Summary
In P. aeruginosa, alginate overproduction, also known as mucoidy, is negatively regulated by the transmembrane protein MucA, which sequesters the alternative sigma factor AlgU. MucA is degraded via a proteolysis pathway that frees AlgU from sequestration, activating alginate biosynthesis. Initiation of this pathway normally requires two signals: peptide sequences in unassembled outer-membrane proteins (OMPs) activate the AlgW protease, and unassembled lipopolysaccharides bind periplasmic MucB, releasing MucA and facilitating its proteolysis by activated AlgW. To search for novel alginate regulators, we screened a transposon library in the non-mucoid reference strain PAO1, and identified a mutant that confers mucoidy through overexpression of a protein encoded by the chaperone-usher pathway gene cupB5. CupB5-dependent mucoidy occurs through the AlgU pathway and can be reversed by overexpression of MucA or MucB. In the presence of activating OMP peptides, peptides corresponding to a region of CupB5 needed for mucoidy further stimulated AlgW cleavage of MucA in vitro. Moreover, the CupB5 peptide allowed OMP-activated AlgW cleavage of MucA in the presence of the MucB inhibitor. These results support a novel mechanism for conversion to mucoidy in which the proteolytic activity of AlgW and its ability to compete with MucB for MucA is mediated by independent peptide signals.
Keywords: Pseudomonas aeruginosa, alginate, MucA, MucB, CupB5, signal transduction
Introduction
Pseudomonas aeruginosa is an important opportunistic human pathogen, capable of thriving in different environmental niches. Activation of the AlgU-regulated extracytoplasmic stress response results in the production of copious amounts of an exopolysaccharide called alginate, forming a capsule that protects the bacterium from the host-immune system. Alginate production by P. aeruginosa is a particular problem for patients with cystic fibrosis, whose dehydrated mucus-filled lungs provide an environment especially suited for bacterial infection (Folkesson et al., 2012). Nonmucoid strains initially colonize the lungs, and alginate overproduction caused by mutations in mucA results in a mucoid phenotype and a chronic lung infection (Ciofu et al., 2005; Hogardt et al., 2007; Mena et al., 2008). Among many roles in pathogenesis, alginate can promote adherence to lung epithelial cells, reduce the efficiency of phagocytosis, and protect bacteria from antibiotics (Govan and Deretic, 1996; Pier et al., 2001; Leid et al., 2005).
The extracytoplasmic stress-response pathways in P. aeruginosa, Escherichia coli, and other Gram-negative bacteria involve regulated intramembrane proteolysis and share common features. For example, outer-membrane and periplasmic stress initiates a proteolytic cascade that ultimately degrades a transmembrane anti-sigma factor and releases the sigma factor to up-regulate stress-response genes (Alba et al., 2002; Kanehara et al., 2002; Walsh et al., 2003; Akiyama et al., 2004; Flynn et al., 2004; Chaba et al., 2007). In P. aeruginosa, proteolysis of wild type MucA, the anti-sigma factor, releases the AlgU sigma factor, which enhances transcription of the genes responsible for alginate biosynthesis from the algD promoter (Martin et al., 1993; Wood et al., 2006).
In P. aeruginosa, degradation of MucA is normally initiated by cleavage by the AlgW protease at site 1 near the periplasmic face of the inner membrane, continues with cleavage by the MucP protease at site 2 near the cytoplasmic face of the membrane, and is completed in the cytoplasm by AAA+ proteases such as ClpXP (Akiyama et al., 2004; Alba et al., 2002; Chaba et al., 2007; Flynn et al., 2004; Kanehara et al., 2002; Qiu et al., 2007; Qiu et al., 2008b; Walsh et al., 2003). Two signals are required to initiate this proteolytic cascade. The C-terminal peptides of proteins that accumulate in the periplasm during stress, including outer-membrane proteins (OMPs), bind to the PDZ domains of the trimeric AlgW protease and allosterically activate site-1 cleavage of MucA, but MucB competes for MucA binding and can inhibit this cleavage (Cezairliyan and Sauer, 2009). Recent studies show that lipopolysaccharides (LPS), a key component of the outer membrane, can bind MucB and cause release of MucA (Lima et al., 2013).
Here, we identify CupB5 as a novel activator of the alginate-production pathway. CupB5, an adhesive protein, is a member of one of the chaperone/usher pathways (CupA-E) that mediate assembly of pili and play roles in bacterial pathogenesis, adhesion, and biofilm formation (Hull et al., 1981; Marklund et al., 1992; Soto and Hultgren, 1999; Vallet et al., 2001; Giraud et al., 2011; Mikkelsen et al., 2009). These secretion systems, which are best characterized in Gram-negative bacteria (Sauer et al., 2000; Waksman and Hultgren, 2009), contain a periplasmic chaperone that maintains proteins to be secreted in an unassembled state and an usher that transports these proteins through the outer membrane. We find that overexpression of CupB5 in the absence of its chaperone induces a mucoid phenotype in P. aeruginosa and provide evidence that a peptide signal in CupB5 enhances OMP-activated cleavage of MucA by AlgW, even in the presence of MucB, allowing initiation of AlgU signaling.
Results
Overexpression of CupB5 induces mucoidy
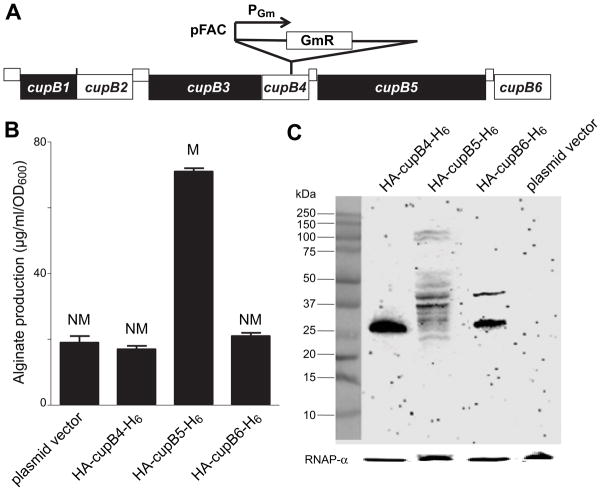
To identify new alginate regulatory genes, we performed mariner-transposon mutagenesis (Wong and Mekalanos, 2000) in the nonmucoid P. aeruginosa reference strain PAO1. We isolated a mucoid variant (PAO1-VE22) and identified the site of a single-copy insertion by inverse PCR and Southern-blot analysis (data not shown). The transposon insertion was six base pairs before the TAG stop codon at the 3′ end of cupB4 (Fig. 1A) and introduced a σ70-dependent PGm promoter that drove expression of the gentamicin resistance cassette and downstream genes (Rubin et al., 1999). Alginate production by PAO1-VE22 (31 ± 1.5 μg/ml/OD600) was substantially higher than that of PAO1 (8.5 ± 0.02 μg/ml/OD600) after 24 h growth at 37°C. To test if altered expression of cupB4, cupB5, or cupB6 was responsible for the mucoidy of PAO1-VE22, we cloned each individual coding sequence with an N-terminal HA epitope and C-terminal H6 tag in pHERD20T under control of an arabinose-inducible promoter (Qiu et al., 2008a). Plasmids expressing the tagged variant of CupB5, but not of CupB4 or CupB6, induced mucoid conversion and high-level alginate production in PAO1 (Fig. 1B). Western-blot analysis indicated that the plasmid-expressed CupB4, CupB5, and CupB6 variants were detectable when overexpressed in PAO1, albeit with fragmentation of CupB5 (Fig. 1C). We conclude that enhanced expression of CupB5 or a CupB5 fragment induces mucoidy and increased alginate in PAO1-VE22.
Fig. 1. Overproduction of CupB5 causes mucoid conversion in PAO1.
(A) Organization of the cupB1-cupB6 gene cluster (www.pseudomonas.com), and location of the transposon insertion site in PAO1-VE22. (B) Alginate production, measured using the carbazole assay, and colony phenotypes (M, mucoid; NM, nonmucoid) were assayed following transformation of P. aeruginosa strain PAO1 with plasmid HERD20T or variants expressing HA-CupB4-H6, HA-CupB5-H6, or HA-CupB6-H6, and overnight growth of colonies on PIA plates (carbenecillin 300 μg ml−1, 0.1% L-arabinose) at 37°C. Values are averages ± SEM (n=3). (C) Cell lysates of PAO1 carrying appropriate plasmids were electrophoresed on SDS polyacrylamide gels, and Western-blots using an anti-HA monoclonal antibody were used to detect HA-CupB4-H6, HA-CupB5-H6, or HA-CupB6-H6. The rightmost lane shows that HA cross-reactive proteins were absent in PAO1 transformed with the pHERD20T empty-vector.
Genetic requirements for CupB5-induced mucoidy
Deletion of five single genes required for the AlgU response (algW, mucP, clpX, clpP, or algU) in PAO1 prevented mucoidy and high-level alginate production when HA-CupB5-H6 was overproduced from a plasmid in these strains (Fig. 2A). In-frame deletion of algW in PAO1-VE22 also prevented alginate overproduction and mucoidy, a phenotype that was complemented by plasmid expression of AlgW but not AlgW lacking its PDZ domain (Fig. 2B). Consistently, when AlgU-dependent promoters were fused to lacZ and introduced into PAO1-VE22 or PAO1-VE2, a strain in which AlgW is activated by overproduction of MucE (Qiu et al., 2007), β-galactosidase levels were increased markedly compared to the same promoter fusions in PAO1 (Fig. S1). CupB5 is normally an extracellular protein (Ruer et al., 2008; Vallet et al., 2001). However, its overexpression in the absence of corresponding levels of the CupB4 chaperone and usher could result in periplasmic accumulation that acts directly or indirectly through AlgW to activate alginate production.
Fig. 2. Genetic requirements for CupB5-mediated alginate overproduction.
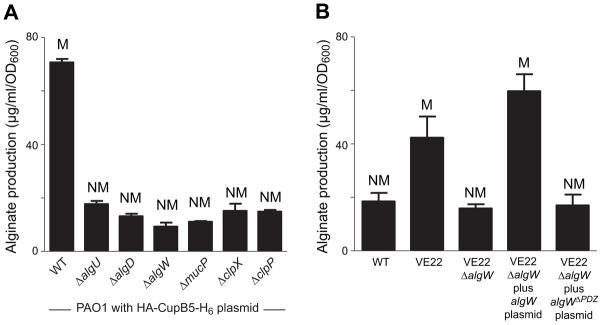
(A) Wild-type (WT) or mutant PAO1 strains were transformed with pHERD20 encoding HA-CupB5-H6, grown on PIA plates with 0.1% L-arabinose for 24 h at 37°C to induce HA-CupB5-H6 overexpression, and mucoid phenotypes and alginate production were determined. The algD gene encodes an enzyme required for alginate production. The algU, algW, mucP, clpX, and clpP genes are required for induction of algU-dependent alginate production. (B) Alginate production and mucoid phenotypes of different strains were measured after growth for 24 h at 37°C on PIA plates plus 300 μg m−1 carbenecillin and 0.1% L-Ara. In both panels, values are averages ± SEM (n=3).
CupB5 contains an internal signal for alginate overproduction
Because C-terminal sequences terminating with an aromatic residue and α-carboxylate can activate AlgW, we initially tested if the C-terminal residues of CupB5 (Asn1016-Ile1017-Trp1018) were responsible for activating AlgW and inducing mucoidy. However, overexpression of CupB5 variants with a C-terminal H6 tag or containing a deletion of the Asn1016-Ile1017-Trp1018 sequence and a C-terminal H6 tag still induced mucoidy in PAO1 (Table S1). Thus, the extreme C-terminal residues of CupB5 play little or no role in mediating alginate overproduction. Moreover, overproduction of variants containing just the N-terminal 491 residues of CupB5 also induced mucoidy (Table S1), indicating that a major inducing signal is located in this portion of the protein.
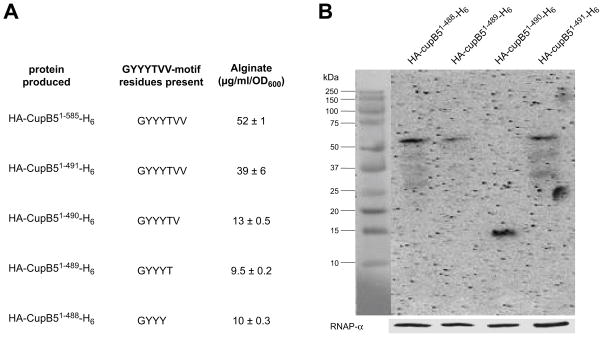
Deletion analyses revealed that the Thr489-Val490-Val491 sequence of CupB5 appeared important for alginate overproduction. Specifically, overexpression of CupB51–491 produced substantially more alginate than overexpression of CupB51–489 or CupB51–488 (Fig. 3A), and Western blotting showed that these proteins were present at a similar intracellular levels (Fig. 3B). Cells expressing CupB51–490 also produced less alginate (Fig. 3A), but this protein appeared to be severely truncated by degradation (Fig. 3B). Based on these results, we hypothesized that the CupB5 activating signal included some or all of the Thr489-Val490-Val491 sequence.
Fig. 3. Delineation of the CupB5 signal that activates alginate production.
(A) CupB5 variants with C-terminal truncations were cloned into pHERD20T, expressed in strain PAO1, and alginate production following growth in the presence of 1% L-arabinose was assayed. Proteins containing CupB5 residues 1–585 and 1–491 contain the complete GYYYTVV491 sequence motif. The remaining proteins contain only a portion of this motif. (B) Western-blot detection using anti-HA monoclonal antibody of truncated CupB5 variants.
CupB5 peptides stimulate OMP-activated AlgW cleavage of MucA and partially relieve MucB inhibition in vitro
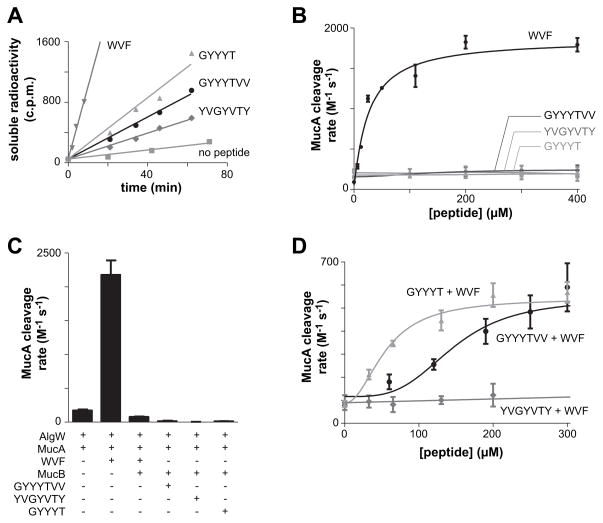
We initially tested if synthetic peptides including the Thr489-Val490-Val491 CupB5 sequence might stimulate alginate production in a manner analogous to OMPs by activating AlgW cleavage of 35S-MucA. One peptide (GYYYTVV) corresponded to the seven C-terminal residues of CupB51–491, another (YVGYVTY) was a sequence-scrambled variant, and a third (GYYYT) corresponded to the C-terminus of CupB51–489, which did not stimulate alginate production in vivo. We assayed the kinetics of AlgW cleavage of 35S-MucA in vitro by the production of acid-soluble peptide fragments in the presence of different concentrations of the CupB5 peptides. Each CupB5 peptide stimulated AlgW cleavage of 35S-MucA to a small degree, but at far lower rates than a peptide (WVF) corresponding to the C-terminal residues of MucE, a potent activator of AlgW (Fig. 4A & 4B). Thus, the CupB5 peptides do not substitute for OMP-like peptides in robust activation of AlgW cleavage of MucA.
Fig. 4. CupB5 peptides relieve MucB inhibition of MucA cleavage by AlgW.
(A) Cleavage of 35S-labelled MucA (20 μM) by AlgW (0.5 μM trimer) was assayed by the time-dependent release of acid-soluble peptides in the presence of either the OMP-like WVF peptide (400 μM), the wild-type GYYYT or GYYYTVV CupB5 peptides (400 μM), the sequence-scrambled YVGYVTY peptide (400 μM), or a buffer control. (B) Second-order rate constants for MucA cleavage were determined by incubating 35S-labelled MucA (20 μM) and AlgW (0.5 μM trimer) with different concentrations of WVF or CupB5 peptides and measuring the time-dependent increase in acid-soluble products. Rates were divided by the MucA and AlgW concentrations to determine activity. Lines are fits to the equation activity = basal + stimulated/(1+(Kact/[peptide])h). Values are averages ± SEM (n=2). (C) Second-order rate constants for cleavage of 35S-MucA (20 μM) by AlgW (0.5 μM trimer) were determined as described in panel B in the presence of different combinations of WVF peptide (110 μM), MucB (25 μM dimer), and wild-type or scrambled CupB5 peptides (405 μM). Values are averages of two or more independent trials ± SEM. (D) Second-order rate constants for cleavage of 35S-MucA (20 μM) by AlgW (0.5 μM trimer) and WVF peptide (150 μM) were determined in the presence of increasing concentrations of the GYYYT, GYYYTVV, or sequence-scrambled YVGYVTY peptides. Lines are fits to the Hill equation: rate = basal + Vmax/(1 + (Ks/[peptide])h). For GYYYT, Ks was 55 ± 13 μM and h was 2.1 ± 0.9. For GYYYTVV, Ks was 143 ± 31 μM and h was 3.5 ± 2.2.
As expected from previous studies (Cezairliyan and Sauer, 2009), addition of MucB suppressed cleavage of 35S-MucA by WVF-activated AlgW (Fig. 4C). Moreover, addition of just the GYYYTVV, YVGYVTY, or GYYYT CupB5 peptides did not result in AlgW cleavage of MucA when MucB was present (Fig. 4C). Next, we tested if the wild-type or scrambled CupB5 peptides could relieve MucB inhibition of AlgW cleavage of 35S-MucA in the presence of the WVF activating peptide. Importantly, titration of GYYYTVV or GYYYT into reactions containing MucA, MucB, AlgW, and WVF peptide resulted in a dose-dependent increase in the rate of MucA cleavage (Fig. 4D). Almost no increase in MucA cleavage was observed with the scrambled YVGYVTY peptide (Fig. 4D), demonstrating sequence specificity. Interestingly, activation of MucA cleavage by GYYYT occurred at lower peptide concentrations and was less cooperative as judged by the Hill constant than activation by the GYYYTVV peptide (Fig. 4D). Thus, the C-terminal valine residues of GYYYTVV are not necessary for activation of MucA cleavage in the presence of MucB in vitro. Although the GYYYTVV and GYYYT peptides activated AlgW cleavage of MucA to similar levels in the presence of WVF peptide and MucB, these levels were approximately one-third of the MucA cleavage rate observed with WVF and AlgW alone (compare Figs. 4B and 4D). We conclude that the CupB5 peptides can partially relieve MucB inhibition of WVF-activated AlgW cleavage of MucA.
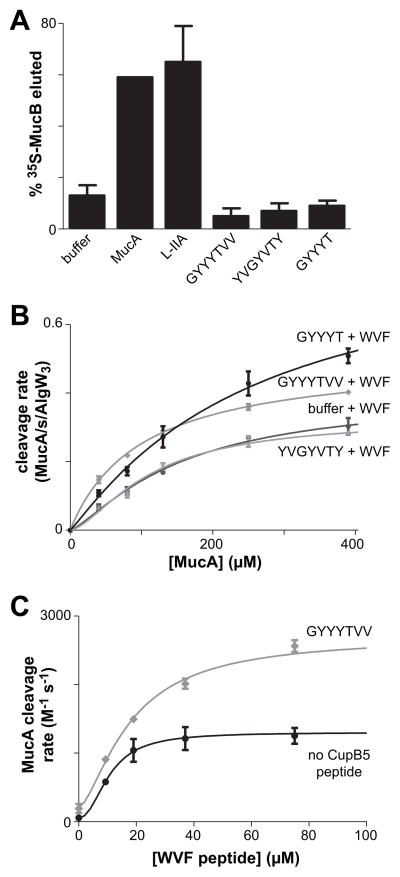
Lipopolysaccharides bind MucB and competitively displace MucA (Lima et al., 2013). Because MucB normally binds peptide sequences in MucA, it seemed plausible that CupB5 peptides might also compete for MucA binding to MucB. We tested this possibility by assaying the ability of the CupB5 peptides to elute 35S-MucB from MucA that had been covalently bound to agarose resin (Fig. 5A). In this assay, the L-IIA lipopolysaccharide fragment and free MucA efficiently competed with resin-bound MucA for MucB binding. However, incubation of 35S-MucB•MucA resin with the GYYYTVV, YVGYVTY, or GYYYT peptides resulted in no more 35S-MucB release than observed in the buffer control (Fig. 5A). This result strongly suggests that the CupB5 peptides do not relieve MucB inhibition by directly competing for its binding to MucA. Binding of the L-IIA lipopolysaccharide fragment converts MucB dimers to tetramers (Lima et al., 2013). By contrast, incubation of MucB with GYYYTVV peptide resulted in no substantial change in elution during gel-filtration chromatography (Fig. S2).
Fig. 5. CupB5 peptides do not compete with MucB for MucA binding but stimulate WVF-activated AlgW cleavage of MucA.
(A) Release of 35S-MucB from MucA-agarose following incubation with buffer, MucA (155 μM), L-IIA (20 mg/mL), or CupB5 peptides (~300 μM). For the CupB5 peptides, values are averages ± SEM (n=3). For L-IIA, the value is the average ± SEM (n=2). (B) Rates of cleavage by AlgW (0.5 μM trimer) and WVF peptide (150 μM) were measured at different concentrations of 35S-MucA in the presence or absence of CupB5 peptides (300 μM). Values are averages ± SEM (n=2) and were fit to the Hill form of the Michaelis-Menten equation, rate = Vmax/(1 + (KM/[substrate])h). (C) Rates of cleavage of 35S-MucA (50 μM) by AlgW (0.5 μM trimer) were assayed at difference concentrations of WVF peptide in the presence or absence of CupB5 peptides (300 μM). Values are averages ± SEM (n=2) and were fit to the equation rate = basal + Vmax/(1 + (Kact/[peptide])h). Without CupB5 peptide, h was 2.2 ± 1, Kact was 11 ± 2 μM, and Vmax was 1250 ± 150 M−1 s−1. With GYYYTVV peptide, h was 1.5 ± 0.8, Kact was 18 ± 7 μM, and Vmax was 2490 ± 490 M−1 s−1.
Because CupB5 peptides did not strongly activate AlgW cleavage of MucA in the absence of the WVF peptide or release MucA from MucB, we speculated that they might act in concert with the WVF peptide to allow AlgW to bind and degrade MucA more efficiently and thus to compete better with MucB for the MucA substrate. This model predicts that MucA should be degraded at a faster rate in the presence of both the WVF and CupB5 peptides. Indeed, when we assayed the rate of cleavage of different concentrations of 35S-MucA by AlgW, faster cleavage was observed when both the WVF and wild-type CupB5 peptides were present compared to only the WVF peptide or the WVF peptide and scrambled CupB5 control peptide (Fig. 5B). We also measured the rate of AlgW cleavage of a fixed concentration of MucA in the presence of increasing concentrations of WVF peptide with or without additional GYYYTVV peptide (Fig. 5C). When GYYYTVV peptide was present, the maximum rate of MucA cleavage increased almost two-fold. We conclude that the CupB5 peptides and WVF peptide act synergistically to enhance AlgW cleavage of MucA, allowing better competition with MucB.
In E. coli, RseA, RseB, and DegS are homologs of MucA, MucB, and AlgW. A YYF peptide efficiently activates DegS cleavage of RseA in the absence of RseB or in the presence of both RseB and lipopolysaccharides (Walsh et al., 2003; Cezairliyan and Sauer, 2007; Lima et al., 2013). The CupB5 peptides did not substantially stimulate cleavage of 35S-RseA by YYF-activated DegS in the presence of RseB (Fig. S3), suggesting that they do not interact with DegS. This result is not surprising, as DegS and AlgW share only 44% sequence homology.
Suppressing and enhancing alginate production
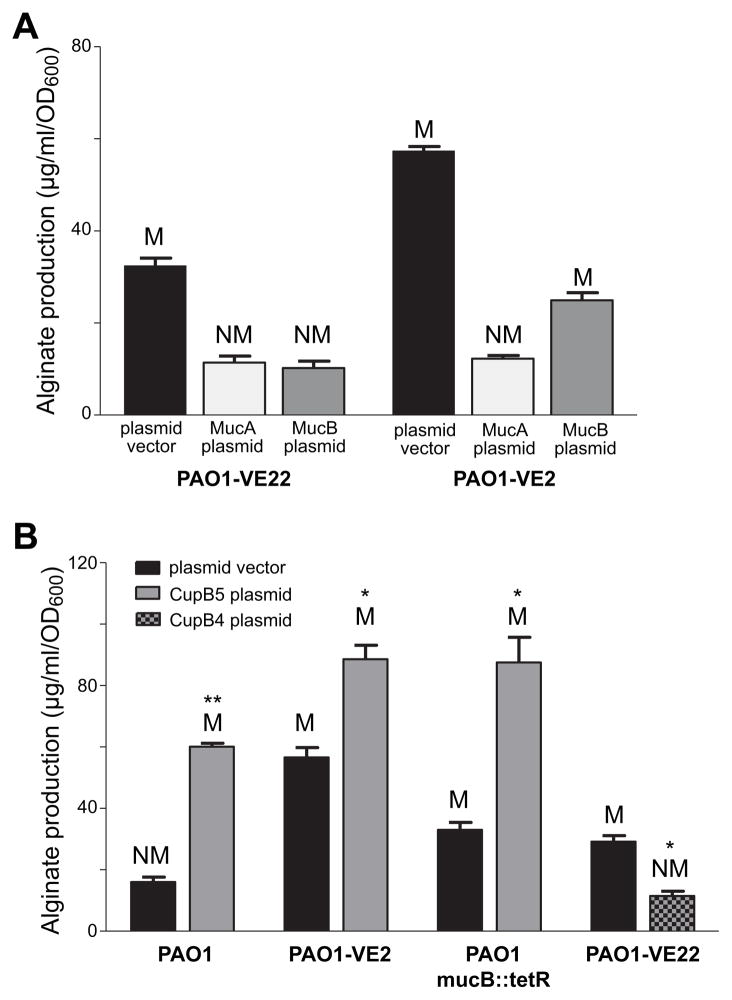
If CupB5 allows AlgW to compete more efficiently with MucB for MucA binding and cleavage, then overproduction of MucB in strain PAO1-VE22 would be expected to decrease alginate production. Indeed, plasmid-mediated MucB overproduction had this effect, as did plasmid-mediated overproduction of MucA (Fig. 6A). The latter result suggests that with enough additional uncleaved MucA, AlgU is efficiently inhibited even when more MucA is degraded than usual. Even the CupB5-induced elevated rate of AlgW cleavage of MucA in this strain must be too slow to free enough AlgU to fully enhance transcription of alginate biosynthesis genes.
Fig. 6. Inducing and suppressing alginate production in P. aeruginosa strains.
(A) Strains overproducing CupB5 (PAO1-VE22) or MucE (PAO1-VE2) were transformed with plasmid HERD20T or variants overexpressing MucA or MucB, grown at 37°C for 24 h on PIA plates supplemented with 0.1% L-arabinose, and alginate production and colony phenotypes (M, mucoid; NM, nonmucoid) were assayed. (B) Alginate production and mucoid phenotypes were assayed in strains PAO1, PAO1-VE2 (mucE overexpressed), PAO1 mucB::tetR (mucB inactivated), and PAO1-VE22 (cupB5 overexpressed) transformed with the HERD20T plasmid vector or variants expressing HA-CupB4-H6 or HA-CupB5-H6. Values are averages ± SEM (n=3). Statistical significance was determine using the Student’s t-test (*P<0.05; **<0.005).
Plasmid-expressed MucB or MucA also decreased alginate production in PAO1-VE2, a strain expressing higher MucE levels (Fig. 6A). The rate of AlgW cleavage of MucA in PAO1-VE2 is apparently too low to fully degrade the excess MucA, resulting in greater inhibition of AlgU and reduced alginate production. Excess MucB presumably outstrips the endogenous anti-MucB signals in PAO1-VE2 and mass action results in enhanced binding to MucA, reducing the rate of MucA cleavage by activated AlgW.
To determine how simultaneous overexpression of MucE and CupB5 affected alginate levels,, we transformed PAO1-VE2 with a plasmid expressing HA-CupB5-H6. Notably, alginate levels in this strain were higher than in the empty-vector control (Fig. 6B), suggesting that activation of AlgW through MucE and through CupB5 occur synergistically. Our biochemical results predict that MucB should not be genetically required for CupB5-mediated pathway activation. To test this prediction, we used a transposon-insertion strain, PAO1-mucB::tetR, which displayed an elevated level of basal alginate production and mucoid phenotype (Fig. 6B). When we transformed PAO1-mucB::tetR with a plasmid expressing HA-CupB5-H6, alginate production increased compared to the empty-vector control (Fig. 6B), indicating that CupB5 overexpression induces pathway activation by a MucB-independent mechanism. Finally, we tested if expression of the CupB4 chaperone, which should facilitate export of CupB5 rather than its periplasmic accumulation, would decrease alginate production. Indeed, when we transformed PAO1-VE22 with a plasmid expressing HA-CupB4-H6, alginate production was suppressed and the mucoid phenotype of the parent strain was reversed (Fig. 6B). This result supports a model in which interactions between accumulated periplasmic CupB5 and periplasmic AlgW leads to induction of alginate production and mucoidy.
Discussion
Based on results from a transposon screen in P. aeruginosa, we discovered that overexpression of CupB5, a protein normally secreted through the outer membrane by a chaperone-usher system, activates alginate production and results in mucoid colonies. CupB5-mediated mucoidy depends on known components of the AlgU stress-response pathway, including the AlgU transcription factor and the AlgW, MucP, and ClpXP proteases. Peptides from a region of CupB5 important for elevated alginate production in vivo stimulate cleavage of MucA by OMP-activated AlgW in vitro and partially relieve MucB inhibition of AlgW cleavage of MucA. Overexpression of either MucB or MucA in strains that overexpress CupB5 also reduces alginate production, supporting a model in which excess periplasmic CupB5 stimulates proteolytic cleavage of MucA by AlgW.
We found that expression of HA-CupB51–491-H6 in PAO1 increased alginate production in vivo and demonstrated that a GYYYTVV peptide (corresponding to CupB5 residues 485–491) antagonized MucB inhibition in vitro. A peptide with the same composition as GYYYTVV but a scrambled sequence did not inhibit MucB. However, we also found that expression of HA-CupB51–489-H6 failed to increase alginate production in vivo, whereas a GYYYT peptide (corresponding to CupB5 residues 485–489) did antagonize MucB inhibition in vitro. This discrepancy between results in vivo and in vitro could arise because the GYYYT peptide in HA-CupB51–489-H6 is altered by proteolysis or is masked by tertiary structure, aggregation, or binding to another cellular component. We also note that expression of full-length HA-CupB5-H6 and truncated HA-CupB51–585-H6 resulted in somewhat higher alginate levels than expression of HA-CupB51–491-H6. This variation could arise from differences in expression levels, cellular interactions, or possibly as a consequence of additional peptide sequences in the longer proteins that facilitate pathway activation.
We initially thought that CupB5 peptides might bind MucB and compete for binding to MucA in a manner analogous to LPS. However, neither GYYYTVV nor GYYYT peptides compete with MucA-agarose for MucB binding and saturating concentrations of these peptides only partially relieve MucB inhibition in vitro. Moreover, unlike LPS, GYYYTVV did not convert MucB dimers into tetramers. By themselves, CupB5 peptides activate AlgW cleavage in vitro far less well than the OMP-like WVF peptide. However, CupB5 peptides stimulate MucA cleavage by WVF-activated AlgW, suggesting that OMP-like and CupB5 peptides bind to distinct sites in AlgW but have a common effect of stabilizing the proteolytically active enzyme.
The GYYYTVV sequence is conserved in CupB5 proteins from different strains of P. aeruginosa but not in CupB5 proteins from other Pseudomonas species (Fig. S3), and some Pseudomonas species have no CupB5 ortholog. Thus, CupB5 activation of AlgW is specific to P. aeruginosa. It remains to be determined if P. aeruginosa utilizes other proteins in this manner or if AlgW homologs in other bacteria are activated by more than one kind of peptide signal.
Activation of the AlgU pathway requires one molecular signal (C-terminal OMP peptides) to activate site-1 cleavage of MucA by the AlgW protease and a second molecular signal (LPS) to relieve MucB inhibition of this cleavage. Overproduction of CupB5 can partially bypass this second requirement by helping OMP-bound AlgW compete with MucB for MucA binding and proteolysis. Expression of CupB5 under non-stress conditions increases alginate production similarly to non-stress expression of MucE (Qiu et al., 2007). In these experiments, a signal that directly antagonizes MucB inhibition could arise indirectly as a consequence of other molecules whose concentrations increase as a consequence of CupB5 or MucE expression or might always be present at some level in the periplasm. For example, some level of unassembled OMPs and LPS is probably always present in the periplasm. Notably, however, simultaneous expression of CupB5 and MucE results in higher levels of alginate, indicating that these signals activate the alginate pathway through synergistic mechanisms
Although CupB5 peptides, OMP peptides, and LPS molecules function by different mechanisms to activate AlgW or relieve MucB inhibition, the molecular logic that gives rise to all three signals is similar. For example, stresses that compromise the transport to and outer-membrane assembly of OMPs and LPS, or that slow usher-mediated CupB5 secretion, would all lead to the periplasmic accumulation of these molecules and activation of the AlgU pathway. Indeed, we found that alginate production was suppressed in strains that overproduced the CupB4 chaperone, allowing export of CupB5. Certain stresses might only activate AlgW through OMP signals, whereas others require an enhanced response via CupB5 or possibly other proteins secreted through the outer membrane. Clearly, however, the AlgU system integrates multiple signals, each reporting on potential dysfunction in outer-membrane assembly, structure, and/or secretion. Dysfunction in just one pathway should give rise to a buffered AlgU response, whereas dysfunction affecting multiple pathways would elicit a robust AlgU response.
Experimental procedures
Bacteria strains, plasmids, and growth conditions
Bacteria strains and plasmids used in this study are shown in Table S2. Escherichia coli strains were grown at 37°C in Lennox broth (LB) or LB agar. P. aeruginosa strains were grown at 37°C in LB, Pseudomonas isolation broth (PIB, Alpha Bioscience) or on Pseudomonas isolation agar (PIA) plates (Difco). When required, the concentration of carbenicillin, tetracycline or gentamicin added to LB broth or plates was 100 μg ml−1, 20 μg ml−1, or 15 μg ml−1, respectively, and 300 μg ml−1, 150 μg ml&−1, or 200 μg ml−1 when added to PIB or PIA plates, respectively.
Transposon mutagenesis
Transposon mutagenesis was carried out via biparental conjugations using E. coli SM10 λpir carrying plasmid pFAC as the donor strain (Wong and Mekalanos, 2000) and PAO1 as the recipient strain. After incubating PAO1 and E. coli cells on LB agar plates at 37°C for 6 h, bacteria were collected, re-suspended in PBS and then plated onto PIA supplemented with gentamicin (200 μg ml−1). Mucoid colonies were identified. The chromosomal DNA of mucoid mutants was isolated using the QIAamp genomic DNA Extraction kit (Qiagen, USA). Approximately, 2 μg of DNA was digested with SalI overnight at 37°C, followed by purification and self-ligation using Fast-Link DNA ligase (Epicentre, USA). The circular closed DNA was used as a template for inverse PCR using GM3OUT and GM5OUT primers (Qiu et al., 2008b). The PCR products were purified and sequenced. Finally, Southern-blot hybridization was also conducted to monitor the copy number of transposon insertions using the Gmr gene as the probe (Head and Yu, 2004).
Genetic construction, transformation and conjugation
All DNA cloning was performed using PCR products digested with appropriate restriction enzymes and ligated into the E. coli to Pseudomonas shuttle vector pHERD20T (Qiu et al., 2008a). All constructs of pHERD20T were sequenced to verify the correct DNA sequences. DNA sequencing was carried out by the Marshall University Genomics Core Facility. The primer sequences used for PCR cloning will be provided upon request. The transformation of One Shot TOP10 (Invitrogen, USA) was performed according to the supplier’s instruction. The transfer of pHERD20T and its derivatives was performed via triparental conjugation using the helper plasmid pRK2013 (Figurski and Helinski, 1979).
Protein preparation and Western blotting
For extraction of total cellular protein, PeriPreps™ Periplasting Kit (Epicentre, USA) was used. Bacteria were inoculated in PIB medium and incubated in shake culture for several hours, until the OD600 was ~0.6. At this point 0.1% L-arabinose was added to induce protein expression. Two hours after induction, bacteria were collected for cell lysis. The protein concentration was measured using Bio-Rad Dc protein assay reagents (Bio-Rad). Equivalent proteins were mixed with 2×sample loading buffer and analyzed on 10–20% pre-cast SDS-PAGE gels (Bio-Rad). Proteins were then transferred to a PVDF membrane (GE) by electro-blotting for immunodetection. A primary antibody, rat Anti-HA (Roche, USA), was used at a dilution of 1:5000. Goat anti-rat immunoglobulin G (heavy and light chains) conjugated with horseradish peroxidase (Pierce, USA) diluted 1: 5000 was used as the secondary antibody. Finally, the proteins on the membrane were visualized using the Amersham ECL kit (GE, USA).
Alginate assay
P. aeruginosa strains were grown at 37°C on triplicate PIA plates for 24 h. Bacteria were collected, suspended in PBS, and the OD600 was measured and adjusted to ~0.5 by addition of PBS. The suspensions were analyzed for the amount of uronic acid in comparison with a standard curve made with D-mannuronic acid lactone (Sigma-Aldrich, USA) as described (Damron et al., 2009).
β-galactosidase assay
Pseudomonas strains carrying the lacZ vector pLP170 containing PalgU, and PalgD were cultured on PIA plates. After 24 h, bacteria were harvested, resuspended in PBS, and OD600 was measured and adjusted to ~0.3. Cells were then permeabilized using toluene, and β-galactosidase activity was measured at OD420 and OD550. The results in Miller Units were calculated according to the follow formula: Miller Units=1000×[OD 420−(1.75 × OD 550)]/[Reaction time (m) × Volume (ml) × OD 600] (Miller, 1972). The reported values represent an average of three independent experiments ± SEM.
Proteins and peptides
Purifications of P. aeruginosa MucA, MucB and AlgW and E. coli RseA, RseB and DegS were performed as described after plasmid-mediated overexpression in E. coli strain X90(DE3) (Sohn et al., 2007; Cezairliyan and Sauer, 2009). Cultures containing 100 μg/mL ampicillin were grown to an OD600 of ~0.6, and protein expression was induced with 100 mM IPTG for two h before harvesting. Cells overexpressing MucB, AlgW, RseB or DegS were resuspended in native lysis buffer [P. aeruginosa proteins: 50 mM sodium phosphate (pH 8), 500 mM KCl, 20 mM imidazole; E. coli proteins: 50 mM sodium phosphate (pH 8), 300 mM NaCl, 10 mM imidazole] and lysed by sonication. Cells overexpressing MucA or RseA were resuspended in denaturing lysis buffer [50 mM sodium phosphate (pH 8), 8 M guanidine hydrochloride (pH 8)] and allowed to lyse at room temperature for 30 min. For all preparations, lysates were cleared by centrifugation and applied to Ni-NTA columns pre-equilibrated in native lysis buffer or denaturing lysis buffer. Columns were washed with lysis buffer and proteins eluted with elution buffer [P. aeruginosa proteins: 50 mM sodium phosphate (pH 8), 500 mM KCl, 500 mM imidazole; E. coli proteins: 50 mM sodium phosphate (pH 8), 300 mM NaCl, 300 mM imidazole]. Proteins were then dialyzed overnight against storage buffer [P. aeruginosa proteins: 50 mM sodium phosphate (pH 7.4), 200 mM KCl, 10% glycerol; E. coli proteins: 50 mM sodium phosphate (pH 8), 200 mM NaCl, 1 mM EDTA] and then again against fresh buffer before being stored frozen at −80°C. Prior to dialysis and storage, MucB and RseB were chromatographed on a size-exclusion column and only fractions corresponding to the dimer were collected. To obtain radiolabeled 35S-MucA,35S-RseA, or 35S-MucB, cells were grown in a defined rich medium (TekNova) lacking methionine and 35S-methionine was added upon induction. Purification of 35S-MucA and 35S-RseA then proceeded as described above for unlabeled MucA and RseA. Cells expressing 35S-MucB were lysed by the addition of lysozyme and three freeze-thaw cycles, and purification then proceeded as described above. All peptides were synthesized by GenScript and purified by reverse-phase HPLC using a C18 column. Purified products were lyophilized, stored at −20°C, then resuspended in storage buffer and used immediately. The L-IIA LPS fragment was a gift from Santiago Lima (MIT) (Lima et al., 2013).
Cleavage assays
Assays were performed as described (Sohn et al., 2007; Cezairliyan and Sauer, 2009). Briefly, cleavage reactions were performed at 25°C in reaction buffer [P. aeruginosa proteins: 50 mM sodium phosphate (pH 7.4), 200 mM KCl, 10% glycerol; E. coli proteins: 150 mM sodium phosphate (pH 8.3), 380 mM NaCl, 10% glycerol, 4 mM EDTA]. Reactions were initiated by the addition of protease and quenched after different times by addition of cold 10% trichloroacetic acid. Following centrifugation, acid-soluble reaction products were quantified by scintillation counting and compared to total radioactivity from control samples. Cleavage activity for peptide-activation assays and experiments involving MucB are reported as a second-order rate constant calculated by dividing the rate of substrate cleavage by the substrate and enzyme concentrations. Activity at different substrate concentrations for the Michaelis-Menten curve was calculated by dividing the rate of substrate cleavage by the enzyme concentration.
MucA-resin assay
Dry cyanogen-bromide activated Sepharose B4 resin (66 mg) was washed with 1 M HCl, equilibrated with coupling buffer [25 mM sodium phosphate (pH 8.3), 50 mM NaCl], and incubated with purified MucA (900 μg) for 1.5 hours. After removal of free protein, reactive groups on the resin were blocked by incubation in 0.5 M ethanolamine (pH 8.3) for 1 h. The MucA resin was washed, resuspended in coupling buffer, and stored at 4 °C. For assays, MucA resin was drained, incubated with a four-fold molar excess of 35S-MucB, washed again, incubated with ligand for 30 min, added to a pre-wetted 96-well Multiscreen HTS-HV Filter Plate (Millipore) over a 96-well polypropylene receiving plate (Greiner), and centrifuged at 1500 × g for 1 min. Elution of 35S-MucB was measured by scintillation counting and normalized by dividing by the total initial 35S-MucB radioactivity.
Supplementary Material
Acknowledgments
This work was supported by the National Aeronautics and Space Administration West Virginia Space Grant Consortium (NASA WVSGC), the Cystic Fibrosis Foundation (CFF-YU11G0), and NIH grant AI-16892. HDY was supported by NIH P20RR016477 and P20GM103434 to the West Virginia IDeA Network for Biomedical Research Excellence, and is also the co-founder of Progenesis Technologies, LLC. T.R.W. was supported through the NASA WVSGC Graduate Research Fellowship. We thank Dongru Qiu for construction of PAO1ΔalgD.
References
- Akiyama Y, Kanehara K, Ito K. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J. 2004;23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev. 2002;16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher JC, Yu H, Mudd MH, Deretic V. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun. 1997;65:3838–3846. doi: 10.1128/iai.65.9.3838-3846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezairliyan BO, Sauer RT. Inhibition of regulated proteolysis by RseB. Proc Natl Acad Sci USA. 2007;104:3771–3776. doi: 10.1073/pnas.0611567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezairliyan BO, Sauer RT. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol. 2009;72:368–379. doi: 10.1111/j.1365-2958.2009.06654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaba R, Grigorova IL, Flynn JM, Baker TA, Gross CA. Design principles of the proteolytic cascade governing the sigmaE-mediated envelope stress response in Escherichia coli: keys to graded, buffered, and rapid signal transduction. Genes Dev. 2007;21:124–136. doi: 10.1101/gad.1496707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofu O, Riis B, Pressler T, Poulsen HE, Hoiby N. Occurrence of hypermutable Pseudomonas aeruginosa in cystic fibrosis patients is associated with the oxidative stress caused by chronic lung inflammation. Antimicrob Agents Chemother. 2005;49:2276–2282. doi: 10.1128/AAC.49.6.2276-2282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damron FH, Qiu D, Yu HD. The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J Bacteriol. 2009;191:2285–2295. doi: 10.1128/JB.01490-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Levchenko I, Sauer RT, Baker TA. Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev. 2004;18:2292–2301. doi: 10.1101/gad.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol. 2012;10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- Giraud C, Bernard CS, Calderon V, Yang L, Filloux A, Molin S, et al. The PprA-PprB two-component system activates CupE, the first non-archetypal Pseudomonas aeruginosa chaperone-usher pathway system assembling fimbriae. Environ Microbiol. 2011;13:666–683. doi: 10.1111/j.1462-2920.2010.02372.x. [DOI] [PubMed] [Google Scholar]
- Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head NE, Yu H. Cross-sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: biofilm formation, virulence, and genome diversity. Infect Immun. 2004;72:133–144. doi: 10.1128/IAI.72.1.133-144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogardt M, Hoboth C, Schmoldt S, Henke C, Bader L, Heesemann J. Stage-specific adaptation of hypermutable Pseudomonas aeruginosa isolates during chronic pulmonary infection in patients with cystic fibrosis. J Infect Dis. 2007;195:70–80. doi: 10.1086/509821. [DOI] [PubMed] [Google Scholar]
- Hull RA, Gill RE, Hsu P, Minshew BH, Falkow S. Construction and expression of recombinant plasmids encoding type 1 or D-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect Immun. 1981;33:933–938. doi: 10.1128/iai.33.3.933-938.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K, Ito K, Akiyama Y. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev. 2002;16:2147–2155. doi: 10.1101/gad.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J Immunol. 2005;175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- Lima S, Guo MS, Chaba R, Gross CA, Sauer RT. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science. 2013;340:837–841. doi: 10.1126/science.1235358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund BI, Tennent JM, Garcia E, Hamers A, Baga M, Lindberg F, et al. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol Microbiol. 1992;6:2225–2242. doi: 10.1111/j.1365-2958.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- Martin DW, Holloway BW, Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J Bacteriol. 1993;175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathee K, McPherson CJ, Ohman DE. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN) J Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, Perez JL, et al. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol. 2008;190:7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen H, Ball G, Giraud C, Filloux A. Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS One. 2009;4:e6018. doi: 10.1371/journal.pone.0006018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. beta-galactosidase assay. In: Miller JH, editor. Experiments in molecular genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- Pier GB, Coleman F, Grout M, Franklin M, Ohman DE. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect Immun. 2001;69:1895–1901. doi: 10.1128/IAI.69.3.1895-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol. 2008a;74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Eisinger VM, Head NE, Pier GB, Yu HD. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology. 2008b;154:2119–2130. doi: 10.1099/mic.0.2008/017368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Eisinger VM, Rowen DW, Yu HD. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2007;104:8107–8112. doi: 10.1073/pnas.0702660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin EJ, Akerley BJ, Novik VN, Lampe DJ, Husson RN, Mekalanos JJ. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc Natl Acad Sci U S A. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruer S, Ball G, Filloux A, de Bentzmann S. The ‘P-usher’, a novel protein transporter involved in fimbrial assembly and TpsA secretion. EMBO J. 2008;27:2669–2680. doi: 10.1038/emboj.2008.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruer S, Stender S, Filloux A, de Bentzmann S. Assembly of fimbrial structures in Pseudomonas aeruginosa: functionality and specificity of chaperone-usher machineries. J Bacteriol. 2007;189:3547–3555. doi: 10.1128/JB.00093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer FG, Barnhart M, Choudhury D, Knight SD, Waksman G, Hultgren SJ. Chaperone-assisted pilus assembly and bacterial attachment. Curr Opin Struct Biol. 2000;10:548–556. doi: 10.1016/s0959-440x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Schurr MJ, Yu H, Martinez-Salazar JM, Boucher JC, Deretic V. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J, Grant RA, Sauer RT. Allosteric activation of DegS, a stress sensor PDZ protease. Cell. 2007;131:572–583. doi: 10.1016/j.cell.2007.08.044. [DOI] [PubMed] [Google Scholar]
- Soto GE, Hultgren SJ. Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol. 1999;181:1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci USA. 2001;98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol. 2009;7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- Wong SM, Mekalanos JJ. Genetic footprinting with mariner-based transposition in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2000;97:10191–10196. doi: 10.1073/pnas.97.18.10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LF, Leech AJ, Ohman DE. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: roles of σ22 (AlgT) and the AlgW and Prc proteases. Mol Microbiol. 2006;62:412–426. doi: 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.