highly specific cell death proteases. They are produced as inactive zymogens and need proteolytic cleavage for activation (Kumar, 2007). Active caspases can be detected using the cleaved Caspase-3 antibody (Cas3*) which cross-reacts with cleaved Drosophila caspases (Fan and Bergmann, 2010; Srinivasan et al., 1998; Yu et al., 2002). Once caspases are activated, they cleave a large number of cellular proteins, which triggers the death of the cell. In surviving cells, caspases are inhibited by inhibitor of apoptosis proteins (IAPs), the most important one in Drosophila being Drosophila IAP1 (Diap1) (Goyal, 2001; Hay et al., 1994; Wang et al., 1999). Many IAPs including Diap1 carry a C-terminal RING E3 ligase domain capable of auto-ubiquitylation and degradation of the IAP (Yang et al., 2000). The IAP-antagonists Reaper (Rpr), Hid, and Grim induce cell death by stimulating the RING activity and degradation of Diap1 (Hays et al., 2002; Holley et al., 2002; Ryoo et al., 2002; Wing et al., 2002; Yoo et al., 2002). Caspases are released from Diap1 inhibition and can now induce apoptosis. Overexpression of the IAP antagonists rpr, hid or grim induces a strong apoptotic response. For example, expression of hid or rpr under the control of the eye-specific promoter GMR causes a strong eye-ablation phenotype (Grether et al., 1995; White et al., 1996) (Figure 1A – C). Hid is unique among the IAP antagonist, because it is negatively regulated by EGFR/Ras/MAPK signaling through inhibitory MAPK phosphorylation and transcriptional downregulation (Bergmann et al., 1998; Kurada and White, 1998).
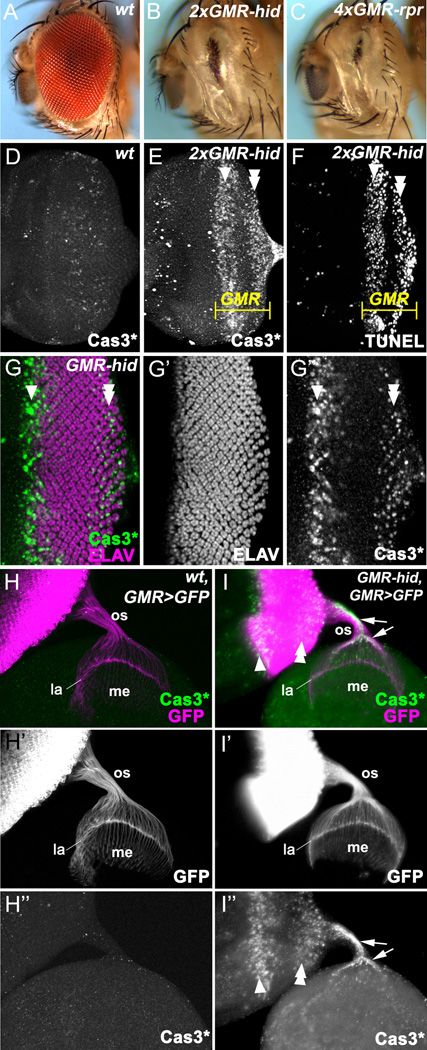
Figure 1. Photoreceptor neurons survive and differentiate in GMR-hid.
Here, and in the following figures, anterior is to the left. White arrowheads in this and subsequent figures indicate the first apoptotic wave induced by GMR-hid, while white double arrowheads indicate the second apoptotic wave.
(A–C) Adult eyes of wild type flies (A), of flies carrying two copies of GMR-hid(B), or four copies of GMR-rpr(C).
(D–F) Wild-type (D) and 2×GMR-hid eye discs (E,F) labeled with cleaved Caspase-3 (Cas3*) antibodies (D,E) and TUNEL assay (F). The GMR expression domain is indicated in yellow. In contrast to a few scattered dying cells in wild type (D)GMR-hid induces two waves of cell death separated by an apoptosis-free zone (E,F).
(G–G’’)GMR-hid eye disc labeled with Cas3* and the neuronal marker ELAV. Despite expression of hid, photoreceptor differentiation appears largely normal.
(H–I) Eye-brain complexes double labeled with GFP and Cas3* (H,I) or GFP (H’,I’) or Cas3* (H’’, I’’). In wild type (GMR-GAL4 UAS-CD8GFP, referred to as GMR>GFP), GFP-positive photoreceptor neurons (R1-R8) project their axons through the optic stalk (os) to the optic lobe. R1-R6 axons terminate at the lamina and form the lamina plexus (la) while R7 and R8 axons project to the medulla (me) (H, H’). Cleaved caspases are not present in wt optic stalks (H’’). The projection pattern of photoreceptor axons in GMR-hid eye discs appears normal (I, I’). Strikingly, strong Cas3*-labeling was also observed in the optic stalk (I’’; arrows) indicating that they contain cleaved caspases. Two apoptotic waves are shown in the GMR-hid eye disc in (I’’).
See also Figure S1.
Despite our detailed knowledge of the biochemical pathways of apoptosis, we know very little about the regulatory mechanisms that control the susceptibility of cells towards apoptosis in the context of a developing organism. It has been widely observed that cells at different developmental stages and of different types can differ in their sensitivity to apoptotic stimuli. As an example, human intestinal cells exhibit segment-specific sensitivities to apoptosis probably due to differential levels of Bcl-2 family of proteins, key regulators of apoptosis in mammals (Gauthier et al., 2001). Furthermore, evading apoptosis is a hallmark of cancer (Hanahan and Weinberg, 2000). Intriguingly, even among different tumor types and depending on their cell cycle status, the susceptibility of tumor cells to apoptosis-inducing anticancer therapies varies (reviewed in (Smith et al., 2000)). However, most of these studies were done in isolated cell lines and only analyzed one or two particular cell types. Thus, a detailed understanding of the survival requirements of all cells in a given tissue at different time points is still lacking.
The developing Drosophila eye tissue, the eye imaginal disc, is an ideal system to study distinct cellular apoptotic responses as the timing of specification of every cell type is known, and many cellular markers exist to follow them. In early larval stages, the cells in the eye imaginal disc proliferate continuously to produce the cell mass required for the production of the eye. During mid-third instar larval stage, an indentation known as the morphogenetic furrow (MF) forms at the posterior edge of the eye disc. All cells in the MF are post-mitotic. The MF moves across the eye disc from posterior to the anterior. In and posterior to the MF, cells are assembled into ommatidia, the functional units of the eye. This begins in the MF with the specification of the first photoreceptor neuron, the R8 cell. By definition, this occurs in ommatidial column 0 (Wolff and Ready, 1993). Still in the MF, the R8 cell induces specification of two pairs of photoreceptor neurons, R2, R5 and R3, R4, forming the five-cell precluster. This specification step requires EGFR signaling (Freeman, 1996; Yang and Baker, 2001). While the MF moves on to the anterior, the remaining unspecified cells re-enter the cell cycle and synchronously divide one more time in the second mitotic wave (SMW) to generate sufficient cells for further specification and differentiation. The SMW occurs in columns 2–4. After the SMW, all cells arrest in G1 and are post-mitotic from now on. The next photoreceptor neurons to be specified are R1 and R6 (column 5), followed by R7 (column 6). The specification of these cell types requires EGFR signaling. Due to their position in the photoreceptor cluster, R7 and R8 are referred to as inner photoreceptors, while R1-R6 are outer photoreceptors. The last cell types specified during larval stages are lens-secreting cone cells (columns 11–15). Thus, the portion of the larval eye disc located posterior to the MF represents a developing field with all developmental states. It is composed of mitotic cells in the SMW, post-mitotic yet unspecified cells, and differentiating cells including photoreceptor neurons and cone cells. Therefore, cells in the MF are developmentally the youngest, while cells towards the posterior edge of the disc are becoming increasingly older. During pupal stages, pigment and bristle cells are added. Finally, a wave of apoptosis removes all interommatidial cells that have not been incorporated into the ommatidia. In the end, each ommatidium is composed of 19 different cells (Brachmann and Cagan, 2003).
Here, we conduct a comprehensive analysis of the apoptosis susceptibilities of the different cell types in the posterior half of the eye imaginal disc. We report that these cell types respond differently to expression of the IAP antagonist hid. Dividing cells in the SMW are extremely sensitive to hid-induced apoptosis. Post-mitotic cells increase their apoptosis resistance in different ways dependent on the status of the specification/differentiation process. Interestingly, as soon as they are specified, photoreceptor neurons accumulate Diap1 and become apoptosis-resistant, while unspecified cells keep Diap1 levels low by Cullin-3-mediated degradation. These cells require EGFR signaling for suppression of hid-induced apoptosis. Finally, at later stages of photoreceptor differentiation, outer photoreceptors are re-sensitized to apoptotic signals by down-regulation of Diap1, potentially in a RING-dependent manner. Surprisingly, the developmentally oldest cell, R8, has the highest apoptotic resistance independently of Diap1. In summary, these data give an impression of how mechanistically complex cellular susceptibilities towards apoptosis are regulated in a developing tissue.
RESULTS
Photoreceptor neurons survive and differentiate in GMR-hid
To test how the different cell types in the posterior eye disc respond to apoptotic signals, we expressed the IAP-antagonist hid in all cells posterior to the MF using the GMR promoter (GMR-hid), resulting in a strong eye ablation phenotype (Figure 1B) (Grether et al., 1995). GMR-driven expression of hid causes massive apoptosis in the posterior half of 3rd instar larval eye imaginal discs as observed by cleaved Caspase3-(Cas3*-) labeling and TUNEL (Figure 1D – F) (Fan and Bergmann, 2008, 2010; Srivastava et al., 2007; Udan et al., 2003). However, although hid is expressed in all cells posterior to the MF (Figure 4B,D), GMR-hid-induced cell death is not uniform, but occurs in two distinct waves separated by an apoptosis-free zone (Figure 1E,F) suggesting that not all cells respond to hid in the same manner. The first apoptotic wave is present between ommatidial columns 3–6, the apoptosis-free zone is between columns 7–15, and the second apoptotic wave begins at column 16. Surprisingly, despite the strong apoptotic phenotype and eye ablation of GMR-hid, photoreceptor differentiation as judged by the neuronal marker ELAV appears normal in larval GMR-hid eye discs (Figure 1G,G’). hid-expressing photoreceptor neurons are even able to project axons into the optic lobe (Figure 1H,H’,I,I’), although strong Cas3*-labeling is detectable in the optic stalk (arrows, Figure 1I’’). Cleaved caspases are not present in wild-type axons (Figure 1H’’) suggesting that they do not fulfill a non-apoptotic function in axons. Thus, although photoreceptor neurons express hid and contain cleaved caspases, they are initially surviving and are able to initiate neuronal differentiation.
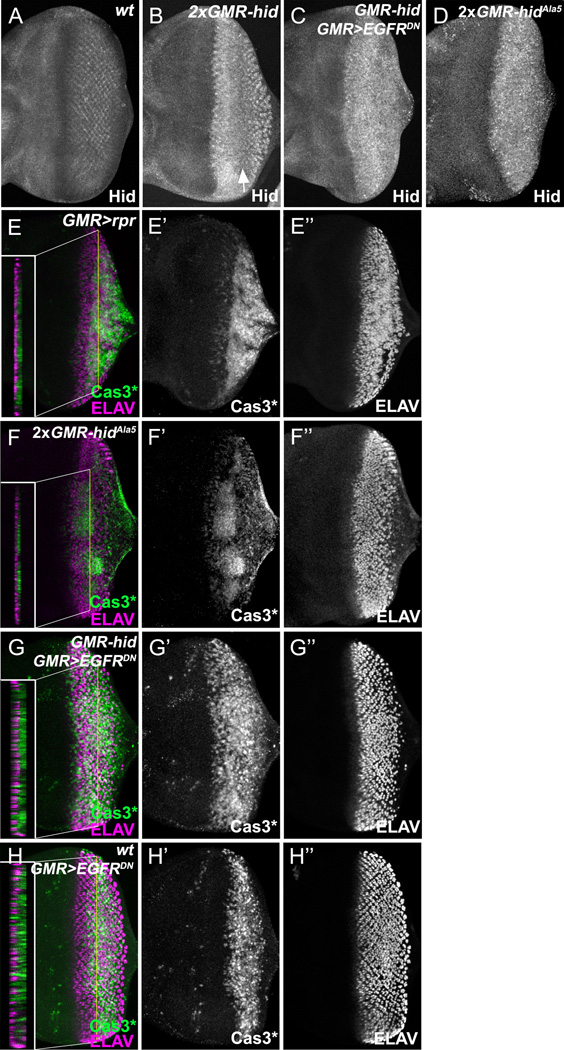
Figure 4. EGFR signaling protects unspecified cells in the apoptosis-free zone.
(A–D) Late 3rd instar eye discs labeled with anti-Hid antibody. Compared to wild type (A), Hid is increased posterior to the MF in GMR-hid eye discs with a reduction of protein levels in the apoptosis-free zone (B, arrow). Reduction of Hid in the apoptosis free zone does not occur in GMR-hid;GMR>EGFRDN(C) and GMR-hidAla5 eye discs (D) suggesting that EGFR/MAPK signaling destabilizes Hid protein.
(E–H) Late 3rd instar eye discs labeled with Cas3* and ELAV. ELAV labels apically located photoreceptor neurons. The apoptosis free zone is missing in EGFR/MAPK-unresponsive GMR-GAL4 UAS-rpr(GMR>rprE) and GMR-hidAla5(F) eye discs, as well as in GMR-hid;GMR>EGFRDN(G). Sections indicate non-overlap of ELAV and Cas3* labelings suggesting that unspecified cells at the basal side of the discs, but not photoreceptor neurons, require EGFR signaling for survival. In the sections, apical is to the left, basal to the right.
(H) Expression of GMR>EGFRDN in otherwise wild-type eye discs induces cell death in the region of the disc which corresponds to the apoptosis-free zone.
Due to the initial resistance of photoreceptor neurons towards hid-induced apoptosis (Figure 1G), GMR-hid imaginal discs are only slightly reduced in overall size at the end of third larval instar stage compared to wild-type eye discs. We found that ~21 ommatidial columns form in GMR-hid eye discs compared to ~24 columns in wild-type eye discs (Cagan and Ready, 1989; Wolff and Ready, 1993). Yet, the resulting adult GMR-hid eye is almost completely ablated (Figure 1B) (Grether et al., 1995). Therefore, we analyzed the size of the eye disc after puparium formation (APF). 2 hours APF, the size of the pupal GMR-hid eye disc is slightly reduced compared to wild-type (Figure S1A,B). The density of the photoreceptor clusters at the posterior end is reduced. 4 hours APF, the second apoptotic wave covers half of the eye disc and the density of the photoreceptor clusters is further declining (Figure S1C,D). The disc is now clearly reduced in size. Interestingly, although the size of the eye disc is decreasing, GMR-hid eye-antennal discs start eversion normally. We were unable to recover any GMR-hid eye discs after eversion. Thus, in early pupal stages most cells are eliminated in GMR-hid. These observations indicate that although photoreceptor neurons are initially resistant to hid-induced apoptosis, they appear to become sensitive to apoptosis in later stages.
These observations prompt several important questions: Which cells are dying in the first and second apoptotic waves? Why is cell death blocked in the apoptosis-free zone? How do photoreceptors survive in the presence of cleaved caspases? How are they becoming sensitive to apoptotic signals in later stages?
Proliferating SMW cells die in the first apoptotic wave of GMR-hid
The first apoptotic wave and the second mitotic wave (SMW) are spatially very close (Figure 2A,A’; Figure S2). In the SMW, all cells except the specified photoreceptors R8, R2/R5, and R3/R4 are proliferating (Baker and Yu, 2001). The photoreceptor neurons are not affected in the first apoptotic wave (see below and Figure 3). Instead, although the proliferation and apoptosis markers BrdU and Cas3* do not overlap (Figure 2A’; Figure S2) because apoptotic cells do not enter S phase, it appears that proliferating SMW cells are subject to cell death in the first apoptotic wave. This is inferred from the reduced number of cells that label positively for the mitotic marker phosphorylated histone 3 (PH3) in GMR-hid imaginal discs compared to wild-type eye discs (arrows, Figure 2B,C). On average, GMR-hid eye imaginal discs contain 29±5 PH3-positive cells derived from the SMW compared to 66±4 in wild-type discs (20 discs counted for each phenotype). This reduction of mitotic cells is rescued by overexpression of the caspase inhibitor P35 in GMR-hid eye discs (Figure 2D,E). Thus, in the first apoptotic wave of GMR-hid, proliferating cells in the SMW are dying suggesting that proliferating cells are very susceptible to hid-induced cell death.
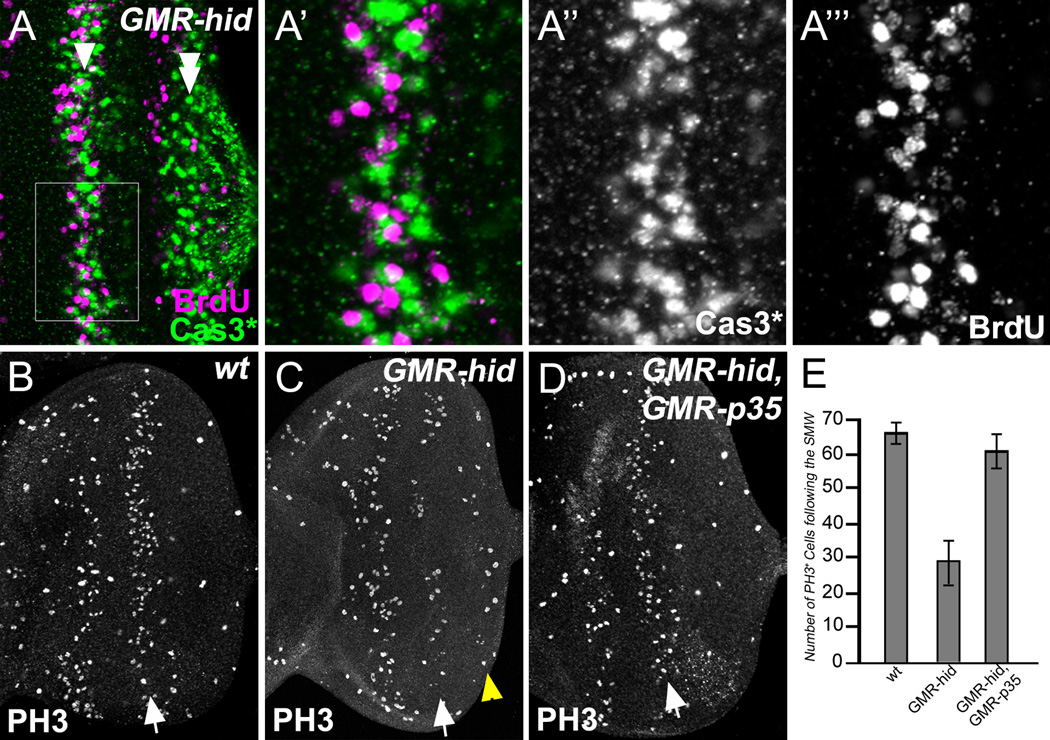
Figure 2. Proliferating SMW cells die in the first apoptotic wave.
(A–A’’’) Late 3rd instar GMR-hid eye disc labeled with Cas3* and BrdU. (A’) shows an enlarged view of the outlined region in (A). The first apoptotic wave (Cas3*-positive, green) and the second mitotic wave (SMW, BrdU-positive, magenta) are spatially very close.
(B–D) Late 3rd instar eye discs labeled with PH3 antibodies to visualize mitotic cells. Compared to wild type (B), the number of mitotic cells in the SMW (arrows) in GMR-hid(C) is strongly reduced. This reduction is rescued in GMR-hid;GMR-p35 eye discs (D). Yellow arrowhead in (C) indicates the wave of compensatory proliferation (Fan and Bergmann, 2008).
(E) Quantification of number of PH3-positive cells in the SMW in various genetic backgrounds. PH3-positive cells in the SMW were counted in wild type, GMR-hid, and GMR-hid;GMR-p35 eye discs (mean ± SD).
See also Figure S2.
Figure 3. Outer photoreceptor neurons R1-R6 die in the second apoptotic wave.
(A) Wild-type late 3rd instar eye disc labeled with Yan (a marker for unspecified cells).
(B–B’’) Late 3rd instar GMR-hid eye disc labeled with Cas3* and Yan. (B’,B’’) show enlarged views of the outlined region in (B). Compared to wild type (A), some unspecified cells are missing in GMR-hid as indicated by arrows (B’’).
(C) Wild-type late 3rd instar eye disc labeled with ELAV (a marker for all photoreceptor neurons R1-R8).
(D–D’’) GMR-hid late 3rd instar eye disc labeled with ELAV and Cas3*. Arrows highlight examples of ELAV-positive photoreceptor neurons that are also Cas3*-positive.
(E–E’’’) Late 3rd instar GMR-hid eye disc labeled with Cas3*, ELAV and the R8-specific marker Senseless (Sens). (E’-E’’’) show enlarged views of the outlined region in (E). Although many photoreceptor clusters are Cas3*-positive in the second apoptotic wave, most R8 cells survive to the end of larval stages. An example with only two photoreceptor neurons left in an ommatidium is indicated by white arrows. One of the two surviving neurons in this ommatidium is R8.
(F–F’’’) Late 3rd instar GMR-hid eye disc labeled with Cas3*, ELAV and Rough (R3,4,2,5-specific). (F’-F’’’) show enlarged views of the outlined region in (F). Most Rough-positive cells are excluded from the second apoptotic wave because they are either immediately eliminated by hid, or they lose the Rough marker as soon as they become apoptotic. Arrows point to a few Rough-positive cells which also label for Cas3*.
(G–H) Late 3rd instar GMR-hid eye discs labeled with ELAV and Seven-up (Svp, R2,5,1,6-specific) (G-G’’) or with TUNEL and Svp (H). (G’-G’’) show enlarged views of the outlined region in (G). Most Svp-positive cells are excluded from the second apoptotic wave because they are either immediately eliminated by hid, or they lose the Svp marker as soon as they become apoptotic. Arrows point to a few Svp-positive cells left in the area corresponding to the second apoptotic wave.
See also Figure S3.
Outer photoreceptor neurons R1-R6 and unspecified cells die in the second apoptotic wave of GMR-hid
Next, using several cell-type-specific markers, we determined the cell types that undergo cell death in the second apoptotic wave in GMR-hid imaginal discs. In this part of the larval eye disc, there are unspecified (interommatidial) cells, photoreceptor neurons and cone cells. Using Yan as a marker for unspecified cells (Figure 3A,B) (Rebay and Rubin, 1995), we found that some unspecified cells are missing in the second apoptotic wave (arrows, Figure 3B’’) suggesting that they are dying. Labeling with ELAV as neuronal marker revealed that the density of photoreceptor clusters and the number of photoreceptors per cluster is progressively reduced towards the posterior end (compare Figure 3C with Figure 3D’). Cas3* labeling overlapped with photoreceptor clusters (Figure 3D,D’’; arrows) suggesting that photoreceptor neurons are also eliminated in the second apoptotic wave.
To obtain a better understanding of photoreceptor cell death in the second apoptotic wave we analyzed individual or subgroups of photoreceptor neurons. Markers for outer photoreceptor neurons, Rough (R2,5,3,4-specific) and Seven-up (Svp, R3,4,1,6-specific), are initially induced in GMR-hid eye discs (Figure 3F,G), but are subsequently eliminated in the second apoptotic wave (Figure 3F–H; see also Figure S3A,B for expression of Rough and Svp in wild type discs). Senseless (Sens) is a marker for R8 photoreceptor neurons, the first photoreceptor to be specified (Nolo et al., 2000). Interestingly, while in many cases almost the entire photoreceptor cluster labels positive for Cas3*, this does not include the R8 cell, and only a few of them are eliminated in the second apoptotic wave during larval stages (Figure 3E–E’’’).
The last photoreceptor neuron to be specified is R7. Labeling with the R7 marker Prospero (Pros) demonstrates that the pattern of R7 cells in GMR-hid is normal (Figure S3C,E). We did not find any Pros-positive cells that are also labeled with Cas3* (Figure S3E,F). Therefore, the majority of R7 neurons, if not all, survive at this stage. Similarly, labeling with Cut, a marker for cone cells, shows that most of them also survive in the second apoptotic wave in GMR-hid (Figure S3D,G,H). Thus, these examples demonstrate that hid-induced cell death affects differentiating cells differently depending on position and developmental age. The outer photoreceptor neurons R1-R6 are largely eliminated by GMR-hid in the second apoptotic wave. R7 and cone cells are the developmentally youngest cells and most of them are still alive at the end of larval stages. Most remarkable, however, is R8, the developmentally oldest cell, which still survives hid-induced apoptosis by the end of larval stages. This suggests that R8 contains a very strong intrinsic program that protects it from apoptosis.
EGFR signaling protects unspecified cells in the apoptosis-free zone
To explain the apoptosis-free zone in GMR-hid (Figure 1G, Figure 3B), we first examined the distribution of Hid protein. Although Hid is strongly induced posterior to the MF in GMR-hid eye discs compared to wild type (Figure 4A,B), a clear reduction of Hid protein is present in the apoptosis-free zone in GMR-hid (Figure 4B, arrow). Because EGFR signaling is known to negatively regulate Hid activity (Bergmann et al., 1998; Kurada and White, 1998), we considered that the reduction of Hid protein in the apoptosis-free zone is the result of EGFR activity. Consistently, the reduction of Hid levels is lost when a dominant negative allele of EGFR (EGFRDN) is expressed in GMR-hid eye discs (Figure 4C). Furthermore, EGFR signaling stimulates MAPK to phosphorylate Hid. Therefore, we tested if MAPK phosphorylation destabilizes Hid protein in the apoptosis free zone. A MAPK-unresponsive mutant of Hid (HidAla5) is indeed stable in the apoptosis-free zone (Figure 4D). These data imply that EGFR/MAPK-dependent regulation of Hid protein levels contributes to the apoptosis-free zone.
The apoptosis-free zone in GMR-hid contains photoreceptor neurons at the apical side and unspecified cells at the basal side of the disc. Although it is known that EGFR signaling protects cells between columns 7 and 15 (Baker and Yu, 2001), it is unclear which of these cell types require EGFR signaling for survival, because Baker and Yu (2001) analyzed egfr mutant clones (Baker and Yu, 2001) in which photoreceptor differentiation is blocked. To reinvestigate this question, we examined eye discs expressing the EGFR/MAPK-unresponsive GMR-rpr and GMR-hidAla5 transgenes (Bergmann et al., 1998; Kurada and White, 1998). In these discs, Cas3*-labeling now covers the entire posterior half of the disc including the area corresponding to the apoptosis-free zone in GMR-hid (Figure 4E’,F’). The majority of photoreceptor neurons as indicated by ELAV still survives in EGFR/MAPK-unresponsive GMR-rpr and GMR-hidAla5 eye discs (Figure 4E’’,F’’). Consistently, sections through the areas which correspond to the apoptosis-free zone show that ELAV labeling is largely excluded from Cas3* labeling which occurs mainly at the basal side of the discs (Figure 4E,F and sections therein). Moreover, expression of EGFRDN results in loss of the apoptosis-free zone in GMR-hid eye discs (Figure 4G’), while expression of EGFRDN in otherwise wild-type background induces cell death specifically in the area which corresponds to the apoptosis-free zone in GMR-hid (Figue 4H’). Taken together, these observations indicate that photoreceptor neurons still survive under the conditions of reduced EGFR signaling, while unspecified cells die. Therefore, EGFR signalling is required to protect unspecified cells in the apoptosis-free zone in GMR-hid eye discs.
Intriguingly, while EGFR signalling is required for photoreceptor differentiation (Freeman, 1996; Yang and Baker, 2001), these cells do not rely completely on EGFR signalling for survival and appear to have developed other mechanisms to protect themselves from apoptosis (see below). The opposite statement can be made for unspecified cells. While EGFR signalling is not sufficient to induce photoreceptor differentiation of unspecified cells, it does protect them from hid-induced apoptosis.
A transient increase of Diap1 reduces apoptosis susceptibility of photoreceptor neurons
As shown in Figure 4D, GMR-hid is expressed in all cells posterior to the MF. However, photoreceptor neurons are protected from apoptosis in the first apoptotic wave and apoptosis-free zone independently of EGFR although they contain cleaved caspases (Figure 1G’, I’’). We therefore investigated how photoreceptor neurons survive despite the presence of cleaved caspases. Because Diap1 is known to inhibit cleaved effector caspases (Ditzel et al., 2008; Li et al., 2011), we examined its protein level in 3rd instar eye discs using Diap1 antibodies. Interestingly, as soon as photoreceptor neurons are specified as judged by ELAV labeling, we detect a strong increase of Diap1 levels in photoreceptor neurons (Figure 5A,B). Diap1 levels remain low in the unspecified interommatidial cells at levels comparable to the tissue anterior to the MF (Figure 5A’,B’). A detailed high-resolution analysis suggests that all eight photoreceptor neurons accumulate Diap1 protein while Diap1 levels in interommatidial cells remain low (Figure S4A–C). In each ommatidium, the photoreceptor axons form a bundle projecting towards the optic lobes of the brain. The brightest signals of Diap1 expression appear to be in axon bundles (Figure 5A’’,B,B’) implying that they inhibit cleaved caspases which are observed here at high levels (Figure 1I’’). Consistent with a protective function of Diap1, loss of one copy of diap1 enhances the GMR-hid-induced eye ablation phenotype (Hay et al., 1995) (Figure 5I,J) through increased cell death in both apoptotic waves and a reduction in the size of the apoptosis free zone (Figure S4D,E). The specificity of the Diap1 antibody used was confirmed in diap1 mosaics (Figure S4F).
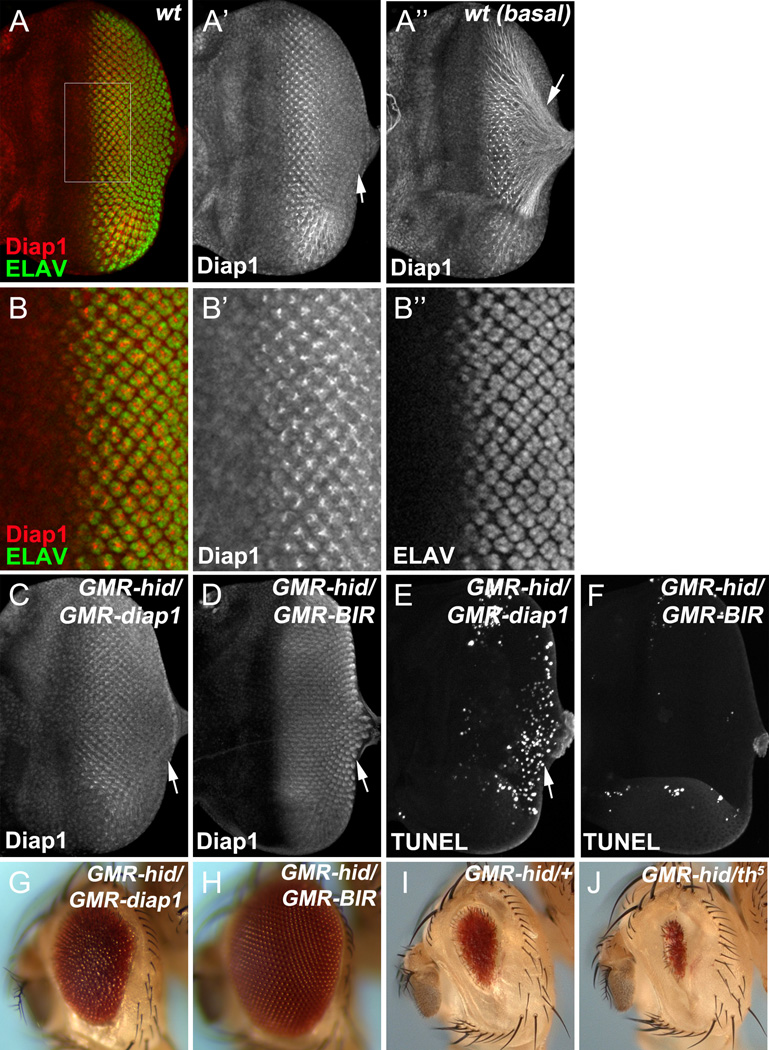
Figure 5. A transient increase of Diap1 protects photoreceptor neurons from apoptosis.
(A, B) Wild type late 3rd instar eye disc labeled with Diap1 and ELAV. (B) shows an enlarged view of the outlined region in (A). (A’’) shows the basal side of the same disc in (A). Diap1 strongly accumulates in ELAV-positive photoreceptor neurons (A, B) and in basally projecting axonal bundles (arrow, A’’ ). In the posterior edge of the disc, levels of Diap1 are gradually reduced (A’, arrow).
(C–F) Late 3rd instar eye discs labeled by Diap1 (C, D) or TUNEL (E, F). Overexpression of full length Diap1, GMR-diap1, in the GMR-hid eye disc shows the same gradual decrease of Diap1 towards the posterior end of the disc (arrow, C) as endogenous Diap1. In contrast, expression of a stabilized form of Diap1, GMR-BIR, in the GMR-hid eye disc maintains accumulated Diap1 levels at the posterior end of the disc (D, arrow). Consequently, GMR-diap1 suppresses the first apoptotic wave in GMR-hid, but not the second (posterior) one indicated by TUNEL-positive cells (E)GMR-BIR suppresses both apoptotic waves in GMR-hid(F).
(G–J) Adult eyes of GMRhid/+;GMR-diap1/+ flies (G)GMRhid/+;GMR-BIR/+ flies (H)GMRhid/+ flies (I), or GMRhid/+ ;th5/+ flies (J). Compared to GMR-diap1, GMR-BIR shows a stronger suppression of GMR-hid (compare H to G). Loss one copy of diap1 by using a null mutant (th5) enhances eye ablation of GMR-hid (compare J to I).
See also Figure S4.
Interestingly, we also observed that protein levels of Diap1 are gradually reduced in developmentally older photoreceptor neurons towards the posterior end of eye discs (Figure 5A’; arrow). We hypothesized that this reduction of Diap1 protein re-sensitizes older photoreceptors to hid-induced apoptosis such that they die in the second apoptotic wave. To test this hypothesis, we overexpressed Diap1 using GMR (GMR-diap1) (Hay et al., 1995). However, while GMR-diap1 is sufficient to suppress the first apoptotic wave, it did not suppress the second apoptotic wave (Figure 5E) causing only a partial suppression of GMR-hid (Hay et al., 1995) (Figure 5G). Nevertheless, we noted that even overexpressed Diap1 protein is subject to the same mechanism that induces down-regulation of Diap1 in older photoreceptor neurons (Figure 5C, arrow). Because RING mutants of Diap1 can suppress GMR-hid-induced apoptosis (Goyal et al., 2000), we tested expression of a RING domain-deleted form of Diap1 (GMR-BIR) (Hay et al., 1995) in GMR-hid background. Consistently, loss of the RING domain resulted in a stabilization and accumulation of Diap1 in developmentally older photoreceptor neurons (Figure 5D, arrow). As a consequence, GMR-BIR completely suppressed GMR-hid-induced apoptosis (Figure 5F) and its adult eye ablation phenotype (Figure 5H). These data suggest that the protein levels of Diap1 determine the apoptosis susceptibility of photoreceptor neurons. While young photoreceptor neurons at the MF increase Diap1 levels, older photoreceptors down-regulate these levels, likely by a RING-dependent mechanism. Therefore, reduced Diap1 levels may account for increased susceptibility to apoptotic signals in older photoreceptor neurons, explaining the formation of the second apoptotic wave in GMR-hid eye discs (Figure 3).
Control of Diap1 protein levels by Cullin-3 E3 ligase
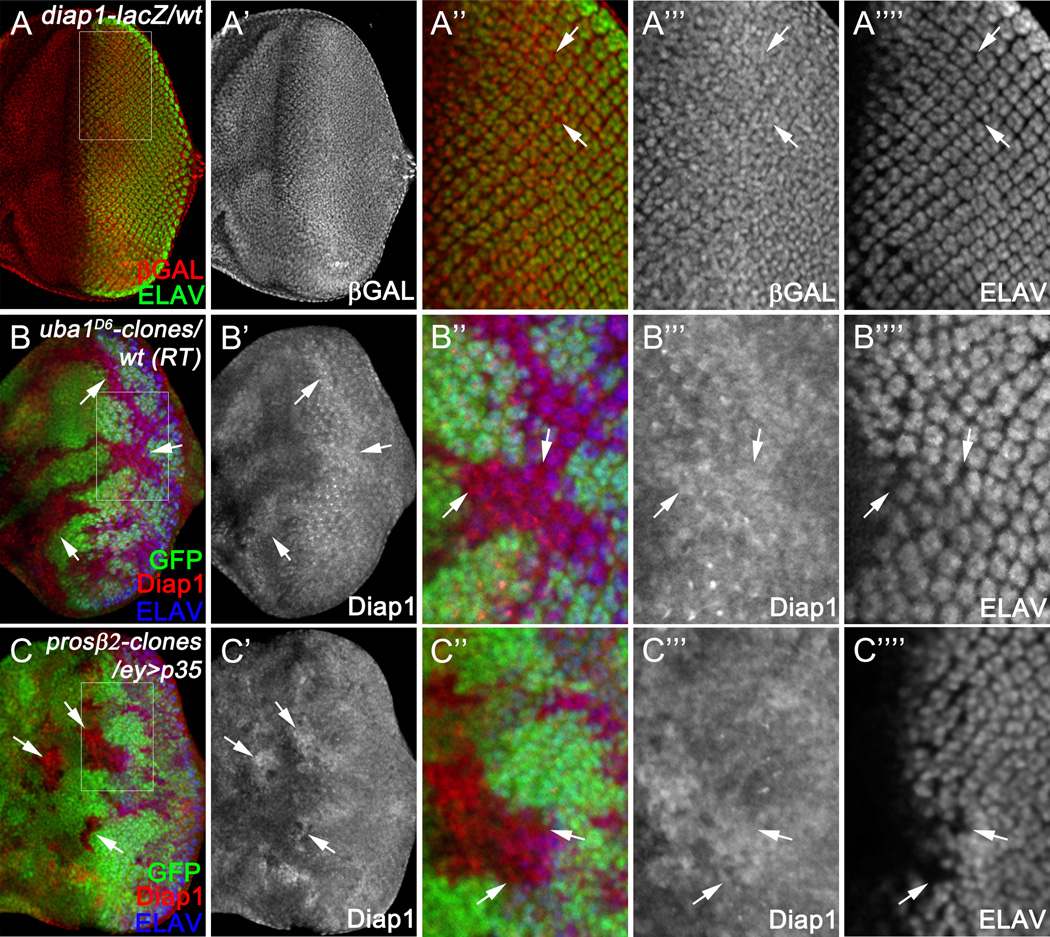
Next, we asked how Diap1 levels are transiently increased in differentiating photoreceptor neurons. We first examined whether the increased levels of Diap1 are due to transcriptional regulation. In situ hybridization of diap1 suggested that, unlike the protein level, transcription of diap1 does not show a neuron-specific pattern (Figure S5) consistent with previous reports (Hays et al., 2002; Udan et al., 2003)). To further investigate this at the single cell resolution, we used diap1-lacZ, an enhancer trap indicating transcriptional expression of diap1 (Ryoo et al., 2002). Both photoreceptor neurons and interommatidial, unspecified cells show comparable expression levels of P-galactosidase (Figure 6A) suggesting that the high protein levels of Diap1 in photoreceptor neurons are due to post-transcriptional regulation.
Figure 6. Ubiquitin-dependent degradation of Diap1 in unspecified cells.
(A) Late 3rd instar eye disc labeled with ELAV. Transcription of diap1 is indicated by the diap1-lacZ reporter (labeled for βGAL). (A’’) shows an enlarged view of the outlined region in (A)diap1-lacZ (βGAL) in unspecified, i.e. interommatidial, cells (examples indicated by arrows) is indistinguishable from that in ELAV-positive photoreceptor neurons.
(B–C) Late 3rd instar eye discs labeled with Diap1 and ELAV. uba1(B) or prosβ2(C) mutant clones are labeled by lack of GFP. (B’’, C’’) show enlarged views of the outlined regions in (B, C), respectively. UAS-p35 was expressed under the control of ey-GAL4 (ey>p35) to block apoptosis in prosβ2 mutant clones (C). Expression of Diap1 is increased in uba1 (B) or prosβ2 mutant clones (C) specifically in ELAV-negative unspecified, i.e. interommatial, cells (arrows, B’’, C’’).
See also Figure S5.
To further investigate how Diap1 is regulated post-transcriptionally, we focused our analysis on ubiquitin-dependent protein degradation (reviewed in (McCarthy, 2013)). In this process, ubiquitin is transferred and attached to the substrate protein through E1-activating enzymes, E2-conjugating enzymes, and E3-ubiquitin ligases consecutively. Poly-ubiquitylated proteins are then degraded by the 26S proteasome. As Uba1 is the only E1-activating enzyme in Drosophila (Lee et al., 2008; Lee et al., 2011; Pfleger et al., 2007), we analysed Diap1 levels in uba1 mutant tissues. Diap1 is increased in uba1 mutant clones located both anterior and posterior to the MF (Figure 6B). Posterior to the MF, this increase of Diap1 is prominent in unspecified, interommatidial cells (Figure 6B”; arrows). Similarly, an increase of Diap1 was observed in prosβ2 mutant tissues, a key component of the 26S proteasome (Figure 6C). These data suggest that in posterior eye tissue, Diap1 levels are kept low specifically in unspecified interommatidial cells through an ubiquitin-dependent protein degradation mechanism maintaining these cells in an apoptosis-susceptible state if they are not protected otherwise (EGFR). Diap1 protein in young photoreceptor neurons, however, is not subject to this degradation mechanism. These cells accumulate Diap1 and increase their apoptotic resistance.
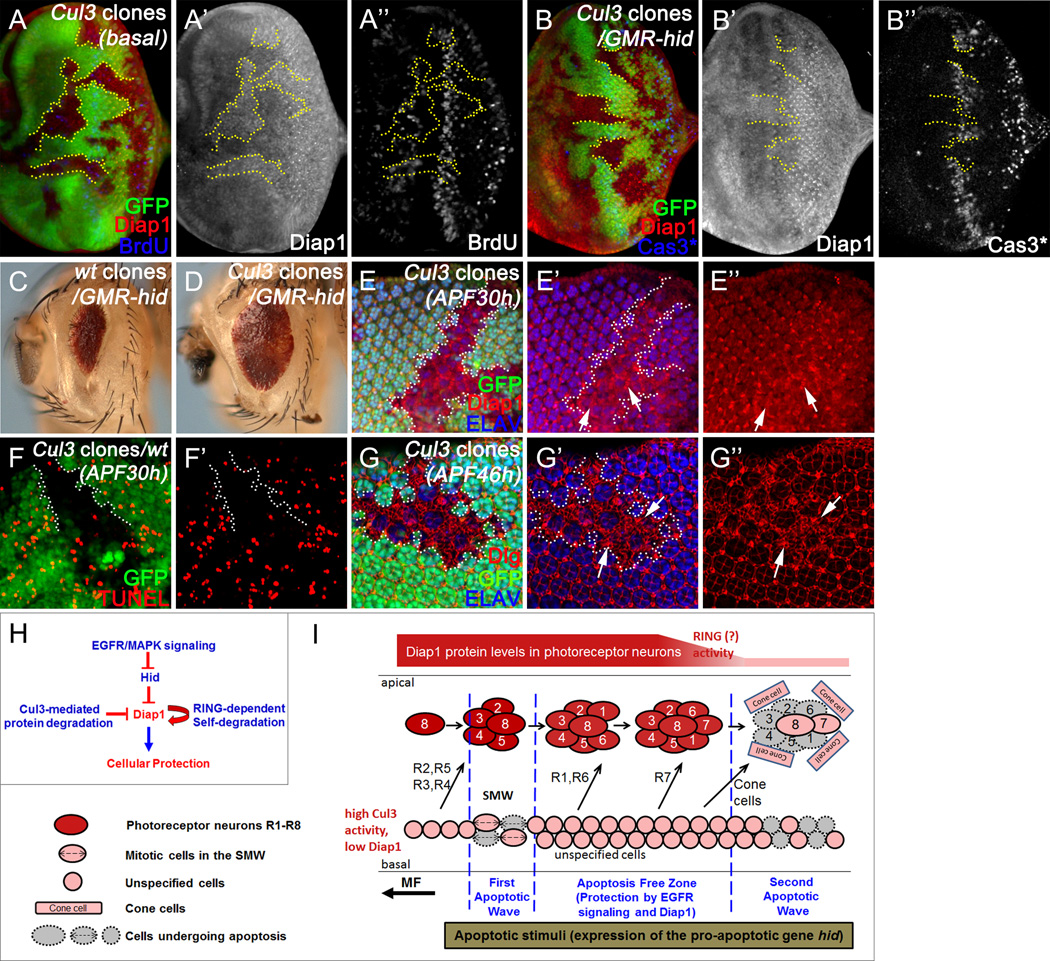
We sought to identify the E3 ubiquitin ligase that targets Diap1. Cullin (Cul) family proteins are the largest known class of ubiquitin E3 ligases (reviewed in (Petroski and Deshaies, 2005)). Both Cul1 and Cul3 have been implicated in regulation of Ci, a key transcription factor in the Hedgehog signaling pathway, in developing Drosophila eye tissues (Ou et al., 2002). We therefore analyzed whether Diap1 can be regulated by Cul1 and Cul3. While Cul1 mutants do not affect Diap1 protein levels (Christiansen et al., 2013), they are increased in Cul3 mutant tissues (Figure 7A,A’). As controls, such an increase is not due to increased cell proliferation (Figure 7A’’) or increased numbers of cells (Figure S6A). Consistent with the role of Cul3 in protein degradation, in situ hybridization analysis shows that diap1 transcripts are not increased in Cul3 mutant tissues (Figure S6B).
Figure 7. Cullin-3-mediated degradation of Diap1 in unspecified cells.
(A–B) Late 3rd instar eye discs labeled by Diap1 and BrdU (A; wt), or Diap1 and Cas3* (B;GMR-hid)Cul3 clones are labeled by lack of GFP. Diap1 protein is increased in Cul3 clones (A’, B’). The proliferation pattern does not change in Cul3 clones (A’’). When Cul3 clones are generated in the GMR-hid eye disc (B), hid-induced apoptosis is suppressed in Cul3 mutant clones (B’’).
(C, D) Adult mosaic eyes of GMR-hid flies with wild type control clones (C) or with Cul3 mutant clones (D)hid-induced eye ablation was partially suppressed by Cul3 mutant clones.
(E–G) Pupal eye discs at 30-hour (E, F) or 46-hour (G) after pupal formation (APF30h or APF46h) labeled with Diap1 and ELAV (E), TUNEL (F), or Dlg and ELAV (G)Cul3 clones are labeled by lack of GFP. Diap1 is increased specifically in Cul3-mutant interommatidial cells (arrows, E-E’’), but not in photoreceptor neurons. Consequently, developmental apoptosis of interommatidial cells is blocked in Cul3 mutant clones (F, F’). This results in additional interommatial cells in Cul3 clones at APF46h (G-G’’).
(H) Several mechanisms modulate the apoptosis susceptibility of various cell types in the eye imaginal disc. First, EGFR/MAPK signaling directly inhibits Hid activity in unspecified cells; Second, Cul3-mediated degradation keeps Diap1 levels low in unspecified cells. Third, H99-independent stimulation of the RING-E3 ligase domain contributes to apoptotic sensitivity of older photoreceptor neurons.
(I) Schematic outline of distinct cellular apoptosis susceptibilities in the developing Drosophila eye tissue. Anterior is to the left. In response to expression of Hid, two apoptotic waves are induced in the SMW (First Apoptotic Wave) and the posterior portion of the developing eye (Second Apoptotic Wave). Unspecified cells located at the basal side of the disc between the two apoptotic waves (the apoptosis-free zone) are protected by EGFR signaling. Moreover, the protein levels of Diap1 are transiently high (indicated by intense red color) in differentiating photoreceptor neurons protecting them from apoptosis at this stage. Later in development, Diap1 levels are down-regulated presumably by RING-dependent self-degradation (indicated by pale red). Therefore, developmentally older photoreceptor neurons become susceptible to apoptosis and die in the second apoptotic wave together with unspecified cells. In addition, R8, R7 and cone cells do not die in the second apoptotic wave suggesting that there are unknown mechanisms that renders these cells resistant to apoptosis.
See also Figure S6.
We also asked whether the increase of Diap1 in Cul3 mutants is functional and can suppress apoptosis. To address this, Cul3 mutant clones were generated in GMR-hid eye discs. GMR-hid-induced Cas3* activity is suppressed in Cul3 clones (Figure 7B). Consequently, GMR-hid-induced adult eye ablation phenotype is suppressed by Cul3 mosaic tissues (Figure 7C,D). We further examined whether Cul3 can regulate developmental apoptosis. We observed that, in mid-pupal eye discs, expression of Diap1 is specifically increased in Cul3 mutant interommatidial cells (arrows, Figure 7E), but not in photoreceptor neurons. Such an increase does inhibit developmental apoptosis of interommatidial cells at this stage (Figure 7F). As a result, many additional interommatidial cells survive in Cul3 mutant clones (Figure 7G, arrows). Therefore, Cul3 mediates Diap1 degradation in unspecified cells to render them susceptible towards apoptosis during development of the Drosophila eye.
We also note that there are more interommatidial cells in Cul3 clones than what would be expected if only apoptosis was suppressed (Figure 7G). Because we do not see a strong effect on proliferation in Cul3 clones (Figure 7A’’), this may not be due to increased proliferation. Instead, it appears that Cul3 clones partially affect photoreceptor specification resulting in an increased number of interommatidial cells.
DISCUSSION
In this paper, using the GMR-hid apoptotic model in Drosophila, we reveal striking differences in the apoptotic susceptibilities of different cell types in the Drosophila retina over a developmental period of only 24 hours (Figure 7H,I). Proliferating cells in the SMW are very susceptible to apoptotic stimuli. In contrast, post-mitotic cells have acquired different mechanisms which can confer – at least transiently - resistance to apoptosis. As previously shown, unspecified cells rely on EGFR signaling for survival (Baker and Yu, 2001). The signal for EGFR activation, Spitz, is likely coming from photoreceptor neurons which require EGFR signaling for specification (Baker and Yu, 2001). Interestingly, although photoreceptor cells require EGFR signaling for specification (Tio and Moses, 1997), the survival of photoreceptor neurons does not solely depend on EGFR signaling. Instead, photoreceptor cells dramatically increase the protein levels of Diap1 at the onset of specification. In contrast, unspecified cells at the basal side of the disc degrade Diap1 by a Cul3-dependent mechanism. The survival of inner photoreceptors R8 and possibly R7 appears to be independent of EGFR signaling and increased Diap1 levels.
Dynamic Diap1 protein pattern
The protein level of Diap1 is under very dynamic control in developing eye imaginal discs (Figure 7H,I). Its level is relatively low in proliferating cells anterior to the MF, in SMW cells and in unspecified cells posterior to the MF. Under normal conditions, these low levels are sufficient to ensure survival of the cells, however, they are very susceptible to apoptotic signals. Our genetic analysis indicates that a Cul3-mediated degradation process maintains Diap1 at low levels in unspecified cells posterior to the MF (Figure 7H,I). Loss of Cul3 results in accumulation of Diap1 causing survival of many additional interommatidial cells in pupal eye discs. Developmentally, it is likely that this Cul3-mediated Diap1 degradation keeps interommatidial cells susceptible to apoptosis so that they can be eliminated by apoptosis if they have not been incorporated into ommatidia at around 28 hours APF (Brachmann and Cagan, 2003).
The control of Diap1 levels by Cul3 adds another level of complexity to Diap1 regulation. For example, Diap1 uses its own RING E3 domain for auto-ubiquitylation and degradation (see below) (Hays et al., 2002; Holley et al., 2002; Ryoo et al., 2002; Wing et al., 2002; Yoo et al., 2002). The kinase IKKε phosphorylates Diap1 and promotes its degradation for proper cell fate specification (Kuranaga et al., 2006). Furthermore, Diap1 is transcriptionally and post-translationally controlled by Hippo signaling to prevent apoptosis during tissue growth (Harvey et al., 2003; Udan et al., 2003; Wu et al., 2008; Zhang et al., 2008).
Interestingly, Cul3 is produced as soma-specific (Cul3S) and testis-specific (Cul3T) isoforms (Arama et al., 2007). Cul3T has been shown to regulate caspase activity in a non-apoptotic process called individualization in developing sperm. However, Cul3T does not target Diap1 for degradation, but another IAP, termed dBruce (Kaplan et al., 2010). Therefore, in the testis, Cul3T regulates dBruce for a non-apoptotic function of caspases, while in the eye disc, Cul3S controls the apoptotic susceptibility of unspecified cells by triggering Diap1 degradation.
While unspecified cells keep Diap1 levels low by the above-described Cul3S-dependent process, photoreceptor neurons accumulate high levels of Diap1 immediately after they are specified and thus survive the first apoptotic wave. A Cul3S-dependent degradation process of Diap1 does not seem to operate in photoreceptor neurons. However, Cul3S is ubiquitously expressed in eye imaginal discs (Ou et al., 2002), raising the question how Cul3S-activity and thus Diap1 degradation is restricted to unspecified cells. One possibility is the specific expression of the substrate recognition protein in unspecified cells. Cul3 utilizes BTB-domain containing proteins to recruit substrates for ubiquitylation and degradation (reviewed in (Petroski and Deshaies, 2005)). In Drosophila, several BTB proteins including Kelch10 (Arama et al., 2007), Roadkill (Baker et al., 2009), Kelch (Hudson and Cooley, 2010), Insomniac (Pfeiffenberger and Allada, 2012; Stavropoulos and Young, 2011), and Diablo (Strutt et al., 2013) have been shown to mediate Cul3-dependent degradation of various protein substrates. However, none of these BTB proteins is required for regulation of Diap1 by Cul3 (data not shown). Another possibility is that an inhibitor of Cul3 is specifically expressed in photoreceptor neurons keeping Cul3 activity low in these cells. An example of such a Cul3 inhibitor is Soti in the testis (Kaplan et al., 2010). Soti competes with dBruce for binding to Kelch10 which regulates activation of caspases during sperm individualization. However, loss of Soti in the eye disc does not affect Diap1 levels (data not shown). Therefore, future work is needed to dissect the specificity of Cul3S-mediated degradation of Diap1 in unspecified cells.
While photoreceptor neurons accumulate high levels of Diap1 starting in the MF, later in development towards the posterior edge of the disc, Diap1 levels are down-regulated. As a result, older photoreceptor neurons, especially R1-R6, become sensitive to apoptotic signals and are eliminated in the 2nd apoptotic wave in GMR-hid. This down-regulation of Diap1 appears to be the result of RING-dependent self-degradation of Diap1 because a RING-depleted mutant of Diap1 is not subject to this type of down-regulation. That raises the question how the RING E3 ligase activity is stimulated at the posterior edge of the discs. It was previously shown that the IAP antagonists Reaper, Hid and Grim stimulate RING activity (Hays et al., 2002; Holley et al., 2002; Ryoo et al., 2002; Wing et al., 2002; Yoo et al., 2002). However, loss of these genes (H99 mutant) does not block the down-regulation of Diap1 at the posterior edge of the disc (data not shown), suggesting that another, yet unknown mechanism or factor accounts for stimulation of the RING domain of Diap1 at the posterior edge of the eye disc.
Another question addresses the functional significance of the Diap1 accumulation in specifying photoreceptor neurons. There is no definitive answer to this question either, but there is a possible hint. In strong diap1 mutant clones which are protected from apoptosis by expression of the caspase inhibitor p35, photoreceptor neurons fail to differentiate (Figure S4F) suggesting that Diap1 may have a role in early specification of photoreceptor neurons. This function of Diap1 does not seem to require the RING domain as mutant alleles lacking the RING domain allow photoreceptor differentiation (Figure S4G).
R8, R7 and cone cells
R8, R7 and cone cells survive in the second apoptotic wave during larval stages. R7 and cone cells are developmentally the youngest cells and their early specification status may permit their survival during larval stages. However, R8 is exceptional. It is the first photoreceptor neuron to be specified and thus has been exposed to Hid activity for the longest time. Yet, compared to R1–6, R8 is more apoptosis-resistant and survives to the end of larval stages (Figure 3E). Because R8 also does not require EGFR signaling for either specification or survival at any time during development (Baker and Yu, 2001; Yang and Baker, 2001) and it down-regulates Diap1, the survival of R8 appears to require a different mechanism. A possible survival mechanism may be exerted by the R8 specification gene senseless (sens) (Nolo et al., 2000). sens has been shown in a different cellular context (salivary glands in embryos) to have a strong anti-apoptotic activity (Chandrasekaran and Beckendorf, 2003). It is currently unknown if sens has a similar protective function in R8.
Relevance for cancer
Cancer cells have developed many strategies to evade apoptosis. One such strategy is the accumulation of IAPs in many tumor types (LaCasse et al., 2008). It is therefore very important to identify the mechanisms that control IAP levels, both transcriptionally and post-translationally. Our work here in Drosophila shows that the lack of a Cul3-mediated degradation process can lead to accumulation of Diap1 and survival of interommatidial cells. It is conceivable that similar Cullin-dependent processes control IAP levels in mammals including humans and that cancer cells may inactivate them to up-regulate IAPs.
In conclusion, the cellular steady-state level of Diap1 appears to be a key modulator of cellular apoptosis susceptibility. Multiple mechanisms are employed to regulate Diap1, hence the cellular susceptibility to apoptosis varies from cell to cell depending on the developing context. As several IAP family proteins exist in mammals as caspase inhibitors (reviewed in (Silke and Meier, 2013)), it is likely that cellular sensitivity and susceptibility to apoptosis can also be modulated at the level of IAP proteins in mammals.
EXPERIMENTAL PROCEDURES
Fly Strains and Crosses
Both transgenic lines GMR-hid10 (Grether et al., 1995) and GMR-hid326 (Fan and Bergmann, 2008) with insertions on the 2nd and 3rd chromosome, respectively, gave similar waves of cell death in developing eye discs. 4xGMR-rpr was obtained by combining CyO;2xGMR-rpr (Kurada and White, 1998) and homozygous GMR-rpr46 (White et al., 1996). GMR-hidAla5 (Bergmann et al., 1998), UAS-EGFRDN (Freeman, 1996), UAS-rpr (Ryoo et al., 2002), th33-1s (Goyal et al., 2000), GMR-diap1, GMR-BIR and th5 (Hay et al., 1995), GMR-p35 and UAS-p35 (Hay et al., 1994), uba-1D6 (Lee et al., 2008), Cul3gft2 and Cul1[EX] (Ou et al., 2002) are as described. GMR-Gal4, ey-GAL4, elavC155-GAL4, UAS-CD8GFP, ey-Flp, UAS-Flp, diap1-lacZ (thj5c8) and Prosβ2 EP3067were obtained from the Bloomington Stock Center.
Mosaic Analysis
The following genotypes were generated to analyze uba-1, Prosβ2, or Cul3 mutant clones in mosaic eye discs: (1) ey-Flp/+; uba-1D6 FRT80B/ P[ubiGFP] FRT80B; (2) ey-GAL4/UAS-Flp UAS-p35; Prosβ2EP3067 FRT80B/ P[ubiGFP] FRT80B; (3) ey-Flp/+; Cul3gft2FRT40A/ P[ubiGFP] FRT40A; to analyze Cul3 clones in GMR-hid background: ey-Flp/+; Cul3gft2FRT40A/ P[ubiGFP] FRT40A; GMR-hid/+. Crosses were raised at 25°C unless otherwise specified. For each genotype, at least 30 discs were analyzed. Representative data are shown.
Immunohistochemistry
Eye-antennal imaginal discs from wandering 3rd instar larvae or pupae at indicated stages were dissected, fixed (with 4% paraformaldehyde for 30min at room temperature), and then labeled with primary and secondary antibodies as described (Fan et al., 2005). Guinea Pig anti-Diap1 (SK14) was kindly provided by P. Meier (Tenev et al., 2002). Guinea Pig anti-Sens was kindly provided by H. Bellen (Nolo et al., 2000). Rabbit anti-Svp is a gift from R. Schulz. Commercial antibodies used are mouse anti-BrdU (BD), rabbit anti-PH3 (Upstate), rabbit anti-cleaved Caspase-3 (Cell Signaling), rat anti-ELAV, mouse anti-Rough, mouse anti-Pros, mouse anti-Cut and mouse anti-Yan (DHSB, U. of Iowa). Secondary antibodies were donkey Fab fragments conjugated to FITC, Cy3 or Cy5 from Jackson ImmunoResearch.
BrdU Labeling, TUNEL and EdU Double Labeling
For BrdU labeling to detect proliferating cells, larval discs were incubated with 0.5 mg/ml BrdU (Sigma) in Schneider’s media for 1 hour at room temperature. Discs were then fixed in 1% paraformaldehyde with 0.01% Tween 20 at 4°C overnight followed by RQ1-DNase (1 mg/ml, Promega) digestion for 2h at 37°C and antibody staining thereafter.
For TdT-mediated dUTP nick end labeling (TUNEL) and EdU double labeling, larval discs were first incubated with 10µM EdU (Molecular Probes) similar as the BrdU labeling described above and then fixed in 4% paraformaldehyde for 30min at room temperature. Fixed discs were further incubated in 100mM Na-Citrate with 0.1% TritonX-100 for 30 min at 65°C followed by detection of dying cells using an in situ cell death detection kit (Roche). EdU labeling was done after TUNEL using a Click-iT® EdU imaging kit (Molecular Probes).
In Situ Hybridization
For in situ hybridization to detect diap1 transcripts, Drosophila cDNA clone GH15248 (BDGP ESTs, Drosophila Genomics Resource Center) was used as a template to generate digoxigenin (DIG)-labeled sense and anti-sense RNA probes (Roche). Labeled probes were detected with NBT/BCIP (Roche) or Tyramide Signal Amplification (TSA, PerkinElmer) as previously described (Chotard et al., 2005).
Imaging
Larval or pupal disc images were taken with either a Zeiss LSM700 or an Olympus FV500 confocal microscope. Adult eyes were photographed by a Zeiss AxioImager equipped with a Zeiss AxioCam HR.
Quantification of mitotic cells
PH3-positive cells in the SMW were counted in wild type, GMR-hid, and GMR-hid;GMR-p35 eye discs (mean ± SD). For each genetic background, 20 representative eye discs were counted.
Supplementary Material
Highlights.
Mitotic cells are extremely sensitive to apoptotic signals.
Post-mitotic, yet undifferentiated, cells require EGFR signaling for survival.
Photoreceptor neurons accumulate the apoptosis inhibitor Diap1 for survival.
Cullin-3 degrades Diap1 in undifferentiated cells.
ACKNOWLEDGMENTS
We would like to thank Hugo Bellen, Cheng-Ting Chien, Georg Halder, Bruce Hay, Andrew Jarman, Pascal Meier, Hyung Don Ryoo, Bob Schulz, Hermann Steller, Steven Stowers, Kristin White, the Bloomington Stock Center, the Drosophila Genomics Resource Center in Indiana, and the Developmental Studies Hybridoma Bank (DSHB) in Iowa, for fly stocks and reagents. We thank Zhihong Chen for assistance on pupal eye dissection and Hillary Graves for help with data acquisition. AB is supported by Grant R01 GM068016 from the National Institute of General Medical Sciences (NIGMS). The content is solely the responsibility of the authors and does not represent the official views of the NIGMS or the NIH. YF is supported by Marie Curie Career Integration Grant (CIG) 630846 from the European Union’s Seventh Framework Programme (FP7) and the Birmingham Fellowship, University of Birmingham, UK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures (Figures S1–S6).
REFERENCES
- Arama E, Bader M, Rieckhof GE, Steller H. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS biology. 2007;5:e251. doi: 10.1371/journal.pbio.0050251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NE, Bhattacharya A, Firth LC. Regulation of Hh signal transduction as Drosophila eye differentiation progresses. Developmental biology. 2009;335:356–366. doi: 10.1016/j.ydbio.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NE, Yu SY. The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell. 2001;104:699–708. doi: 10.1016/s0092-8674(01)00266-5. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Agapite J, McCall K, Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–341. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Cagan RL. Patterning the fly eye: the role of apoptosis. Trends Genet. 2003;19:91–96. doi: 10.1016/S0168-9525(02)00041-0. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Developmental biology. 1989;136:346–362. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran V, Beckendorf SK. senseless is necessary for the survival of embryonic salivary glands in Drosophila. Development (Cambridge, England) 2003;130:4719–4728. doi: 10.1242/dev.00677. [DOI] [PubMed] [Google Scholar]
- Chotard C, Leung W, Salecker I. glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron. 2005;48:237–251. doi: 10.1016/j.neuron.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Christiansen AE, Ding T, Fan Y, Graves HK, Herz HM, Lindblad JL, Bergmann A. Non-cell autonomous control of apoptosis by ligand-independent Hedgehog signaling in Drosophila. Cell death and differentiation. 2013;20:302–311. doi: 10.1038/cdd.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel M, Broemer M, Tenev T, Bolduc C, Lee TV, Rigbolt KT, Elliott R, Zvelebil M, Blagoev B, Bergmann A, et al. Inactivation of effector caspases through nondegradative polyubiquitylation. Mol Cell. 2008;32:540–553. doi: 10.1016/j.molcel.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. Distinct mechanisms of apoptosis-induced compensatory proliferation in proliferating and differentiating tissues in the Drosophila eye. Developmental cell. 2008;14:399–410. doi: 10.1016/j.devcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell death and differentiation. 2010;17:534–539. doi: 10.1038/cdd.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Soller M, Flister S, Hollmann M, Muller M, Bello B, Egger B, White K, Schafer MA, Reichert H. The egghead gene is required for compartmentalization in Drosophila optic lobe development. Developmental biology. 2005;287:61–73. doi: 10.1016/j.ydbio.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier R, Laprise P, Cardin E, Harnois C, Plourde A, Reed JC, Vezina A, Vachon PH. Differential sensitivity to apoptosis between the human small and large intestinal mucosae: linkage with segment-specific regulation of BCL-2 homologs and involvement of signaling pathways. J Cell Biochem. 2001;82:339–355. doi: 10.1002/jcb.1172. [DOI] [PubMed] [Google Scholar]
- Goyal L. Cell death inhibition: keeping caspases in check. Cell. 2001;104:805–808. doi: 10.1016/s0092-8674(01)00276-8. [DOI] [PubMed] [Google Scholar]
- Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. Embo J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes & development. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development (Cambridge, England) 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Hays R, Wickline L, Cagan R. Morgue mediates apoptosis in the Drosophila melanogaster retina by promoting degradation of DIAP1. Nature cell biology. 2002;4:425–431. doi: 10.1038/ncb794. [DOI] [PubMed] [Google Scholar]
- Holley CL, Olson MR, Colon-Ramos DA, Kornbluth S. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nature cell biology. 2002;4:439–444. doi: 10.1038/ncb798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson AM, Cooley L. Drosophila Kelch functions with Cullin-3 to organize the ring canal actin cytoskeleton. The Journal of cell biology. 2010;188:29–37. doi: 10.1083/jcb.200909017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan Y, Gibbs-Bar L, Kalifa Y, Feinstein-Rotkopf Y, Arama E. Gradients of a ubiquitin E3 ligase inhibitor and a caspase inhibitor determine differentiation or death in spermatids. Developmental cell. 2010;19:160–173. doi: 10.1016/j.devcel.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Kumar S. Caspase function in programmed cell death. Cell death and differentiation. 2007;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- Kurada P, White K. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998;95:319–329. doi: 10.1016/s0092-8674(00)81764-x. [DOI] [PubMed] [Google Scholar]
- Kuranaga E, Kanuka H, Tonoki A, Takemoto K, Tomioka T, Kobayashi M, Hayashi S, Miura M. Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell. 2006;126:583–596. doi: 10.1016/j.cell.2006.05.048. [DOI] [PubMed] [Google Scholar]
- LaCasse EC, Mahoney DJ, Cheung HH, Plenchette S, Baird S, Korneluk RG. IAP-targeted therapies for cancer. Oncogene. 2008;27:6252–6275. doi: 10.1038/onc.2008.302. [DOI] [PubMed] [Google Scholar]
- Lee TV, Ding T, Chen Z, Rajendran V, Scherr H, Lackey M, Bolduc C, Bergmann A. The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously. Development (Cambridge, England) 2008;135:43–52. doi: 10.1242/dev.011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TV, Fan Y, Wang S, Srivastava M, Broemer M, Meier P, Bergmann A. Drosophila IAP1-mediated ubiquitylation controls activation of the initiator caspase DRONC independent of protein degradation. PLoS Genet. 2011;7:e1002261. doi: 10.1371/journal.pgen.1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang J, Shi Y. Structural mechanisms of DIAP1 auto-inhibition and DIAP1-mediated inhibition of drICE. Nature communications. 2011;2:408. doi: 10.1038/ncomms1418. [DOI] [PubMed] [Google Scholar]
- McCarthy N. Ubiquitylation: Mediation by degradation. Nature reviews. 2013;13:146. doi: 10.1038/nrc3471. [DOI] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- Ou CY, Lin YF, Chen YJ, Chien CT. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes & development. 2002;16:2403–2414. doi: 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nature reviews. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pfeiffenberger C, Allada R. Cul3 and the BTB adaptor insomniac are key regulators of sleep homeostasis and a dopamine arousal pathway in Drosophila. PLoS Genet. 2012;8:e1003003. doi: 10.1371/journal.pgen.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Harvey KF, Yan H, Hariharan IK. Mutation of the Gene Encoding the Ubiquitin Activating Enzyme Uba1 Causes Tissue Overgrowth in Drosophila. Fly. 2007;1:95–105. doi: 10.4161/fly.4285. [DOI] [PubMed] [Google Scholar]
- Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–866. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nature cell biology. 2002;4:432–438. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- Silke J, Meier P. Inhibitor of apoptosis (IAP) proteins-modulators of cell death and inflammation. Cold Spring Harbor perspectives in biology. 2013;5 doi: 10.1101/cshperspect.a008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Gao G, Zhang X, Wang G, Dou QP. Regulation of tumor cell apoptotic sensitivity during the cell cycle (Review) International journal of molecular medicine. 2000;6:503–507. doi: 10.3892/ijmm.6.5.503. [DOI] [PubMed] [Google Scholar]
- Srinivasan A, Roth KA, Sayers RO, Shindler KS, Wong AM, Fritz LC, Tomaselli KJ. In situ immunodetection of activated caspase-3 in apoptotic neurons in the developing nervous system. Cell death and differentiation. 1998;5:1004–1016. doi: 10.1038/sj.cdd.4400449. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Scherr H, Lackey M, Xu D, Chen Z, Lu J, Bergmann A. ARK, the Apaf-1 related killer in Drosophila, requires diverse domains for its apoptotic activity. Cell death and differentiation. 2007;14:92–102. doi: 10.1038/sj.cdd.4401931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos N, Young MW. insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron. 2011;72:964–976. doi: 10.1016/j.neuron.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H, Searle E, Thomas-Macarthur V, Brookfield R, Strutt D. A Cul-3-BTB ubiquitylation pathway regulates junctional levels and asymmetry of core planar polarity proteins. Development (Cambridge, England) 2013;140:1693–1702. doi: 10.1242/dev.089656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenev T, Zachariou A, Wilson R, Paul A, Meier P. Jafrac2 is an IAP antagonist that promotes cell death by liberating Dronc from DIAP1. EMBO J. 2002;21:5118–5129. doi: 10.1093/emboj/cdf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tio M, Moses K. The Drosophila TGF alpha homolog Spitz acts in photoreceptor recruitment in the developing retina. Development (Cambridge, England) 1997;124:343–351. doi: 10.1242/dev.124.2.343. [DOI] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nature cell biology. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- White K, Tahaoglu E, Steller H. Science. Vol. 271. New York, NY: 1996. Cell killing by the Drosophila gene reaper; pp. 805–807. [DOI] [PubMed] [Google Scholar]
- Wing JP, Schreader BA, Yokokura T, Wang Y, Andrews PS, Huseinovic N, Dong CK, Ogdahl JL, Schwartz LM, White K, et al. Drosophila Morgue is an F box/ubiquitin conjugase domain protein important for grim-reaper mediated apoptosis. Nature cell biology. 2002;4:451–456. doi: 10.1038/ncb800. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Pattern formation in the Drosophila retina. II. NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Developmental cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Xu D, Woodfield SE, Lee TV, Fan Y, Antonio C, Bergmann A. Genetic control of programmed cell death (apoptosis) in Drosophila. Fly (Austin) 2009;3 doi: 10.4161/fly.3.1.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Baker NE. Role of the EGFR/Ras/Raf pathway in specification of photoreceptor cells in the Drosophila retina. Development (Cambridge, England) 2001;128:1183–1191. doi: 10.1242/dev.128.7.1183. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Science. Vol. 288. New York, NY: 2000. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli; pp. 874–877. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, Feldman RM, Clem RJ, Muller HA, Hay BA. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nature cell biology. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Yu SY, Yoo SJ, Yang L, Zapata C, Srinivasan A, Hay BA, Baker NE. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development (Cambridge, England) 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Developmental cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.