Abstract
Dysfunctions of the mitochondria and the ubiquitin–proteasome system, as well as generation of reactive oxygen species (ROS), are linked to many aging-related neurodegenerative disorders. However, the order of these events remains unclear. Here, we show that the initial impairment occurs in mitochondria under proteasome inhibition. Fluorescent redox probe measurements revealed that proteasome inhibition led to mitochondrial oxidation followed by cytosolic oxidation, which could be prevented by a mitochondrial-targeted antioxidant or antioxidative enzyme. These observations demonstrated that proteasome dysfunction causes damage to mitochondria, leading them to increase their ROS production and resulting in cytosolic oxidation. Moreover, several antioxidants found in foods prevented intracellular oxidation and improved cell survival by maintaining mitochondrial membrane potential and reducing mitochondrial ROS generation. However, these antioxidant treatments did not decrease the accumulation of protein aggregates caused by inhibition of the proteasome. These results suggested that antioxidative protection of mitochondria maintains cellular integrity, providing novel insights into the mechanisms of cell death caused by proteasome dysfunction.
Impairment of the ubiquitin–proteasome system (UPS), mitochondrial dysfunction, and generation of reactive oxygen species (ROS) have been strongly associated with cell death–mediated aging and the pathogenesis of neurodegenerative disorders, e.g., Alzheimer's and Parkinson's disease1,2,3. However, the order in which these events occur, as well as the intracellular mechanisms that connect proteasome and mitochondrial impairment, remain unclear.
The UPS is a major protein degradation pathway that is essential for cellular protein homeostasis4. The UPS eliminates misfolded or damaged proteins, which are selectively polyubiquitinated, recognized, and then degraded by proteasomes. Over the course of aging, proteasomal activity declines as a result of decreased expression and oxidative modification of proteasomal proteins5,6. Previous studies demonstrated that genetic mutations in UPS components result in several rare familial neurodegenerative diseases, providing a direct link between UPS dysfunction and neurodegeneration7,8. Accumulation of protein aggregates caused by proteasome dysfunction is considered to be a hallmark of aging and the most likely cause of neurodegenerative diseases. Proteasome inhibition leads to cell death; although the mechanism is incompletely understood, recent work has suggested that a shortage of free amino acids (normally produced by proteasomal degradation) is one possible cause of the cell death that occurs when the proteasome is inhibited9. Several reports have indicated that protein aggregates can generate ROS through interactions with redox-active metal ions10,11,12. Although moderate levels of ROS function as signals to promote cell proliferation and survival, excessive levels of ROS induce oxidative modifications of proteins, thereby promoting protein aggregation13,14. Compounding the problem, oxidative modifications of the proteasome reduce its stability and function, leaving it less able to remove protein aggregates15,16.
Mitochondria fulfill a number of essential cellular functions. Although their primary role is generation of ATP through oxidative phosphorylation, they also control programmed cell death, a key feature of neurodegenerative diseases3,17. Mitochondria are also the major site of intracellular ROS production, which occurs via electron leakage as a byproduct of ATP generation by oxidative phosphorylation. Mitochondrial antioxidative enzymes such as manganese SOD (MnSOD) and glutathione peroxidase detoxify ROS, but excess ROS generation can overwhelm the capacity of these defenses, leading to mitochondrial damage. Mitochondria themselves are susceptible to ROS: in particular, the components of the electron transport chain (ETC) and mitochondrial DNA (mtDNA) are vulnerable to oxidative damage3,18. mtDNA has a 10-fold higher mutation frequency relative to nuclear DNA19. Mitochondrial function is impaired over the course of aging20,21, and injured mitochondria produce higher levels of ROS via further electron leakage from the impaired ETC. This vicious cycle leads to age-related accumulation of ROS, inducing alterations in the mitochondrial genome and generating oxidized proteins that cause further mitochondrial dysfunction.
In this study, we investigated the changes that occur in the intracellular redox state following induction of proteasome dysfunction and assessed redox maintenance by redox modulators, by monitoring the intracellular redox state using our newly developed redox sensor probe, Redoxfluor22,23. Following proteasome inhibition, the cytosolic milieu was in an oxidized state, but the normal redox state could be restored by treating cells with two natural antioxidants, resveratrol and sesamin, or with mitochondrial-targeted antioxidants and antioxidative enzymes. Antioxidant treatments increased cell viability, but did not decrease the level of protein aggregation. Following proteasome inhibition, mitochondrial redox state was oxidized (concomitant with the mitochondrial accumulation of ubiquitinated proteins) before similar changes occurred in the cytosol. Taken together, our results show that mitochondria are the primary initial sites of oxidative damage following proteasome dysfunction. Furthermore, antioxidative protection of mitochondria maintained cellular integrity even in the presence of protein aggregates caused by proteasome dysfunction.
Results
Inhibition of proteasome-mediated protein degradation leads to intracellular oxidation
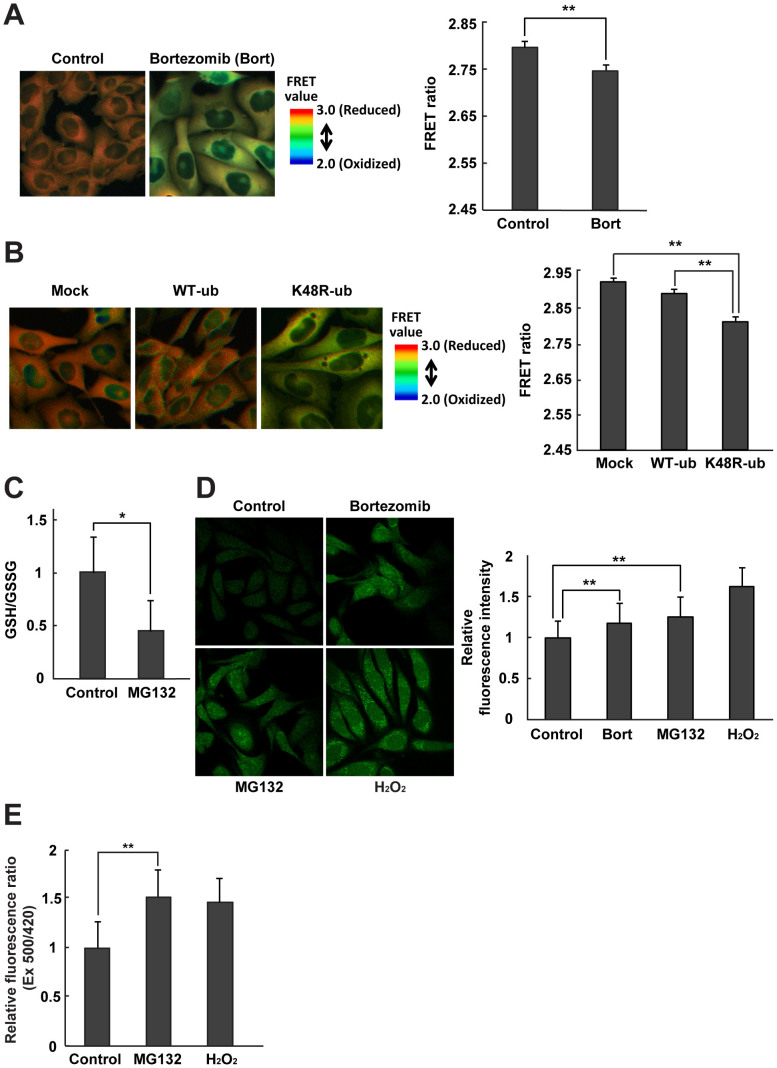
In order to assess the intracellular redox state in cells after proteasome-inhibition, we used Chinese Hamster Ovary (CHO) cells stably expressing the redox-sensing protein, Redoxfluor22. Redoxfluor possesses tandem redox-sensing regions derived from the yeast oxidative stress–responsive transcription factor Yap1p, flanked by cerulean (CYP) and citrine (YFP), and exhibits fluorescence resonance energy transfer (FRET) corresponding to its redox-dependent structural change22. Redoxfluor directly interacts with ROS and glutathione, and efficiently reacts with thioredoxin in the presence of peroxiredoxin (Tsa1p)22,24; consequently, it displays the integrated redox state as a FRET ratio. Visualization of the FRET ratio under control conditions revealed orange-colored areas in the cytosolic region (Fig. 1A, left panel). By contrast, treatment with the proteasome inhibitor bortezomib for 8 h changed the FRET ratio color from orange to yellow, indicating oxidation of the cytosolic redox state. Bortezomib also dramatically increased the level of polyubiquitinated proteins relative to the control condition (Supplementary Fig. S1). Quantitation of the FRET ratios confirmed the statistical significance of the result (Fig. 1A, right panel).
Figure 1. Proteasome and ubiquitination inhibition leads to cellular oxidation.
FRET ratio images of CHO cells expressing Redoxfluor are shown (left panels), and the FRET ratio is quantitated (right panels), (A) after 8 h treatment with 1 μM bortezomib (bort), and (B) 24 h after transfection with HA-WT ubiquitin or K48R ubiquitin. (C) The ratio of GSH/GSSG after 8 h treatment with 10 μM MG132. (D) Fluorescence images (left panel) and quantitation (right panel) of relative fluorescence intensity representing cytosolic ROS detection by CM-H2DCFDA and (E) cytosolic H2O2–dependent fluorescence ratio of Hyper after 8 h treatment with the proteasome inhibitor bortezomib or MG132. H2O2 was used as a positive control. Quantitated values are shown as means ± s.e.m. of three independent measurements. **: p < 0.01, *: p < 0.05.
Ubiquitination is one of the crucial steps of UPS, in which proteins targeted for proteolysis are tagged with ubiquitin, leading to polyubiquitination. Among the seven lysine residues in ubiquitin, Lys48 plays a vital role in polyubiquitination, and Lys48-linked polyubiquitination is a major signal for targeting of proteins to the proteasome25. Therefore, mutation of the Lys48 residue to arginine (K48R) interferes with the polyubiquitination of damaged proteins and their proteasomal targeting. Overexpression of K48R HA-ubiquitin in CHO cells lowered the FRET ratio of Redoxfluor, indicating that this mutation had led to an oxidized redox state by inhibiting K48-linked polyubiquitination. By contrast, overexpression of HA-ubiquitin did not affect the FRET ratio (Fig. 1B).
To confirm the intracellular oxidization detected by Redoxfluor, we examined the effect of proteasome inhibition by MG132 on the glutathione redox state. The GSH/GSSG ratio was reduced after treatment with the drug (Fig. 1C). Similarly, the intracellular fluorescence intensity of a ROS probe, CM-H2DCFDA, increased in the presence of bortezomib or MG132, although the increase was slightly lower than that observed upon the addition of H2O2 (Fig. 1D). It remains controversial whether this probe can be used to measure ROS production in cells26. Therefore, we also used the hydrogen peroxide sensor HyPer to detect the increase of intracellular hydrogen peroxide under proteasome inhibition (Fig. 1E). Cytosolic HyPer exhibited a substantial increase in the fluorescent ratio (Ex 500/420), suggesting that MG132 treatment increased the level of H2O2 level. These results demonstrated that impairment of the UPS led to intracellular oxidation, reflected in both the GSH/GSSG ratio and the levels of ROS (including H2O2).
Antioxidants, as redox modulators, counteract the intracellular oxidation caused by proteasome inhibition
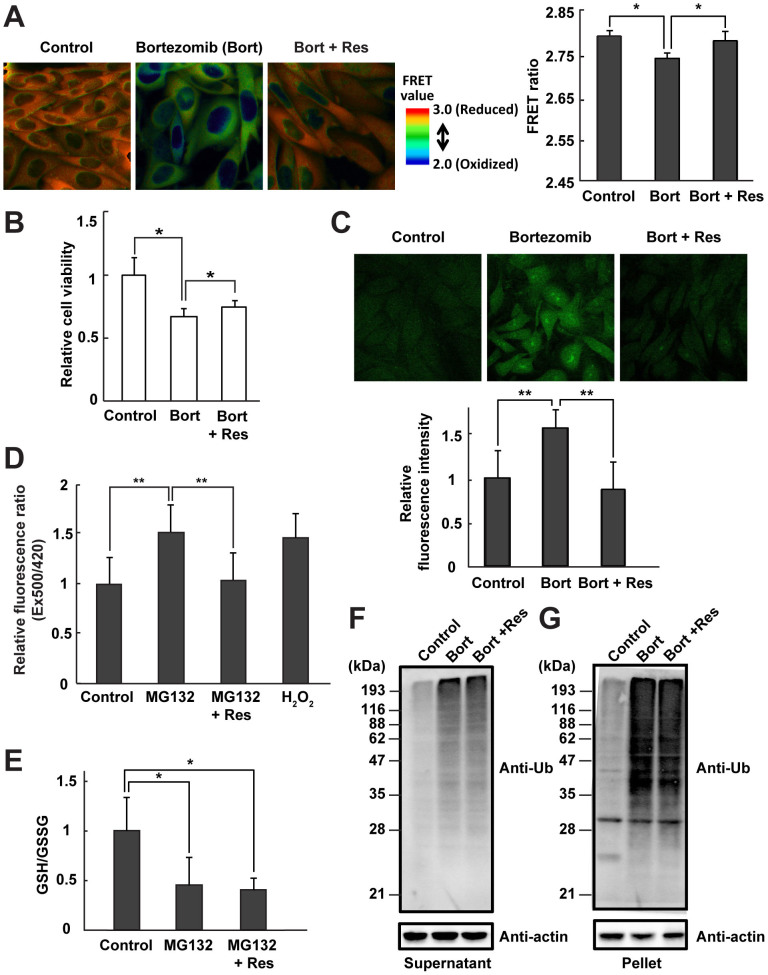
Next, we examined the redox-modulating effect of the antioxidant resveratrol under proteasome inhibition. CHO cells that were simultaneously treated with bortezomib and resveratrol exhibited the same cytosolic orange FRET image observed in control cells (Fig. 2A, left panel). The quantitated FRET ratio of cells treated with bortezomib and resveratrol was higher than that of cells treated with the proteasome inhibitor alone (Fig. 2A, right panel), indicating that resveratrol promoted recovery from an oxidized state to a normal cytosolic redox state. In addition, antioxidant treatment partially but significantly improved the cell viability of both bortezomib-treated and MG132-treated cells (Fig. 2B and Supplementary Fig. S2A).
Figure 2. Resveratrol functions as a redox modulator under proteasome inhibition.
(A) FRET analysis of CHO cells expressing Redoxfluor. Fluorescence images are shown (left panel) and the relative fluorescence intensity is quantitated (right panel). (B) Relative cell viability as determined by the MTT assay. (C) Cytosolic ROS detection using CM-H2DCFDA. Fluorescence images are shown (upper panel), and fluorescence levels are quantitated (lower panel). (D) Cytosolic H2O2 detection using Hyper. H2O2 (200 μM) was used as a positive control. (E) The ratio of GSH/GSSG, and (F, G) immunoblotting of polyubiquitinated proteins with anti-ubiquitin in 1% NP-40–soluble (F) and -insoluble (G) CHO cell fractions. All treatments were performed for 8 h: DMSO (control); 1 μM bortezomib (bort); 10 μM MG132; or simultaneous treatment with the proteasome inhibitors, bortezomib or MG132, and 10 μM resveratrol (Res). Quantitated values are shown as the means ± s.e.m. of three independent measurements. **: p < 0.01, *: p < 0.05.
In proteasome inhibitor–treated cells, intracellular production of ROS, including H2O2, as detected by CM-H2DCFDA or HyPer, was also suppressed by the addition of resveratrol (Fig. 2C and D and Supplementary Fig. S2B), in accordance with the improved viability of these cells. However, resveratrol did not remedy the oxidized glutathione redox state (GSH/GSSG) (Fig. 2E), suggesting that intracellular glutathione contributed to attenuation of intracellular ROS levels even in the presence of resveratrol. Furthermore, resveratrol did not decrease the accumulation of polyubiquitinated proteins in the detergent-soluble and -insoluble fractions including protein aggregates (Fig. 2F and G). Although Redoxfluor can also detect the glutathione redox state22, it is more sensitive to the ROS level than the glutathione redox state in the cytosol, resulting in an apparent discrepancy in assessments of the redox state obtained using glutathione redox state vs. Redoxfluor. Together, these data indicated that resveratrol can function as a redox modulator to prevent ROS production, maintain intracellular redox homeostasis, and promote cell viability in the presence of proteasome inhibitors.
Another antioxidant, sesamin, exhibited similar effects on redox maintenance and cell viability under proteasome inhibition (Supplementary Fig. S3A and B). Although sesamin was less effective than resveratrol in reducing cytosolic ROS levels in bortezomib-treated cells, the reduction was statistically significant (Fig. 2C and Supplementary Fig. S3C). By contrast to resveratrol and sesamin, ascorbic acid did not serve as a redox modulator under proteasome inhibition (Supplementary Fig. S4).
Proteasome inhibition causes a decrease in mitochondrial membrane potential and an increase in mitochondrial ROS production
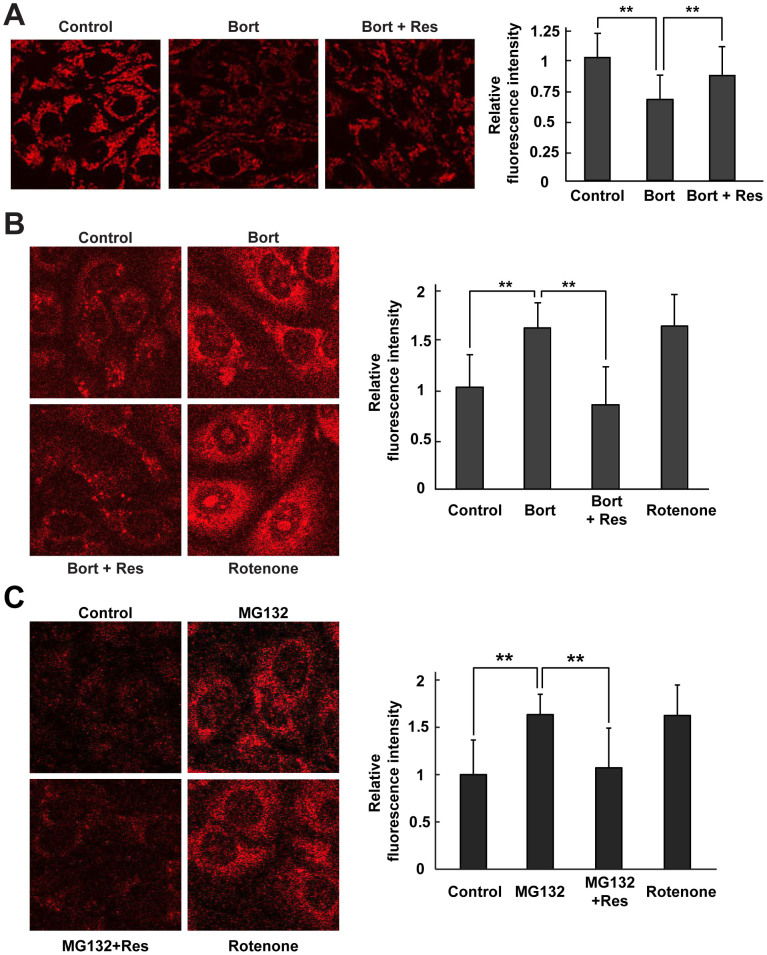
The results described above indicated that proteasome inhibition caused cytosolic oxidation (as detected by Redoxfluor) and an increase in cytosolic and nuclear ROS levels (as detected by HyPer). Because mitochondria are considered to be the major intracellular source of ROS18, we investigated whether the observed cytosolic oxidation was due to impaired mitochondria. Mitochondrial membrane potential, a consequence of the electrochemical proton gradient maintained for the purpose of ATP synthesis, is an important indicator of functional mitochondria. Therefore, we monitored mitochondrial membrane potential in CHO cells under proteasome inhibition using a mitochondrial-targeted probe, Mitotracker Red CMXRos, which accumulates in mitochondria depending on its membrane potential. In cells treated with the proteasome inhibitor alone, the fluorescence intensity of the probe decreased, indicating a loss in mitochondrial membrane potential (Fig. 3A). However, simultaneous treatment with the proteasome inhibitor and resveratrol prevented the reduction in mitochondrial membrane potential.
Figure 3. Resveratrol protects mitochondrial membrane integrity and function in cells under proteasome inhibition.
(A) Mitochondrial membrane potential was detected using the mitochondrial membrane potential (Δψm)-sensitive fluorochrome Mitotracker® Red CMXRos. (B, C) Mitochondrial ROS were detected using the MitoSOX Red probe 8 h after treatment with bortezomib alone or bortezomib + resveratrol (B) or with MG132 alone or MG132 + resveratrol (C). Fluorescence images are shown (left panels), and the relative fluorescence intensity is quantitated (right panels). Rotenone was used as positive control. Quantitated values are shown as means ± s.e.m. of three independent measurements. **: p < 0.01.
Next, we measured mitochondrial ROS production under proteasome inhibition using MitoSOX Red, a fluorogenic superoxide-detecting dye that specifically targets mitochondria. Proteasome inhibition by bortezomib or MG132 led to increased production of mitochondrial superoxide anion in CHO cells; a similar effect was also caused by addition of rotenone, which increases mitochondrial superoxide anion production by inhibiting complex I of the ETC (Fig. 3B and C). Because these treatments resulted in loss of mitochondrial membrane potential, as shown in Fig. 3A, we interpret the elevated concentration of mitochondrial superoxide anion under proteasome inhibition as an indication of electron leaks from the ETC. In bortezomib-treated cells, resveratrol effectively prevented an increase in mitochondrial production of superoxide anion. A similar effect was also detected using sesamin (Supplementary Fig. S3D and E). Thus, resveratrol and sesamin reversed the effects of proteasome inhibition–mediated intracellular oxidation by maintaining membrane potential and preventing mitochondrial ROS production.
Proteasome inhibition induces mitochondrial aberration prior to cytosolic oxidation
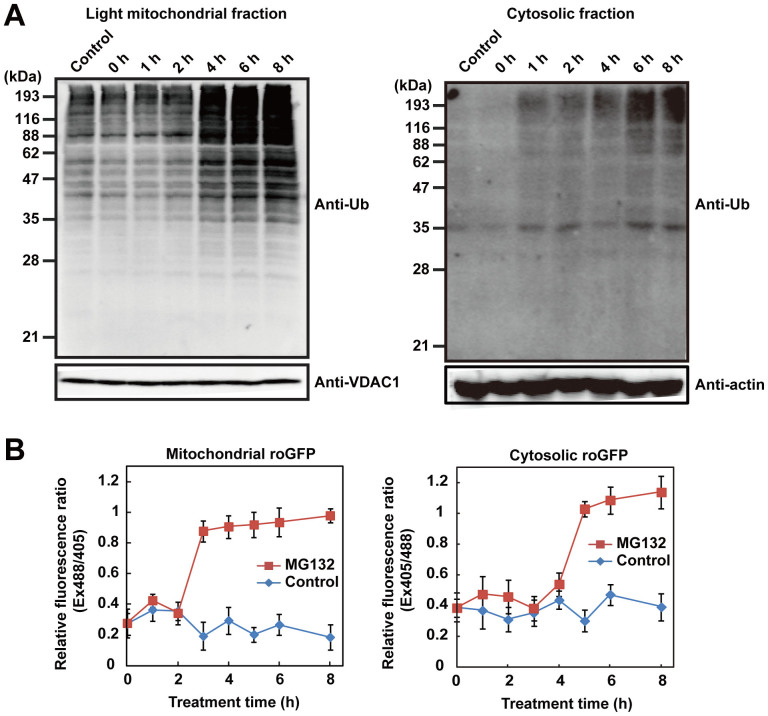
The results described so far demonstrate that proteasome inhibition causes mitochondrial damage, reflected by a loss of membrane potential and mitochondrial ROS generation. To uncover the relationship between mitochondrial damage and proteasome inhibition, we examined mitochondrial accumulation of polyubiquitinated proteins. Mitochondrial proteins that undergo degradation by the UPS are released from the outer mitochondrial membrane into the cytosol27,28,29, and proteasome inhibition causes accumulation of ubiquitinated proteins in the outer mitochondrial membrane30. Therefore, we monitored accumulation of polyubiquitinated protein in the light mitochondrial fraction (LMF), which includes the outer mitochondrial membrane, following proteasome inhibition. Polyubiquitinated proteins in the LMF were sharply elevated 4 h after treatment with MG132, and gradually increased further thereafter (Fig. 4A, left panel). We validated the subcellular fractionation by detecting compartment-specific marker proteins. The LMF was considerably enriched in an outer mitochondrial membrane marker protein, VDAC1, but did not contain a peroxisome-localized catalase. However, low levels of ER-localized ERp72 and cytosolic GAPDH were present as contaminants in the LMF (Supplementary Fig. S6A). In the cytosolic fraction, polyubiquitinated proteins were elevated at 6 h after MG132 treatment, 2 h later than these proteins accumulated in the LMF (Fig. 4A, right panel). Furthermore, treatment with proteasome inhibitors increased the proportion of HA-tagged ubiquitin co-localized on mitochondria (detected using MitoTracker Deep Red), indicating that polyubiquitinated proteins accumulated in the mitochondria (Supplementary Fig. S6B). Under normal conditions, HA-tagged ubiquitin was diffusely distributed throughout the cell, although the concentration appeared to be somewhat elevated in the nucleus.
Figure 4. Proteasome inhibition induces mitochondrial oxidation and ubiquitinated protein accumulation before cytosol is oxidized.
(A) Time course of accumulation of polyubiquitinated proteins in the light mitochondria fraction (LMF) and (B) time-lapse measurement of the redox state in the mitochondria (left panels) and cytosol (right panels) using mitochondria or cytosolic-targeted roGFP after treatment with DMSO (control) or 10 μM MG132. VDAC1, an outer membrane mitochondrial protein, was used as a mitochondrial loading control, and actin was used as a cytosolic loading control.
Next, we monitored the mitochondrial redox state following proteasome inhibition using mitochondria-targeted roGFPs because we could not prepare a functional construct of mitochondria-targeted Redoxfluor so far. We assessed the redox state by ratiometric measurement of roGFP fluorescence intensity excited at 405 and at 488 nm31,32. In this assay, a higher ratio (405/488 in roGFP1/the cytosolic roGFP, 488/405 in roGFP2/the mitochondrial one) indicated an oxidized state, whereas a lower ratio indicated a reduced state. The mitochondrial redox state was oxidized 3–4 h after MG132-treatment (Fig. 4B, left panel), coincident with the timing of mitochondrial accumulation of polyubiquitinated proteins (Fig. 4A). By contrast, the cytosolic redox state assessed using cytosolic roGFP was oxidized 5–6 h post-treatment, also coincident with the timing of cytosolic accumulation of polyubiquitinated proteins (Fig. 4A), which as noted above occurred later than the mitochondrial oxidation (Fig. 4B, right panel). Neither cytosolic nor mitochondrial redox states changed in the absence of proteasome inhibition. These data indicated that proteasome inhibition caused mitochondrial oxidation and accumulation of polyubiquitinated proteins before similar changes occurred in the cytosol.
Mitochondrial ROS are responsible for the cytosolic oxidation and cell death observed after proteasome inhibition
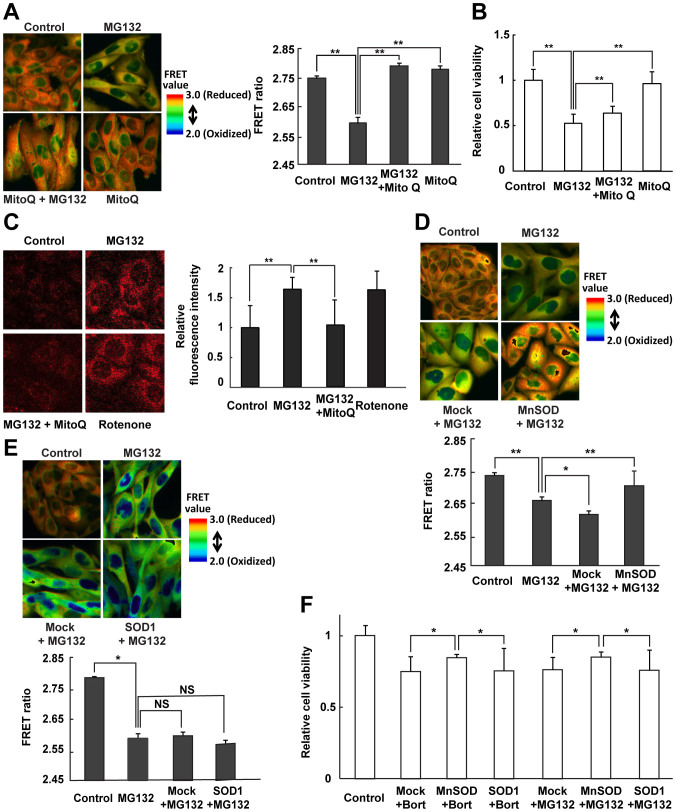
Proteasome inhibition induced mitochondrial oxidation prior to cytosolic oxidation, whereas the antioxidants derived from foods suppressed cytosolic oxidation and cell death by maintaining mitochondrial integrity and preventing ROS generation (Figs. 2–4). Thus, resveratrol appears to counter mitochondrial ROS production by acting as an ROS scavenger, thereby maintaining the cytosolic redox state. Resveratrol has been reported to function not only as a direct ROS scavenger, but also as an inducer of antioxidant gene expression33. To ascertain the mechanism by which resveratrol blocks the deleterious consequences of proteasome inhibition, we examined the effect of MitoQ, a mitochondrial-targeted antioxidant in which the ubiquinol moiety of coenzyme Q is conjugated to a lipophilic triphenylphosphonium that blocks mitochondrial ROS generation and preserves mitochondrial function34, in proteasome-inhibited cells. In CHO cells expressing Redoxfluor treated with MG132 exhibited a yellow FRET image, similar to that observed in cells treated with bortezomib (Fig. 5A and Fig. 1A). Although MitoQ treatment alone did not affect the FRET ratio of Redoxfluor in CHO cells, MitoQ maintained more reduced conditions in MG132-treated cells. Similar to resveratrol or sesamin treatment, MitoQ treatment also prevented mitochondrial ROS generation, as detected by MitoSOX, and improved the cell viability of MG132-treated cells as effectively as resveratrol (Fig. 5B and C). These data indicated that, in MitoQ-treated cells, cell viability and the cytosolic redox state recovered after proteasome inhibition because mitochondrial ROS generation was prevented.
Figure 5. Antioxidative protection of mitochondria improves cytosolic oxidation under proteasome inhibition.
(A, D, E) FRET ratio images of CHO cells expressing Redoxfluor (left panel in A, upper panels in D, E) and quantitation of the FRET ratio (right panel in A, lower panels in D, E) in cells under proteasome inhibition 8 h after treatment with 10 μM MG132, or after simultaneous treatment with 10 μM MG132 and 1 μM MitoQ (A), and 24 h after transfection with (D) MnSOD and (E) SOD1. (C) Mitochondrial ROS detection using the MitoSOX Red probe. Fluorescence images are shown (left panel), and the relative fluorescence intensity is quantitated (right panel). (B, F) Relative cell viability as determined by the MTT assay. Assay was performed at 8 h after treatment with MG132 alone or MG132 + MitoQ (B) at 24 h after the transfection (F). Quantitated values are shown as means ± s.e.m. of three independent measurements. **: p < 0.01, *: p < 0.05, NS: not significant.
Next, we examined the effect of the expression of mitochondrial or cytosolic SOD on redox state under proteasome inhibition. MnSOD, which is exclusively localized in mitochondrial matrix, provides the primary mitochondrial antioxidative defense against ROS35. MnSOD overexpression increased the FRET ratio of Redoxfluor in MG132-treated CHO cells relative to that of mock-transfected cells (Fig. 5D). Overexpressed MnSOD co-localized with the specific mitochondrial marker MitoTracker Deep Red (Supplementary Fig. S5B). By contrast, overexpression of SOD1, which is mainly localized in the cytosol, had no significant effect on the FRET ratio of Redoxfluor in MG132-treated CHO cells (Fig. 5E). The expression of transfected SOD1 was confirmed by immunostaining (Supplementary Fig. S5D). Similar to redox modulators such as resveratrol and MitoQ, overexpressed MnSOD restored cell viability, whereas overexpressed SOD1 did not (Fig. 5F). These data suggested that suppression of superoxide anions in mitochondria, not in the cytosol, maintains a reduced cytosolic redox state under proteasome inhibition. Taken together, these data demonstrate that proteasome inhibition caused mitochondrial ROS generation followed by cytosolic oxidation, and that suppression of ROS generation in mitochondria rescued both the cytosolic redox state and cell viability.
Discussion
We used a newly developed redox sensor, Redoxfluor22, which was stably expressed in CHO cells, to assess the intracellular redox state. Redoxfluor integrates multiple different redox indicators: ROS, glutathione, and thioredoxin22,23,24. In a previous study, we showed that a peroxisome-deficient cell line had a reduced redox state relative to the wild-type cell line22. In this study, we screened various conditions and chemicals that changed the intracellular redox state and affected cellular viability. Many previous studies have shown that proteasome inhibition causes ROS generation and oxidization of the intracellular glutathione redox state36,37,38,39. In addition, disturbance of the normal polyubiquitination machinery causes abnormal protein degradation associated with increased oxidative damage40,41. Redoxfluor enabled us to visualize oxidation in the cytosol caused by inhibition of either the proteasome or K48-linked polyubiquitination (Fig. 1A and B). Furthermore, several redox modulators could return the oxidation state to normal and could substantially rescue cellular viability, although the glutathione redox state remained oxidized (Fig. 2 and Supplementary Fig. S2). The physiological redox state, as represented by the FRET ratio of Redoxfluor, correlated positively with cell viability.
We revealed the sequential events following inhibition of the UPS (Fig. 6). Mitochondrial impairment occurred soon after proteasome inhibition, as judged by accumulation of polyubiquitinated proteins in mitochondria, resulting in loss of mitochondrial membrane potential and the generation of ROS, e.g., superoxide anion, from the ETC. Mitochondrial superoxide anion is swiftly converted into H2O2. Because the cytosolic redox capacity is larger than the mitochondrial one, H2O2 diffused into the cytosol were eliminated by cytosolic antioxidant systems until the ROS levels generated by mitochondria exceeded the cytosolic redox defense. After that, mitochondria-generated ROS caused oxidation in the cytosol, concomitant with the decrease in cell viability. Thus, the delay in cytosolic oxidation observed in Fig. 4B might be due to the high capacity of the cytosolic antioxidant system. The sequence of events that occurred after proteasome inhibition is supported by the following observations: i) Time-lapse detection of local redox changes and ubiquitinated protein accumulation revealed that the mitochondrial redox state was oxidized concomitant with mitochondrial accumulation of polyubiquitinated proteins, prior to similar changes in the cytosol (Fig. 4). ii) Mitochondrial-specific antioxidative treatments (MitoQ treatment or MnSOD overexpression) suppressed mitochondrial ROS generation and rescued the cytosolic redox state and cell viability (Fig. 5 and Supplementary Fig. S5C). However, mitochondrial antioxidative treatments resulted in only partial rescue of cell viability, suggesting that proteasome inhibition causes cell death by alternative mechanisms, e.g., failure of amino-acid recycling9.
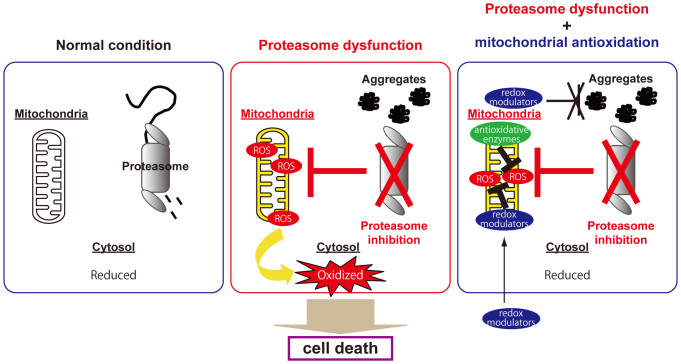
Figure 6. Mitochondrial integrity is essential for preventing cytosolic oxidative stress caused by proteasome inhibition.
The cytosol is normally maintained in a reducing state (left panel). Proteasomal dysfunction first damages mitochondrial integrity; then, the resulting ROS produced by impaired mitochondria lead to cytosolic oxidation, and eventually to cell death (middle panel). Reversal of mitochondrial oxidation by redox modulators, e.g., resveratrol and antioxidative enzymes, prevents cytosolic oxidation and eventual cell death without reducing the level of protein aggregation in cells under proteasome inhibition (right panel).
Proteasome inhibition causes the accumulation of polyubiquitinated protein aggregates, which is associated with aging and neurodegenerative diseases1,6,42. In neuronal mitochondria, proteasome inhibition induces rapid accumulation of polyubiquitinated proteins30, apparently resulting in mitochondrial impairment. We found that the level of polyubiquitinated proteins in the LMF increased suddenly, concomitant with mitochondrial oxidation, 3–4 h after addition of the proteasome inhibitor; the increase of polyubiquitinated proteins and oxidation were observed in the cytosol ~2 h later (Fig. 4A). Antioxidative treatments targeting the mitochondria diminished mitochondrial ROS production, restored the cytosolic redox state, and rescued cell viability; however, these antioxidative treatments did not reduce the level of polyubiquitinated proteins (Figs. 2, 3 and 5). Thus, under proteasome dysfunction, antioxidative protection of mitochondria was sufficient to prevent cell death even in the presence of accumulated polyubiquitinated proteins (Fig. 6).
Using proteasome-inhibited CHO-cells stably expressing Redoxfluor, we screened various chemicals and treatment strategies for the ability to return the cytosolic oxidative state to normal levels and restore cell viability. Among the compounds that worked were resveratrol and sesamin, which are found in many natural products (Fig. 2 and Supplementary Fig. S3). Natural compounds such as these have attracted a great deal of attention for their potential ability to prevent neurodegenerative diseases and aging. Resveratrol and sesamin induce the expression of antioxidant enzymes via the activation of the oxidative stress-responding transcription factor Nrf233,43,44,45,46,47, as well as by inducing molecular chaperones48, thereby preventing the accumulation of protein aggregates. However, our results showed that resveratrol did not attenuate formation of protein aggregates, but did restore the cytosolic redox state and substantially improved cell viability (Fig. 2). Although the mechanism by which these natural compounds function as redox modulators needs to be clarified in more detail, we suspect that resveratrol primary acts as a ROS scavenger in mitochondria. Our data suggest that in general, these natural compounds act as redox modulators in cell death caused by UPS perturbation, which is related to aging and neurodegenerative diseases.
In conclusion, mitochondrial dysfunction represents the onset of critical events that happen after proteasome impairment, leading to intracellular oxidative stress and eventually to cell death. Using the physiological redox sensor Redoxfluor, we found that resveratrol and sesamin function as redox modulators to restore not only the intracellular redox state but also cell viability after proteasome inhibition (Fig. 2 and Supplementary Fig. S2). Meanwhile, antioxidative protection of mitochondria, including treatment with these natural redox modulators, maintained cellular integrity under proteasome inhibition in spite of the accumulation of protein aggregates (Fig. 6). These novel insights into the mechanism of cell death caused by proteasome dysfunction will lead to further advancements in understanding the molecular mechanisms governing aging and neurodegenerative disorders, some of which are caused by proteasome dysfunction, in relationship to the subcellular redox state.
Methods
Cell culture, construction of plasmids, and transfection
CHO cells were grown in Ham's F-12 medium supplemented with 10% fetal bovine serum and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) under humidified air containing 7% CO2 at 37°C. MnSOD and SOD1 expression plasmids were constructed as follows: Total RNA was extracted from HeLa cells using an RNeasy Mini Kit (Qiagen, Tokyo, Japan) and reverse-transcribed using ReverTraAce enzyme (TOYOBO, Osaka, Japan). The resulting cDNA was used as a template for the amplification of MnSOD (forward: 5′-ATGTTGAGCCGGGCAGT-G-3′, reverse: 5′-TTACTTTTTGCAAGCCATGTATCTTTCAGTTACAT-3′) and SOD1 (forward: 5′-CCGGAATTCGCCACCATGTACCCATACGATGTTCCAGATTACGCTGCGACGA AGGCCGTG-3′, reverse: 5′- ATTTGCGGCCGCTTATTGGGCGATCCCAATTACA CC-3′). The PCR-amplified products were inserted into pCDNA 3.1. HA (hemagglutinin)-tagged wild-type and K48R ubiquitin expression plasmids, as well as cytosolic roGFP1 and mitochondrial-targeted roGFP2 were kindly provided by Dr. Kazuhiro Nagata (Kyoto Sangyo University, Japan)49 and Dr. S. James Lemington (Oregon University)31, respectively. The plasmid encoding cytosolic HyPer (pHyPer-cyto) was obtained from Evrogen. Cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen, Tokyo, Japan).
Reagents and antibodies
Bortezomib was purchased from Selleck Chemicals. MG132, resveratrol, and sesamin were obtained from Wako Pure Chemical (Osaka, Japan). MitoQ was a kind gift from Dr. Michael P. Murphy (MRC Mitochondrial Biology Unit, Cambridge, UK). Anti-ubiquitin (Santa Cruz Biotechnology, Dallas, TX), anti-MnSOD (Life Span BioSciences, Seattle, WA), anti-VDAC1 and anti-catalase (both from Abcam, Cambridge, MA), anti-HA (Covance, Denver, PA), anti-β-actin (Sigma-Aldrich, Tokyo, Japan), anti-GAPDH (Cell signaling technology, Beverly, MA), and anti-ERp72 (Enzo Life Sciences, Farmingdale, NY) antibodies were used for immunostaining.
FRET analysis using Redoxfluor, measurement of cytosolic hydrogen peroxide using HyPer, and measurement of redox state using roGFP
FRET measurements were performed as previously described22. Fluorescence images of Redoxfluor, HyPer, and roGFP were acquired using an IX70 fluorescence microscope (Olympus, Tokyo, Japan) equipped with a CoolSNAP HQ2 CCD camera (Photometrics, Tucson, AZ). HyPer and roGFP fluorescence was excited using the XF1076 filter (400AF30, Omega Optical, Brattleboro, VT) for excitation of the peak around 420 nm (400 nm for roGFP), and the U-MNIBA cube (BP 470–490, Olympus) for excitation of the peak around 500 nm (480 nm for roGFP); the corresponding YFP and GFP emissions were observed using the U-MNIBA cube (BP 515–550), respectively. Acquired images were analyzed using the MetaMorph imaging software (Universal Imaging Corp., Downingtown, PA).
Quantitation of intracellular glutathione by LC-MS
Intracellular glutathione measurements were performed as previously described24. Briefly, a chilled methanol extraction of cells was freeze-dried and resuspended in 1% acetonitrile, and then applied to a Hydrosphere C18 column (YMC) on a Prominence nano HPLC system (Shimadzu, Kyoto, Japan) in line with a 4000 QTRAP mass spectrometer (AB Sciex Instruments, Foster City, CA).
MTT assay
Cells were grown in a 96-well microplate for 24 h, and then incubated with 5 mg/ml MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) for 4 h at 37°C (Nacalai Tesque, Kyoto, Japan). Next, 100 μl of solubilization solution (isopropanol with 0.04 mol/l HCl) was added to dissolve precipitated formazans. The plate was sealed and incubated overnight in a humidified chamber at 37°C; the next day, absorbance was read at 570 nm. Relative cell numbers were expressed as a percentage of untreated controls.
Fluorescence microscopy
All confocal fluorescence images were obtained using an LSM 510 META confocal microscope (Carl Zeiss, Tokyo, Japan). For detection of mitochondrial membrane potential and ROS, cells were labeled at 37°C for 10 min (30 min for CM-H2DCFDA (5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester)) with the following fluorescent probes: 0.4 μM Mitotracker® Red CMXRos (chloromethyl-X-rosamine), 5 μM MitoSOX Red probe, or 10 μM CM-H2DCFDA. After the incubation, these fluorescent probes were washed out with Hank's balanced salt solution. ImageJ was used to quantitate fluorescence intensity. For immunocytochemistry, cells overexpressing MnSOD were stained with Mitotracker Deep Red for 30 min in the dark, and then fixed with 4% paraformaldehyde at room temperature (r.t.) for 20 min. Cells were permeabilized with 0.2% Triton X-100 for 10 min at r.t., and then incubated in blocking buffer (1% glycerol, 1% bovine serum albumin, 1% goat serum, and 0.2% Triton X-100) for 1 h. Cells were incubated with a rabbit anti-rat MnSOD antibody for 2 h, and then with an Alexa Fluor 488–conjugated goat anti-rabbit IgG secondary antibody for 1 h. All fluorescent probes were obtained from Invitrogen.
Mitochondrial fractionation
Cells were resuspended in homogenization medium (HM) (0.25 M sucrose, 1 mM EDTA, 20 mM HEPES-NaOH [pH 7.4]) and homogenized by gently passing the sample 10 times through a 25-gauge needle. The homogenate was centrifuged at 1,000 × g and then the carefully removed supernatant was centrifuged at 3,000 × g. Crude light mitochondria fraction (LMF) was prepared by centrifuging the resultant supernatant at 17,000 × g for 15 min. Crude LMF was further purified by 19–27% continuous ioxadiol density-gradient ultracentrifugation in a swinging-bucket rotor at approximately 167,000 × g for 2 h at 4°C.
Author Contributions
M.O., J.H. and Y.S. designed the experiments. S.M. and M.T. performed the experiments and analyzed the data. S.M., J.H. and Y.S. wrote the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr. Kazuhiro Nagata (Kyoto Sangyo University) and Dr. S. James Lemington (Oregon University) for providing us with plasmids, and Dr. Michael P. Murphy (MRC, Mitochondrial Biology Unit, Cambridge, UK) for his kind gift of MitoQ. This work was supported by a Grant-in-Aid for Exploratory Research (23651235) and a Grant-in-Aid for Scientific Research (B) (26292052) from JSPS and the Uehara Memorial Foundation to Y.S.; a grant from the Takeda Science Foundation and a Grant-in-Aid for Scientific Research (C) (26440050) to J.H.; a Grant-in-Aid for Young Scientists (B) (24780100) from JSPS to M.O.; and a scholarship from MEXT to S.M.
References
- Keller J. N., Hanni K. B. & Markesbery W. R. Impaired proteasome function in Alzheimer's disease. J Neurochem 75, 436–439 (2000). [DOI] [PubMed] [Google Scholar]
- Balaban R. S., Nemoto S. & Finkel T. Mitochondria, oxidants, and aging. Cell 120, 483–495 (2005). [DOI] [PubMed] [Google Scholar]
- Lin M. T. & Beal M. F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–795 (2006). [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Orian A. & Schwartz A. L. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22, 442–451 (2000). [DOI] [PubMed] [Google Scholar]
- Keller J. N., Huang F. F. & Markesbery W. R. Decreased levels of proteasome activity and proteasome expression in aging spinal cord. Neuroscience 98, 149–156 (2000). [DOI] [PubMed] [Google Scholar]
- Carrard G., Bulteau A. L., Petropoulos I. & Friguet B. Impairment of proteasome structure and function in aging. Int J Biochem Cell Biol 34, 1461–1474 (2002). [DOI] [PubMed] [Google Scholar]
- Kitada T. et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998). [DOI] [PubMed] [Google Scholar]
- Lee J. T., Wheeler T. C., Li L. & Chin L. S. Ubiquitination of alpha-synuclein by Siah-1 promotes alpha-synuclein aggregation and apoptotic cell death. Hum Mol Genet 17, 906–917 (2008). [DOI] [PubMed] [Google Scholar]
- Suraweera A., Munch C., Hanssum A. & Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell 48, 242–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabner B. J., El-Agnaf O. M., German M. J., Fullwood N. J. & Allsop D. Protein aggregation, metals and oxidative stress in neurodegenerative diseases. Biochem Soc Trans 33, 1082–1086 (2005). [DOI] [PubMed] [Google Scholar]
- Allsop D., Mayes J., Moore S., Masad A. & Tabner B. J. Metal-dependent generation of reactive oxygen species from amyloid proteins implicated in neurodegenerative disease. Biochem Soc Trans 36, 1293–1298 (2008). [DOI] [PubMed] [Google Scholar]
- Jomova K., Vondrakova D., Lawson M. & Valko M. Metals, oxidative stress and neurodegenerative disorders. Mol Cell Biochem 345, 91–104 (2010). [DOI] [PubMed] [Google Scholar]
- Rhee S. G. Cell signaling. H2O2, a necessary evil for cell signaling. Science 312, 1882–1883 (2006). [DOI] [PubMed] [Google Scholar]
- Avery S. V. Molecular targets of oxidative stress. Biochem J 434, 201–210 (2011). [DOI] [PubMed] [Google Scholar]
- Droge W. Free radicals in the physiological control of cell function. Physiol Rev 82, 47–95 (2002). [DOI] [PubMed] [Google Scholar]
- Martindale J. L. & Holbrook N. J. Cellular response to oxidative stress: signaling for suicide and survival. J Cell Physiol 192, 1–15 (2002). [DOI] [PubMed] [Google Scholar]
- Danial N. N. & Korsmeyer S. J. Cell death: critical control points. Cell 116, 205–219 (2004). [DOI] [PubMed] [Google Scholar]
- Sohal R. S., Sohal B. H. & Orr W. C. Mitochondrial superoxide and hydrogen peroxide generation, protein oxidative damage, and longevity in different species of flies. Free Radic Biol Med 19, 499–504 (1995). [DOI] [PubMed] [Google Scholar]
- Mecocci P. et al. Oxidative damage to mitochondrial DNA shows marked age-dependent increases in human brain. Ann Neurol 34, 609–616 (1993). [DOI] [PubMed] [Google Scholar]
- Turner C. & Schapira A. H. Mitochondrial dysfunction in neurodegenerative disorders and ageing. Adv Exp Med Biol 487, 229–251 (2001). [DOI] [PubMed] [Google Scholar]
- Trifunovic A. & Larsson N. G. Mitochondrial dysfunction as a cause of ageing. J Intern Med 263, 167–178 (2008). [DOI] [PubMed] [Google Scholar]
- Yano T. et al. A novel fluorescent sensor protein for visualization of redox states in the cytoplasm and in peroxisomes. Mol Cell Biol 30, 3758–3766 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku M. & Sakai Y. Assessment of physiological redox state with novel FRET protein probes. Antioxid Redox Signal 16, 698–704 (2012). [DOI] [PubMed] [Google Scholar]
- Oku M., Hoseki J., Ichiki Y. & Sakai Y. A fluorescence resonance energy transfer (FRET)-based redox sensor reveals physiological role of thioredoxin in the yeast Saccharomyces cerevisiae. FEBS Lett 587, 793–798 (2013). [DOI] [PubMed] [Google Scholar]
- Pickart C. M. Targeting of substrates to the 26S proteasome. FASEB J 11, 1055–1066 (1997). [DOI] [PubMed] [Google Scholar]
- Rota C., Chignell C. F. & Mason R. P. Evidence for free radical formation during the oxidation of 2′-7′-dichlorofluorescin to the fluorescent dye 2′-7′-dichlorofluorescein by horseradish peroxidase: possible implications for oxidative stress measurements. Free Radic Biol Med 27, 873–881 (1999). [DOI] [PubMed] [Google Scholar]
- Xu S., Peng G., Wang Y., Fang S. & Karbowski M. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol Biol Cell 22, 291–300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E. B. & Rutter J. Mitochondrial quality control by the ubiquitin-proteasome system. Biochem Soc Trans 39, 1509–1513 (2011). [DOI] [PubMed] [Google Scholar]
- Tanaka A. et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol 191, 1367–1380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Kanthasamy A., Anantharam V. & Kanthasamy A. G. Mitochondrial accumulation of polyubiquitinated proteins and differential regulation of apoptosis by polyubiquitination sites Lys-48 and -63. J Cell Mol Med 13, 1632–1643 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson G. T. et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem 279, 13044–13053 (2004). [DOI] [PubMed] [Google Scholar]
- Dooley C. T., Dore T. M., Hanson G. T., Jackson W. C., Remington S. J. & Tsien R. Y. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem 279, 22284–22293 (2004). [DOI] [PubMed] [Google Scholar]
- de la Lastra C. A. & Villegas I. Resveratrol as an antioxidant and pro-oxidant agent: mechanisms and clinical implications. Biochem Soc Trans 35, 1156–1160 (2007). [DOI] [PubMed] [Google Scholar]
- Murphy M. P. & Smith R. A. Targeting antioxidants to mitochondria by conjugation to lipophilic cations. Annu Rev Pharmacol Toxicol 47, 629–656 (2007). [DOI] [PubMed] [Google Scholar]
- Fukai T. & Ushio-Fukai M. Superoxide dismutases: Role in redox signaling, vascular function, and diseases. Antioxid Redox Signal 15, 1583–1606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa L., Gomes E. & Rockwell P. Reactive oxygen species induced by proteasome inhibition in neuronal cells mediate mitochondrial dysfunction and a caspase-independent cell death. Apoptosis 12, 1389–1405 (2007). [DOI] [PubMed] [Google Scholar]
- Keller J. N., Hanni K. B. & Markesbery W. R. Possible involvement of proteasome inhibition in aging: implications for oxidative stress. Mech Ageing Dev 113, 61–70 (2000). [DOI] [PubMed] [Google Scholar]
- Ling Y. H., Liebes L., Zou Y. & Perez-Soler R. Reactive oxygen species generation and mitochondrial dysfunction in the apoptotic response to Bortezomib, a novel proteasome inhibitor, in human H460 non-small cell lung cancer cells. J Biol Chem 278, 33714–33723 (2003). [DOI] [PubMed] [Google Scholar]
- Vali S. et al. Insights into the effects of α-synuclein expression and proteasome inhibition on glutathione metabolism through a dynamic in silico model of Parkinson's disease: validation by cell culture data. Free Radic Biol Med 45, 1290–1301 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun D. H., Gray D. A., Halliwell B. & Jenner P. Interference with ubiquitination causes oxidative damage and increased protein nitration: implications for neurodegenerative diseases. J Neurochem 90, 422–430 (2004). [DOI] [PubMed] [Google Scholar]
- Hyun D. H. Effect of the overexpression of mutant ubiquitin (K48R) on the cellular response induced by 4-hydroxy-2,3-trans-nonenal, an end-product of lipid peroxidation. Neurosci Lett 477, 115–120 (2010). [DOI] [PubMed] [Google Scholar]
- McNaught K. S. P., Belizaire R., Isacson O., Jenner P. & Olanow C. W. Altered proteasomal function in sporadic Parkinson's disease. Exp Neurol 179, 38–46 (2003). [DOI] [PubMed] [Google Scholar]
- Leonard S. S. et al. Resveratrol scavenges reactive oxygen species and effects radical-induced cellular responses. Biochem Biophys Res Commun 309, 1017–1026 (2003). [DOI] [PubMed] [Google Scholar]
- Lorenz P., Roychowdhury S., Engelmann M., Wolf G. & Horn T. F. W. Oxyresveratrol and resveratrol are potent antioxidants and free radical scavengers: Effect on nitrosative and oxidative stress derived from microglial cells. Nitric Oxide - Biol and Ch 9, 64–76 (2003). [DOI] [PubMed] [Google Scholar]
- Rubiolo J. A., Mithieux G. & Vega F. V. Resveratrol protects primary rat hepatocytes against oxidative stress damage: activation of the Nrf2 transcription factor and augmented activities of antioxidant enzymes. Eur J Pharmacol 591, 66–72 (2008). [DOI] [PubMed] [Google Scholar]
- Osawa T., Namiki M. & Kawakishi S. Role of dietary antioxidants in protection against oxidative damage. Basic Life Sci 52, 139–153 (1990). [DOI] [PubMed] [Google Scholar]
- Hamada N. et al. Involvement of heme oxygenase-1 induction via Nrf2/ARE activation in protection against H2O2-induced PC12 cell death by a metabolite of sesamin contained in sesame seeds. Bioorg Med Chem 19, 1959–1965 (2011). [DOI] [PubMed] [Google Scholar]
- Putics A., Vegh E. M., Csermely P. & Soti C. Resveratrol induces the heat-shock response and protects human cells from severe heat stress. Antioxid Redox Signal 10, 65–75 (2008). [DOI] [PubMed] [Google Scholar]
- Morito D. et al. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRDeltaF508. Mol Biol Cell 19, 1328–1336 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information