Abstract
We have previously identified 55 nonribosomal proteins in PAB1-mRNP complexes in Saccharomyces cerevisiae using mass spectrometric analysis. Because one of the inherent limitations of mass spectrometry is that it does not inform as to the size or type of complexes in which the proteins are present, we consequently used analytical ultracentrifugation with fluorescent detection system (AU-FDS) to determine which proteins are present in the 77S monosomal translation complex that contains minimally the closed-loop structure components (eIF4E, eIF4G, and PAB1), mRNA, and the 40S and 60S ribosomes. We assayed by AU-FDS analysis 33 additional PAB1-mRNP factors but found that only five of these proteins were present in the 77S translation complex: eRF1, SLF1, SSD1, PUB1, and SBP1. eRF1 is involved in translation termination, SBP1 is a translational repressor, and SLF1, SSD1, and PUB1 are known mRNA binding proteins. Many of the known P body/stress granule proteins that associate with the PAB1-mRNP were not present in the 77S translation complex, implying that P body/stress granules result from significant protein additions after translational cessation. These data inform that AU-FDS can clarify protein complex identification that remains undetermined after typical immunoprecipitation and mass spectrometric analyses.
Keywords: protein synthesis, analytical ultracentrifugation, translation proteome, PAB1 mRNP complexes
Introduction
The eukaryotic mRNA-protein (mRNP) complexes are involved in a wide range of biological processes. These include complexes involved in the synthesis of mRNA in the nucleus, the transport of the mRNA into the cytoplasm, the synthesis of protein from the mRNA, the subsequent degradation of the mRNA, and, in response to stresses, the sequestration of the mRNA in quiescent nontranslating complexes. Global approaches to identifying the components of these many mRNP complexes have identified a rich assortment of proteins, many of them present in the majority of these studies.1–6 The primary limitation of these previous approaches, which generally utilized mass spectrometric and immunoprecipitation analysis, is that they principally inform only about possible components in mRNP complexes and not which factors are present in which functionally relevant complexes. In addition, no information is provided as to the size of the complexes that are visualized.
We have previously utilized the novel technique of analytical ultracentrifugation with fluorescent detection system (AU-FDS) to unambiguously identify the size and components of the monosomal translation complex from Saccharomyces cerevisiae.7 This 77S translating complex had not previously been isolated or visualized using such traditional techniques as sucrose gradient analysis or gel permeation chromatography. We demonstrated, using GFP fusions to unique yeast proteins, that the 77S complex contains the 40S and 60S ribosomal subunits, mRNA, and the closed-loop structural components, eIF4E, eIF4G, and PAB1, the latter of which are required for a variety of translational processes. Other translation initiation factors were notably absent from this complex. This 77S monosomal translating complex was also responsive to translational stresses and was shown to be distinct from the free 80S ribosome that is not bound to mRNA.7
We have consequently tackled the proteomic analysis of the eukaryotic mRNP by determining which known mRNP complex proteins are components of the translating ribosome. Based on our previous mass spectrometric determination of the PAB1-mRNP proteome, we studied a large group of proteins that were likely, presumed, or possibly associated with translating complexes.6 Out of 33 such proteins, we found only five to be present in the 77S translating complex. Many known P body/stress granule mRNP components were not present in the 77S complex, implying a significant protein addition and structural arrangement that occurs on formation of such mRNA degradation/translationally quiescent complexes. Our studies in combination with a recent study that analyzed which mRNP complex components are present in P body/stress granules5 begin the absolute determination of which components of the mRNP exist in which exact functional complexes.
Results
Methodology for determining presence in the 77S monosomal translation complex
Previously, we have used mass spectrometric analysis to identify 55 nonribosomal proteins that specifically associated with Flag-PAB1.6 Within this group, all but 10 had functions related to some role in which PAB1 may be involved: splicing, mRNA export, nucleolar/RNA biogenesis, translation, mRNA decay, or RNA binding. Because PAB1 has so many potential of roles in the cell, these 45 remaining proteins do not necessary have to be present all in one complex with PAB1. To clarify these putative interactions, we used AU-FDS in combination with GFP fusions to many of these putative interactors to identify which of these proteins existed in the 77S monosomal translation complex. For this analysis, we limited our studies to those proteins that might be involved in some aspect of PAB1 function involving protein synthesis or mRNA degradation, and, therefore, we did not generally analyze the nucleolar/RNA biogenesis factors (18 proteins), splicing factors (3), mRNA transport proteins (3), and the proteins not apparently related to PAB1 (10). As controls, we did include in our study two nucleolar proteins (RRP5 and LHP1) and UBP3, a ubiquitin protease, all of which were shown to interact with PAB1.6 We also analyzed one mRNA export protein (GBP2) that may have some role in the translation process and stress granule formation.8
Two criteria were used to verify if any of the putative PAB1-mRNP interacting proteins were in the 77S monosomal translation complex. First, Flag-PAB1-containing complexes were purified by specific binding to Flag agarose beads and the resultant complexes subjected to AU-FDS analysis. The abundance of a specific GFP fusion protein present in the 77S complex was compared to the abundance of the 77S complex as determined following Flag immunoprecipitation of extracts from an isogenic strain containing the GFP fusion but lacking Flag-PAB1 (see Fig. 1). All GFP fusions were present at their chromosomal location and do not result in any obvious growth defects.7,9 Only those GFP fusion proteins that displayed significant presence in a 77S translation complex were considered likely components of the monosomal translation complex.
Figure 1.
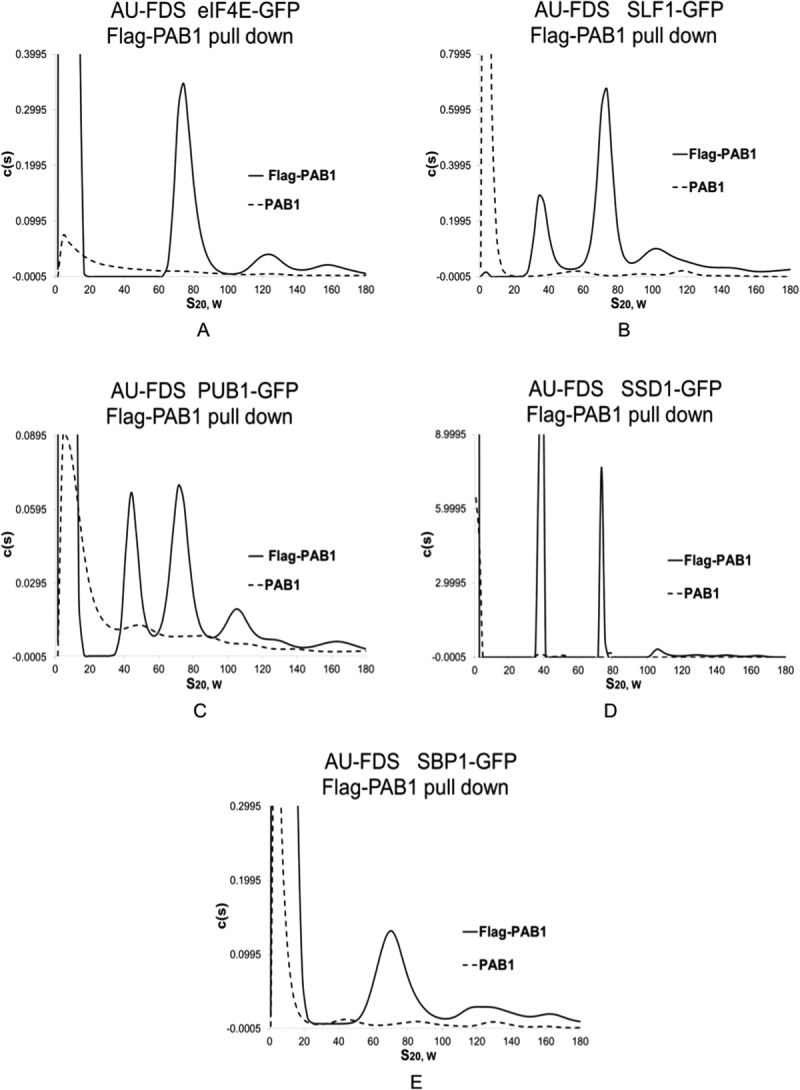
Identification of new proteins in the 77S monosomal translation complex. Cells were pregrown to mid-log phase on medium containing 2% glucose prior to cell lysis and purification of Flag-PAB1-containing complexes. Equivalent levels of protein extracts were used in the respective Flag pull downs. Experiments displayed together in a particular part of the figure were conducted in the same centrifugation analysis. All strains were isogenic (Mata ura3 leu2 his3 in which the GFP fusion to the protein as indicated at its chromosomal location is marked with HIS3)9 and carried either Flag-PAB1 or not, as indicated. Flag-PAB1 was expressed from either YC504 or YC776.7 A. Strain containing eIF4E-GFP; B. SLF-GFP; C. PUB1-GFP; D. SSD1-GFP; E. SBP1-GFP.
The second criterion that we used was that the putative 77S monosomal translation complex containing the specific GFP fusion was subjected to the stress of glucose deprivation that represses translation and causes an approximate four-fold reduction in 77S translation complex abundance (Fig. 2).7 Any GFP fusion protein that met both these criteria, that is, significant presence in a 77S complex and 77S complex abundance sensitivity to glucose deprivation, was considered to be part of the monosomal translating complex.
Identification of new components of the 77S translation complex
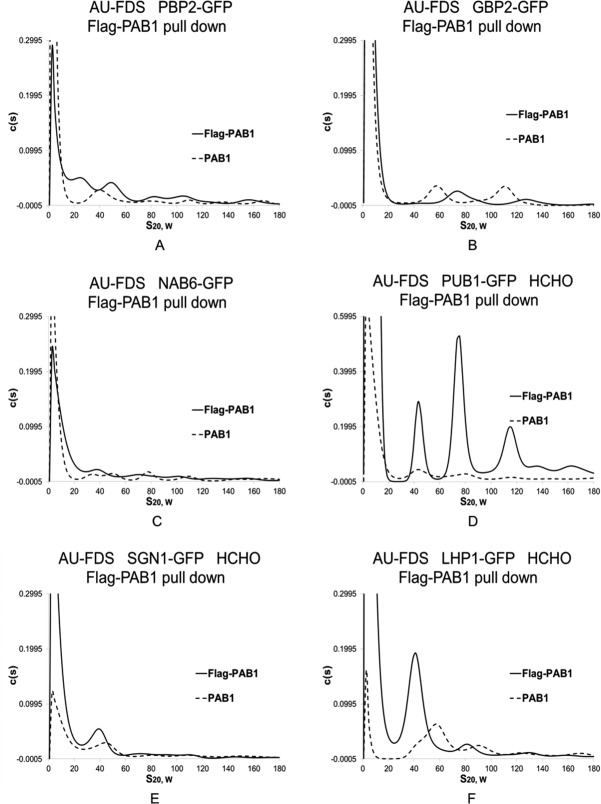
Previously, we had established that the following proteins were present in the 77S monosomal translating complex: eIF4E, eIF4G1, eIF4G2, and PAB1. Figure 1(A) displays a typical AU-FDS analysis for eIF4E-GFP versus the no Flag-PAB1 control, and Figure 2(A) displays the glucose sensitivity of eIF4E-GFP presence in the 77S translating complex. We had also previously found that initiation factors that are components of the 43S translation complex, eIF2, eIF3, and eIF5, were not part of the 77S translating complex.7 Of the new proteins that we analyzed based on our previous mass spectrometric analysis of the PAB1-mRNP proteome, only four proteins were present in a 77S complex: SLF1, PUB1, SSD1, and SBP1 [Fig. 1(B–E), respectively]. The average S value for the complex for each of these proteins was found to be for SLF1 as 75.9S (1.0S Standard Error of the Mean: SEM), PUB1 as 76.6S (1.1S SEM), SSD1 as 78.6S (2.1S SEM), and SBP1 as 76.6S (0.70S SEM), (average of six determinations for each protein). In contrast, the two nucleolar proteins, UBP3, the mRNA export proteins, a number of RNA binding factors, and the mRNA decay proteins were not present in a 77S complex: CBC1, GCD11, GCD6, GBP2, HRP1, NAB3, NAB6, LHP1, PBP1, PBP2, RRP5, SGN1, SMB1, SUP35 (eRF3), TMA46, UBP3, UPF1, XRN1, and yGR250c [see Fig. 3(A–C) for typical analyses conducted for PBP2, GBP2, and NAB6; Table I summarizes the data; and Supporting Information Figs. 3 display profiles for the additional factors].
Figure 2.
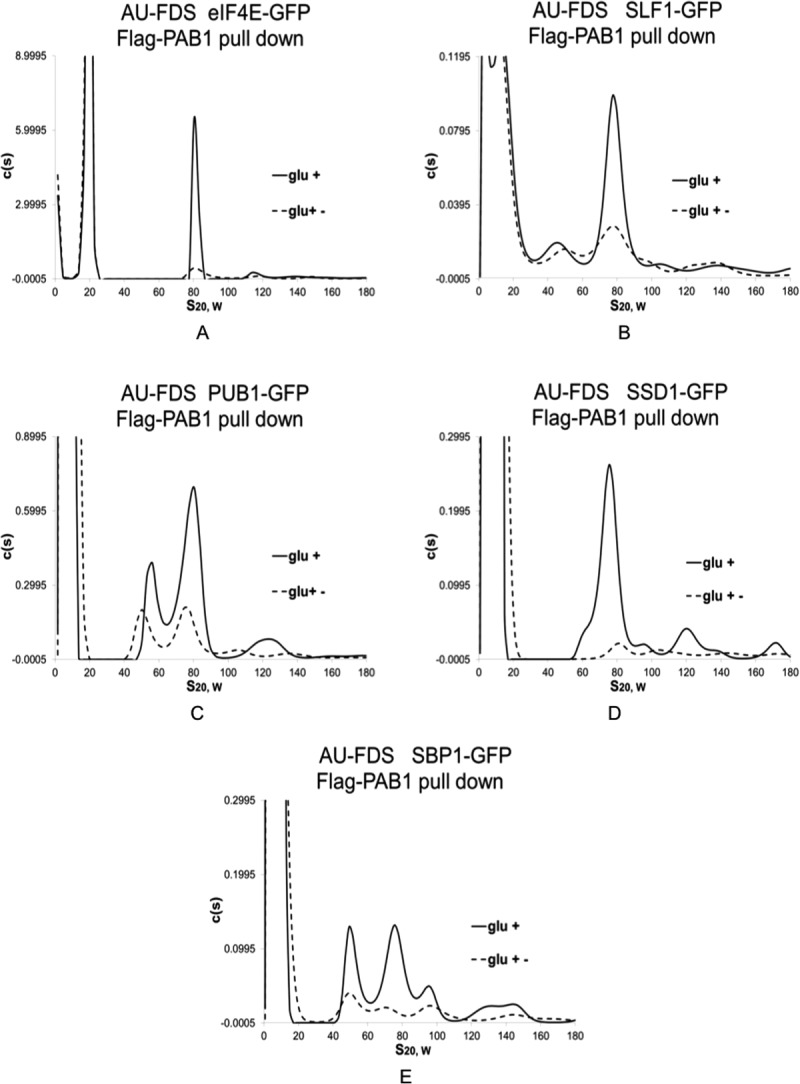
Effect of glucose depletion on the 77S monosomal translation complex. Glucose growth conditions (glc +) were the same as described in Figure 1, and glucose depletion conditions (glc +−) occurred after pregrowth in glucose-containing medium followed by growth for 10 min in medium lacking glucose. The eIF4E-GFP data was taken from Ref.7. A–E: strains are the same as described in Figure 1.
Figure 3.
AU-FDS analysis of other PAB1-interacting factors. AU-FDS analysis and glucose growth conditions were conducted as described in Figure 1. Flag-PAB1-containing strains carried either plasmid YC504 or YC776 whereas PAB1-containing strains lacked the Flag-PAB1 plasmid. Formaldehyde treatment of cells prior to cell lysis for 30 min was as described.16 A–C. Glucose grown cells; D–F. Glucose grown cells treated with formaldehyde (HCHO). A. Strain carrying PBP2-GFP; B. GBP2-GFP; C. NAB6-GFP; D. PUB1-GFP; E. SGN1-GFP; and F. LHP1-GFP.
Table I.
Proteins Analyzed for Presence in the 77S Translation Complex
| Protein | Presence in 77S translation complex | Presence in P body/stress granules | Cellular function |
|---|---|---|---|
| RPS4B/RPS30A | Y | Na | Translation/Ribosome |
| RPL7A/RPL6B | Y | N | Translation/Ribosome |
| PAB1 | Y | Y | Translation/Closed-loop |
| eIF4E | Y | Y | Translation/Closed-loop |
| eIF4G1 | Y | Y | Translation/Closed-loop |
| eIF4G2 | Y | Y | Translation/Closed-loop |
| eRF1 | Y | Na,b | Translation termination |
| eRF3 | N | Na,b | Translation termination |
| DBP5 | N | c | Translation termination |
| eIF3b | N | Na | Translation initiation |
| eIF2α | N | Na | Translation initiation |
| eIF4A | N | N | Translation initiation |
| eIF5 | N | N | Translation initiation |
| GCD6 | N | c | Translation initiation |
| GCD11 | N | c | Translation initiation |
| SBP1 | Y | Y | Translation repressor |
| PAT1 | N | Y | Translation repressor |
| DHH1 | N | Y | Translation repressor |
| SCD6 | N | c | Translation repressor |
| SLF1 | Y | Y | Translation |
| TMA46 | N | c | Translation |
| SSD1 | Y | c | RNA binding |
| PUB1 | Y | Y | RNA binding |
| NAB3 | N | N | RNA binding |
| NAB6 | N | Y | RNA binding |
| CBC1 | N | c | RNA binding |
| PBP1 | N | Y | RNA binding |
| PBP2 | N | c | RNA binding |
| PUF3 | N | Y | RNA binding |
| SGN1 | N | c | RNA binding |
| CCR4 | N | Y | mRNA degradation |
| LSM1 | N | Y | mRNA degradation |
| DCP1 | N | Y | mRNA degradation |
| DCP2 | N | Y | mRNA degradation |
| XRN1 | N | Y | mRNA degradation |
| UPF1 | N | Y | mRNA degradation |
| yGR250c | N | Y | mRNA degradation |
| GBP2 | N | Y | mRNA export |
| HRP1 | N | Y | mRNA export |
| SMB1 | N | c | Splicing |
| RRP5 | N | c | Nucleolar |
| LHP1 | N | c | Nucleolar |
| UBP3 | N | N | Ubiquitin protease |
Protein presence in the 77S translation complex was taken from data herein or in Ref.7. Presence of the protein in P body/stress granules following glucose deprivation was determined from Refs.6, 8, and10–13.
proteins that were not present in glucose depleted stress granules but which were found in robust heat shock stress granules.14,15
proteins that infrequently associate with P bodies or stress granules but are not clearly identified with them.5,8
location being unknown.
To ensure that our analyses were not missing proteins weakly associated with the 77S complex that might dissociate on our Flag immunoprecipitations, a number of our strains were pretreated with formaldehyde to cross-link translation complexes.16,17 Prior treatment of cells with formaldehyde has been shown to stabilize translation complexes and to not interfere with their subsequent characterization.10,16,18 We, therefore, conducted our Flag-PAB1 pull down experiments in the presence of formaldehyde for the following GFP tagged proteins: LHP1, yGR250c, SGN1, SMB1, RRP5, NAB6, CBC1, NAB3, PBP1, and HRP1. None of these proteins were found to be enriched in the 77S complex in the experimental Flag-PAB1 pull down sample as compared to that of the Flag control [for typical analyses see SGN1 and LHP1, Fig. 3(E,F), compared to PUB1, Fig. 3(D); also see Supporting Information Fig. 3]. These data suggest that our studies were not missing the identification of transiently associated factors with the 77S complex.
For each of the four new proteins found to be present in a 77S complex, the 77S complex abundance decreased on the stress of glucose depletion [typical analyses displayed in Fig. 2(B–E)]. The average decrease in 77S complex abundance for each of these proteins following glucose depletion compared favorably with that observed previously for the total 77S complex, eIF4E, eIF4G1, and RPS4B and RPL6B (Table II).7 These concurrences establish that the 77S complex in which SSD1, SBP1, PUB1, and SLF1 are present is the monosomal translating complex.
Table II.
Effect of Glucose Depletion on the 77S Complex Isolated from Flag-PAB1 Pull-Downs
| GFP fusion | Mean abundance of 77S complex following glucose depletion (%) | SEM |
|---|---|---|
| AU-A260 (total) | 22 | 6.8 |
| eIF4E | 17 | 3.7 |
| eIF4G1 | 8.4 | 1.1 |
| RPS4B | 19 | 5.6 |
| RPL6B | 17 | 3.9 |
| eRF1 | 34 | 7.3 |
| SBP1 | 16 | 6.6 |
| SSD1 | 8.3 | 0.35 |
| SLF1 | 14 | 3.4 |
| PUB1 | 30 | 2.7 |
The abundance of the 77S complex using Flag-PAB1 pull-downs was assessed from glucose or glucose depleted cultures split prior to depletion of glucose. All proteins indicated were GFP fusions. Western analysis was conducted on the resultant Flag pull-downs and demonstrated that equivalent levels of Flag-PAB1 had been isolated when comparing pull-downs from glucose grown cultures to those from glucose depleted cultures.35 AU-A260 identification of the 77S complex reflects the concentration of the total 77S complex.7 Data for AU-A260, eIF4E, and RPS4B were taken from Ref.7.
The identification of each of these proteins in the 77S monosomal translating complex agrees with previous studies. SBP1 is known to be involved in translational repression and to bind eIF4G.11,19,20 SSD1 and PUB1 are RNA binding proteins that control expression of a number of yeast genes,21 and they may be bound to the mRNA during translation. SLF1 is known to interact with eIF4E.4
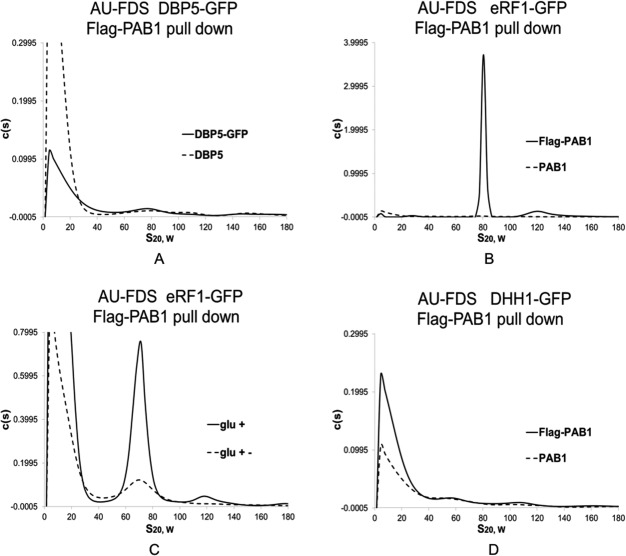
Translation termination factor eRF1 is a component of the 77S complex but DBP5 is not
Although we observed that eRF3 was not a component of the 77S translating complex, other studies have suggested termination factors might be associated with this translation complex.22 Correspondingly, we analyzed by AU-FDS for the presence of eRF1 and DBP5. DBP5 aids in release of the polypeptide from the translating ribosome, whereas eRF1 binds to the stop codon, prevents additional tRNA binding, and binds eRF3 that aids in polypeptide release.23 DBP5 was not found to be in a 77S complex [Fig. 4(A)] whereas eRF1 did migrate in a 77S complex [Fig. 4(B) in which eRF1-GFP migrated in a 76.9S peak, 0.95S SEM for five determinations]. On glucose deprivation, the level of eRF1 dropped to 34% of glucose growth conditions [Fig. 4(C); Table II], confirming that eRF1 is present in translating complexes, as predicted from previous in vitro studies.22
Figure 4.
eRF1 is present in the 77S monosomal translation complex. Growth conditions (glc + and glc +−) and AU-FDS analyses were conducted as described in Figures 1 and 2. Flag-PAB1 and PAB1 designate strains carrying Flag-PAB1 plasmid or not as indicated in Figure 1. A. DBP5-GFP; B, C. eRF1-GFP; D. DHH1-GFP.
Translational repressor proteins are not generally present in translational complexes
Because SBP1 is a translational repressor and is a component of mRNA translation complexes, we determined if other known translational repressors associate with the 77S translation complex. We, therefore, analyzed by AU-FDS the GFP fusions to DHH1, PAT1, and SCD6. PAT1 and DHH1 are known to act with SBP1 in promoting translational arrest and movement of mRNP complexes into P body and stress complexes.11,19,20 SCD6 interacts with eIF4G and presumably aids in translational repression.19 We found, however, that none of these proteins were present in the 77S monosomal translation complex [for example, see Fig. 4(D), DHH1, and Supporting Information Figs. 1 and 2 for PAT1 and SCD6]. Relatedly because of links between DHH1 and decapping, deadenylation, and mRNA degradation factors, we also analyzed CCR4, DCP1, DCP2, and LSM1, but none of these proteins were found to be present in the 77S translating complex (Supporting Information Fig. 4). Because DHH1, PAT1, GBP2, HRP1, NAB6, PBP1, UPF1, XRN1, CCR4, DCP1, DCP2, LSM1, and yGR250c all are present in P bodies or stress granules but not in translation complexes, our data also implies that a significant proportion of the P body/stress granule components become part of these latter complexes only following stress and are not significant components of active translation complexes prior to the introduction of the stress condition. These results further suggest a major reconstruction and addition of factors to the PAB1-mRNP following stress and translational cessation.
SBP1 and SLF1 do not bind to the free 80S ribosome and associate in translational complexes independently of contacts to PAB1
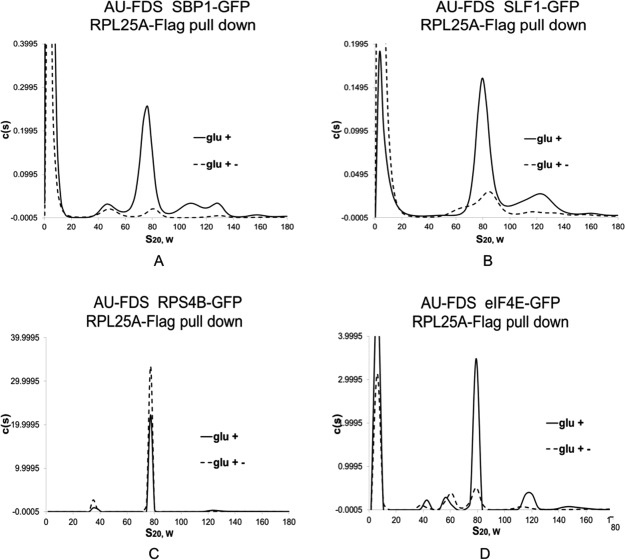
Both the SBP1 and SLF1 proteins had previously been implicated to be part of translational complexes. We, therefore, chose these two proteins for further study. We had shown initially that the closed-loop components, eIF4E, eIF4G, and PAB1, do not appreciably associate with the free 80S ribosome.7 We consequently determined if SBP1 and SLF1 were solely associated with the 77S translation complex or were also capable of binding the free 80S ribosome. To conduct this analysis, we used the ability of RPL25A-Flag to pull down both the 77S translating complex and the free 80S ribosome.7 In this case, if a significant pool of SBP1 or SLF1 were part of the free 80S complex, RPL25A-Flag would immunoprecipitate these complexes. Importantly, such a pool of SBP1 or SLF1 would not be sensitive to the stress of glucose deprivation and their abundance in the 80S complex would not decrease following such a stress.7
Following immunoprecipitation with RPL25A-Flag, we found that the 77S complex abundance containing either SBP1-GFP or SLF1-GFP remained sensitive to the stress of glucose removal [Fig. 5(A,B), respectively]. The reduction in the 77S complex for SBP1-GFP was to 8.7% (SEM of 0.40%) of the glucose grown sample and for SLF1-GFP was to 29% (SEM of 3.0%) of the control, which is similar to the overall reduction in abundance for proteins observed following Flag-PAB1 pull downs (Table II). In contrast, the abundance of RPS4B-GFP, which is present in both the 77S and 80S complexes, was undiminished by glucose deprivation [Fig. 5(C)] while the abundance of eIF4E-GFP, which is only in the 77S complex, was reduced following this stress [Fig. 5(D)].7 These data imply that there is not a significant pool of SBP1 or SLF1 in the 80S free ribosomal pool.
Figure 5.
SBP1 and SLF1 are not present in the free 80S ribosome. Glucose growth (glc +) and depletion (glc +−) conditions were conducted as described in Figure 2. RPL25A-Flag pull downs were conducted as described previously using strains carrying JC288 (RPL25A-Flag URA3)7 Data for RPS4B-GFP and eIF4E-GFP are taken from Ref.7. Strains in panels A–D carry the GFP fusion as indicated.
Our second analysis with SBP1 and SLF1 was to determine whether their associations with the 77S complex were dependent on the presence of PAB1. Previously, we had shown that 13 proteins interact with PAB1-mRNP complexes dependent on particular PAB1 domains.6 These data implied that these interactions required the presence of PAB1. SLF1 association with PAB1-mRNP complexes was reduced if the RRM1 domain of PAB1 was removed, whereas SBP1 interacted with the PAB1-mRNP complexes irrespective of deletion of any particular PAB1 domain.6
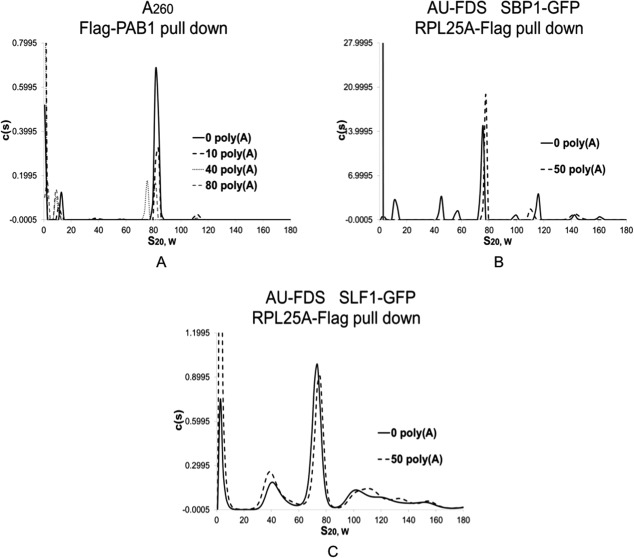
To screen for SLF1 and SBP1 presence in the 77S complex based on the presence of PAB1, we used a competition assay24 to selectively remove PAB1 from the poly(A) tail prior to conducting our pull downs and AU-FDS analysis. Previous analysis had shown that prior addition of poly(A) blocked the ability of PAB1 to associate with either SLF1 or SBP1.25 For the present study, treatment of crude extracts with increasing concentrations of free poly(A) prior to conducting the Flag pull down experiment was found to specifically remove PAB1 from the poly(A) tail, as evidenced by the reduced ability of Flag-PAB1 to purify the 77S complex when increasing concentrations of free poly(A) were added prior to the Flag pull down [Fig. 6(A); average reduction using 50 µg of poly(A) was to 14% of that observed with the addition of no poly(A) for three determinations]. However, when 50 µg of poly(A) was added to extracts and RPL25A-Flag pull downs were then conducted in the presence of either SBP1-GFP or SLF1-GFP in order to detect proteins still associated with the 77S complex, no diminishment in 77S complex abundance was detected [Fig. 6(B,C)]. These results imply that prior removal of PAB1 from 77S complexes did not impair the ability of SBP1 or SLF1 to associate with the 77S translating complex, indicating that SBP1 and SLF1 can associate with the 77S complex through interactions independent of PAB1.
Figure 6.
SBP1 and SLF1 contact the 77S complex independent of PAB1. PAB1 was competed off the 77S complex by the addition of poly(A) for 30 min prior to Flag pull downs.6,24 The micrograms of poly(A) are indicated in the Figure and was chosen to be 50 µg in panels B and C since 40 µg gave similar results as did 80 µg. AU-FDS and AU-260 analysis and growth conditions were as described in Figure 1. Flag pull downs were conducted with strains either containing Flag-PAB1 or RPL25A-Flag as indicated. A. Strain AS319/YC504 that lacks a GFP tagged protein7; B. SBP1-GFP; C. SLF1-GFP.
Discussion
AU-FDS defines the existence of multiple mRNP complexes
Two major studies have conducted global proteomic analysis to determine which factors are present in mRNP complexes in yeast,5,6 whereas other general proteomic analyses have yielded information on the PAB1-mRNP proteome.1–4 These studies have principally relied on immunoprecipitation of various tagged PAB1 molecules bound to the mRNA poly(A) tail or purification of mRNA-containing proteins previously cross-linked to RNA. Regardless of method, there are very significant overlaps in the identified proteins in these sets of mRNP proteins, validating all of these approaches. It is very unlikely, however, that all of these proteins exist in one complex considering their known variations in location within the cell.9 One recent study did analyze the subcellular location of many of these mRNP complex proteins and extended the analysis to determining subcellular location following the stress of glucose depletion.5 These results established major shifts in subcellular locations following glucose depletion, implying that many different mRNP complexes exist in the cell.
Because of the inherent limitations of immunoprecipitation and mass spectrometric analysis in informing on the character and size of protein complexes, we used AU-FDS to specifically and precisely determine the size of the complexes in which many of these mRNP proteins exist. Our data established that only a subset of 33 identified mRNP proteins exist in the 77S monosomal translation complex. Our criterion for presence in mRNP complexes relied on coimmunoprecipitation with PAB1.6 The assumption was that PAB1 was bound to the mRNA tail and would be found in complexes associated with mRNA. One caveat to this assumption is that it is known that PAB1 can bind poly(A) sequences not present in poly(A) tails and consequently may be either bound to other RNA than mRNA or to mRNA not specifically functioning in a translational role.26 Therefore, in general, our results indicate that the PAB1-mRNP proteome is quite diverse in character with only a small number of the factors it contains actually being present in translating complexes.
We also analyzed 21 known P body/stress granule components and found that only seven were present in translating complexes: eIF4E, eIF4G1, eIF4G2, PAB1, SBP1, SLF1, and PUB1. None of these proteins are considered proteins primarily involved in mRNA degradation. In contrast, of the other proteins not present in translating complexes that are present in P body/stress granules, six proteins are significantly involved in mRNA degradation (XRN1, UPF1, CCR4, LSM1, DCP1, and DCP2)5,27 and three are translational repressors (DHH1, PAT1, and SCD6). Two conclusions can, therefore, be drawn. The first is that many of the mRNP proteins involved in P body/stress granules associate in such bodies only after translation ceases. Second, these proteins' abilities to associate in P body/stress granules may be specifically related to translational cessation and processing and decay of the mRNA. These data imply, furthermore, that a major reconstruction of the mRNP takes place following cessation of translation in which numerous new factors associate into these mRNP complexes.
New components of the translating ribosome
The character and components of the translating ribosomal complex are poorly understood other than that it contains the 40S and 60S ribosome and key translational closed-loop factors such as PAB1, eIF4E, and eIF4G. Previously, other proteins' presence in translating ribosomes was based primarily on sucrose gradient analysis of polysomal material followed by western analysis using antibody directed against a particular protein. This type of analysis is unsatisfactory, for a protein's presence in the polysomal material is based on comigration and can not correlate with individual ribosomal peaks due to the lack of resolution in conducting the western analysis on only 15 or so fractions across the sucrose gradient. Additional analyses are often conducted wherein polysomes might be disrupted with EDTA to demonstrate the movement of a particular protein out of the polysomal fractions. This analysis is also unsatisfactory as EDTA can disrupt many macromolecular interactions in addition to polysomal associations.
We have endeavored to overcome these limitations by using AU-FDS analysis to detect the 77S monosomal translation complex and determine which other proteins are components of this complex. We consequently established that eRF1, SSD1, SBP1, PUB1, and SLF1 are components of the 77S monosomal translation complex. The termination factor eRF1 has previously been suggested to be part of the translation initiation complex based on in vitro reconstruction experiments.22 Because other termination factors, DBP5 and eRF3, were not found in the 77S translation complex, either these factor interactions with the 77S translation complex are much weaker or more transient during translation termination or their roles occur with monosomal complexes not responsive to the stress of glucose depletion, as observed for eRF3 (Supporting Information Fig. 3).
The role for the remaining four translation-associated factors is less clear. SBP1, an abundant RNA binding protein, has been identified as functioning in mRNA degradation and translational repression.20 Models explaining SBP1 function suggest that SBP1 protein could directly bind mRNA and inhibit the function of translation initiation factors, or SBP1 protein could directly bind the mRNA and facilitate the full assembly of a translational repression complex. In that SBP1 is presumed to bind eIF4G and by this means carry out translational repression,11,19 SBP1 may function to help remove eIF4G from the mRNA or occlude its function. Its specific localization to both 5′ and 3′ UTRs,5 suggests that it is binding sequences in such regions. An alternative possibility is that it localizes to the 5′ and 3′ sequences of the mRNA because of its interaction with closed-loop structure components which, of course, are particularly localized at the 5′ end (through eIF4E) and the 3′ end (through PAB1). Our data demonstrating that SBP1 associates with the 77S complex irrespective of the presence of PAB1 suggests that its interactions with the RNA or eIF4G but not with PAB1 are more critical to its functional role.
SLF1 has been reported as a RNA binding protein that associates with polyribosomes.28 Using whole genome mass spectrometric analysis, it was reported that SLF1 interacted with eIF4E,4 which is consistent with our results. We also established that SLF1 does not interact in the 77S complex solely through its interaction with PAB1. Previously, we had implicated the RRM1 domain of PAB1 as important to SLF1 association with the PAB1-mRNP using mass spectrometric analyses.6 Those results were, however, less direct than the current studies. Particularly, the SLF1 mass spectrometric data that demonstrated specific PAB1 interactions were limited by the low number of SLF1 peptides detected using these mass spectrometric analyses.
PUB1 is a cytoplasmic mRNA binding protein that stabilizes transcripts containing AU-rich elements (AREs) or stabilizer elements (STEs). Nuclear poly(A) binding protein 2 (NAB2) is also known to interact with PUB1, and NAB2 functions together with PUB1 to modulate mRNA stability. These data suggest a model where nuclear events are coupled to the control of mRNA turnover in the cytoplasm.29 Several lines of evidence also suggest that PUB1 may be involved in mRNA metabolism. Both mammalian homologues of PUB1, HuR and the TIA-1/TIAR, are involved in translational regulation. While HuR acts as a translational enhancer or repressor,30 the TIA-1 and TIAR proteins are involved in ARE-mediated translational repression.31 Global mRNA turnover in isogenic PUB1 and pub1Δ strains, as determined by gene expression analysis, also demonstrated that 573 genes exhibit a significant reduction in half-life in a pub1Δ strain. Subsequently, the binding specificity of PUB1 was examined using affinity purification followed by microarray analysis to comprehensively distinguish between direct and indirect targets. PUB1 was found to significantly bind 368 cellular transcripts.32 PUB1 was also found to bind to discrete subsets of cellular transcripts and to post-transcriptionally regulate their expression at multiple levels.31
Our demonstration that PUB1 is found in the 77S translation complex is hence consistent with PUB1 binding to translating mRNA and therefore being part of translation complexes. Whether this implies that PUB1 is only present in the 77S translation complex because mRNA are also in the complex or that it plays a specific functional role in the translation process is unclear. If PUB1 is adventitiously present in the 77S complex because of its binding to so many mRNA, it would suggest that many other mRNA binding proteins should also be present in the 77S translation complex. Yet, several other mRNA binding proteins such as NAB3, NAB6, SGN1, PBP1, PBP2, and PUF3 (Table II) were not shown to be present in the 77S complex.
SSD1 is a protein with a role in maintenance of cellular integrity and interacts with components of the TOR pathway. Systematic global screens have identified about 200 genes that show genetic or physical interactions with SSD1.33 These genes show a striking enrichment for post-translational modifiers, including 19 kinases and nine histone deacetylases, and genes involved in the cell cycle and cell morphogenesis.21 SSD1 mutants also display sensitivity to high osmolarity, caffeine, fungicides, and numerous other compounds, which suggests a role for this protein in the maintenance of cell wall integrity,34 but its mechanism of action in this function remains obscure. Therefore, these roles for SSD1 are consistent with SSD1 association with specific mRNAs, a significant number of which encode cell wall remodeling proteins.21 SSD1 may, therefore, like PUB1, be present in translating complexes because it binds a number of mRNA.
In summary, of the new factors we have established as being components of the translating complexes, PUB1, SSD1, and SLF1 appear present during active translation because they bind mRNA and may not have separable roles on translation in general. SBP1 and eRF1, in contrast, have known global translational functions either in termination or repression of translation.
Materials and Methods
AU-FDS analysis
Analytical ultracentrifugation with fluorescent detection was conducted as described.7 All yeast strains were isogenic, each carrying a different gene fused to the coding sequence of GFP as indicated in the text.9 All strains were transformed with either plasmid YC504 (Flag-PAB1 TRP1) or YC766 (Flag-PAB1 URA3), and isolation of PAB1-containing complexes by Flag-agarose chromatography was conducted as described.7 Growth of strains on glucose-containing and –deprived medium has been described.7 It should also be noted that peak profiles as determined by AU-FDS analysis can vary between different analyses [see, for example, SBP1 in Figs. 1(E), 2(E), and Supporting Information Fig. 3(A)]. The cause for this is probably SEDFIT mathematical fitting of the resultant sedimentation profiles.
Other techniques
Analysis of free 80S ribosomes was conducted as previously described.7 Briefly, plasmid JC288 (RPL25A-Flag-URA3) was transformed into strains containing SLF1-GFP and SBP1-GFP. Following purification of RPL25A-Flag complexes by Flag-agarose chromatography, AU-FDS analysis was used to detect the resultant complexes. The poly(A) competition assays were conducted by adding upwards to 80 µg of poly(A) to extracts for 30 min prior to conducting the Flag-PAB1 pull downs.25 Formaldehyde treatment of cells prior to cell lysis was conducted exactly as previously described.16
Acknowledgments
We wish to thank the Roy Parker laboratory for generously providing strains containing GFP-tagged proteins. This is Scientific Contribution Number 2550.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information Figures.
References
- 1.Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Gavin AC, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Höfert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 3.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sørensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 4.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrín-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell SF, Jain S, She M, Parker R. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 2013;20:127–133. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richardson R, Denis CL, Zhang C, Nielsen MO, Chiang YC, Kierkegaard M, Wang X, Lee DJ, Andersen JS, Yao G. Mass spectrometric identification of proteins that interact with specific domains of the poly(A) binding protein. Mol Genet Genomics. 2012;287:711–730. doi: 10.1007/s00438-012-0709-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Zhang C, Chiang Y-C, Toomey S, Power MP, Granoff ME, Richardson R, Xi W, Lee DJ, Chase S, Laue TM, Denis CL. Use of the novel technique of analytical ultracentrifugation with fluorescence detection system identifies a 77S monosomal translation complex. Protein Sci. 2012;21:1253–1268. doi: 10.1002/pro.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huh WK, Flavo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 10.Hoyle NP, Castelli LM, Campbell SG, Holmes LEA, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nissan T, Rajyarguru P, She M, Song S, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Grousl T, Ivanov P, Frydlova I, Vasicova P, Janda F, Votobva J, Malinska K, Malcova I, Novakova L, Janoskova D, Valasek L, Hasek J. Robust heat shock induces eIF2α-phosphorylation-indpendent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J Cell Sci. 2009;122:2078–2088. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- 15.Grousl T, Ivanov P, Malcova I, Pompach P, Frydlova I, Slaba R, Senoihrabkova L, Novakova L, Hasek J. Heat shock-induced accumulation of translation elongation and termination factors precedes assembly of stress granules in S. cerevisiae. PLoS One. 2013;8:e57083. doi: 10.1371/journal.pone.0057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen KH, Szamecz B, Valasek L, Jivotovskaya A, Shin B-S, Hinnebusch AG. Functions of eIF3 downstream of 48S assenbkt unoact /aug recognition and GCN4 translational control. EMBO J. 2004;23:1166–1177. doi: 10.1038/sj.emboj.7600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valásek L, Szamecz B, Hinnebusch AG, Nielsen KH. In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol. 2007;429:163–183. doi: 10.1016/S0076-6879(07)29008-1. [DOI] [PubMed] [Google Scholar]
- 18.Jivotovskaya AV, Valásek L, Hinnebusch AG, Nielsen KH. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol Cell Biol. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajyarguru P, She M, Parker R. Scd6 targets eIF4G to repress translation: RGG motif portiens as a class of eIF4G-binding proteins. Mol Cell. 2012;45:244–254. doi: 10.1016/j.molcel.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segal SP, Dunckley T, Parker R. Sbp1p affects translational repression and decapping in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:5120–5130. doi: 10.1128/MCB.01913-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baierlaein C, Krebber H. Translation termination: new focus and insights. RNA Biol. 2010;7:548–550. doi: 10.4161/rna.7.5.12686. [DOI] [PubMed] [Google Scholar]
- 24.Yao G, Chiang YC, Zhang C, Lee DJ, Laue TM, Denis CL. PAB1 self-association precludes its binding to poly(A), thereby accelerating CCR4 deadenylation in vivo. Mol Cell Biol. 2007;27:6243–6253. doi: 10.1128/MCB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao G. 2006. Analysis of the role of poly(A) binding protein (PAB1) in the mRNA degradation process in yeast. Ph.D Dissertation, University of New Hampshire.
- 26.Mullin C, Duning K, Barnekow A, Richter D, Kremerskothen J, Mohr E. Interaction of rat poly(A)-binding protein with poly(A)- and non-poly(A) sequences is preferentially mediated by RNA recognition motifs 3+4. FEBS Lett. 2004;576:437–441. doi: 10.1016/j.febslet.2004.09.054. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Chiang YC, Denis CL. CCR4, a 3'-5' poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobel SG, Wolin SL. Two yeast La motif-containing proteins are RNA-binding proteins that associate with polyribosomes. Mol Biol Cell. 1999;10:3849–3862. doi: 10.1091/mbc.10.11.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Apponi LH, Kelly SM, Harreman MT, Lehner AN, Corbett AH, Valentini SR. An interaction between two RNA binding proteins, Nab2 and Pub1, links mRNA processing/export and mRNA stability. Mol Cell Biol. 2007;27:6569–6579. doi: 10.1128/MCB.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.López de Silanes I, Lal A, Gorospe M. HuR: post-transcriptional paths to malignancy. RNA Biol. 2005;2:11–13. doi: 10.4161/rna.2.1.1552. [DOI] [PubMed] [Google Scholar]
- 31.Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, Anderson P. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duttagupta R, Tian B, Wilusz CJ, Khounh DT, Soteropoulos P, Ouyang M, Dougherty JP, Peltz SW. Global analysis of Pub1p targets reveals a coordinate control of gene expression through modulation of binding and stability. Mol Cell Biol. 2005;25:5499–5011. doi: 10.1128/MCB.25.13.5499-5513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, Stark C, Ho Y, Botstein D, Andrews B, Boone C, Troyanskya OG, Ideker T, Dolinski K, Batada NN, Tyers M. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibeas JI, Yun DJ, Damsz B, Narasimhan ML, Uesono Y, Ribas JC, Lee H, Hasegawa PM, Bressan RA, Pardo JM. Resistance to the plant PR-5 protein osmotin in the model fungus Saccharomyces cerevisiae is mediated by the regulatory effects of SSD1 on cell wall composition. Plant J. 2001;25:271–280. doi: 10.1046/j.1365-313x.2001.00967.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang C. 2011. Identifying novel proteins in translation complexes by using analytical ultracentrifugation with fluorescent detection system. Ph.D Dissertation, University of New Hampshire.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information Figures.