Abstract
Objective
The purpose of this study was to explore whether non-HLA genetic markers can improve type 1 diabetes (T1D) prediction in a prospective cohort with high-risk HLA-DR,DQ genotypes.
Methods
The Diabetes Autoimmunity Study in the Young (DAISY) follows prospectively for development of T1D and islet autoimmunity (IA) children at increased genetic risk. A total of 1709 non-Hispanic White DAISY participants have been genotyped for 27 non-HLA single nucleotide polymorphisms and one microsatellite.
Results
In multivariate analyses adjusting for family history and HLA-DR3/4 genotype, PTPN22 (rs2476601) and two UBASH3A (rs11203203 and rs9976767) SNPs were associated with development of IA (HR=1.87, 1.55 and 1.54 respectively, all p≤0.003), while GLIS3 and IL2RA showed borderline association with development of IA. INS, UBASH3A and IFIH1 were significantly associated with progression from IA to diabetes (HR=1.65, 1.44 and 1.47 respectively, all p≤0.04), while PTPN22 and IL27 showed borderline association with progression from IA to diabetes. In survival analysis, 45% of general population DAISY children with PTPN22 rs2476601 TT or HLA-DR3/4 and UBASH3A rs11203203 AA developed diabetes by age 15, compared to 3% of children with all other genotypes (p<0.0001). Addition of non-HLA markers to HLA-DR3/4,DQ8 did not improve diabetes prediction in first-degree relatives.
Conclusion
Addition of PTPN22 and UBASH3A SNPs to HLA-DR,DQ genotyping can improve T1D risk prediction.
Keywords: Type 1 diabetes, islet autoimmunity, non-HLA genetic markers, prediction
Introduction
The HLA region on chromosome 6p21 is considered the major susceptibility locus for type 1 diabetes (odds ratio > 6) with an estimated 30-50% of the genetic risk for diabetes attributed to this region 1. With the advent of genome wide association studies (GWAS), more than 50 non-HLA susceptibility gene markers have been associated with type 1 diabetes 2-5. A majority of these loci appear to have effects in the immune system. INS 6 and PTPN22 7show the strongest association (odds ratio ∼2), notably weaker compared to the HLA region.
Class II HLA genotypes in combination with islet autoantibodies can predict diabetes risk in first-degree relatives (FDR) of persons with type 1 diabetes. We have previously published on the association of INS, PTPN22 and UBASH3A with islet autoimmunity (IA) and type 1 diabetes in the Diabetes Autoimmunity Study in the Young (DAISY) 8, 9. In an article published in Pediatric Diabetes in 2012 9, we reported on the association of UBASH3A with both IA and type 1 diabetes with a cumulative risk for diabetes of 22% by age 10 for those general population DAISY children having UBASH3A AA genotype with HLA-DR3/4,DQB1*0302. In this study, we have genotyped an additional 8 non-HLA single nucleotide polymorphisms (SNPs) in 7 genes (ERBB3 (rs2292239), CLEC16A (rs12708716), IL27 (rs4788084), CTRB (rs7202877), C14orf (rs4900384), GSDM (rs2290400), HORMAD2 (rs5753037), UBASH3A (rs9976767)), and further explored the independent predictive value of novel non-HLA markers on the risk of IA and progression from IA to diabetes, controlling for the effects of HLA-DR,DQ genotypes. We have also developed a genetic risk model, adding non-HLA markers (UBASH3A AA, PTPN22 TT) to high risk HLA-DR3/4 in order to refine diabetes risk prediction, and report a risk of diabetes by age 15 years of 45% for those DAISY general population children in the high risk genetic stratum. Finally, we tested a set of SNPs, previously found to significantly discriminate diabetes in the BABYDIAB cohort 10, in the DAISY study.
Methods
Study population
Since 1993, DAISY has followed two cohorts of young children at increased risk of type 1 diabetes: FDR of type 1 diabetes patients and general population children found through a newborn screening to carry high-risk HLA-DR,DQ genotypes. The details of screening and follow-up have been previously published 11. Briefly, 31,881 newborns from the general population of Denver, Colorado have been screened for HLA-DR,DQ genotypes that carry susceptibility to type 1 diabetes. All children with DR3/4,DQB1*0302, DR3/3 and DR4/4,DQB1*0302 and a sample of those with DR4/DRx, DQB1*0302 or DR3/DRx (where DRx ≠ DR3 or DR4) were invited to participate in DAISY. Although general population children were included in DAISY only if they had the above susceptibility HLA genotypes, non-diabetic offspring and siblings of patients with type 1 diabetes were invited to participate regardless of their HLA genotype. A total of 1709 non-Hispanic White (NHW) participants (858 general population children and 851 FDR children, including 477 multiple siblings) were genotyped for 27 non-HLA single nucleotide polymorphisms and one microsatellite. Of those, 116 developed persistent IA and 66 of these progressed to diabetes during the 10-year mean prospective follow-up. Informed consent was obtained from the parents of each study subject. The Colorado Multiple Institutional Review Board approved all study protocols.
Islet Autoantibodies
Measurement of islet autoantibodies to insulin, GAD65, IA-2 and ZnT8 was performed in the Clinical Immunology Laboratory at the Barbara Davis Center using previously described radio-immunoassays 12. IA was defined as presence of one or more of the autoantibodies to insulin, GAD65, IA-2 or ZnT8 on at least 2 consecutive visits 3-12 months apart, and still positive at last visit.
Genotyping
INS-23Hph1 (rs689), CTLA-4 T17A (rs231775), and PTPN22 R620W (rs2476601) polymorphisms were genotyped using a linear array (immobilized probe) method essentially as described in Mirel et al. 13. The following SNPs were genotyped in the laboratory of Dr. Cisca Wijmenga using Illumina GoldenGate Beadexpress assays (veracode 48-plex): IL2RA (rs12251307), SH2B3 (rs3184504), PTPN2 (rs1893217), C10orf59 (rs10509540), IL18RAP (rs917997), BACH2 (rs11755527) and TAGAP (rs1738074).
Taqman SNP genotyping assays (Applied Biosystems, CA USA) were utilized to obtain genotype information on the following SNPs as described previously 8: CD69 (rs4763879), GAB3 (rs2664170), GLIS3 (rs7020673), IL10 (rs3024496), SIRPG (rs2281808), PRKD2 (rs425105), UBASH3A (rs11203203), IFIH1 (rs1990760) and SLC30A8 (rs13266634).
The following SNPs were genotyped by utilizing the Taqman SNP genotype based OpenArray platform [Applied Biosystems, CA USA]: ERBB3 (rs2292239), CLEC16A (rs12708716), IL27 (rs4788084), CTRB (rs7202877), C14orf (rs4900384), GSDM (rs2290400), HORMAD2 (rs5753037), UBASH3A (rs9976767). Custom designed arrays were loaded using the OpenArray AccuFill system and cycling was performed on a GeneAmp 9700 PCR system, all gDNA template load and run parameters according to manufacturer protocol. Genotypes were analyzed using the OpenArray SNP genotyping analysis software v.1.0.3 and Taqman Genotyper Software 2.0.
CCR5 genotypes were determined using a fluorescent-based method. PCR fragments were generated using primers that differentiate between the wild type genotype (CCR5/CCR5) at 225bp and the homozygous mutant (Δ32/Δ32) at 193bp. Reactions (25 μl) were assembled using FailSafe PCR PreMix J, 2.5 U MasterAmp Taq polymerase (Epicentre), 10 nmol each primer and 100 ng of genomic template. The PCR product was amplified via 35 PCR cycles of 94°C for 30 sec, 57°C for 35 sec, 72°C for 1 min, and a final extension of 72°C for 45 min. Products were diluted 1:60 and separated by capillary electrophoresis on an ABI3100-Avant Genetic Analyzer (Applied Biosystems). Alleles were identified using GeneMapper v3.5 (Applied Biosystems).
Statistical analysis
Analyses were performed in SAS version 9.2 and PRISM software. Cox proportional hazard models were used to test the effect of each genetic polymorphism on time to development of IA and progression from IA to diabetes. Multivariate model with Weibull distribution (outcome: IA) and Cox PH model (outcome: diabetes) included family history of diabetes (yes/no) and the presence of the HLA-DR3/4-DQB1*0302 genotype (yes/no); independently significant non-HLA polymorphisms were identified by backward selection at a critical level of 0.05. Each SNP in the model was defined according to the number of risk allele present (0, 1 or 2) and was treated as a continuous variable in the model. In the Cox regression model, we checked proportionality by including time-dependent covariates and all p values were non-significant. Since our analyses were based on a priori hypotheses, P values were not corrected for multiple testing. We performed survival analysis of progression to IA and diabetes with PRISM software, using the log-rank test and an alpha level for significance set at 0.05. Only one of the two UBASH3A SNPs (rs11203203) was included in the model since both UBASH3A SNPs are in linkage disequilibrium (D'=1.0, r2=0.63). Based on initial significant results, survival analyses were stratified by high and low genetic risk groups. High risk genetic group included all subjects with UBASH3A AA in addition to HLA-DR3/4 as well as all subjects with PTPN22 TT (whether they were HLA-DR3/4 or not, since this group is small), while low risk genetic group included all other genotypes. Finally, survival analyses including nine out of the 12 genes recently tested in a model by Winkler et al 10 were performed. Receiver operator curve (ROC) analysis was performed with area under the curve (AUC) calculated for the 9 gene variants. The SNPs were in Hardy-Weinberg equilibrium except for C10orf59, PRKD2 and GAB3, which were therefore excluded from the multivariate and survival analyses. To determine whether inclusion of multiple siblings per family in this cohort affected our findings, we performed analyses accounting for the clustering of patients within a family (using the robust sandwich estimate for statistical inference) and found similar results (data not shown).
Results
In multivariate analyses adjusting for family history and HLA-DR3/4 high-risk genotype, PTPN22 (rs2476601) and two UBASH3A (rs11203203 and rs9976767) SNPs were associated with development of IA (HR=1.87, 1.55 and 1.54 respectively, all p≤0.003), while GLIS3 and IL2RA showed borderline association with development of IA (Table 1). On the other hand, INS, UBASH3A and IFIH1 were significantly associated with progression from IA to diabetes (HR=1.65, 1.44 and 1.47 respectively, all p≤0.04), while PTPN22 and IL27 showed borderline association with progression from IA to diabetes. Some of these SNPs might be close to reach statistical significance due to smaller numbers, especially in the group looking at progression to diabetes. The other non-HLA markers tested did not predict development of IA or diabetes. There were no significant interactions between any of the SNPs and HLA-DR3/4-DQB1*0302.
Table 1. Non-HLA gene polymorphisms as potential predictors of islet autoimmunity (IA) and progression from IA to type 1 diabetes (T1D) in DAISY non-Hispanic white population a.
| Gene | SNP | Risk Allele | Islet Autoimmunity (N=1709)b | Progression from IA to T1D (N=116)c | ||
|---|---|---|---|---|---|---|
| HR d (95% CI) | p-value | HR (95% CI) | p-value | |||
| ERBB3 | rs2292239 | T | 1.12 (0.85-1.48) | 0.43 | 1.32 (0.81-2.15) | 0.26 |
| CLEC16A | rs12708716 | G | 0.94 (0.70-1.27) | 0.69 | 0.94 (0.63-1.40) | 0.77 |
| IL27 | rs4788084 | T | 0.98 (0.74-1.29) | 0.87 | 1.45 (0.98-2.14) | 0.06 |
| CTRB | rs7202877 | G | 1.01 (0.65-1.59) | 0.96 | 0.75 (0.39-1.41) | 0.37 |
| C14orf | rs4900384 | G | 0.77 (0.56-1.06) | 0.11 | 0.64 (0.40-1.05) | 0.08 |
| GSDM | rs2290400 | T | 0.91 (0.70-1.19) | 0.51 | 0.97 (0.66-1.42) | 0.86 |
| HORMAD2 | rs5753037 | T | 0.92 (0.69-1.23) | 0.59 | 1.16 (0.79-1.71) | 0.44 |
| BACH2 | rs11755527 | G | 1.01 (0.76-1.35) | 0.95 | 0.88 (0.59-1.32) | 0.54 |
| C10orf59 | rs10509540 | G | 0.84 (0.69-1.03) | 0.10 | 0.96 (0.72-1.26) | 0.74 |
| CD69 | rs4763879 | A | 1.10 (0.84-1.44) | 0.49 | 0.93 (0.65-1.35) | 0.72 |
| GAB3 | rs2664170 | G | 0.91 (0.71-1.15) | 0.42 | 0.92 (0.67-1.26) | 0.61 |
| GLIS3 | rs7020673 | C | 0.77 (0.60-1.00) | 0.05 | 0.77 (0.55-1.09) | 0.14 |
| IFIH1 | rs1990760 | C | 1.07 (0.81-1.40) | 0.65 | 1.47 (1.02-2.12) | 0.04 |
| IL10 | rs3024496 | A | 1.27 (0.97-1.65) | 0.08 | 1.21 (0.85-1.73) | 0.29 |
| IL18RAP | rs917997 | A | 1.16 (0.84-1.60) | 0.36 | 0.94 (0.57-1.54) | 0.80 |
| IL2RA | rs12251307 | A | 0.61 (0.37-1.02) | 0.06 | 1.15 (0.58-2.29) | 0.69 |
| INS | rs689 | A | 1.29 (0.93-1.79) | 0.12 | 1.65 (1.05-2.59) | 0.03 |
| PRKD2 | rs425105 | C | 0.87 (0.61-1.26) | 0.47 | 0.83 (0.49-1.38) | 0.47 |
| PTPN2 | rs1893217 | G | 1.33 (0.95-1.84) | 0.09 | 1.11 (0.70-1.77) | 0.65 |
| PTPN22 | rs2476601 | T | 1.87 (1.32-2.66) | 0.001 | 1.59 (0.97-2.61) | 0.06 |
| SH2B3 | rs3184504 | A | 0.99 (0.76-1.31) | 0.97 | 0.80 (0.56-1.15) | 0.23 |
| SIRPG | rs2281808 | T | 0.85 (0.64-1.13) | 0.25 | 1.03 (0.72-1.48) | 0.87 |
| TAGAP | rs1738074 | A | 0.90 (0.68-1.19) | 0.47 | 1.20 (0.83-1.73) | 0.32 |
| UBASH3A | rs11203203 | A | 1.55 (1.19-2.02) | 0.001 | 1.44 (1.01-2.04) | 0.04 |
| UBASH3A | rs9976767 | G | 1.54 (1.16-2.05) | 0.003 | 1.47 (0.97-2.21) | 0.07 |
| SLC30A8 | rs13266634 | T | 0.95 (0.70-1.27) | 0.71 | 1.07 (0.70-1.65) | 0.75 |
| CTLA4 | rs231775 | G | 1.20 (0.93-1.56) | 0.16 | 0.95 (0.66-1.36) | 0.76 |
| CCR5 | microsatellite | Del32 | 0.93 (0.60-1.47) | 0.77 | 1.04 (0.55-1.98) | 0.89 |
Multivariate analyses, adjusted for HLA-DR3/4,DQB1*0302 and family history of type 1 diabetes
Total N=1709 (116 subjects with IA)
Total N=116 (66 subjects with T1D)
HR = hazard ratio
Backward multivariate regression analyses including all SNPs were performed for both development of IA and progression from IA to type 1 diabetes (Table 2). SNPs that remained significantly associated with IA, adjusting for family history and HLA-DR3/4-DQB1*0302, included PTPN22, UBASH3A, INS and GLIS, while the final model for progression from IA to diabetes only included UBASH3A.
Table 2. Predictors of islet autoimmunity and progression to type 1 diabetes a.
| Predictor | Islet Autoimmunity | Progression to diabetes | ||
|---|---|---|---|---|
| HR b (95% CI) | p-value | HR (95% CI) | p-value | |
| HLA-DR3/4, DQB1*0302 | 3.96 (2.22-7.06) | <0.0001 | 5.93 (2.83-12.43) | <0.0001 |
| Cohort (FDR) c | 2.09 (1.20-3.64) | 0.009 | 2.68 (1.21-5.96) | 0.02 |
| rs689 (INS) | 1.95 (1.20-3.19) | 0.008 | NA d | |
| rs2476601 (PTPN22) | 1.93 (1.16-3.22) | 0.01 | NA | |
| rs9976767 (UBASH3A) | 1.63 (1.12-2.37) | 0.01 | 2.11 (1.14-3.89) | 0.02 |
| rs7020673 (GLIS3) | 0.65 (0.45-0.93) | 0.02 | NA | |
Multivariate model including all variables with an a level of <0.05
HR = hazard ratio
FDR = first-degree relatives
NA = not significant for progression from IA to diabetes
Based on the results of backward multivariate regression analyses, we performed survival analyses with those significant variables (HLA-DR3/4-DQB1*0302, PTPN22, UBASH3A, INS and GLIS) in order to refine diabetes risk prediction. There was no further improvement in prediction by including INS or GLIS, so the final high-risk stratum includes all subjects with UBASH3A AA in addition to HLA-DR3/4 as well as all subjects with PTPN22 TT (whether they were HLA-DR3/4 or not, since this group is small), while the low risk group has all other genotypes. Cumulative incidence of development of IA showed a higher risk of IA by age 15 years for the high (26%) compared to the low risk group (5%) in the general population (N=843) (Figure 1A). Risk of diabetes by age 15 years was also higher in subjects with high (45%) compared to those with low risk (3%) (p<0.0001) (Figure 1B). In comparison, survival analysis stratified by HLA-DR3/4,DQB1*0302 showed a risk of persistent IA and diabetes by age 15 years for HLA-DR3/4 of “only” 12% and 15% respectively (Figure 1C and 1D). In the DAISY general population, the positive predictive value for diabetes is slightly better with this genetic risk stratum than HLA-DR3/4 alone (17.4 vs. 6.4) for similar negative predictive value (98.6 vs. 99.2), while sensitivity was lower (42 vs. 75%) and specificity was better (95 vs. 74%) compared to HLA-DR3/4 alone.
Figure 1. Progression to Islet Autoimmunity (IA) and Diabetes in DAISY NHW general population children (N=843)*: IA by genetic risk strata (1A), Diabetes by genetic risk strata (1B), IA by HLA-DR3/4 (1C) and Diabetes by HLA-DR3/4 (1D).
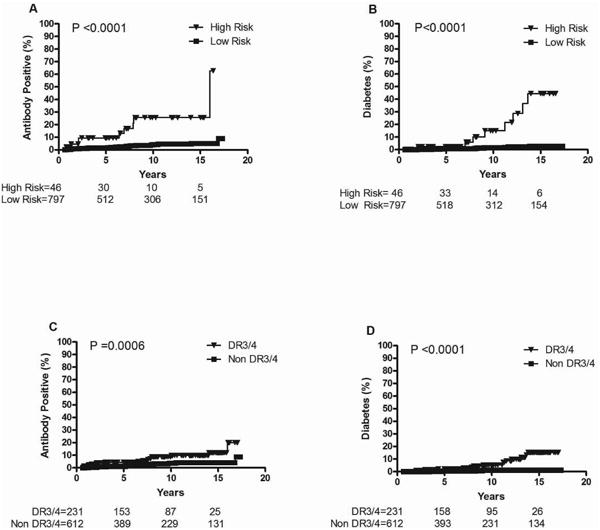
High risk: all subjects with UBASH3A AA in addition to HLA-DR3/4 as well as all subjects with PTPN22 TT; Low risk: all other genotypes
Follow-up time was defined as the age of the child at the 1st of the 2 consecutive positive visits for affected children and age of the child at the last visit for unaffected children.
Non DR3/4 refers to not having the highest risk HLA DR3/4,DQB1*0302 genotype.
*15 subjects not included due to missing either UBASH3A or PTPN22 genotyping
Addition of non-HLA markers to HLA-DR3/4,DQ8 did not improve diabetes prediction in DAISY FDR (Figure 2). The cumulative risk of IA among FDR reached 51% in the high risk group by age 15 (Figure 2A), while the cumulative risk for diabetes was 29% (Figure 2B). Cumulative incidence of IA and diabetes by age 15 years showed similar risk for FDR with HLA-DR3/4 only (39% and 35% respectively) (Figure 2C and 2D).
Figure 2. Progression to Islet Autoimmunity (IA) and Diabetes in DAISY NHW first-degree relatives (N=816)*: IA by genetic risk strata (1A), Diabetes by genetic risk strata (1B), IA by HLA-DR3/4 (1C) and Diabetes by HLA-DR3/4 (1D).
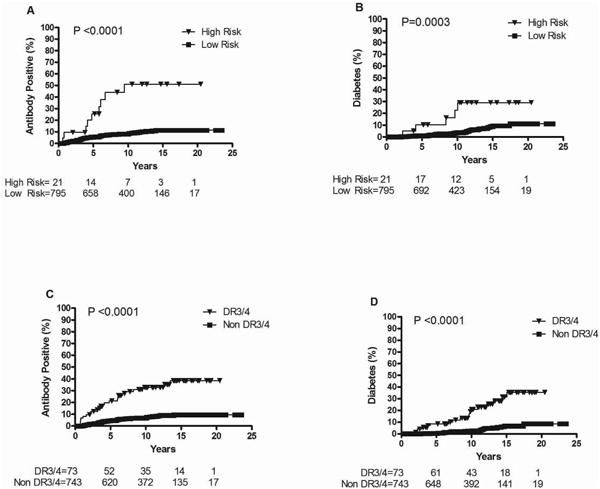
High risk: all subjects with UBASH3A AA in addition to HLA-DR3/4 as well as all subjects with PTPN22 TT; Low risk: all other genotypes
Follow-up time was defined as the age of the child at the 1st of the 2 consecutive positive visits for affected children and age of the child at the last visit for unaffected children.
Non DR3/4 refers to not having the highest risk HLA DR3/4,DQB1*0302 genotype.
*35 subjects not included due to missing either UBASH3A or PTPN22 genotyping
Nine (ERBB3, PTPN2, IFIH1, PTPN22, KIAA0350/CLEC16A, CTLA4, SH2B3, IL18RAP, IL10) out of the 12 genes recently tested in a model by Winkler et al.10 have been genotyped in DAISY. For all 9 gene SNPs, a score of 2 was given if the child was homozygous for the susceptible allele, 1 if heterozygous and 0 if homozygous for the non-susceptible allele. The sum of the scores for the 9 genes was assigned as the combined risk score for each child. Although the distribution of the combined risk scores did not reach statistical significance between children who developed diabetes compared to autoantibody negative children, threshold points could be observed at SNP-risk allele score of <5 and >9, which were used to define low (<5), intermediate (6-9) and high (>9) risk categories (Figure 3). These thresholds showed a similar stratification of diabetes risk in DAISY than in BABYDIAB, although the survival analyses only trended towards significance in the general population (p=0.06), likely due to smaller numbers (Figure 4). Receiver operator curve (ROC) analysis was performed, but area under the curve (AUC) was not statistically significant (AUC 0.55, 95% CI 0.45-0.65).
Figure 3. Non-HLA gene SNP-risk allele score distribution in DAISY general population and first-degree relatives.
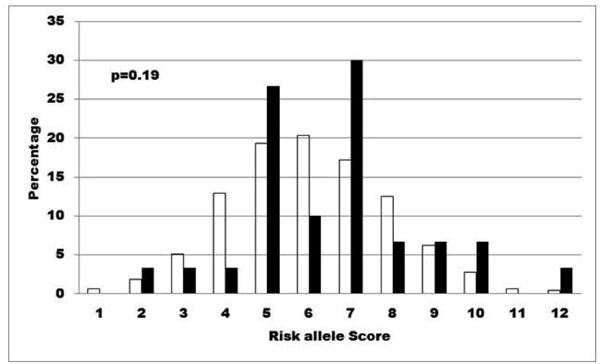
Distribution of risk allele scores derived from 9 (ERBB3, PTPN2, IFIH1, PTPN22, KIAA0350/CLEC16A, CTLA4, SH2B3, IL18RAP, IL10) type 1 diabetes susceptibility genes in nondiabetic autoantibody negative children (unfilled bars) compared to children who progressed to diabetes (filled bars) in DAISY
N=1332 antibody negative children and N=37 children with type 1 diabetes (antibody positive subjects who have not developed diabetes and subjects missing data for one or more SNPs are not included)
Figure 4. Progression to Diabetes in DAISY general population children (left) and DAISY first-degree relatives (right).
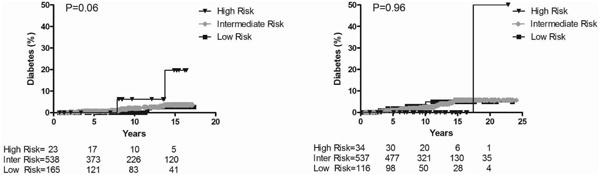
SNP-risk allele score categories are low <5, intermediate 5-9 and high >9
Inter: intermediate
N=726 general population children and N=687 first-degree relatives (subjects missing data for one or more SNPs were not included)
Discussion
High-density SNP analysis, GWAS and follow-up meta-analyses have added to the list of non-HLA loci associated with type 1 diabetes (more than 50 to date) 2-4, 14. Still the strongest signals by far are associated with the HLA region (OR>6) with the next highest non-HLA signals in INS and PTPN22 15. The prospective DAISY study has now genotyped 28 previously confirmed non-HLA loci to test the robustness of these associations with the advantage of evaluating the effect of candidate SNPs on the prospectively observed development of diabetes phenotypes (development of IA and progression from IA to diabetes). In addition to the non-HLA genes most strongly associated with type 1 diabetes in previous studies (PTPN22 and INS) 16, 17, we recently found a robust association of IA and diabetes for UBASH3A 9. While PTPN22, INS and UBAH3A seem to be the main non-HLA risk factors in the DAISY cohort, some SNPs might not reach statistical significance due to smaller numbers, especially in the group looking at progression to diabetes.
This is the first study to describe a genetic risk stratum for diabetes in a prospective cohort following general population children screened at birth for high-risk HLA-DR,DQ genotypes. The risk definition includes HLA class II, PTPN22 and UBASH3A. This genetic risk stratum significantly improves prediction of type 1 diabetes in DAISY general population children with a risk of diabetes by age 15 years of 45% for those subjects in the high risk compared to 3% for those in the low risk stratum. If confirmed in another population, these prediction models could be used for screening high-risk general population children into potential clinical research trials. We have previously published on two SNPs (rs2040410 and rs7454108) that are 98.6% sensitive and 99.7% specific for HLA DR3/4-DQ8 18. A new genetic stratum including a total of 4 SNPs (rs2476601, rs11203203, rs2040410 and rs7454108) could potentially be applicable for screening of type 1 diabetes risk, followed by diabetes antibody testing in those subjects found at high genetic risk. Although the negative predictive value is good, the positive predictive value remains low. These results should be confirmed in additional cohorts with long-term follow-up such as The Environmental Determinants of Diabetes in the Young (TEDDY) and ideally in a general population without HLA susceptibility genotypes for type 1 diabetes.
Winkler et al recently published on improved prediction of diabetes by including 12 non-HLA risk genes in children with high-risk HLA genotypes 10. Stratified survival analyses showed risk ranging from 0% by age 14 years for children in the low-risk category to 7.1% for children in the high-risk category. Interestingly, our risk score distribution (Figure 3) seems to be shifted to the left compared to the paper by Winkler et al,, which might be due to population stratification (although we limited our study to non-Hispanic White subjects) or to higher population frequencies of the three SNPs (CD25 rs11594656, IL2 rs4505848, COBL rs4948088) not included in our study. One of the strengths and similarities for these two studies is that they are prospective studies in which timing of IA and diabetes onset are closely monitored and time to event analyses are possible. However, there are several important differences between the BABYDIAB article and our study. First, BABYDIAB population only includes first-degree relatives. Second, the number and type of SNPs included were different and HLA high-risk genotypes were defined according to the TEDDY criteria in BABYDIAB 19, while for the DAISY study, only high-risk HLA DR3/4-DQ8 genotype was considered as a categorical variable.
Another study looked at the joint effects of HLA, INS, PTPN22 and CTLA4 genes and found that multiple susceptibility loci confer a very high risk of diabetes, but only a small proportion of the population carries all high risk alleles 20. When assessing the predictive utility of these genetic risk markers by ROC curve, multiple susceptibility genotypes seemed to improve disease prediction only marginally compared to HLA genotype alone 20. ROC analyses did not improve disease prediction in this DAISY study. Limitations of ROC analysis include the fact that it does not take into account time to event, which is actually one of the strength of this prospective DAISY cohort study.
So far, prediction of type 1 diabetes has mainly been based on family history, age of onset of proband, autoantibody number and levels, and genetic susceptibility markers such as INS and HLA-DR3/4-DQB1*0302 21-25. Addition of PTPN22 and UBASH3A SNPs to HLA-DR,DQ genotyping can help improve prediction of type 1 diabetes.
Acknowledgments
This research was supported by NIH grants R37 DK32493, RO1 DK32083, DK050979, N01 AI15416. A.K.S. was supported by the JDRF Grant 11-2010-206 Early Career Patient-oriented Diabetes Award.
The authors declare that there is no duality of interest associated with this manuscript.
Abbreviations
- GWAS
genome wide association studies
- FDR
first-degree relatives
- DAISY
Diabetes Autoimmunity Study in the Young
- IA
islet autoimmunity
- NHW
non-Hispanic White
- SNPs
single nucleotide polymorphisms
- TEDDY
The Environmental Determinants of Diabetes in the Young
References
- 1.Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: Molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet. 1996;59:1134–1148. [PMC free article] [PubMed] [Google Scholar]
- 2.Burren OS, Adlem EC, Achuthan P, Christensen M, Coulson RM, Todd JA. T1DBase: update 2011, organization and presentation of large-scale data sets for type 1 diabetes research. Nucleic Acids Res. 2011;39(Database issue):D997–1001. doi: 10.1093/nar/gkq912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009 doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper JD, Smyth DJ, Smiles AM, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40(12):1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smyth DJ, Plagnol V, Walker NM, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359(26):2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell GI, Horita S, Karam JH. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. diab. 1984;33:176–183. doi: 10.2337/diab.33.2.176. [DOI] [PubMed] [Google Scholar]
- 7.Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36(4):337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 8.Steck AK, Wong R, Wagner B, et al. Effects of non-HLA gene polymorphisms on development of islet autoimmunity and type 1 diabetes in a population with high-risk HLA-DR,DQ genotypes. diab. 2012;61(3):753–758. doi: 10.2337/db11-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rewers M, Bugawan TL, Norris JM, et al. Newborn screening for HLA markers associated with IDDM: Diabetes Autoimmunity Study in the Young (DAISY) diabetol. 1996;39:807–812. doi: 10.1007/s001250050514. [DOI] [PubMed] [Google Scholar]
- 10.Yu L, Rewers M, Gianani R, et al. Antiislet autoantibodies usually develop sequentially rather than simultaneously. J Clin Endocrinol Metab. 1996;81:4264–4267. doi: 10.1210/jcem.81.12.8954025. [DOI] [PubMed] [Google Scholar]
- 11.Mirel DB, Valdes AM, Lazzeroni LC, Reynolds RL, Erlich HA, Noble JA. Association of IL4R haplotypes with type 1 diabetes. diab. 2002;51(11):3336–3341. doi: 10.2337/diabetes.51.11.3336. [DOI] [PubMed] [Google Scholar]
- 12.Winkler C, Krumsiek J, Lempainen J, et al. A strategy for combining minor genetic susceptibility genes to improve prediction of disease in type 1 diabetes. Genes Immun. 2012 doi: 10.1038/gene.2012.36. [DOI] [PubMed] [Google Scholar]
- 13.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med. 2009;360(16):1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 15.Reddy MP, Wang H, Liu S, et al. Association between type 1 diabetes and GWAS SNPs in the southeast US Caucasian population. Genes Immun. 2011;12(3):208–212. doi: 10.1038/gene.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howson JM, Walker NM, Smyth DJ, Todd JA. Analysis of 19 genes for association with type I diabetes in the Type I Diabetes Genetics Consortium families. Genes Immun. 2009;10(Suppl 1):S74–S84. doi: 10.1038/gene.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson K, Wong R, Barriga KJ, et al. rs11203203 is associated with type 1 diabetes risk in population pre-screened for high-risk HLA-DR,DQ genotypes. Pediatr Diabetes. 2012;13(8):611–615. doi: 10.1111/j.1399-5448.2012.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barker JM, Triolo TM, Aly TA, et al. Two single nucleotide polymorphisms identify the highest-risk diabetes HLA genotype: potential for rapid screening. diab. 2008;57(11):3152–3155. doi: 10.2337/db08-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Environmental Determinants of Diabetes in the Young (TEDDY) study: study design. Pediatr Diabetes. 2007;8(5):286–298. doi: 10.1111/j.1399-5448.2007.00269.x. [DOI] [PubMed] [Google Scholar]
- 20.Bjornvold M, Undlien DE, Joner G, et al. Joint effects of HLA, INS, PTPN22 and CTLA4 genes on the risk of type 1 diabetes. diabetol. 2008;51(4):589–596. doi: 10.1007/s00125-008-0932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orban T, Sosenko JM, Cuthbertson D, et al. Pancreatic islet autoantibodies as predictors of type 1 diabetes in the Diabetes Prevention Trial-Type 1. Diab care. 2009;32(12):2269–2274. doi: 10.2337/dc09-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walter M, Albert E, Conrad M, et al. IDDM2/insulin VNTR modifies risk conferred by IDDM1/HLA for development of Type 1 diabetes and associated autoimmunity. diabetol. 2003;46(5):712–720. doi: 10.1007/s00125-003-1082-z. [DOI] [PubMed] [Google Scholar]
- 23.Bonifacio E, Hummel M, Walter M, Schmid S, Ziegler AG. IDDM1 and multiple family history of type 1 diabetes combine to identify neonates at high risk for type 1 diabetes. Diab care. 2004;27(11):2695–2700. doi: 10.2337/diacare.27.11.2695. [DOI] [PubMed] [Google Scholar]
- 24.Mrena S, Virtanen SM, Laippala P, et al. Models for predicting type 1 diabetes in siblings of affected children. Diab care. 2006;29(3):662–667. doi: 10.2337/diacare.29.03.06.dc05-0774. [DOI] [PubMed] [Google Scholar]
- 25.Steck AK, Johnson K, Barriga KJ, et al. Age of Islet Autoantibody Appearance and Mean Levels of Insulin, but Not GAD or IA-2 Autoantibodies, Predict Age of Diagnosis of Type 1 Diabetes: Diabetes Autoimmunity Study in the Young. Diab care. 2011;34(6):1397–1399. doi: 10.2337/dc10-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]


