Abstract
Extracellular binding proteins or antagonists are important factors that modulate ligands in the transforming growth factor (TGF-β) family. While the interplay between antagonists and ligands are essential for developmental and normal cellular processes, their imbalance can lead to the pathology of several disease states. In particular, recent studies have implicated members of the differential screening-selected gene in neuroblastoma (DAN) family in disease such as renal fibrosis, pulmonary arterial hypertension, and reactivation of metastatic cancer stem cells. DAN family members are known to inhibit the bone morphogenetic proteins (BMP) of the TGF-β family. However, unlike other TGF-β antagonist families, DAN family members have roles beyond ligand inhibition and can modulate Wnt and vascular endothelial growth factor (VEGF) signaling pathways. This review describes recent structural and functional advances that have expanded our understanding of DAN family proteins with regards to BMP inhibition and also highlights their emerging roles in the modulation of Wnt and VEGF signaling pathways.
Keywords: Keywords DAN, BMP, TGF-β, Wnt, VEGF, extracellular antagonists
Introduction
The Transforming Growth Factor-β (TGF-β) superfamily is a large collection of secreted ligands that play numerous roles in a variety of biological processes. Ligands can be subdivided into the Activin, Bone Morphogenetic Protein (BMP), and TGF-β subclasses. Functionally, ligands orchestrate proliferation and differentiation protocols that help dictate the organization of tissues, organs, and the body plan.1,2 In the adult, ligands regulate tissue homeostasis, direct wound healing processes, and are deeply embedded in reproduction and the immune response. A variety of mechanisms have evolved to achieve proper cellular signaling outcomes. For example, ligand modulation occurs when secreted protein antagonists directly neutralize the signaling capacity of the TGF-β ligands.2–4 However, misregulation of ligand signaling within adults is causative of numerous human diseases, such as cancer and fibrosis.5–9
Members of the differential screening-selected gene in neuroblastoma (DAN) family form a diverse group of antagonists that were originally identified as BMP inhibitors. The family consists of seven members: Nbl1 (the founding member, also known as Dan and DAND1), SOST (Sclerostin), uterine sensitization-associated gene-1 (USAG-1/Wise), Coco (DAND5), Gremlin (Drm), Cerberus (Cer1), and protein related to Dan and Cerberus (PRDC/Gremlin-2).4,10,11 DAN family members are expressed during development where they have a profound role in limb bud formation and digitation, kidney formation and morphogenesis, and left-right axis specification.12–16 However, increased DAN protein levels in adults are often associated with a number of severe disease-states, including pulmonary and renal fibrosis and cancer.5,7,8,17–19 Recently, DAN family proteins have been directly implicated in the reactivation of metastatic breast cancers within lung tissue.5 Therefore, due to their critical roles in human development and disease, there is significant need to structurally and functionally evaluate the members composing the DAN family of protein antagonists.
In addition to the inhibition of BMP signaling, certain DAN family members can modulate other signaling pathways. For example, the Xenopus form of Cerberus can directly inhibit nodal signaling, a specific Activin subclass member that controls left-right body symmetry.20 More intriguingly, specific members of the DAN family harbor the ability to interact with the Wnt and VEGF (vascular endothelial growth factor) signaling cascades. SOST, for example, has been shown to be a strong Wnt antagonist (for specific Wnt ligands) by directly binding the Wnt co-receptors LRP5 and LRP6, with specific mutations in SOST leading to bone dysplasia.21–23 Additionally, Gremlin has recently been show to function as an agonist of the VEGF signaling cascade, where it directly stimulates angiogenesis through its interactions with vascular endothelial growth factor receptor 2 (VEGFR2).24–26
In the past, reviews have elegantly highlighted the importance of DAN family members in biology and disease.4,27 However, our understanding of the molecular and structural underpinnings defining how DAN family proteins control and regulate BMPs, as well as other signaling cascades, is underdeveloped and in need of further characterization. Therefore, this review will focus on the emerging structural and biochemical studies that have provided new insight for understanding how DAN family proteins specifically function to regulate BMP, Wnt, and VEGF signaling.
Overview of TGF-β Signaling
In humans, the TGF-β superfamily has expanded to include a very diverse portfolio of ligands, composing ∼40 unique members. For downstream signaling to occur, the dimeric ligand has to sequester both two Type I and two Type II receptors, with an overall stoichiometry of 1 : 2 : 2. This leads to phosphorylation of the Type I receptors by the Type II receptors, followed by phosphorylation and activation of intracellular SMAD (mothers against decapentaplegic homolog) transcription factors (Fig. 1). For a more detailed description of ligand and receptor binding and signaling, which is not discussed thoroughly here, please refer to the following reviews.28–30
Figure 1.
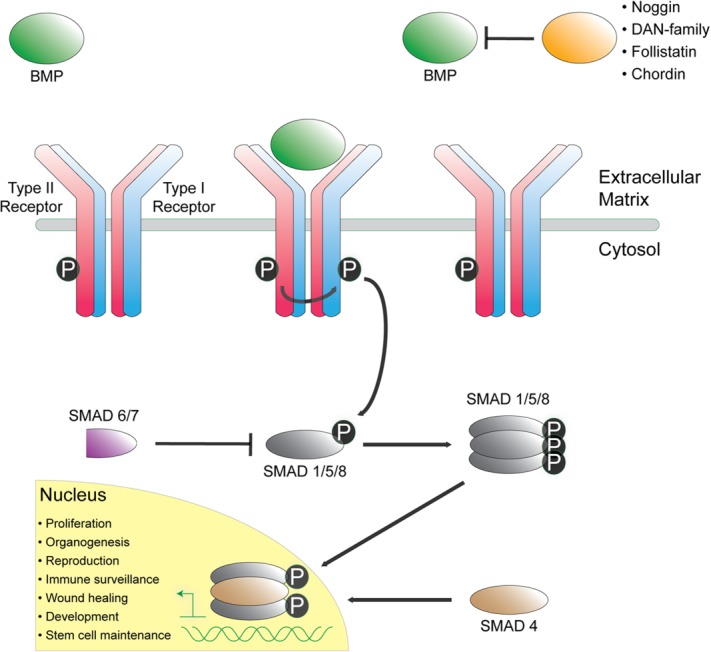
Overview of BMP/ TGF-β signaling. Secreted ligands (green) signal by binding and activating two of each Type I (blue) and Type II (red) receptors. Upon binding, the Type I receptor is phosphorylated by the Type II receptor, leading to Type I kinase domain activation and subsequent phosphorylation of intracellular SMAD transcription factors (gray). Activated SMAD molecules oligomerize and accumulate in the nucleus to combine with coactivators and corepressors to regulate gene expression. Several structurally diverse extracellular antagonists (orange) bind and sequester ligands to inhibit and block ligand–receptor interaction and activation.
Structural studies in the early 1990s revealed that ligands adopt a propeller-like dimer with a pair of β-strands extending distally in opposite directions.28,29 The overall fold has been historically described as two hands shaking, where the set of anti-parallel β-strands are referred to as Finger 1 (F1, β1/β2) and Finger 2 (F2, β3/β4), and the intervening helix is called the wrist region [Fig. 2(A)]. An unpaired cystine at the dimer interface forms an intermolecular disulfide bond that links the opposing monomer chains. The other conserved cysteines form a cystine-knot motif that is present in numerous other growth factors. Within this motif, a ring is formed from linking adjacent strands (β2 and β4) through two proximally spaced disulfide bonds [Fig. 2(A)]. An additional disulfide bond linking β1 to β3 is threaded through this ring to complete the knot fold. The overall curvature of the fingers, as a result, forms very prominent convex and concave surfaces [Fig. 2(B)]. With these surfaces, the ligands display multiple large hydrophobic interfaces, which play critical roles in defining their ability to sequester and bind cognate receptors and antagonists [Fig. 2(C)].30–33
Figure 2.
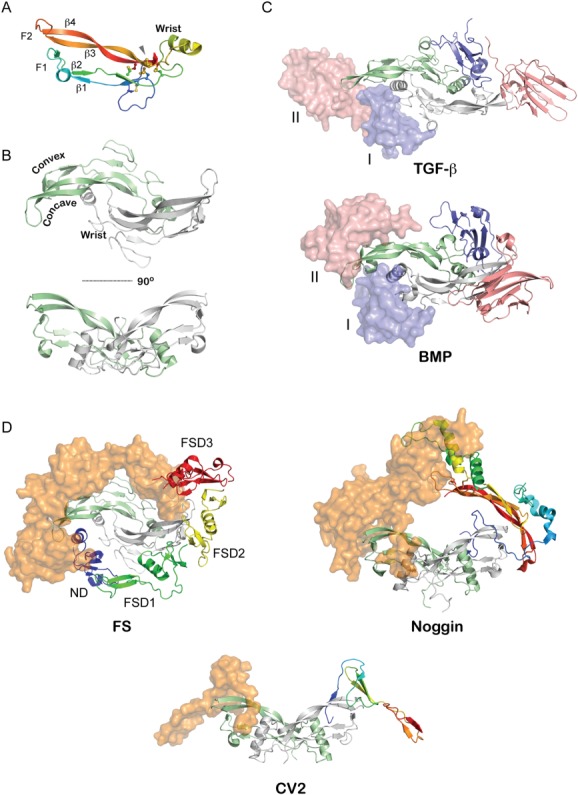
Structure of TGF-β ligands and their associated complexes. A: Representative structure of a TGF-β ligand monomer (Myostatin from PDB 3HH2).37 Intramolecular disulfide bonds are shown as sticks. Labels indicate various β-stands in the ligand as well as identify the finger-wrist-finger architecture of the ligands. B: Structure of the mature TGF-β ligand homodimer (PDB 3HH2) with one monomer colored in light green and another in gray.37 The monomers are linked via an intermolecular disulfide bond. This architecture exposes extended convex and concave surfaces on the protein, which have a strong hydrophobic character and define the ability of the protein to interact with its cognate receptors. C: Structure of TGF-β ligands bound to their Type I and Type II receptors (PDB 2PJY).93 The ligand is represented in ribbon (green, gray) with the receptors represented in both surface and ribbon representation (pink, blue). The receptors for the TGF-β subclass (top) come into contact during binding since the Type I receptors binds more towards the fingertips of the ligand while those for the BMP subclass (bottom) do not, where the Type I receptor binds more significantly to the exposed convex epitope of BMP ligands (PDB 2GOO).94 D: Representative structures of various BMP-antagonist complexes. Ligands are indicated by ribbon diagrams (green, gray), while antagonists are shown as rainbow colored ribbons (one half) and orange surface representations. (left) FS-Myostatin complex (PDB 3HH2).37 Labels indicate various domains of FS. (middle) Noggin-BMP7 complex (PDB 1M4U).34 (right) CV2-BMP2 complex (PDB 3BK3).39 A single VWC domain was solved in complex from the larger multidomain CV2 protein.
Secondary to the discovery of the TGF-β superfamily of ligands, researchers uncovered numerous factors that could bind and neutralize their activity. In the past years, developmental biology has had a defining role identifying several protein modulators that function to bind and block ligand signaling, including Noggin, Chordin, and members of the Follistatin (FS) and DAN family of protein antagonists2,3 (Fig. 1). Interestingly, despite the significant structural conservation across the TGF-β superfamily, their extracellular antagonists are extremely structurally diverse, where size, domain layout, and the overall and local folds are highly variable. Furthermore, these antagonists exhibit significant structural diversity even within their corresponding families.
In 2002, the structure of Noggin bound to BMP7 was solved, making it the first structure of any TGF-β ligand-antagonist complex.34 This structure showed that Noggin directly blocks both the Type I and Type II receptor binding interfaces on BMP7 [Fig. 2(D)]. Similar to the ligands, Noggin exists as a disulfide linked homodimer, where each monomer contains a growth factor like cystine-knot motif and a series of finger-like β-strands. In 2005, the structure of the FS-Activin A complex was determined [Fig. 2(D)].35 FS, being much larger and structurally unique in comparison to Noggin, utilizes multiple domains for binding, where two molecules of FS form a donut-like structure around the ligand. Despite this difference, both FS and Noggin directly bind and block all four receptor binding epitopes of the ligand. Stemming from this work, our lab has resolved a number of FS-ligand structures, which has revealed that these antagonists can undergo conformational shifts to accommodate differences in ligand structure and their corresponding exposed surfaces.35–38 Most recently, the structure of a single domain of Crossveinless-2 (CV2) bound to BMP2 was resolved, which utilizes a prescribed “clip” mechanism to pinch together the ligand fingers and inhibit receptor sequestration [Fig. 2(D)].39 Taken together, these structures highlight the diversity for both the structures and mechanisms utilized to antagonize TGF-β signaling. Unfortunately, a significant number of TGF-β and BMP antagonists remain structurally and functionally unclassified and under represented in the Protein Data Bank (PDB), including the DAN family of BMP antagonists.
DAN Family of BMP Antagonists
The founding member of the DAN family, Nbl1 (also known as Dan or DAND1), was identified as a potential tumor suppressor in neuroblastoma cell lines and for its role in cell cycle regulation.40 Nbl1 was shown to exist as a 165 amino acid long glycoprotein containing a cysteine-rich domain (CRD) with flanking, non-cysteine containing, N- and C-termini [Fig. 3(B)]. In the following years, several other proteins containing this CRD (subsequently referred to as the DAN domain) were identified, accounting for seven total members [Fig. 3(A)].3,4
Figure 3.
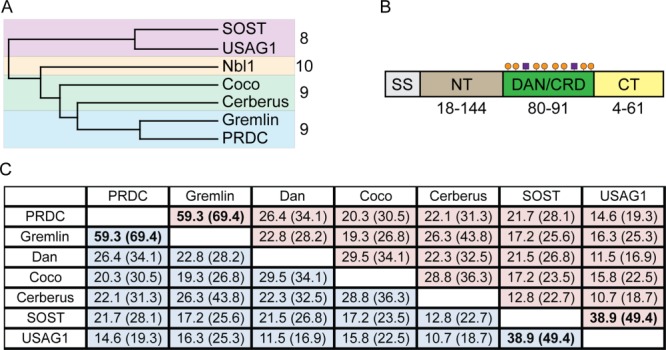
DAN-family of protein antagonists. A: Phylogenetic tree based upon amino acid conservation across the family. The family can be separated into three groups based upon extended amino acid and cysteine conservation. Different colors (red, yellow, blue) indicate the different subgroups. Numbers indicate the number of cysteines within each group of proteins. B: Overall DAN family architecture. Different colors represent the different regions found in DAN-family proteins. SS is the signal sequence (gray), NT is the N-terminus (brown), DAN/CRD is the functional DAN-domain or cystine-rich domain (green), and CT is the C-terminus (yellow). Orange circles represent the conserved eight cysteines in all family members that help define conservation within the family as well as their core cystine-knot and fold. Purple boxes indicate the location of variable cysteines. Numbers below the diagram represent the range of amino acids found in these varying regions across the family. For example, in the NT, DAN only contains 18 amino acids while Cerberus has 144. These numbers help to show that the family maintains the majority of its conservation within the DAN domain while exhibiting a large amount of variability outside of this domain. C: Amino acid sequence identity table. Identity of only the DAN domain is in parentheses. Numbers in bold represent the highest scores across the family, occurring between (1) PRDC and Gremlin and (2) SOST and USAG-1.
Despite sharing the DAN domain, conservation across this family is exceptionally low, with only a handful (<5%) of residues being completely conserved. By cross-comparison, the family can be subdivided into three main groups based upon both extended amino acid and cysteine conservation: (1) Gremlin, PRDC, Cerberus, and Coco (containing nine cysteines); (2) SOST and USAG-1 (containing 8 cysteines); and (3) Nbl1 (containing 10 cysteines) [Fig. 3(A)]. In addition, the N- and C-termini are extraordinarily variable both in length and composition. On average, the N-terminus is typically longer than the C-terminus. One exception is Nbl1, which has the longest C-terminus with 58 residues and the shortest N-terminus with 10 residues. Two pairs of family members stand out as the most similar; Gremlin and PRDC, which are 65% identical, and SOST and USAG-1, which are 45% identical [Fig. 3(C,D)]. The remaining members only share <30% identity with the next closest member. This sequence diversity parallels the wide range of function associated with the family.
Although Nbl1 was the first member discovered, bioinformatics suggests that Gremlin was the first to evolve.41 While phenotypic roles vary broadly within the family, it had been speculated that each DAN family member functioned through high affinity inhibition of BMP ligands. However, the extent and specificity for each member to bind to BMPs is incomplete and often controversial. For the known, strong BMP antagonists in the DAN family, including Gremlin, PRDC, and Coco (Grem1, Grem2, and Grem3, respectively), their target affinity and ability to inhibit BMP signaling and downstream SMAD 1/5/8 phosphorylation is robust, showing preference for BMP2, BMP4, and BMP7.3–5,42–48 In addition, Nbl1 also shows preference for BMP2 and BMP4, as well as GDF5 (growth and differentiation factor 5 or BMP14), although with reduced affinity.49–51 For the less robust BMP antagonists within the DAN family, results and conclusions are often conflicting, bringing distortion to our perception of their true activities.
The DAN family members Cerberus, SOST, and USAG-1 have proven difficult to characterize in terms of BMP signal inhibition, resulting in conflicting manuscripts regarding their function. In terms of Cerberus, the Xenopus isoform of the protein shows a strong phenotype towards BMP inhibition, as well as Wnt and Nodal antagonism.20 However, human and mouse Cerberus exhibit a significantly less pronounced phenotype towards BMP inhibition, despite showing strong in vitro binding to both BMP2 and BMP4.52–55 In terms of SOST and USAG-1, there is conflicting in vivo evidence to support SOST in the direct inhibition of BMP ligands, with more compelling evidence existing for USAG-1.56–61 On the other hand, in vitro data shows that both SOST and USAG-1 can inhibit BMP ligands, although less potently when compared to Noggin.56,57,59,62–65 Culminating this multitude of results, SOST, and probably USAG-1, appears to be less specific for BMP2 and BMP4, showing a greater preference for BMP5, BMP6, and BMP7.59,62–65 Although SOST does not inhibit BMP7 signaling when both are applied exogenously, inhibition is observed endogenously during cotransfection.64,65 Therefore, it has been proposed that SOST may function to antagonize BMP signaling through direct interaction with the latent form or prodomain of the ligand.64,65
Until recently, work pinpointing the functional BMP binding epitope within the DAN family has been minimal. Initially, a study on Gremlin suggested that a particular stretch of amino acids within its DAN domain likely defined its ability to antagonize BMP4. Furthermore, this study showed that the termini of Gremlin could be replaced without compromising BMP inhibition.48 Despite this success, deletion mutagenesis provided little insight for identifying the corresponding BMP binding epitope within USAG-1.62
Molecular studies of DAN family proteins started in 2007 with two NMR (nuclear magnetic resonance) structures of SOST [Fig. 4(A)].66,67 SOST is a monomer with a structured DAN domain and highly flexible N- and C-termini. The DAN domain exhibits a finger-wrist-finger architecture with the cystine-knot motif positioned opposite the fingertips [Fig. 4(A)]. As anticipated, the overall architecture of the fold is very similar to other cystine-knot containing proteins and growth factors, including BMP, Noggin, FSH-β (follicle stimulating hormone-β), Artemin, and VEGF.34,68–70 However, a fourth and additional disulfide bond linking Finger 1 to Finger 2 is present in SOST and is characteristic of the DAN-domain [Fig. 4(A)]. The significance of this disulfide bond is unknown but could potentially provide stability to the fingers.
Figure 4.
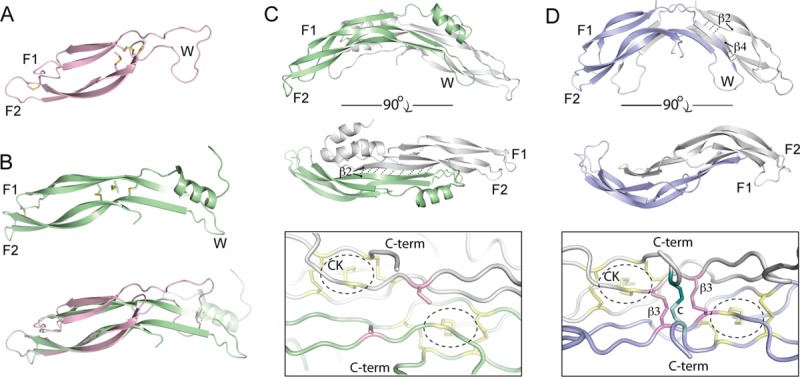
Structures of DAN-family antagonists. A: NMR Structure of SOST (PDB 2K8P) represented in ribbon diagram (pink).66 Disulfide bonds are shown as sticks and the finger-wrist-finger architecture of the proteins are labeled, showing each finger and the wrist region. B: (top) Crystal structure of one PRDC monomer (PDB 4JPH) represented in ribbon (green).43 (bottom) Overlay of the SOST and PRDC monomer structures. The N-terminal helix of PRDC is faded to ease comparison of the core DAN domains. As can be seen, there is substantially more secondary structural content, in the form of β-strands, in the wrist region of PRDC as compared to SOST. These β-strands in PRDC help form the dimer interface of the protein. C: (top) Structure of the PRDC dimer. The opposing monomer chains are shown in different colors (green, gray) with labels indicating the β-strands involved in dimerization and dashed lines showing hydrogen bonds. (bottom) Zoomed in view of the cystine-knot of PRDC near its free, unbound, cysteine. Disulfide bonds that form the cystine-knot are colored yellow and circled. The 9th or unpaired cysteines of PRDC are colored pink. D: (top) Crystal structure of the Norrin dimer (PDB 4MY2).73 The opposing monomer chains are shown in different colors (blue, gray) with labels indicating the finger-wrist-finger architecture and the β-strands and hydrogen bonds important for stabilizing the dimer fold. (bottom) Zoomed in view of the cystine-knot of Norrin near its intermolecular disulfide bonds. The cystine-knot is annotated as above. Norrin forms three intermolecular disulfide bonds; two that are adjacent to the CK and in similar locations to the free cysteine in PRDC (pink) and one in the C-terminus (teal).
Although the structure of SOST was shown to be monomeric, it was believed that DAN family members containing 9 cysteines existed as mature, disulfide-bonded homodimers.11 Since the odd cysteine in DAN family proteins aligns closely with the odd cysteine in BMPs required for intermolecular disulfide bonding, this hypothesis became the accepted dogma.11,71 In 2012, our lab used a combination of size exclusion chromatography (SEC) and analytical ultracentrifugation (AUC) to show that PRDC was in fact a mature homodimer.71 Intriguingly, the study revealed that the PRDC homodimers were not linked via an intermolecular disulfide bond. This result was further recapitulated for Nbl1, which contains 10 cysteines.71 Both the PRDC and Nbl1 homodimers showed significant resistance to reduction (1 mM DTT) and denaturation (6M urea).71 Furthermore, mutation of the odd cysteine in PRDC had little effect on the protein's ability to dimerize and inhibit BMP signaling in vitro and in vivo.71 In support, expression of PRDC and Gremlin (not published) in HEK293 cells did not yield a dimeric form that was reducible upon addition of DTT.71 Taken together, it is likely that PRDC, as well as Cerberus, Gremlin, and Coco, do not function as covalently linked dimers.
Recently, we determined the structure of PRDC by X-ray crystallography [Fig. 4(C)].43 This structure revealed that two monomers of PRDC come together to form an extended, zipper-like antiparallel β-sheet in the core of the protein [Fig. 4(C)]. Similar to BMP ligands, the PRDC homodimer forms in a head-to-tail or antiparallel fashion, strongly resembling a growth factor type fold. This dimerization likely causes the global arching seen in the fold of the PRDC dimer, where N-terminal α-helices, not apparent in the structures of SOST, flank the DAN domain [Fig 4(C)].43,66,67 These N-terminal helices in PRDC appear to interact with the opposing monomer, potentially providing stabilizing forces to the dimer. Taken together with the zipper-like dimer interface at the core of the protein, these stabilizing forces likely account for the ability of the PRDC dimer to resist both reduction and denaturation.71 Finally, looking at the concave surface of the PRDC dimer, the odd cysteines of the protein monomers are shown to exist in the free-sulfhydryl state, where these cysteines are substantially outside of the acceptable range for disulfide formation [Fig. 4(C)]. This distance constraint between cysteines likely explains the absence of disulfide linked oligomers during oxidative refolding within mammalian cells and in vitro following bacterial cell production.43,71,72
Upon comparison, the PRDC monomer fold strongly resembles that of SOST [Fig. 4(B)]. However, differences in the two structures help explain why SOST does not form homodimers similar to PRDC. PRDC has additional α-helical content in its N-terminus as well as increased β-strand content within both the wrist region and DAN domain [Fig. 4(B,C)]. These two elements comprise a major portion of the dimer interface in PRDC, which are absent in SOST and likely explained by significant sequence variation (Fig. 5).43 In a different sense, USAG-1 has been shown via crosslinking studies to form noncovalent homodimers, despite sharing high sequence similarity with SOST even within the wrist region.62 Because of these discrepancies, more work discerning the oligomeric state within the DAN family is needed. Further, it will be interesting to determine whether or not the oligomeric state of these proteins correlates with functionality.
Figure 5.
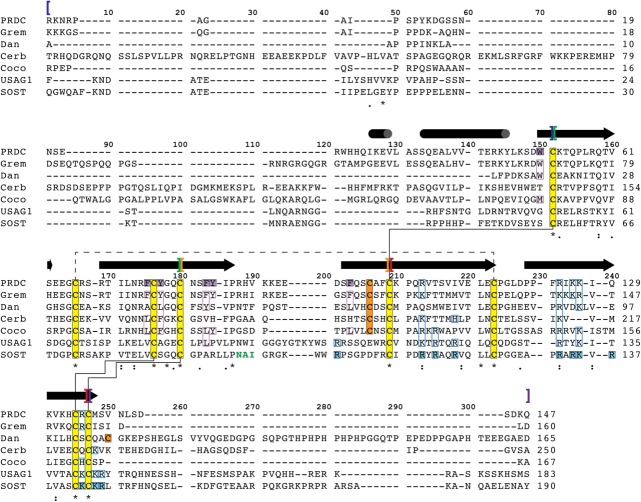
DAN-family protein multiple sequence alignment. Members of the DAN-family were aligned using ClustalW.95 Numbers on the right of the alignment indicated the amino acid number at that position for the corresponding protein (with amino acid 1 being the first translated from the signal sequence). Brackets above the alignment dictate various regions in the proteins, blue being the N-terminus, green being Finger 1, orange being the Wrist, red being Finger 2, and purple being the C-terminus. The black cylinders (helices) and arrows (β-strands) represent secondary structure content based upon the crystal structure of PRDC.43 Yellow filled boxes show the 8 conserved cysteines throughout the family while those in orange filled boxes are additional cysteines dictated for specific family members. Solid black lines between cysteines indicated disulfide bonds forming the cystine-knot in the DAN-domain while a dotted line represents the disulfide bond linking Finger 1 to Finger 2. Purple filled boxes represent those amino acids identified in PRDC to be important for BMP binding while those in purple outlined boxes are seemingly conserved for certain family members.43 Blue-filled boxes represent amino acids identified for SOST to be important for heparin binding while those in blue outlined boxes represent those amino acids that are conserved across the remaining DAN-family.66 The bold green text in the sequence of SOST represents the linear motif identified to be important for binding to the Wnt coreceptors LRP5 and LRP6.79,80
Concurrently with structural characterization, mutagenic studies were performed on PRDC to determine the propensity of various regions to antagonize BMP ligands. These results showed that PRDC derives a significant portion of its BMP affinity along the convex surface of the dimer within the DAN domain.43 This surface, which is partially buried or protected by flanking α-helices, showed the greatest reduction in BMP binding and inhibition when altered. Furthermore, these identified amino acids were large and hydrophobic in nature, in agreement with the hydrophobic tendencies observed for both antagonist and receptor mediated binding to BMP and TGF-β ligands.
Comparing these residues across the entirety of the DAN family shows that a number of these amino acids are, in part, conserved. Conservation is most significant for those DAN family members described as being moderate or strong antagonists, specifically PRDC, Gremlin, Dan, and Coco (Fig. 5). For the weaker or less pronounced antagonists, these residues show limited conservation, especially in the case of SOST and USAG-1, potentially accounting for part of the observed differences in functionality (Fig. 5). However, oligomerization might also play a role in BMP affinity. The homodimerization of certain DAN family members, such as PRDC Nbl1, could provide increased affinity BMP ligands through avidity effects, similar to the interaction of Noggin with BMP7. Therefore, it can be speculated that SOST might have lost part of its ability to inhibit BMP signaling through the evolutionary loss of its ability to dimerize.
On the basis of the structure/function data available for PRDC, a model has been proposed to describe mechanistically how the protein might function to bind and antagonize BMP ligands.43 Since a portion of the BMP binding epitope on PRDC is buried by flanking N-terminal helices, it is speculated that these helices are required to move prior to ligand binding. This is supported by both crystallographic temperature factors and 1-D SAXS (small angle x-ray scattering) data suggesting intrinsic flexibility within these regions. Taken together, PRDC can be thought to exist in a state of flux, where the N-termini helices are capable of sampling nearby space, either shielding (closed conformation) or exposing (open conformation) the underlying BMP binding epitope. While modeling strategies have been utilized to build a hypothetical structure of the PRDC-BMP complex, obtaining reliable results has proven problematic. This is compounded by a lack of data characterizing the overall fold of the complex, the stoichiometry of the interaction, and which receptor binding motifs PRDC functions to block on BMP.
While Noggin retains several similarities to the DAN family in terms of fold and size, it is likely that the structure of a DAN-BMP complex will differ from that of Noggin-BMP7. For example, Gremlin has been shown not to require its N-terminus to antagonize BMP4. In addition, both Gremlin and PRDC have been supported to require amino acids within their DAN domains to achieve BMP inhibition.48 Noggin, however, derives the majority of its affinity for BMP7 from its N-terminus, as seen structurally [Fig. 2(D)].34 Furthermore, Noggin exists in a different dimeric conformation as compared with PRDC, sustaining a head-to-head or parallel conformation supported via an intermolecular disulfide bond. Ultimately, more work needs to be done to validate the current PRDC binding model, where the structure of a DAN-BMP complex would prove most beneficial.
Recently, the crystal structure of Norrin was determined [Fig. 4(D)].73 While Norrin is typically not categorized in the DAN family, it has been shown to inhibit BMP signaling as well as activate Wnt signaling.74–77 Similar to PRDC, Norrin forms an arched head-to-tail dimer, sharing the finger-wrist-finger and cystine-knot architecture, including the disulfide bond linking Finger 1 to Finger 2. While the similarity between Norrin and PRDC is striking, significant differences are apparent between their corresponding wrist regions and dimer interfaces. In Norrin, the β-strand (β2) of one chain forms an anti-parallel interaction with the β2 of the adjacent chain, similar to PRDC, but then extends to form additional anti-parallel contacts with β4 [Fig. 4(C,D)]. This additional H-bonding likely accounts for the increased curvature present in the wrist region of Norrin. Interestingly, Norrin contains three additional cysteines that allow it to form a highly stable disulfide-bonded dimer. Two of these cysteines lie in proximity of the corresponding location of the unpaired cysteine in PRDC, forming two intermolecular disulfide bonds that are spaced a single residue apart [Fig. 4(C,D)].73 Furthermore, a third disulfide bond links the opposing Norrin monomers to bring their C-termini together [Fig. 4(D)]. Future studies are needed to elucidate how Norrin functions to antagonize BMP signaling and if it parallels the DAN family.
An interesting aspect of DAN function is the ability of these antagonists to interact with the extracellular matrix, including heparin and heparan oligosaccharides. It has been shown that SOST, Gremlin, PRDC, and USAG-1 can all bind to heparin or heparan sulfate, a characteristic not shared across the entirety of the family (such as Nbl1 and Cerberus).3,26,43,62,66 NMR studies of SOST showed that the heparin binding epitope was dispersed throughout the DAN domain, extending from the wrist region into Finger 2.66 However, the location of heparin binding does not appear to be conserved across the family through comparison by multiple alignment (Fig. 5). Comparison of the electrostatic surface potential shows a clear distinction between SOST and PRDC (Fig. 6). Using PRDC to model other DAN family members, the distribution of positive charges is shown to be highly variable (similar to BMP ligands), suggesting differences in their heparin binding epitopes, affinity, and potentially their functional use of heparin and heparan binding (Fig. 6).
Figure 6.
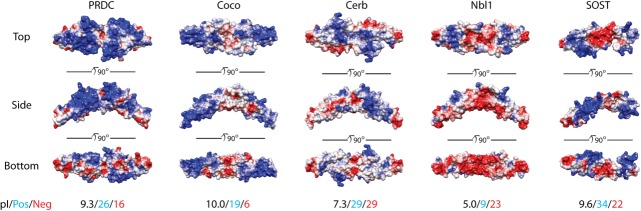
DAN-family electrostatic surface potentials. Surface potentials for several different DAN-family members are shown using top, side, and bottom views. Potentials were calculated using APBS and are colored on a scale of −6 to 6 kbT/ec (red to white to blue).96 Red indicates a negative surface potential while blue represents a positive surface potential. PRDC surface representation is based upon the crystal structure of the dimer. Coco, Cerberus, and Nbl1 were modeled using SwisProt2.0 using PRDC (PDB 4JPH) as an input structure.43,97 These members are plotted as dimers, where Nbl1 is known to exist as a dimer and speculated for Coco and Cerberus. SOST surface potential is based upon the NMR solution of the protein (PDB 2K8P) and is shown as a monomer.66 Beneath the protein surfaces are numbers indicating the pI (black) of each DAN-family protein, the number of positive residues (blue; lysines and arginines), and the number of negative residues (red; glutamates and aspartates) per monomer of each full-length antagonist.
Role in Wnt Signaling
It was originally thought that all members of the DAN family would function to inhibit BMP signaling. The first indication of signaling cross-talk was shown in 1999, where Xenopus Cerberus (xCerb) was found to inhibit Wnt signaling in addition to BMP signaling.20 Furthermore, these activities localized to different regions in xCerb, with xWnt8 binding localizing to the N-terminus and BMP and Nodal binding localizing to the DAN domain. Interestingly, direct binding of Wnt ligands appears unique to xCerb, where the mouse homologue lacks this capability.55
In addition to xCerb, SOST and USAG-1 have been shown to modulate Wnt signaling. Initially, USAG-1 was described in both Wnt activating and inhibitory roles.78 This phenotype was later shown to derive from a direct interaction between USAG-1 and the Wnt co-receptor, LRP6 (low-density lipoprotein-related protein 6).62 In 2001, mutations in the SOST gene were identified in humans with the bone disorder sclerosteosis, with the disease being initially associated with a BMP inhibitory phenotype.21 However, this phenotype has been reclassified, as SOST also shares the ability to antagonize Wnt signaling through a direct interaction with LRP5/LRP6.22,23 Since both BMP and Wnt signaling directly control bone mass, therapeutics targeted at sequestering SOST have become highly regarded, prompting structural studies aimed at understanding the interaction between SOST and LRP5/6.79,80
The ectodomain of LRP5/6 consists of four β-propeller/EGF-like domains. It has been shown that the first repeat contains a peptide recognition motif capable of binding the consensus peptide sequence NXI.80 This sequence is present in both Wnt antagonists Dickkopf-1 (Dkk1) and SOST, which are structurally unrelated. In SOST, the NXI motif is located in a loop within the wrist region of its DAN-domain. Furthermore, this motif is present in USAG-1 but absent in the remaining DAN family members. The NXI motif of SOST is located in the distal tip of the molecule, where it is likely accessible for interaction with LRP5/6. In contrast, the corresponding segment in PRDC, which lacks the NXI motif, forms part of its dimer interface and is partially buried.43 This suggests that access to this peptide segment might differ depending on the monomeric or dimeric state of the protein. While SOST appears to be monomeric, however, USAG-1 has been implicated as a dimer, suggesting that dimerization may not limit the NXI motif from binding LRP5/6.62 As such, it will be interesting to determine what role oligomerization plays in mediating both BMP and Wnt antagonism.
While DAN family members have been implicated in Wnt antagonism, this role is complex as different mechanisms for inhibition likely exist depending on the specific Wnt ligand in question. For example, differing results have been reported in regards to the inhibition of Wnt3a by SOST.79,81,82 Wnt3a binds to the third propeller domain of LRP5/6 while SOST binds to the first propeller domain, possibly implying an allosteric mechanism of regulation.79,83 However, the concentration of SOST required to inhibit Wnt3a versus Wnt1 signaling is drastically different, suggesting that inhibition of Wnt3a by SOST may not be physiological.82 Furthermore, a short peptide derived from SOST containing the NXI motif was capable of antagonizing Wnt1 but not Wnt3a signaling.79,82,83 Taken together, the mechanisms required to antagonize these different Wnt ligands is inherently different. To complicate matters further, USAG-1 and SOST can also bind to the alternative Wnt coreceptor, LRP4.79,84,85 While the NXI motif is necessary for LRP5/6 binding, it does not appear to be involved in the interaction with LRP4, suggesting different mechanisms of mediating Wnt signaling.79
Given that Norrin activates Wnt signaling through direct interaction with the Wnt coreceptors, it raises the possibility that certain DAN family members may share this ability. On the basis of the similarity of the PRDC and Norrin structures, in addition to their conserved dimeric and growth factor like folds, it will be interesting to see whether or not certain DAN family members can also activate canonical Wnt signaling.74–76
Role in VEGF Signaling
Recently, the DAN family member, Gremlin, was shown to promote angiogenesis, pointing to roles in both cancer progression (or inhibition) and tumor survival.24,86,87 Gremlin was also shown to be upregulated in endothelial cells isolated from human lung cancers.88 Interestingly, this proangiogenic activity of Gremlin is not related to its role in BMP or even, possibly, Wnt antagonism, but through its ability to activate the VEGF receptor VEGFR2.25 Binding studies have shown that Gremlin specifically interacts with VEGFR2 (not VEGFR1 or VEGFR3) with an apparent affinity of 47 nM.25 While this is comparatively lower than its affinity for BMP2 or BMP4, it was shown that incubation of Gremlin with BMP did not interfere with VEGFR2 activation, suggesting that alternative epitopes may be utilized to interact with VEGFR2 versus BMP ligands.25 In addition, competition assays have suggested that the binding epitopes on VEGFR2 likely overlap between VEGF and Gremlin. Furthermore, heparin is necessary for Gremlin mediated activation of VEGFR2.26,43 Taken together, Gremlin likely functions to bind BMP ligands and VEGFR2 utilizing alternative and independent epitopes.
To gain insight into the Gremlin-VEGFR2 interaction, we compared the structure of PRDC (70% sequence identify to Gremlin) to the structure of the VEGF ligands. While the overall RMSD is high by direct comparison (9.4 Å), there are several significant similarities between these seemingly different proteins. Overall, both PRDC and VEGF share a similar growth factor like fold, where two monomers are arranged head-to-tail to form a homodimer (Fig. 7). Each monomer contains a centrally located cystine-knot and an N-terminal helix that bridges the two monomers, forming part of their dimer interfaces.70 However, the PRDC dimer is more elongated than the VEGF dimer, with a tip-to-tip distance that is ∼25 Å greater (Fig. 7). Moreover, VEGF dimerization is covalent, supported via two intermolecular disulfide bonds, whereas PRDC is noncovalent with a more compressed dimer interface.43,70
Figure 7.
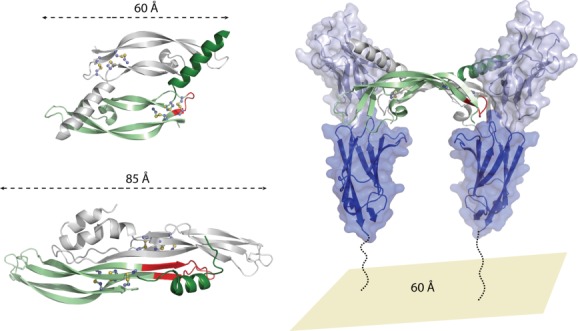
VEGF binding to the VEGFR2 receptor and comparison to PRDC. (top left) Structure of the VEGF-C dimer shown in ribbon representation with one monomer colored green and the opposing colored gray (PDB 2X1X).70 (bottom left) Structure of the PRDC dimer shown in ribbon representation with one monomer colored green and the opposing colored gray.43 For both PRDC and VEGF-C, sticks are shown to represent disulfide bonds. Numbers and lines indicate the length of the proteins from one end to the other. Regions highlighted in red indicated the wrist regions of these proteins. (right) The second and third extracellular domains of VEGFR2 are shown in ribbon and surface representation, colored light and dark blue, respectively. The dotted lines indicate that a substantial portion of the protein is not included in this model (extracellular domains 4–7). For VEGF-C, the wrist region (shown in red) folds out of the when binding to VEGFR2 and is important for mediating the interaction and affinity with this receptor. Number represents the approximate distance between opposing VEGFR2 receptors.
Given that VEGFR2 dimerizes and activates upon VEGF binding, it is easy to speculate that a dimer of Gremlin could perform a similar function.70 However, due to the structural differences between Gremlin and VEGF, it is possible that the spatial assembly of VEGFR2 upon agonist binding is different and/or unique. For VEGF, receptor binding occurs distally at the fingertips and involves contacts with both monomers of the ligand (Fig. 7).70 For Gremlin, the more elongated dimer could position the receptors farther apart during activation, possibly altering signaling outcomes. Interestingly, the kinetics of receptor activation were shown to be different between VEGF and Gremlin with maximum signaling occurring at 12 min versus 21 min, respectively.89 It will be exciting to see how the structures of these complexes differ and if other DAN family members share the capacity to stimulate VEGF signaling.
Conclusions
While the knowledge base for DAN family mediated BMP antagonism is growing, much work is needed to mechanistically understand how this occurs. On the basis of recent observations of cross-talk with both VEGF and Wnt signaling, it will be important to determine what features and epitopes allow for DAN family proteins to differentiate these pathways and which proteins within the family harbor the ability to do so. Furthermore, more work is needed to determine how oligomeric state impacts both BMP antagonism as well as noncanonical signaling roles. Although not discussed at length in this review, Gremlin has been supported to interact with Slit-1 and Slit-2, as well as MMI-1 (macrophage migration inhibitory factor-1).90,91 Additionally, SOST has been shown to bind to the BMP antagonist, Noggin, with low nM affinity.92 As the number of cross-interactors increases for this family, it will be interesting to determine how this impacts cellular signaling outcomes. In final, additional structural and functional work is needed to address these differences, where understanding how DAN family proteins differentiate these multiple protein partners could prove valuable when developing therapeutics to treat diseases associated with misregulated BMP or DAN family signaling.
Glossary
- BMP
bone morphogenetic protein
- DAN
differential screening-selected gene aberrative in neuroblastoma
- GDF
growth and differentiation factor
- Nbl1
neuroblastoma suppression of tumorigenicity 1
- PRDC
protein related to DAN and Cerberus
- TGF-β
transforming growth factor-β
- SOST
Sclerostin
- LRP
low density lipoprotein receptor-related protein
- USAG-1
uterine sensitization-associated gene-1
- VEGF
vascular endothelial growth factor.
References
- 1.Hogan BL. Bone morphogenetic proteins in development. Curr Opin Genet Dev. 1996;6:432–438. doi: 10.1016/s0959-437x(96)80064-5. [DOI] [PubMed] [Google Scholar]
- 2.Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morphogenetic proteins: a critical review. Cell Signal. 2011;23:609–620. doi: 10.1016/j.cellsig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Rider CCC, Mulloy BB. Bone morphogenetic protein and growth differentiation factor cytokine families and their protein antagonists. Biochem J. 2010;429:1–12. doi: 10.1042/BJ20100305. [DOI] [PubMed] [Google Scholar]
- 4.Walsh DW, Godson C, Brazil DP, Martin F. Extracellular BMP-antagonist regulation in development and disease: tied up in knots. Trends Cell Biol. 2010;20:244–256. doi: 10.1016/j.tcb.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Gao H, Chakraborty G, Lee-Lim AP, Mo Q, Decker M, Vonica A, Shen R, Brogi E, Brivanlou AH, Giancotti FG. The BMP inhibitor Coco reactivates breast cancer cells at lung metastatic sites. Cell. 2012;150:764–779. doi: 10.1016/j.cell.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waite KA, Eng C. From developmental disorder to heritable cancer: it's all in the BMP/TGF-β family. Nat Rev Genet. 2003;4:763–773. doi: 10.1038/nrg1178. [DOI] [PubMed] [Google Scholar]
- 7.Koli K, Myllärniemi M, Vuorinen K, Salmenkivi K, Ryynänen MJ, Kinnula VL, Keski-Oja J. Bone morphogenetic protein-4 inhibitor Gremlin is overexpressed in idiopathic pulmonary fibrosis. Am J Pathol. 2006;169:61–71. doi: 10.2353/ajpath.2006.051263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanagita M. Inhibitors/antagonists of TGF-β system in kidney fibrosis. Nephrol Dial Transplant. 2012;27:3686–3691. doi: 10.1093/ndt/gfs381. [DOI] [PubMed] [Google Scholar]
- 9.Massagué J. TGF-β in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu DR, Economides AN, Wang X, Eimon PM, Harland RM. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol Cell. 1998;1:673–683. doi: 10.1016/s1097-2765(00)80067-2. [DOI] [PubMed] [Google Scholar]
- 11.Avsian-Kretchmer O, Hsueh AJW. Comparative genomic analysis of the eight-membered ring cystine knot-containing bone morphogenetic protein antagonists. Mol Endocrinol. 2004;18:1–12. doi: 10.1210/me.2003-0227. [DOI] [PubMed] [Google Scholar]
- 12.Michos O, Panman L, Vintersten K, Beier K, Zeller R, Zúñiga A. Gremlin-mediated BMP antagonism induces the epithelial-mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development. 2004;131:3401–3410. doi: 10.1242/dev.01251. [DOI] [PubMed] [Google Scholar]
- 13.Michos O, Gonçalves A, Lopez-Rios J, Tiecke E, Naillat F, Beier K, Galli A, Vainio S, Zeller R. Reduction of BMP4 activity by Gremlin 1 enables ureteric bud outgrowth and GDNF/WNT11 feedback signalling during kidney branching morphogenesis. Development. 2007;134:2397–2405. doi: 10.1242/dev.02861. [DOI] [PubMed] [Google Scholar]
- 14.Khokha MK, Hsu D, Brunet LJ, Dionne MS, Harland RM. Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat Genet. 2003;34:303–307. doi: 10.1038/ng1178. [DOI] [PubMed] [Google Scholar]
- 15.Zúñiga A, Haramis AP, McMahon AP, Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401:598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]
- 16.Marques S, Borges AC, Silva AC, Freitas S, Cordenonsi M, Belo JA. The activity of the Nodal antagonist Cerl-2 in the mouse node is required for correct L/R body axis. Genes Dev. 2004;18:2342–2347. doi: 10.1101/gad.306504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Zhang Q. Bone morphogenetic protein-7 and Gremlin: new emerging therapeutic targets for diabetic nephropathy. Biochem Biophys Res Commun. 2009;383:1–3. doi: 10.1016/j.bbrc.2009.03.086. [DOI] [PubMed] [Google Scholar]
- 18.Tamminen JA, Parviainen V, Rönty M, Wohl AP. Gremlin-1 associates with fibrillin microfibrils in vivo and regulates mesothelioma cell survival through transcription factor slug. Oncogenesis. 2013;2:e66. doi: 10.1038/oncsis.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurila R, Parkkila S, Isola J, Kallioniemi A, Alarmo E-L. The expression patterns of gremlin 1 and noggin in normal adult and tumor tissues. Int J Clin Exp Pathol. 2013;6:1400–1408. [PMC free article] [PubMed] [Google Scholar]
- 20.Piccolo S, Agius E, Leyns L, Bhattacharyya S, Grunz H, Bouwmeester T, De Robertis EM. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature. 1999;397:707–710. doi: 10.1038/17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunkow ME, Gardner JC, Van Ness J, Paeper BW, Kovacevich BR, Proll S, Skonier JE, Zhao L, Sabo PJ, Fu Y, RS Alisch, L Gillett, T Colbert, P Tacconi, D Galas, H Hamersma, P Beighton, J Mulligan. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am J Hum Genet. 2001;68:577–589. doi: 10.1086/318811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, Harris SE, Wu D. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. 2005;280:19883–19887. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- 23.Ellies DL, Viviano B, McCarthy J, Rey J-P, Itasaki N, Saunders S, Krumlauf R. Bone density ligand, Sclerostin, directly interacts with LRP5 but not LRP5G171V to modulate Wnt activity. J Bone Miner Res. 2006;21:1738–1749. doi: 10.1359/jbmr.060810. [DOI] [PubMed] [Google Scholar]
- 24.Stabile H, Mitola S, Moroni E, Belleri M, Nicoli S, Coltrini D, Peri F, Pessi A, Orsatti L, Talamo F. Bone morphogenic protein antagonist Drm/Gremlin is a novel proangiogenic factor. Blood. 2007;109:1834–1840. doi: 10.1182/blood-2006-06-032276. [DOI] [PubMed] [Google Scholar]
- 25.Mitola S, Ravelli C, Moroni E, Salvi V, Leali D, Ballmer-Hofer K, Zammataro L, Presta M. Gremlin is a novel agonist of the major proangiogenic receptor VEGFR2. Blood. 2010;116:3677–3680. doi: 10.1182/blood-2010-06-291930. [DOI] [PubMed] [Google Scholar]
- 26.Chiodelli P, Mitola S, Ravelli C, Oreste P, Rusnati M, Presta M. Heparan sulfate proteoglycans mediate the angiogenic activity of the vascular endothelial growth factor receptor-2 agonist Gremlin. Arterioscler Thromb Vasc Biol. 2011;31:e116–e127. doi: 10.1161/ATVBAHA.111.235184. [DOI] [PubMed] [Google Scholar]
- 27.Costello CM, Cahill E, Martin F, Gaine S, McLoughlin P. Role of Gremlin in the lung: development and disease. Am J Respir Cell Mol Biol. 2010;42:517–523. doi: 10.1165/rcmb.2009-0101TR. [DOI] [PubMed] [Google Scholar]
- 28.Daopin S, Piez KA, Ogawa Y, Davies DR. Crystal structure of transforming growth factor-β2: an unusual fold for the superfamily. Science. 1992;257:369–373. doi: 10.1126/science.1631557. [DOI] [PubMed] [Google Scholar]
- 29.Schlunegger MP, Grütter MG. An unusual feature revealed by the crystal structure at 2.2Å resolution of human transforming growth factor-β2. Nature. 1992;358:430–434. doi: 10.1038/358430a0. [DOI] [PubMed] [Google Scholar]
- 30.Kirsch T, Sebald W, Dreyer MK. Crystal structure of the BMP-2–BRIA ectodomain complex. Nat Struct Mol Biol. 2000;7:492–496. doi: 10.1038/75903. [DOI] [PubMed] [Google Scholar]
- 31.Greenwald J, Groppe J, Gray P, Wiater E, Kwiatkowski W, Vale W, Choe S. The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol Cell. 2003;11:605–617. doi: 10.1016/s1097-2765(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 32.Greenwald J, Fischer WH, Vale WW, Choe S. Three-finger toxin fold for the extracellular ligand-binding domain of the type II Activin receptor serine kinase. Nat Struct Biol. 1999;6:18–22. doi: 10.1038/4887. [DOI] [PubMed] [Google Scholar]
- 33.Thompson TB. Structures of an ActRIIB:Activin A complex reveal a novel binding mode for TGF-β ligand:receptor interactions. EMBO J. 2003;22:1555–1566. doi: 10.1093/emboj/cdg156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Groppe J, Greenwald J, Wiater E, Rodriguez-Leon J, Economides AN, Kwiatkowski W, Affolter M, Vale WW, Belmonte JCI, Choe S. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature. 2002;420:636–642. doi: 10.1038/nature01245. [DOI] [PubMed] [Google Scholar]
- 35.Thompson TB, Lerch TF, Cook RW, Woodruff TK. The structure of the Follistatin: activin complex reveals antagonism of both type I and type II receptor binding. Dev Cell. 2005;9:535–543. doi: 10.1016/j.devcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Stamler R, Keutmann HT, Sidis Y, Kattamuri C, Schneyer A, Thompson TB. The structure of FSTL3:Activin A complex: differential binding of N-terminal domains influences Follistatin-type antagonist specificity. J Biol Chem. 2008;283:32831–32838. doi: 10.1074/jbc.M801266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cash JN, Rejon CA, McPherron AC, Bernard DJ, Thompson TB. The structure of Myostatin:Follistatin 288: insights into receptor utilization and heparin binding. EMBO J. 2009;28:2662–2676. doi: 10.1038/emboj.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cash JN, Angerman EB, Kattamuri C, Nolan K, Zhao H, Sidis Y, Keutmann HT, Thompson TB. Structure of Myostatin:Follistatin-like 3: N-terminal domains of Follistatin-type molecules exhibit alternate modes of binding. J Biol Chem. 2012;287:1043–1053. doi: 10.1074/jbc.M111.270801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang JL, Qiu LY, Kotzsch A, Weidauer S, Patterson L, Hammerschmidt M, Sebald W, Mueller TD. Crystal structure analysis reveals how the Chordin family member Crossveinless 2 Blocks BMP-2 receptor binding. Dev Cell. 2008;14:739–750. doi: 10.1016/j.devcel.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura Y, Ozaki T, Ichimiya S, Nakagawara A, Sakiyama S. Ectopic expression of DAN enhances the retinoic acid-induced neuronal differentiation in human neuroblastoma cell lines. Biochem Biophys Res Commun. 1998;243:722–726. doi: 10.1006/bbrc.1998.8112. [DOI] [PubMed] [Google Scholar]
- 41.Le Petillon Y, Oulion S, Escande M-L, Escriva H, Bertrand S. Identification and expression analysis of BMP signaling inhibitors genes of the DAN family in amphioxus. Gene Expr Patterns. 2013;13:377–383. doi: 10.1016/j.gep.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 42.Bates TJD, Vonica A, Heasman J, Brivanlou AH, Bell E. Coco regulates dorsoventral specification of germ layers via inhibition of TGFβ signalling. Development. 2013;140:4177–4181. doi: 10.1242/dev.095521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nolan K, Kattamuri C, Luedeke DM, Deng X, Jagpal A, Zhang F, Linhardt RJ, Kenny AP, Zorn AM, Thompson TB. Structure of protein related to dan and cerberus: insights into the mechanism of bone morphogenetic protein antagonism. Structure. 2013;21:1417–1429. doi: 10.1016/j.str.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pearce JJ, Penny G, Rossant J. A mouse Cerberus/Dan-related gene family. Dev Biol. 1999;209:98–110. doi: 10.1006/dbio.1999.9240. [DOI] [PubMed] [Google Scholar]
- 45.Sudo S, Avsian-Kretchmer O, Wang LS, Hsueh AJ. Protein related to DAN and Cerberus is a bone morphogenetic protein antagonist that participates in ovarian paracrine regulation. J Biol Chem. 2004;279:23134–23141. doi: 10.1074/jbc.M402376200. [DOI] [PubMed] [Google Scholar]
- 46.Ideno H, Takanabe R, Shimada A, Imaizumi K, Araki R, Abe M, Nifuji A. Protein related to DAN and Cerberus (PRDC) inhibits osteoblastic differentiation and its suppression promotes osteogenesis in vitro. Exp Cell Res. 2009;315:474–484. doi: 10.1016/j.yexcr.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 47.Topol LZ, Bardot B, Zhang Q, Resau J, Huillard E, Marx M, Calothy G, Blair DG. Biosynthesis, post-translation modification, and functional characterization of Drm/Gremlin. J Biol Chem. 2000;275:8785–8793. doi: 10.1074/jbc.275.12.8785. [DOI] [PubMed] [Google Scholar]
- 48.Sun J, Zhuang F-F, Mullersman JE, Chen H, Robertson EJ, Warburton D, Liu Y-H, Shi W. BMP4 activation and secretion are negatively regulated by an intracellular Gremlin-BMP4 interaction. J Biol Chem. 2006;281:29349–29356. doi: 10.1074/jbc.M603833200. [DOI] [PubMed] [Google Scholar]
- 49.Dionne MS, Skarnes WC, Harland RM. Mutation and analysis of Dan, the founding member of the Dan family of transforming growth factor β antagonists. Mol Cell Biol. 2001;21:636–643. doi: 10.1128/MCB.21.2.636-643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hung WT, Wu FJ, Wang CJ, Luo CW. DAN (NBL1) specifically antagonizes BMP2 and BMP4 and modulates the actions of GDF9, BMP2, and BMP4 in the rat ovary 1. Biol Reprod. 2012;86:158. doi: 10.1095/biolreprod.111.096172. [DOI] [PubMed] [Google Scholar]
- 51.Katsu K, Tokumori D, Tatsumi N, Suzuki A, Yokouchi Y. BMP inhibition by DAN in Hensen's node is a critical step for the establishment of left-right asymmetry in the chick embryo. Dev Biol. 2012;363:15–26. doi: 10.1016/j.ydbio.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 52.Belo JA, Bachiller D, Agius E, Kemp C, Borges AC, Marques S, Piccolo S, De Robertis EM. Cerberus-like is a secreted BMP and Nodal antagonist not essential for mouse development. Genesis. 2000;26:265–270. [PubMed] [Google Scholar]
- 53.Belo JA, Silva AC, Borges AC, Filipe M, Bento M, Gonçalves L, Vitorino M, Salgueiro AM, Texeira V, Tavares AT, S Marques. Generating asymmetries in the early vertebrate embryo: the role of the Cerberus-like family. Int J Dev Biol. 2009;53:1399–1407. doi: 10.1387/ijdb.072297jb. [DOI] [PubMed] [Google Scholar]
- 54.Yu X, He F, Zhang T, Espinoza-Lewis RA, Lin L, Yang J, Chen Y. Cerberus functions as a BMP agonist to synergistically induce Nodal expression during left-right axis determination in the chick embryo. Dev Dyn. 2008;237:3613–3623. doi: 10.1002/dvdy.21769. [DOI] [PubMed] [Google Scholar]
- 55.Chi L, Saarela U, Railo A, Prunskaite-Hyyryläinen R, Skovorodkin I, Anthony S, Katsu K, Liu Y, Shan J, Salgueiro A-M, JA Belo, J Davies, Y Yokouchi, SJ Vainio. A secreted BMP antagonist, Cer1, fine tunes the spatial organization of the ureteric bud tree during mouse kidney development. PLoS ONE. 2011;6:e27676. doi: 10.1371/journal.pone.0027676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Bezooijen RL, Svensson JP, Eefting D, Visser A, van der Horst G, Karperien M, Quax PHA, Vrieling H, Papapoulos SE, Dijke ten P, CW Lowik. Wnt but not BMP signaling is involved in the inhibitory action of Sclerostin on BMP-stimulated bone formation. J Bone Miner Res. 2007;22:19–28. doi: 10.1359/jbmr.061002. [DOI] [PubMed] [Google Scholar]
- 57.van Bezooijen RL, Roelen BA, Visser A, van der Wee-Pals L, de Wilt E, Karperien M, Hamersma H, Papapoulos SE, Dijke ten P, Löwik CW. Sclerostin is an osteocyte-expressed negative regulator of bone formation, but not a classical BMP antagonist. J Exp Med. 2004;199:805–814. doi: 10.1084/jem.20031454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Bezooijen RL, DeRuiter MC, Vilain N, Monteiro RM, Visser A, van der Wee-Pals L, van Munsteren CJ, Hogendoorn PCW, Aguet M, Mummery CL, SE Papapoulos, P TenDjike, CW Lowik. SOST expression is restricted to the great arteries during embryonic and neonatal cardiovascular development. Dev Dyn. 2007;236:606–612. doi: 10.1002/dvdy.21054. [DOI] [PubMed] [Google Scholar]
- 59.Winkler DG, Sutherland MK, Geoghegan JC, Yu C, Hayes T, Skonier JE, Shpektor D, Jonas M, Kovacevich BR, Staehling-Hampton K. Osteocyte control of bone formation via Sclerostin, a novel BMP antagonist. EMBO J. 2003;22:6267–6276. doi: 10.1093/emboj/cdg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanagita M, Okuda T, Endo S, Tanaka M, Takahashi K, Sugiyama F, Kunita S, Takahashi S, Fukatsu A, Yanagisawa M. Uterine sensitization-associated gene–1 (USAG-1), a novel BMP antagonist expressed in the kidney, accelerates tubular injury. J Clin Invest. 2006;116:70–79. doi: 10.1172/JCI25445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanaka M, Endo S, Okuda T, Economides AN, Valenzuela DM, Murphy AJ, Robertson E, Sakurai T, Fukatsu A, Yancopoulos GD. Expression of BMP-7 and USAG-1 (a BMP antagonist) in kidney development and injury. Kidney Int. 2008;73:181–191. doi: 10.1038/sj.ki.5002626. [DOI] [PubMed] [Google Scholar]
- 62.Lintern KB, Guidato S, Rowe A, Saldanha JW, Itasaki N. Characterization of wise protein and its molecular mechanism to interact with both Wnt and BMP signals. J Biol Chem. 2009;284:23159–23168. doi: 10.1074/jbc.M109.025478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kusu N, Laurikkala J, Imanishi M, Usui H, Konishi M, Miyake A, Thesleff I, Itoh N. Sclerostin is a novel secreted osteoclast-derived bone morphogenetic protein antagonist with unique ligand specificity. J Biol Chem. 2003;278:24113–24117. doi: 10.1074/jbc.M301716200. [DOI] [PubMed] [Google Scholar]
- 64.Dijke ten P, Krause C, de Gorter DJJ, Löwik CWGM, van Bezooijen RL. Osteocyte-derived Sclerostin inhibits bone formation: its role in bone morphogenetic protein and Wnt signaling. J Bone Joint Surg Am 90 Suppl. 2008;1:31–35. doi: 10.2106/JBJS.G.01183. [DOI] [PubMed] [Google Scholar]
- 65.Krause C, Korchynskyi O, de Rooij K, Weidauer SE, de Gorter DJJ, van Bezooijen RL, Hatsell S, Economides AN, Mueller TD, Löwik CWGM, P tenDjike. Distinct modes of inhibition by Sclerostin on bone morphogenetic protein and Wnt signaling pathways. J Biol Chem. 2010;285:41614–41626. doi: 10.1074/jbc.M110.153890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Veverka V, Henry AJ, Slocombe PM, Ventom A, Mulloy B, Muskett FW, Muzylak M, Greenslade K, Moore A, Zhang L, J Gong, X Qian, C Paszty, RJ Taylor, MK Robinson, MD Carr. Characterization of the structural features and interactions of Sclerostin: molecular insight into a key regulator of Wnt-mediated bone formation. J Biol Chem. 2009;284:10890–10900. doi: 10.1074/jbc.M807994200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weidauer SE, Schmieder P, Beerbaum M, Schmitz W, Oschkinat H, Mueller TD. NMR structure of the Wnt modulator protein Sclerostin. Biochem Biophys Res Commun. 2009;380:160–165. doi: 10.1016/j.bbrc.2009.01.062. [DOI] [PubMed] [Google Scholar]
- 68.Jiang X, Liu H, Chen X, Chen P-H, Fischer D, Sriraman V, Yu HN, Arkinstall S, He X. Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor. Proc Natl Acad Sci USA. 2012;109:12491–12496. doi: 10.1073/pnas.1206643109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Silvian L, Jin P, Carmillo P, Boriack-Sjodin PA, Pelletier C, Rushe M, Gong B, Sah D, Pepinsky B, Rossomando A. Artemin crystal structure reveals insights into heparan sulfate binding. Biochemistry. 2006;45:6801–6812. doi: 10.1021/bi060035x. [DOI] [PubMed] [Google Scholar]
- 70.Leppänen VM, Prota AE, Jeltsch M, Anisimov A, Kalkkinen N, Strandin T, Lankinen H, Goldman A, Ballmer-Hofer K, Alitalo K. Structural determinants of growth factor binding and specificity by VEGF receptor 2. Proc Natl Acad Sci USA. 2010;107:2425–2430. doi: 10.1073/pnas.0914318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kattamuri C, Luedeke DM, Nolan K, Rankin SA, Greis KD, Zorn AM, Thompson TB. Members of the DAN family are BMP antagonists that form highly stable noncovalent dimers. J Mol Biol. 2012;424:313–327. doi: 10.1016/j.jmb.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kattamuri C, Luedeke DM, Thompson TB. Expression and purification of recombinant protein related to DAN and Cerberus (PRDC) Protein Expr Purif. 2012;82:389–395. doi: 10.1016/j.pep.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ke J, Harikumar KG, Erice C, Chen C, Gu X. Structure and function of Norrin in assembly and activation of a Frizzled 4–Lrp5/6 complex. Genes Dev. 2013;27:2305–2319. doi: 10.1101/gad.228544.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu Q, Wang Y, Dabdoub A, Smallwood PM, Williams J, Woods C, Kelley MW, Jiang L, Tasman W, Zhang K. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell. 2004;116:883–895. doi: 10.1016/s0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- 75.Xu S, Cheng F, Liang J, Wu W, Zhang J. Maternal xNorrin, a canonical Wnt signaling agonist and TGF-β antagonist, controls early neuroectoderm specification in Xenopus. PLoS Biol. 2012;10:e1001286. doi: 10.1371/journal.pbio.1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ye X, Wang Y, Cahill H, Yu M, Badea TC, Smallwood PM, Peachey NS, Nathans J. Norrin, frizzled-4, and Lrp5 signaling in endothelial cells controls a genetic program for retinal vascularization. Cell. 2009;139:285–298. doi: 10.1016/j.cell.2009.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deng C, Reddy P, Cheng Y, Luo C-W, Hsiao C-L, Hsueh AJW. Multi-functional Norrin is a ligand for the LGR4 receptor. J Cell Sci. 2013;126:2060–2068. doi: 10.1242/jcs.123471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Itasaki N, Jones CM, Mercurio S, Rowe A, Domingos PM, Smith JC, Krumlauf R. Wise, a context-dependent activator and inhibitor of Wnt signalling. Development. 2003;130:4295–4305. doi: 10.1242/dev.00674. [DOI] [PubMed] [Google Scholar]
- 79.Holdsworth G, Slocombe P, Doyle C, Sweeney B, Veverka V, Le Riche K, Franklin RJ, Compson J, Brookings D, Turner J, J Kennedy, R Garlish, J Shi, L Newnham, D McMillan, M Muzylak, MD Carr, AJ Henry, T Ceska, MK Robinson. Characterization of the interaction of Sclerostin with the low density lipoprotein receptor-related protein (LRP) family of Wnt co-receptors. J Biol Chem. 2012;287:26464–26477. doi: 10.1074/jbc.M112.350108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bourhis E, Wang W, Tam C, Hwang J, Zhang Y, Spittler D, Huang OW, Gong Y, Estevez A, Zilberleyb I. Wnt antagonists bind through a short peptide to the first β-propeller domain of LRP5/6. Structure. 2011;19:1433–1442. doi: 10.1016/j.str.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 81.Winkler DG, Sutherland MSK, Ojala E, Turcott E, Geoghegan JC, Shpektor D, Skonier JE, Yu C, Latham JA. Sclerostin inhibition of Wnt-3a-induced C3H10T1/2 cell differentiation is indirect and mediated by bone morphogenetic proteins. J Biol Chem. 2005;280:2498–2502. doi: 10.1074/jbc.M400524200. [DOI] [PubMed] [Google Scholar]
- 82.Boschert V, van Dinther M, Weidauer S, van Pee K, Muth EM, Dijke ten P, Mueller TD. Mutational analysis of Sclerostin shows importance of the flexible loop and the cystine-knot for Wnt-signaling inhibition. PLoS ONE. 2013;8:e81710. doi: 10.1371/journal.pone.0081710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bourhis E, Tam C, Franke Y, Bazan JF, Ernst J, Hwang J, Costa M, Cochran AG, Hannoush RN. Reconstitution of a frizzled8 Wnt3a LRP6 signaling complex reveals multiple Wnt and Dkk1 binding sites on LRP6. J Biol Chem. 2010;285:9172–9179. doi: 10.1074/jbc.M109.092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choi HY, Dieckmann M, Herz J, Niemeier A. Lrp4, a novel receptor for Dickkopf 1 and Sclerostin, is expressed by osteoblasts and regulates bone growth and turnover in vivo. PLoS ONE. 2009;4:e7930. doi: 10.1371/journal.pone.0007930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ohazama A, Johnson EB, Ota MS, Choi HY, Choi HJ, Porntaveetus T, Oommen S, Itoh N, Eto K, Gritli-Linde A, J Herz, PT Sharpe. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS ONE. 2008;3:e4092. doi: 10.1371/journal.pone.0004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ravelli C, Mitola S, Corsini M, Presta M. Involvement of αvβ3 integrin in Gremlin-induced angiogenesis. Angiogenesis. 2013;16:235–243. doi: 10.1007/s10456-012-9309-6. [DOI] [PubMed] [Google Scholar]
- 87.Chen MH, Yeh YC, Shyr YM, Jan YH, Chao Y, Li CP, Wang SE, Tzeng CH, Chang PMH, Liu CY, MH Chen, M Hsiao, CY Huang. Expression of Gremlin 1 correlates with increased angiogenesis and progression-free survival in patients with pancreatic neuroendocrine tumors. J Gastroenterol. 2013;48:101–108. doi: 10.1007/s00535-012-0614-z. [DOI] [PubMed] [Google Scholar]
- 88.Mulvihill MS, Kwon YW, Lee S, Fang LT, Choi H, Ray R, Kang HC, Mao JH, Jablons D, Kim IJ. Gremlin is overexpressed in lung adenocarcinoma and increases cell growth and proliferation in normal lung cells. PLoS ONE. 2012;7:e42264. doi: 10.1371/journal.pone.0042264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maiolo D, Mitola S, Leali D, Oliviero G, Ravelli C, Bugatti A, Depero LE, Presta M, Bergese P. Role of nanomechanics in canonical and noncanonical pro-angiogenic ligand/VEGF receptor-2 activation. J Am Chem Soc. 2012;134:14573–14579. doi: 10.1021/ja305816p. [DOI] [PubMed] [Google Scholar]
- 90.Chen B, Blair DG, Plisov S, Vasiliev G, Perantoni AO, Chen Q, Athanasiou M, Wu JY, Oppenheim JJ, Yang D. Cutting edge: bone morphogenetic protein antagonists Drm/Gremlin and Dan interact with Slits and act as negative regulators of monocyte chemotaxis. J Immunol. 2004;173:5914–5917. doi: 10.4049/jimmunol.173.10.5914. [DOI] [PubMed] [Google Scholar]
- 91.Müller I, Schönberger T, Schneider M, Borst O, Ziegler M, Seizer P, Leder C, Müller K, Lang M, Appenzeller F, O Lunov, B Buchele, M Fahrleitner, M Olbrich, H Langer, T Geisler, F Lang, M Chatterjee, JF deBoer, UJ Tietge, J Bernhagen, T Simmet, M Gawaz. Gremlin-1 is an inhibitor of macrophage migration inhibitory factor and attenuates atherosclerotic plaque growth in ApoE-/- Mice. J Biol Chem. 2013;288:31635–31645. doi: 10.1074/jbc.M113.477745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Winkler DG, Yu C, Geoghegan JC, Ojala EW, Skonier JE, Shpektor D, Sutherland MK, Latham JA. Noggin and Sclerostin bone morphogenetic protein antagonists form a mutually inhibitory complex. J Biol Chem. 2004;279:36293–36298. doi: 10.1074/jbc.M400521200. [DOI] [PubMed] [Google Scholar]
- 93.Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, Schwarz PM, Wrana JL, Hinck AP. Cooperative assembly of TGF-β superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell. 2008;29:157–168. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 94.Allendorph GP, Vale WW, Choe S. Structure of the ternary signaling complex of a TGF-β superfamily member. Proc Natl Acad Sci USA. 2006;103:7643–7648. doi: 10.1073/pnas.0602558103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holst M, Saied F. Multigrid solution of the Poisson—Boltzmann equation. J Comput Chem. 1993;14:105–113. [Google Scholar]
- 97.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]