Abstract
Prostate cancer risk–associated variants have been reported in populations of European descent, African-Americans and Japanese using genome-wide association studies (GWAS). To systematically investigate prostate cancer risk–associated variants in Chinese men, we performed the first GWAS in Han Chinese. In addition to confirming several associations reported in other ancestry groups, this study identified two new risk-associated loci for prostate cancer on chromosomes 9q31.2 (rs817826, P = 5.45 × 10−14) and 19q13.4 (rs103294, P = 5.34 × 10−16) in 4,484 prostate cancer cases and 8,934 controls. The rs103294 marker at 19q13.4 is in strong linkage equilibrium with a 6.7-kb germline deletion that removes the first six of seven exons in LILRA3, a gene regulating inflammatory response, and was significantly associated with the mRNA expression of LILRA3 in T cells (P < 1 × 10−4). These findings may advance the understanding of genetic susceptibility to prostate cancer.
Prostate cancer is the second most frequently diagnosed cancer and the sixth leading cause of cancer-related death in men, with an estimated 914,000 new cases and 258,000 deaths per year globally1. Incidence and mortality rates for prostate cancer vary by 25-fold and 10-fold, respectively, around the world1. The highest incidence rates are found in Western developed countries, and the highest mortality rates are found in African-Americans, whereas the lowest incidence and mortality rates are reported in Asians. These differences suggest genetic heterogeneity as well as different environmental exposures in prostate cancer development in various ancestry groups.
Multiple GWAS of prostate cancer have been performed in populations of European descent, and more than 40 prostate cancer susceptibility loci have been identified2–12. In addition, GWAS of prostate cancer in Japanese13,14 and African-American15 populations identified eight and one novel loci for prostate cancer, risk, respectively. However, no GWAS for prostate cancer has been reported in the Chinese population.
In this study, we performed a multistage GWAS of prostate cancer in the Chinese Consortium for Prostate Cancer Genetics (ChinaPCa), with a total of 4,484 prostate cancer cases and 8,934 controls from Han Chinese population. The characteristics of the subjects in each stage are summarized in Supplementary Table 1. In the first stage, 731,458 SNPs were genotyped in 1,497 cases and 1,008 controls using Illumina Human OmniExpress BeadChips. After quality control filtering (Online Methods), 587,294 SNPs in 1,417 cases and 1,008 controls were retained in the subsequent analyses. A principal-component analysis was first performed using EIGENSOFT software16 to assess the genetic background of the subjects. Results from this analysis were consistent with subjects being Han Chinese (Supplementary Fig. 1). The distribution of the top two eigens is shown in Supplementary Figure 2. We performed association analysis for each of these SNPs with prostate cancer risk with PLINK17, assuming an additive model and adjusting for the first eigen. Quantile-quantile plot analysis of the association results showed an inflation factor (λ) of 1.08 (Supplementary Fig. 3). Similar results were found when analysis was adjusted for the top two eigens (λ = 1.08), likely reflecting differences in the ascertainment of cases and controls (Online Methods). Across the genome, one SNP at 8q24 (rs1456315, P = 1.18 × 10−12) exceeded the predefined genome-wide significance level of P < 5.0 × 10−8 (Supplementary Fig. 4). This SNP is located in region 2 of 8q24, which has previously been reported as a prostate cancer susceptibility locus3 (Supplementary Table 2). Suggestive evidence for association was also found for many other regions throughout the genome, although these associations did not reach genome-wide significance. These regions include eight additional prostate cancer risk–associated loci previously reported in populations of European descent and four loci reported in the Japanese population (P < 0.05) (Supplementary Table 2).
To confirm new prostate cancer risk–associated loci suggested in the first stage of the GWAS, we selected a subset of independently associated SNPs for replication on the basis of three criteria: (i) P < 5.0 × 10−3 in the association test (4,323 SNPs met this criterion), (ii) r2 < 0.5 for linkage disequilibrium (LD) between SNPs (166 SNPs met both of these criteria) and (iii) similar allele frequency in the controls to that reported in two additional large GWAS of Chinese populations (difference of ≤0.02)18,19 (43 SNPs met all three criteria) (Online Methods). Forty-three independent SNPs were selected and genotyped in an additional 782 cases and 1,792 controls (replication 1). Association analysis using the same method as in the GWAS stage confirmed two SNPs that associated at P < 1.16 × 10−3 (accounting for 43 independent tests): rs817826 at 9q31.2 (P = 1.05 × 10−3) and rs103294 at 19q13.4 (P = 4.98 × 10−7) (Supplementary Table 3). To further confirm these associations, we genotyped these two SNPs in two additional case-control series (replication 2: 1,102 cases and 4,501 controls; replication 3: 1,183 cases and 1,633 controls). These two SNPs showed significant association in each stage (replication 2: rs817826, P = 3.09 × 10−7 and rs103294, P = 2.62 × 10−3; replication 3: rs817826, P = 2.22 × 10−3 and rs103294, P = 3.65 × 10−4) (Table 1). After combining the results from all four stages using a meta-analysis assuming a fixed effect, associations at these two SNPs exceeded genome-wide significance. For rs817826 at 9q31.2, the rs817826[C] allele was associated with an odds ratio (OR) of 1.41 for prostate cancer risk (95% confidence interval (CI) = 1.29–1.54; P = 5.45 × 10−14). For rs103294 at 19q13.4, the rs103294[C] allele was associated with an OR of 1.28 for prostate cancer risk (95% CI = 1.21–1.36; P = 5.34 × 10−16). There was no evidence for heterogeneity among the four stages at rs817826 (Phet = 0.83, I2 = 0) or rs103294 (Phet = 0.53, I2 = 0).
Table 1.
Summary results of associations with prostate cancer risk at 9q31.2 and 19q13.4 in Chinese men
| Marker | Allelesa | Location | Gene | Study | Genotypesb |
MAFc |
OR (95% CI)d |
Pd | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |||||||
| rs817826 | T/C | 9q31.2 | RAD23B KLF4 | GWAS | 1,128/275/14 | 860/144/4 | 0.107 | 0.075 | 1.43 (1.17–1.77) | 6.82 × 10−4 |
| Replication 1 | 619/154/9 | 1,513/264/14 | 0.110 | 0.082 | 1.39 (1.14–1.70) | 1.05 × 10−3 | ||||
| Replication 2 | 858/226/18 | 3,770/701/27 | 0.119 | 0.084 | 1.47 (1.27–1.71) | 3.09 × 10−7 | ||||
| Replication 3 | 927/234/10 | 1,368/251/12 | 0.109 | 0.084 | 1.33 (1.11–1.59) | 2.22 × 10−3 | ||||
| Combinede | 1.41 (1.29–1.54) | 5.45 × 10−14 | ||||||||
| rs103294 | T/C | 19q13.4 | LILRA3 | GWAS | 701/579/137 | 579/371/58 | 0.301 | 0.242 | 1.28 (1.12–1.45) | 2.94 × 10−4 |
| Replication 1 | 388/311/82 | 1,036/655/97 | 0.304 | 0.237 | 1.40 (1.23–1.60) | 4.98 × 10−7 | ||||
| Replication 2 | 577/430/94 | 2,621/1,609/263 | 0.281 | 0.238 | 1.25 (1.13–1.39) | 2.62 × 10−5 | ||||
| Replication 3 | 602/485/85 | 943/592/90 | 0.279 | 0.238 | 1.25(1.11−1.41) | 3.65 × 10−4 | ||||
| Combinede | 1.28(1.21−1.36) | 5.34 × 10−16 | ||||||||
Major allele/minor allele.
Major homozygotes/heterozygotes/minor homozygotes.
Minor allele frequency (MAF).
ORs, 95% CIs and corresponding P values in an additive model were estimated using a logistic regression model.
Results in an additive model from GWAS and replication stages were combined by meta-analysis using a fixed-effect model under the absence of heterogeneity among studies for both loci (rs817826: P = 0.83, I2 = 0; rs103294: P = 0.53, I2 = 0).
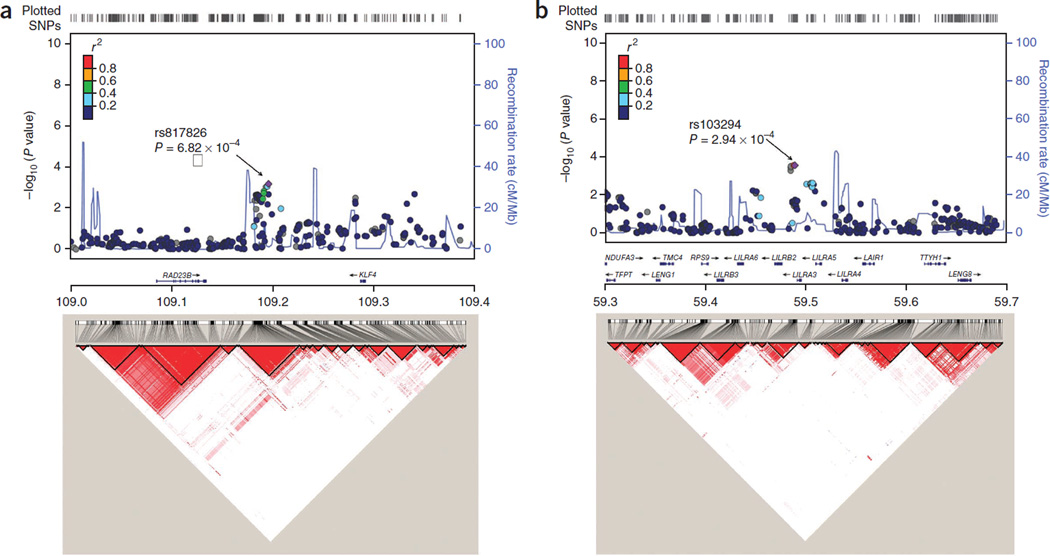
To further characterize prostate cancer associations at these two loci, we imputed known common SNPs in the flanking regions of these SNPs for subjects in the first-stage GWAS on the basis of haplo-type data from the 1000 Genomes Project Han Chinese in Beijing, China (CHB) and Japanese in Tokyo, Japan (JPT) subjects (Phase 1 integrated data version 3, released March 2012) using IMPUTE2.2.2 software (Fig. 1). Flanking regions were determined for each locus on the basis of local LD information (r2 ≥ 0.10) and association results (P < 0.01) from the first-stage data (∼17.5 kb for 9q31.2 and ∼48 kb for 19q13.4). Considering that only a small number of SNPs were successfully imputed in the region of 19q13.4, we genotyped an additional 19 SNPs in this region (Online Methods). None of the additional SNPs in either of these regions showed a stronger association with prostate cancer than the respective index SNP (rs817826 and rs103294). After conditioning on the index SNP at each region, no association was found at P < 0.01 for the remaining SNPs at 9q31.2 and 19q13.4 (Supplementary Tables 4 and 5), suggesting that no additional independent prostate cancer risk–associated loci exist in these two regions.
Figure 1.
Regional association plots. (a,b) Regional plots (top) and LD maps (bottom) at 9q31.2 (a) and 19q13.4 (b). For regional plots, association of individual SNP is plotted as –log10P against chromosomal position. Results for both genotyped and imputed SNPs are shown. Symbol colors represent the LD of the SNP with the most significant SNP at each locus (purple diamond). The right y axis shows the recombination rate estimated from 1000 Genomes Project CHB and JPT data. LD maps were based on D′ values using CHB and JPT genotypes from the 1000 Genomes Project (Phase 1 integrated data version 3, released March 2012).
We also examined associations of these two SNPs with clinical characteristics of prostate cancer in a case-only analysis (Supplementary Table 6). No significant associations were observed with prostate-specific antigen (PSA) levels at diagnosis, Gleason score, tumor, node, metastasis (TNM) stage or aggressiveness in the combined prostate cancer cases from the four stages. Similarly, we did not find any association of these two SNPs with serum PSA levels in the controls (Supplementary Fig. 5).
We tested association of these two SNPs with prostate cancer risk in two populations of European descent: Cancer Genetic Markers of Susceptibility (CGEMS) in the United States and CAncer Prostate in Sweden (CAPS) (Supplementary Table 7). For rs103294 at 19q13.4, the allele frequency differed considerably in the population of European descent from that seen in the Chinese population, and no association was found. For rs817826 at 9q31, however, the allele frequency was similar to that seen in the Chinese population, the reported risk allele showed higher frequency in cases relative to controls in both studies and the association with prostate cancer risk was statistically significant in the combined analysis of these two studies (P = 0.023). Further studies in other ancestry groups are warranted, including evaluation of the broader regions surrounding each implicated prostate cancer risk–associated SNP.
We next performed expression quantitative trait locus (eQTL) analysis to examine whether the two prostate cancer risk–associated SNPs correlate with expression of nearby genes within a 2-Mb region centered on the index SNP, using a publicly available database20 (Online Methods and Supplementary Table 8). The rs103294[C] risk allele at 19q13.4 was consistently associated with increased expression of LILRA3 (encoding leukocyte immunoglobulin-like receptor subfamily A member 3) in T cells from 75 individuals, as measured by 2 probes for the gene (P ≤ 1 × 10−4 after permutation)21 (Supplementary Fig. 6). No consistent association with expression was found in other cell types and tissues for LILRA3 and other surrounding genes. No correlation was observed between rs817826 at 9q31.2 and expression of nearby genes in any measured cell type (fibroblast, lymphoblastoid cell line and T cell)21 or tissue type (adipose, lymphoblastoid cell line and skin)22 (Supplementary Table 8). Considering the small number of samples used in eQTL analysis, additional studies are needed to better understand the association of these SNPs with the mRNA expression of nearby genes.
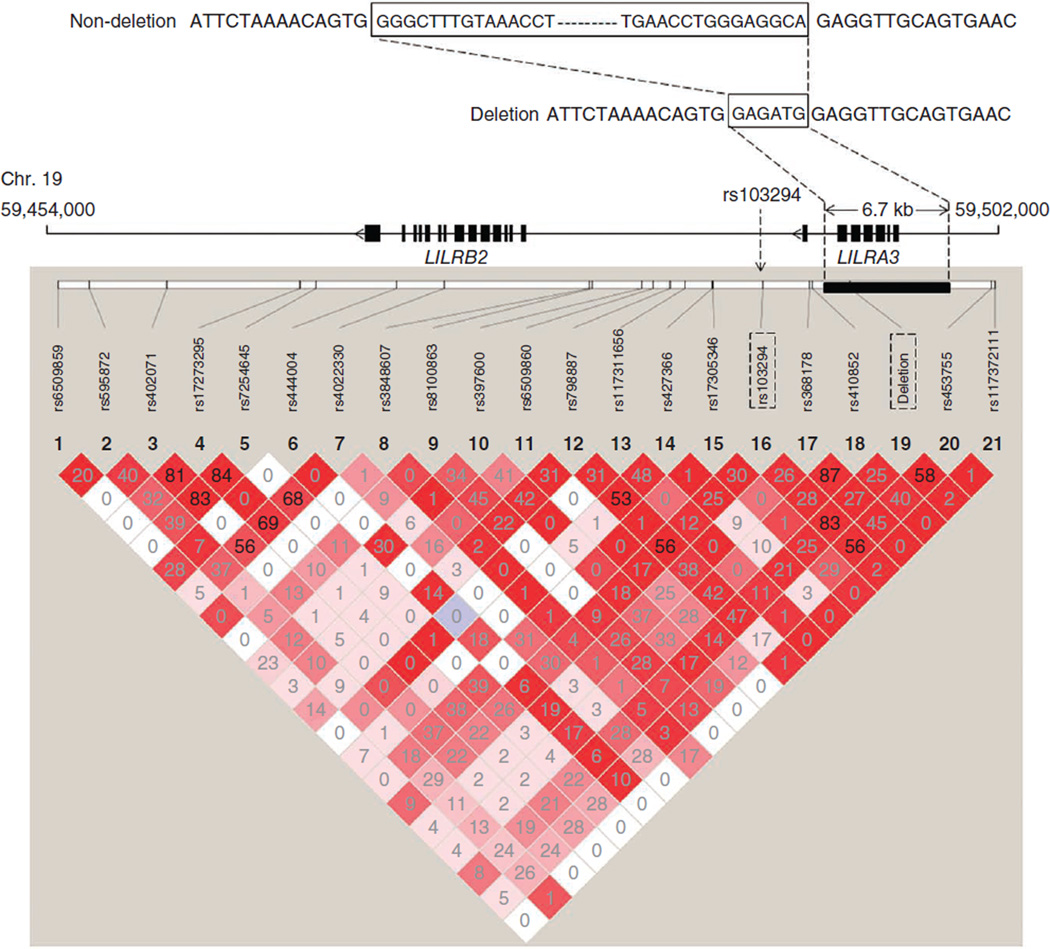
The rs103294 SNP is located within the leukocyte immunoglobulin-like receptor (LIR) gene cluster at 19q13.4. The LD block containing rs103294 overlaps with LILRA3 (Fig. 1). There is a 6.7-kb known germline deletion within the LILRA3 gene that removes the first six of a total of seven exons in the gene23 (Fig. 2). The deletion was reported to be more common in Northeastern Asians (0.56–0.84) than Europeans (0.17) or individuals from other populations (0.10–0.26)24. We measured the deletion status in all the subjects in the GWAS stage using a previously reported method24. Strong LD between the deletion variant and rs103294 (r2 = 0.83) was observed, and the non-deleted allele was on the haplotype containing the rs103294[C] risk allele (Fig. 2). The non-deleted allele was more common in cases (0.307) than controls (0.252) (P = 9.60 × 10−4; Supplementary Table 9). LIR family members, including LILRA3, are expressed on immune cells, where they bind to major histocompatibility complex (MHC) antigens and regulate immune and inflammatory responses25. The mRNA expression of LILRA3 is low in prostate tissues in the UCSC database and was not detectable in our analysis of prostate tissue samples from 80 individuals with benign prostatic hyperplasia (BPH) (Supplementary Table 10). The role of LILRA3 in prostate cancer development is largely unknown, although the eQTL data in T cells, as well as other emerging evidence26, may suggest a potential role of chronic inflammation in prostate carcinogenesis. Nevertheless, further functional characterization of this gene and other genes at 19q13.4 is needed to fully evaluate their contribution to prostate cancer development.
Figure 2.
Germline deletion at LILRA3 and genotyped SNPs flanking the gene. Top, schematic of the LILRA3 deletion that is in strong LD with prostate cancer risk–associated SNP rs103294 at 19q13.4. Bottom, pairwise LD (r2) values calculated based on data from GWAS stage samples.
The rs817826 SNP at 9q31.2 resides in an intergenic region between RAD23B (62 kb centromeric) and KLF4 (91 kb telomeric), and the LD block containing rs817826 does not overlap with these two genes (Fig. 1). A bioinformatics analysis at this region, based on Encyclopedia of DNA Elements (ENCODE) data annotated by the UCSC browser, did not reveal additional functional regions (Supplementary Fig. 7). Additional genetic and functional studies are needed to delineate the mechanism by which the 9q31.2 locus contributes to prostate cancer development.
In summary, we conducted the first GWAS of prostate cancer in Han Chinese and identified two new susceptibility loci at 9q31.2 and 19q13.4. These findings may improve the understanding of prostate cancer susceptibility and provide clues for further functional studies. Our study highlights the importance of GWAS of complex diseases in diverse populations.
URLs
EIGENSOFT, http://genepath.med.harvard.edu/~reich/Software.htm; Genevar, http://www.sanger.ac.uk/resources/software/genevar; HapMap, http://hapmap.ncbi.nlm.nih.gov/; IMPUTE, https://mathgen.stats.ox.ac.uk/impute/impute.html; LocusZoom, http://csg.sph.umich.edu/locuszoom/; PLINK 1.07, http://pngu.mgh.harvard.edu/~purcell/plink/download.shtml; R statistical software, http://www.r-project.org/; UCSC database, http://genome.ucsc.edu/.
ONLINE METHODS
Study subjects
Demographic characteristics and clinical features of study subjects are summarized in Supplementary Table 1. These Chinese subjects were part of the ChinaPCa27,28. Briefly, all of the Chinese subjects are male Han Chinese and were recruited from the southeastern region of China by members of ChinaPCa. All of the cases were hospital based and were pathologically diagnosed as having primary prostate cancer. Cancer-free controls were recruited from the community or selected from subjects undergoing routine physical examination in local hospitals. In the GWAS stage, 1,497 cases and 1,008 controls were mainly recruited from Shanghai and surrounding areas. Subjects in replication 1 were also from Shanghai (782 cases and 1,792 controls). Subjects in replications 2 (1,102 cases and 4,501 controls) and 3 (1,183 cases and 1,633 controls) were mainly recruited from Nanjing and surrounding areas. In addition, we also included a population-based case-control study (2,919 cases and 1,612 controls) from Sweden, CAPS, for replication29. For CAPS, individuals with prostate cancer were identified and recruited from four of the six regional cancer registries in Sweden and the National Prostate Cancer Register. Control subjects were all males without a diagnosis of prostate cancer and were randomly selected from the Swedish Population Registry by frequency matching to the cases on the basis of age (groups of 5-year intervals) and geographic region. After informed consent was obtained, a blood sample was obtained from each subject for DNA extraction. This study was approved by the Institutional Review Board of each participating institution.
Selection of SNPs for the confirmation study
To confirm suggestive association signals identified in the GWAS stage that were not within loci that have already been reported, two steps were used to select a subset of SNPs for first-stage replication (replication 1). First, a CLUMP analysis in PLINK was performed to identify SNPs that were independently associated with prostate cancer risk, requiring association at P < 1 × 10−3 and r2 of <0.5 for LD. Second, to exclude potential false positive SNPs due to unstable estimates in controls, SNP allele frequencies were compared between control subjects from the GWAS stage and subjects from two additional large GWAS of Han Chinese (including some females)18,19. Only SNPs with an allele frequency difference of ≤0.02 between these control subjects were selected. SNPs on the X chromosome were not applied to this step. These two steps led to the identification of 43 candidate SNPs for replication (Supplementary Table 3).
SNP genotyping and quality control in the GWAS stage
DNA samples were extracted from blood samples and were genotyped using Illumina Human OmniExpress BeadChips. A total of 731,458 SNPs were genotyped in 1,497 cases and 1,008 controls. A standard quality control procedure was applied to select samples and SNPs for further analysis. Samples were removed if they (i) had an overall genotyping rate of <95%; (ii) had ambiguous gender; or (iii) were duplicates or showed familial relationships (PI_HAT > 0.025). SNPs were excluded if they had (i) a call rate of <95%; (ii) a minor allele frequency (MAF) of <0.05; or (iii) P < 1 × 10−3 in a Hardy-Weinberg equilibrium test among controls. After quality control analysis, a total of 1,417 cases and 1,008 controls with 587,292 SNPs remained.
SNP genotyping in replication stages
On the basis of results from the GWAS stage, 43 SNPs were selected for replication (Supplementary Table 3). Genotyping of these SNPs was performed using the MassARRAY iPLEX (Sequenom) or TaqMan (Applied Biosystems) systems. Duplicates and negative controls were included in each 96-well plate for quality control. Genotyping was performed by technicians blinded to sample status. The average concordance rate between duplicate samples was >99%.
Fine-mapping study at 19q13.4
To further analyze the prostate cancer risk locus 19q13.4 in detail, we densely genotyped a set of markers within a 48-kb region (chr. 19: 59,454,000–59,502,000) in 1,497 cases and 1,008 controls from the GWAS stage. Tagging SNPs were determined on the basis of the CHB SNP data set in the 1000 Genomes Project (1000 Genomes Phase 1 integrated version, March 2012), requiring a MAF of ≥0.10, a call rate of ≥0.95, a Hardy-Weinberg equilibrium test P value of >0.001 and pairwise r2 of <0.5. SNPs residing in the deleted region were excluded from selection as a tagging SNP. In this analysis, 16 tagging SNPs, including rs 103294, were selected. In addition, we included seven low-frequency (MAF < 0.10) SNPs to increase the coverage density of this region. Genotyping of the total 23 SNPs was performed using the MassARRAY iPLEX system. We successfully genotyped 20 SNPs after excluding 3 SNPs with low call quality (Supplementary Table 5).
We also genotyped the known ∼6.7-kb germline deletion (chr. 19: 59,492,668–59,499,432) of the LILRA3 gene in all subjects from the GWAS stage. A previously reported PCR sequence-specific primer typing method30 was used to detect the presence or absence of this deletion. Primers and PCR conditions were the same as those described previously30. Deletion status was independently determined by two technicians that were blinded to the genotype at rs 103294.
Examination of LILRA3 mRNA expression in prostate tissues
RNA templates were extracted from 80 BPH tissues using the RNeasy Miniprep kit (Qiagen). cDNA was synthesized from 1 µg of RNA template using reverse transcriptase and oligo(dT) primer (Promega). We measured the mRNA levels of LILRA3 using a quantitative RT-PCR assay. Primer sequences are given in Supplementary Table 11. The primers complement exons 6 and 7 and were designed to differentially target deleted and non-deleted LILRA3 mRNA. Expression of ACTB (encoding β-actin) was also examined to normalize the expression of LILRA3.
Statistical analysis
A logistic regression model was used to analyze the association of each SNP with prostate cancer risk, assuming an additive genetic model, which was implemented in PLINK version 1.07 (see URLs)17. ORs and 95% CIs were estimated from logistic regression analysis with adjustment for age and the top eigen. Ancestry and population stratification were determined by principal-component analysis, using data from four populations (CHB, JPT, Utah residents of Northern and Western European ancestry (CEU) and Yoruba from Ibadan, Nigeria (YRI)) of the HapMap 2 project (see URLs) and the cases and controls genotyped in the GWAS stage, implemented in the EIGENSOFT package (see URLs). The first two principal components for each individual were plotted. The logistic regression model was also applied for testing association with prostate cancer risk in replication stages. The relationships of SNPs with prostate cancer clinical features, including PSA levels, Gleason score, clinical stage (T, N or M stage) and aggressiveness, were also evaluated using an additive model for cases in the GWAS and replication stages independently. Associations of SNPs with PSA levels in controls were assessed using linear regression analyses assuming an additive model after log transformation of the original values to approach normalization. Results from the GWAS and replication stages were combined by meta-analysis. Cochran’s Q statistic was used to test for heterogeneity, and the I2 statistic was used to quantify the proportion of the total variation caused by heterogeneity. A random-effect model (DerSimonian-Laird) was adopted if there was an indication of heterogeneity between studies (P for Q ≤ 0.05); otherwise, the fixed-effect model was applied (P for Q > 0.05). A predefined P value of 5.0 × 10−8 was set as a threshold for genome-wide significance. In replication stages, association was assumed to be significant after Bonferroni correction, at P < 1.16 × 10−3 (0.05/43) or 0.025 (0.05/2) for replication 1 and 2, respectively. For regions that met the statistical criteria of genome-wide association with prostate cancer risk, ungenotyped SNPs were imputed using IMPUTE software (see URLs) with 1000 Genomes Project CHB and JPT subjects serving as reference haplotype maps. A posterior probability of >0.90 was applied to call imputed genotypes. Imputed SNPs were excluded if they had (i) a call rate of <95%; (ii) a MAF of <0.05; or (iii) P < 1 × 10−3 in a Hardy-Weinberg equilibrium test in controls. Conditional analysis was then applied to test the independence of SNPs in each region, using the SNPs that were originally determined to be significant as covariates. Regional plots were created using LocusZoom31 (see URLs). SAS 9.2 (SAS Institute) and R 2.9.1 (see URLs) were also used for data analysis.
eQTL analysis
Two identified loci, rs817826 at 9q31.2 and rs103294 at 19q13.4, were tested for correlation with nearby gene expression, as measured by probes using the publicly available eQTL database Genevar20 (see URLs). Genotype and expression data within this database are derived from 3 cell types (fibroblast, lymphoblastoid cell line and T cell) from 75 individuals from Geneva21 and 3 tissue types (166 adipose, 156 lymphoblastoid cell line and 160 skin) from healthy female twins22. The expression probes located within 1 Mb of the 5′ and 3′ end of the specified SNPs were analyzed. Differences in the distribution of normalized expression levels between genotypes were compared using a linear regression model. To avoid false positive associations due to multiple tests, we set a significance threshold of P < 1.0 × 10−3 and also assessed significance using 10,000-fold permutations. Specifically, two probes, ILMN_1786303 on exon 6 and ILMN_1661631 on exon 7 of LILRA3, were used in the measurement of LILRA3 expression.
Supplementary Material
ACKNOWLEDGMENTS
We thank all of the subjects included in this study. This work was partially funded by the National Key Basic Research Program grant 973 (2012CB518300 to Y.S and 2012CB518301 to J.X.), the Key Project of the National Natural Science Foundation of China (81130047 to J.X.), intramural grants from the Fudan University Thousand Talents Program and Huashan Hospital (to J.X.), the US National Institutes of Health (NCI CA129684 to J.X.), the National Natural Science Foundation of China (30945204 to Z.M and 30973009 to D.Y.), the Ministry of Health Special Research Fund for Public Interests (201002007 to L.J.) and the National Science & Technology Pillar Program (2011BAI09B00 to L.J.).
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
Y.S., J.X. and Z.M. directed the study, obtained financial support and were responsible for study design, interpretation of results and manuscript writing. D.Y., M.W., F.L. and C.X. recruited study subjects and managed respective project. G.J. performed statistical analyses, summarized results and drafted the manuscript. X.W., Q.S., Z.C., Z.T., J.Q., F.Z., Zhong Wang (affiliation 20), Y.F., D.H., Q. Wei, J. Guo, D.W., Xin Gao, J. Yuan, Gongxian Wang, Y. Xu, Guozeng Wang, H. Yao, P.D., Y.J., M.S., J. Yang, J.O.-Y., H.J., Y. Zhu, S.R., Z.Z., C.Y., Xu Gao, B.D., Z.H., Y.Y., Q. Wu, H.C., P.P., Y. Zheng, X. Zheng, Y. Xiang, J. Gong, R.N. and X.L. recruited subjects and prepared samples. J.L., X.-O.S., W.Z. and X. Zhang provided the allele frequency data from their GWAS populations. H. Yu, Zhong Wang (affiliation 4), S.T., J.F., Jishan Sun and W.L. performed statistical and bioinformatics analyses and carried out experiments. F.W. and H.G. provided samples and information from CAPS. A.H., J.R., Q.D., H.S., L.J., R.S., D.L., Jielin Sun and S.L.Z. coordinated the project. All of the authors reviewed, approved and contributed to the final version of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Jemal A, et al. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nat. Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 3.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 4.Gudmundsson J, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 5.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 6.Eeles RA, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 7.Gudmundsson J, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat. Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 9.Eeles RA, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudmundsson J, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeager M, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kote-Jarai Z, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat. Genet. 2011;43:785–791. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takata R, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat. Genet. 2010;42:751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 14.Akamatsu S, et al. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nat. Genet. 2012;44:426–429. doi: 10.1038/ng.1104. [DOI] [PubMed] [Google Scholar]
- 15.Haiman CA, et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat. Genet. 2011;43:570–573. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patterson N, Price AL, Reich D. Population structure and eigen analysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng W, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang XJ, et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat. Genet. 2009;41:205–210. doi: 10.1038/ng.310. [DOI] [PubMed] [Google Scholar]
- 20.Yang TP, et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26:2474–2476. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimas AS, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nica AC, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torkar M, et al. Arrangement of the ILT gene cluster: a common null allele of the ILT6 gene results from a 6.7-kbp deletion. Eur. J. Immunol. 2000;30:3655–3662. doi: 10.1002/1521-4141(200012)30:12<3655::AID-IMMU3655>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 24.Hirayasu K, et al. Evidence for natural selection on leukocyte immunoglobulin-like receptors for HLA class I in Northeast Asians. Am. J. Hum. Genet. 2008;82:1075–1083. doi: 10.1016/j.ajhg.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borges L, Hsu ML, Fanger N, Kubin M, Cosman D. A family of human lymphoid and myeloid Ig-like receptors, some of which bind to MHC class I molecules. J. Immunol. 1997;159:5192–5196. [PubMed] [Google Scholar]
- 26.De Marzo AM, et al. Inflammation in prostate carcinogenesis. Nat. Rev. Cancer. 2007;7:256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, et al. Systematic confirmation study of reported prostate cancer risk– associated single nucleotide polymorphisms in Chinese men. Cancer Sci. 2011;102:1916–1920. doi: 10.1111/j.1349-7006.2011.02036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M, et al. Replication and cumulative effects of GWAS-identified genetic variations for prostate cancer in Asians: a case-control study in the ChinaPCa consortium. Carcinogenesis. 2012;33:356–360. doi: 10.1093/carcin/bgr279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng SL, et al. Cumulative association of five genetic variants with prostate cancer. N. Engl. J. Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 30.Hirayasu K, et al. Long-term persistence of both functional and non-functional alleles at the leukocyte immunoglobulin-like receptor A3 (LILRA3) locus suggests balancing selection. Hum. Genet. 2006;119:436–443. doi: 10.1007/s00439-006-0152-y. [DOI] [PubMed] [Google Scholar]
- 31.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.