Abstract
Background aims
The Quantum® Cell Expansion System (Quantum; Terumo BCT, Inc, Lakewood, CO, USA) is a novel hollow fiber-based device that automates and closes the cell culture process, reducing labor intensive tasks such as manual cell culture feeding and harvesting. The manual cell selection and expansion processes for the production of clinical-scale quantities of bone marrow-derived human mesenchymal stromal cells (BM-hMSCs) have been successfully translated onto the Quantum platform previously. The formerly static, manual, in vitro process performed primarily on tissue culture polystyrene substrates may raise the question of whether BM-hMSCs cultured on a hollow fiber platform yields comparable cell quality.
Methods
A rigorous battery of assays was used to determine the genetic stability of BM-hMSCs selected and produced with the Quantum. In this study, genetic stability was determined by assessing spectral karyotype, micronucleus formation and tumorigenicity to resolve chromosomal aberrations in the stem cell population. Cell phenotype, adherent growth kinetics and tri-lineage differentiation were also evaluated. HMSC bone marrow aspirates, obtained from three approved donors, were expanded in parallel using T225 culture flasks and the Quantum.
Results
BM-hMSCs harvested from the Quantum demonstrated immunophenotype, morphology and tri-lineage differentiation capacity characteristics consistent with the International Society of Cell Therapy standard for hMSCs. Cell populations showed no malignant neoplastic formation in athymic mice 60 days post-transplant, no clonal chromosomal aberrations were observed and no DNA damage was found as measured by micronucleus formation.
Conclusions
Quantum-produced BM-hMSCs are of comparable quality and demonstrate analogous genetic stability to BM-hMSCs cultured on tissue culture polystyrene substrates.
Keywords: cell proliferation, chromosome, mesenchymal stromal cell, micronucleus, spectral karyotyping, xenograft
Introduction
Since the discovery of colony-forming unit fibroblasts was reported by Friedenstein et al. in 1966–1970 (1–3), the primary motivation for the therapeutic development of mesenchymal stromal cells (MSCs) continues to be their intrinsic nature for self-renewal and their multi-potency. The demand for research and clinical-grade MSCs necessitates not only that a sufficient quantity of cells be made available but also that these cells should manifest genetic stability for tissue homeostasis. For example, MSCs derived from adult bone marrow aspirates and other tissue sources have the ability to differentiate into adipogenic, chondrogenic, osteogenic and myogenic lineages, which have now been incorporated into various regenerative cell-based clinical trials. Human MSCs (hMSCs) are being studied for numerous immunologic, orthopedic, cardiovascular, dermatologic and neural indications. In cell therapy, one of the development challenges is to expand a population of MSCs that comprise 0.001–0.01% of the nucleated cells in the bone marrow (4). We employed a series of methods to quantify genetic stability to support further the development of stem cell processing from bone marrow aspirates by automated cell expansion systems. The Quantum System (Terumo BCT, Inc, Lakewood, CO, USA) is a functionally closed, automated hollow fiber system designed to grow both adherent and suspension cells.
The objective of this study was to characterize the genetic stability of bone marrow-derived hMSCs (BM-hMSCs) that have been selected and expanded in both the Quantum System and on tissue culture polystyrene surfaces (TCPS). Using this functional approach, we sought to compare the impact of manual and automated expansion methods on chromosome fragmentation and aberrations by assessing hMSC surface biomarker expression, chromosome stability, micronucleus formation, and malignant cell transformation. To detect these events, we used the International Society of Cell Therapy (ISCT) qualifying criteria (5), including immunophenotyping coupled with spectral karyotyping, automated micronucleus analysis and the engraftment of cells in the athymic mouse model. Although considered rare, Wang et al. (6) reported the outgrowth of a transformed cell population from normal BM-hMSC culture that generated solid tumors in immunocompromised mice. Adherent cells were isolated from BM-hMSCs that exhibited contact-independent growth, chromosome aneuploidy and translocations as well as formation of multiple tumors in an immunocompromised murine model. Wang et al. (6) suggested that either the transformed population of hMSCs developed in culture or the abnormal cells existed in the donor and were expanded in culture. In view of the clinical development of adult stem cells, it seems prudent to assess hMSCs for their genetic stability to confirm that suitable conditions exist for ex vivo propagation, particularly as processes become automated. Spectral karyotyping (SKY) is a genome scanning technique based on 24 DNA probes that can detect abnormalities on the average of 3 Mb pairs in length (7). This technique is most useful in identifying chromosomal material lacking band formation, defining subtle rearrangements and identifying translocations unresolved by classic G-banding cytogenetics.
In eukaryotic mitosis, chromosome number and structure are generally conserved for somatic cells until senescence or transformation. However, if mitosis is interrupted or if chromosomes are damaged, the distribution of genetic material in anaphase between the daughter cells may become altered because of acentric fragmentation, delayed chromosome migration, non-disjunction and the mis-repair or unrepair of DNA (8,9). As a result, genetic material that is not incorporated into the daughter nuclei in telophase can replicate in cell progeny and form micronuclei (MN) in the cytoplasm (9,10). Consequently, measuring the frequency of MN is one of several methods by which to quantify the genetic stability of mammalian cells. More recently, the in vitro micronucleus assay has been incorporated into the working guidelines from the International Conference on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use (S2R1 Step4, 09 Nov 2011). In addition, the in vitro MicroFlow (Litron Laboratories, Rochester, NY, USA) technique has been shown to be a practical and robust method for the evaluation of clastogenic effects and aneuploid inducers in human TK6 lymphoblastoid cells (11). Although multiple cell lines have been evaluated with this protocol, this is the first time to our knowledge that the frequency of MN in hMSCs has been assessed by flow cytometry using the in vitro MicroFlow Kit (MNvit; Litron Laboratories).
Based on transplantation studies by Rygaard and Povlsen (12,13), the athymic mouse model remains an essential method for the measurement of tumorigenesis emanating from cell implants in both biologic and drug development. With this classic model, the effects of malignant cell clones can be assessed directly by subcutaneous or ectopic implantation through the measurement of tumor volume while animal health is monitored. Xenograft studies in the athymic mouse provide a convenient proof-of-principle with respect to oncogenesis and neovascularization or the lack thereof.
Methods
Donor exclusion criterion
The genetic stability of hMSCs was characterized by assessing surface biomarker expression, chromosome stability, micronucleus formation and malignant cell transformation after the passage 2 expansion. If the harvested cell population is not characterized as hMSCs according to the ISCT standard (biomarker expression, differentiation, morphology), the donor was excluded from genetic stability characterization. Donor BM3130 was excluded from the study because of contamination of the osteogenic differentiation assay. Donors BM3204 and BM3241 were excluded from the study because of inadequate quantities of cells to perform any of the differentiation assays for the TCPS lineage.
Quantum System and flask BM-hMSC selection and expansion
Initially, six human bone marrow aspirates supplied by normal adult donors, who gave informed consent through a certified clinical collection facility (All-Cells, Alameda, CA, USA), were divided on arrival into Quantum and TCPS lineages for comparison (Figure 1). Fresh human bone marrow aspirates (hBM-APs) were unprocessed before cell culture in both parallel lineages. MSC biomarker screening by flow cytometry from the first passage indicated that hMSCs from three of the donors met the ISCT biomarker expression profile.
Figure 1.
Automated Quantum System and manual TCPS used for isolation and expansion of hMSCs.
HMSCs from these qualified donors (BM3164, male/38 years old; BM3346, female/21 years old; BM3437, male/32 years old) were re-suspended and expanded over 5 passages in alpha minimal essential medium (α-MEM 12–169F; Lonza, Allendale, NJ, USA) supplemented with 10% HyClone fetal bovine serum (FBS SH30070.03; Thermo Scientific HyClone, Logan, UT, USA), Gibco® GlutaMAX™ (35050; Life Technologies, Grand Island, NY, USA) dipeptide 2 mmol/L under mixed gas (5% CO2, 20% O2, balance N2) conditions at 37°C. The average cell seeding densities per cm2 for the TCPS hMSC lineages were 9.50 × 102 (passage 1), 9.53 × 102 (passage 2), 9.50 × 102 (passage 3) and 9.50 × 102 (passage 4). In comparison, the average cell seeding densities per cm2 for the Quantum hMSC lineages were 9.21 × 102 (passage 1), 9.52 × 102 (passage 2), 9.52 × 102 (passage 3) and 9.52 × 102 (passage 4). Statistical analysis of the hMSC seeding densities in passages 1–4 showed that there was no significant difference between the two lineages across all 4 passages (P = 0.492, 95% confidence interval [CI]). Cells were isolated based their ability to adhere to the TCPS surface or the fibronectin-coated hollow fiber membrane of the Quantum System. Non-adherent cells were removed by washing on day 1 after seeding without the use of density gradients. Glucose and lactate metabolism were monitored in the culture media of the Quantum by a RapidLab 860 Blood Gas Analyzer (Siemens Healthcare, Tarrytown, NY, USA); for the purpose of adjusting media, addition rates were doubled to maintain glucose ≥50 mg/dL. On harvest with Gibco® Trypsin-EDTA 0.25% (25200, Life Technologies, Grand Island, NY, USA), the cells were evaluated for concurrence with the ISCT criteria for hMSC phenotype and tri-lineage differentiation.
In the xenograft studies, human colorectal adenocarcinoma cells (HCT-15) from American Type Culture Collection (CCL-225; ATCC, Manassas, VA, USA) were expanded as a positive control in Falcon® T225 flasks (BD Biosciences, San Jose, CA, USA) using RPMI-1640 medium (ATCC, 30–2001) supplemented with 10% FBS, GlutaMAX™ 2 mmol/L under mixed gas (5% CO2, 20% O2, balance N2) conditions at 37°C and harvested with Trypsin-EDTA 0.25%. DNA and cytogenetic analyses of HCT-15 cells were confirmed by ATCC. Flask media was changed every 3–4 days.
hMSC differentiation
After hMSC harvests (passages 1–4), cells from both the Quantum System and flask lineages were seeded into six-well (triplicate) tissue culture plates (16,666/ well) with complete α-MEM supplemented with FBS (10%), 2 mmol/L GlutaMAX™, 1% antibiotics (Penicillin 17–602E/Streptomycin 17–836E; Lonza) and incubated at 37°C in 5% CO2 until confluent for adipogenesis and osteogenesis assays. At ≥90% confluency, un-induced cells were maintained in complete media, and the stimulated cells were incubated in respective differentiation media for 21 days. Adipogenesis induction media included α-MEM, 1% FBS, antibiotics (as listed previously), 1 µmol/L dexamethasone (D2915; Sigma-Aldrich, St Louis, MO, USA), 0.5 mmol/L isobutylmethylxanthine (I5879; Sigma-Aldrich), 100 µmol/L indomethacin (I7378; Sigma-Aldrich) and 100 µg/mL insulin (I9278; Sigma-Aldrich). At the conclusion of adipogenic induction, cells were washed with phosphate-buffered saline (PBS, 17–516; Lonza), fixed with 4% formalin (HT50-1–2; Sigma-Aldrich) for 30 min, washed with de-ionized water, permeabilized with 60% isopropanol for 2–5 min, stained with 0.3% oil red O (O0625; Sigma-Aldrich), washed three times with de-ionized water and counterstained with hematoxylin (GHS116; Sigma-Aldrich) to record the presence of lipid vacuoles associated with adipocytes by phase-contrast microscopy (Olympus CKX 41 with Q-Capture Pro 6.0 software; Olympus Corp, Tokyo, Japan). At the conclusion of osteogenic induction, cells were washed with PBS, fixed with 4% formalin (HT50-1–2; Sigma-Aldrich) for 10 min, washed with PBS, stained with 2% alizarin red S (A5533; Sigma-Aldrich), washed three times with PBS and counterstained with hematoxylin (GHS116; Sigma-Aldrich) to record the presence of calcium deposits associated with osteocytes by phase-contrast microscopy. Osteogenesis induction media included 0.1 µmol/L dexamethasone (D2915; Sigma-Aldrich), 0.05 mmol/L ascorbic acid (A8960; Sigma-Aldrich) and 10 mmol/L β-glycerophosphate (G9422; Sigma-Aldrich).
To evaluate chondrogenesis induction, 2.0 million cells were plated at high density in 100 mL per well (triplicate) from both the Quantum and the flask hMSC lineages in a six-well tissue culture plate with complete media and allowed to attach for 90 min at room temperature. After room temperature incubation, the complete media were replaced with chondrogenic base media (4 mL/well) and incubated at 37°C with 5% CO2. After 24 h, the base media were replaced in the stimulated wells with chondrogenic stimulation media. Chondrogenesis base media included α-MEM, 2 mmol/L GlutaMAX™, 1% antibiotics (as listed previously), 1% ITS+ (recombinant insulin, human transferrin, selenite; I-2771-5ML; Sigma-Aldrich), 13 µmol/L ascorbic acid (A8960; Sigma-Aldrich), 1 mmol/L sodium pyruvate (P5280; Sigma-Aldrich) and Pierce 0.0625% bovine serum albumin (23209; Thermo Scientific). Chondrogenesis induction media included chondrogenesis base media plus 0.1 µmol/L dexamethasone (D2915; Sigma-Aldrich) and 0.01 mg/mL transforming growth factor-β3 (100-36E; Pepro-Tech, Rocky Hill, NJ, USA). At the conclusion of induction, cells were fixed with 4% formalin for 1 h, washed with PBS, treated with 2% acetic acid (A8976; Sigma-Aldrich) and stained with 1% alcian blue (RT 26026-13; EMS, Hatfield, PA, USA) to record the matrix deposition of sulfated glycosaminoglycan associated with chondrocytes by phase-contrast microscopy.
hMSC immunophenotyping
The combination of positive (CD73+, CD90+, CD105+) and negative (CD14+, CD19+, CD34+, CD45+, HLA-DR+) cell surface antigen expression is currently used to identify multipotent MSCs, in part, by flow cytometry per ISCT guidelines. In addition, cell viability was confirmed by staining cells with 7-amino-actinomycin D (7-AAD). Post-harvest samples were prepared for multi-color immunofluorescent analysis by concentrating 1.1 million cells via centrifugation at 300g for 7 min and re-suspension in flow blocking solution (2% normal goat serum (100–109; Gemini Bio-Products, West Sacramento, CA, USA)/PBS) for 10 min at ambient temperature, centrifuged, decanted, re-suspended in 1.1 mL of flow wash-stain solution (2% FBS, 0.06% sodium azide (S8032; Sigma-Aldrich), PBS) and aliquoted. Non-specific isotype control stain panel included mIgG1-FITC (555909; BD Pharmingen, Grand Island, NY, USA), mIgG1-PE (555749; BD Pharmingen), mIgG1-APC (554681; BD Pharmingen) and mIgG2a–FITC (555573; BD Pharmingen). HMSC antigen detection and cell viability stain panel included anti-CD14-FITC (555397; BD Pharmingen), anti-CD19-APC (555415; BD Pharmingen), anti-CD34-APC (555824; BD Pharmingen), anti-CD45-FITC (555482; BD Pharmingen), anti-CD73-PE (550257; BD Pharmingen), anti-CD90-FITC (MCA90F; Serotec, Kidlington, UK), anti-CD105-APC (17–1057; eBioscience, San Diego, CA, USA) and 7-AAD (559925; BD Pharmingen). MSC biomarker data from passages 1–4 were acquired and analyzed with paired isotype controls on a dual-laser FACSCalibur (BD Biosciences, San Jose, CA, USA) equipped with FACSComp and Cell-Quest Pro 5.2 software.
SKY
Cells from parallel TCPS and Quantum expansions that exhibited ISCT morphology, surface biomarker expression and tri-lineage differentiation for hMSCs were subjected to SKY. The plastic-adherent hMSCs were seeded at the density of 1.5 × 106 to T75 flasks with α-MEM supplemented with 10% FBS and 2 mmol Gibco GlutaMAX™. After 24 h, colcemid (0.05 µg/mL) was used for metaphase arrest for 2 h, 30 min, following cell harvesting, hypotonization in 0.075M KCl for 25 min and fixation in 3:1 methanol:glacial acetic acid. CytoClear (5 µL/mL of fixative; ProCell, Genial Genetics, Chester, UK) was added to remove persistent cytoplasm. Cell suspensions were dropped onto slides and subjected to the protocol for SKY as described (7). The human SKY probe cocktail and reagents for hybridization and immunochemical detection were provided by Applied Spectral Imaging (ASI, Vista, CA, USA). Three fluorophores (fluorescein isothiocyanate [FITC], rhodamine and Texas red) and two haptens (biotin and digoxigenin) were used to generate the 24-chromosome painting probe. The probe was denatured for 10 min at 80°C and incubated at 37°C for 45 min to reduce non-specific background labeling. The specimens were incubated in 2× saline sodium citrate (SSC) at 37°C for 30 min and dehydrated in ethanol series, then chromosomal DNA was denatured in 70% formamide at 70°C for 1–2 min and dehydrated. The denatured probe was applied to the selected area, which was covered with a coverslip and sealed with rubber cement. Hybridization was allowed to occur for 48 h at 37°C in a dry chamber. To wash the unbound reagents, three consecutive 5-min incubations were performed in 50% formamide/2× SSC and 2× SSC at 45°C, followed by a 2-min wash in 4× SSC/0.1% Tween 20 at room temperature. Biotin was detected with avidin/Cy5, and digoxigenin was detected with mouse anti-digoxigenin antibody plus sheep anti-mouse/Cy5.5 (ASI reagents). Chromatin was counter-stained with 0.15 µg/mL 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI) in VECTA-SHIELD mounting medium (Vector Laboratories, Burlingame, CA, USA). At least 50 metaphases per specimen were imaged using the SD200 SpectraCube system with the SKY-1 optical filter (Chroma Technology, Brattleboro, VT, USA), mounted on an Olympus BX60 microscope (Olympus Corp, Tokyo, Japan). SKYView version 1.6.2 software (ASI) was used for complete analyses of approximately 40 metaphases per specimen. Analyses have used the spectral images representing the hybridization profiles and the classified image, which is a pseudo-colored image with distinct colors for easy interpretation of the spectral content for each chromosome that was generated by conversion of the emission spectra to the display colors through assignment of blue, green and red colors to specific sections of the emission spectrum. Electronically inverted DAPI images (band images) also were used to identify deletions and intra-chromosomal rearrangements and to assign the chromosome breakpoints. Karyotype results were reported following nomenclature guidelines (14).
In vitro micronucleus assay by flow cytometry
The Litron Laboratories in vitro MicroFlow Kit (MNvit) was used to prepare and stain hMSC samples of 5.0 × 105 each for the presence of MN using ethidium monoazide (EMA) and SYTOX Green (15–17), components of the MIcroFlow Kit. EMA crosses the compromised outer membrane of apoptotic and necrotic cells and covalently binds to DNA through photo-activation. Positive control hMSCs were incubated with mitomycin C (MMC) (C4287; Sigma-Aldrich) at 1.5 µmol/L (0.5 µg/mL) for 1.5–2.0 cell cycles to induce the formation of MN. MMC is a direct-acting alkylating agent used as MN induction agent (18). After treatment, cells were trypsinized, counted by Vi-CELL XR cell counter (Beckman Coulter, Brea, CA, USA), centrifuged (300g for 7 min) and washed, and the cytoplasmic membranes were digested with a non-ionic detergent to liberate nuclei and MN. The lysis solutions contained SYTOX Green (a pan nuclei acid dye) and RNase. Acquisition and analysis of the samples were accomplished using templates provided with the MicroFlow kits and a FACSCalibur flow cytometer equipped with CellQuest Pro 5.2 software.
Tumorigenesis in athymic (nu/nu) mouse model
HMSC xenograft studies were conducted in accordance with an approved protocol under Institutional Animal Care & Use Committee guidelines at the University of Colorado–Denver, Anschutz Medical Campus, Office of Laboratory Animal Resources, Center for Comparative Medicine. In each of the three 60-day experiments, 32 athymic female (nu/nu) mice from Charles River Laboratories (Wilmington, MA, USA) were quarantined for 1 → 2 weeks before entry into study and subsequently weighed, randomly assigned and sorted into four equal arms (media control, positive control American type culture collection HCT-15 tumor cells, flask hMSC lineage cells and Quantum hMSC lineage cells). Host animals were implanted with HCT-15 tumor cells (on the order of 107) 1 week before subcutaneous implantation of donor-derived passage 2 → 3 hMSCs (1–5 × 106) in 0.2 mL base media to minimize the possibility of cross-contamination and to verify host animal engraftment. The athymic (nu/nu) mouse model was chosen because it is known to be deficient in T cells but not deficient in B cells or natural killer (NK) cells to maintain a viable host animal at the University of Colorado–Denver, Anschutz Medical Campus, Office of Laboratory Animal Resources controlled barrier facility where certain endemic biologic agents are present. NK cells are not considered to be a factor in this study because they need to have prior recognition of specific tumor cell histocompatibility complex (class I) to exert their lytic function on transformed cells (19). Cancer cells also have been shown to express ligands that inhibit NKG2D receptors, which reduce the cytotoxic effect of NK cells (18). Mice were housed in autoclaved, individually ventilated cages on Aspen Chip bedding (Harlan Teklad, Indianapolis, IN, USA) with a nestlet, provided reverse osmosis hyperchlorinated water via an automatic watering system and provided an ad lib irradiated diet (2918; Harlan Laboratories, Indianapolis, IN, USA). Sentinel testing is performed quarterly by Charles River Laboratories. The mice used in this study did not present with any detectable microbial infection and were housed at 22.2 ± 1.1°C with a minimum of 10–15 air changes per hour and 30–70% humidity. Animal condition, weight and neoplasm volume (neoplasm volume = width2 × length/2) were recorded twice weekly using a calibrated Scout balance (Ohaus Corp, Parsippany, NJ, USA), ProMax calipers (F. V. Fowler, Newton, MA, USA) and LABCAT software (IPA, Lawrenceville, NJ, USA) (20,21). Animals were either removed from the study with respect to animal health or at the conclusion of the experiment per protocol.
Statistics
Statistical analysis was performed with the central value functions in MiniTab 16 statistical software (MiniTab, State College, PA, USA). Student t test calculations were used to quantify the P value probability of differences between Quantum and TCPS (flask) mean values at a CI of 95%.
Results
hMSC expansion and phenotype summary
In our study, the cells derived from three human bone marrow aspirates exhibited comparable morphology, attachment, tri-lineage differentiation and biomarker phenotype consistent with ISCT criteria for MSCs (5). At passage 2, the Quantum and flask lineages generated an average of 17.0 and 21.5 doublings, respectively, and at passage 4, the Quantum and flask lineages generated an average of 22.5 and 27.9 doublings, respectively, which may have been the result of bi-directional cell loading techniques or media addition protocols that are under development (Table I). Statistical analysis of these data from passage 0–4 suggests that there was no significant difference between the two stem cell population doublings over the course of the study even though there were differences in media volume/ cm2 (P = 0.055, 95% CI). Additionally, Rojewski et al. (22) previously demonstrated the expansion of hMSCs in a fibronectin-coated Quantum.
Table I.
Summary of mesenchymal stromal cell expansion and International Society of Cell Therapy characterization from human bone marrow aspirates at passage 0–4.
| Passage 0 | |||||||
| Cells loaded |
Cells harvested |
Number of doublings |
|||||
| Donor BM3164 | Quantum | 4.22E+04 | 1.24E+08 | 11.5 | |||
| TCPS | 5.08E+03 | 4.99E+07 | 13.3 | ||||
| Donor BM3346 | Quantum | 1.04E+04 | 6.86E+07 | 12.7 | |||
| TCPS | 1.46E+03 | 4.03E+07 | 14.7 | ||||
| Donor BM3437 | Quantum | 2.02E+04 | 5.41E+07 | 11.4 | |||
| TCPS | 2.43E+03 | 2.76E+07 | 13.5 | ||||
| Passage 1 | |||||||
| Cells loaded |
Cells harvested |
Cumulative number of doublings |
Morphology: day 1 attachment debris low spindle/ fibroblastic (yes/no) |
Differentiation: + chondrogenic, + osteogenic, + adipogenic, inconclusive i/ or n/a) (yes/no) |
Phenotype: ≤2% positive (CD14, CD19, CD34, CD45, HLA-DR), ≥95% positive (CD73, CD90, CD105) (yes/no) |
||
| Donor BM3164 | Quantum | 2.00E+07 | 9.43E+07 | 13.8 | Yes | No (+A, iO, +C) | Yes except |
| 2.00E+07 | 9.90E+07 | 13.8 | CD105 = 43.3% | ||||
| TCPS | 1.71E+06 | 4.72E+07 | 18.1 | Yes | No (+A, iO, iC) | Yes | |
| Donor BM3346 | Quantum | 2.00E+07 | 9.28E+07 | 14.9 | Yes | Yes (+A, +O, +C) | Yes except |
| 2.00E+07 | 9.08E+07 | 14.9 | CD14 = 2.6% | ||||
| TCPS | 1.71E+06 | 1.27E+07 | 17.7 | Yes | No (+A, +O, n/a C) | Yes except CD14 = 4.8% |
|
| Donor BM3437 | Quantum | 1.80E+07 | 1.17E+08 | 14.1 | Yes | Yes (+A, +O, +C) | Yes |
| 1.80E+07 | 8.67E+07 | 13.7 | |||||
| TCPS | 1.71E+06 | 4.46E+07 | 18.2 | Yes | Yes (+A, +O, +C) | Yes | |
| Passage 2 | |||||||
| Donor BM3164 | Quantum | 2.00E+07 | 1.04E+08 | 16.2 | Yes | Yes (+A, +O, +C) | Yes |
| 2.00E+07 | 8.06E+07 | 15.8 | |||||
| 2.00E+07 | 1.06E+08 | 16.2 | |||||
| TCPS | 3.43E+06 | 2.11E+07 | 20.7 | Yes | Yes (+A, +O, +C) | Yes | |
| Donor BM3346 | Quantum | 2.00E+07 | 1.59E+08 | 17.9 | Yes | Yes (+A, +O, +C) | Yes |
| 2.00E+07 | 1.40E+08 | 17.7 | |||||
| 2.00E+07 | 1.54E+08 | 17.8 | |||||
| TCPS | 6.43E+06 | 1.26E+08 | 22.0 | Yes | Yes (+A, +O, +C) | Yes | |
| Donor BM3437 | Quantum | 2.00E+07 | 1.38E+08 | 16.9 | Yes | Yes (+A, +O, +C) | Yes |
| 2.00E+07 | 1.78E+08 | 17.2 | |||||
| 2.00E+07 | 1.77E+08 | 17.2 | |||||
| TCPS | 6.43E+06 | 8.17E+07 | 21.8 | Yes | Yes (+A, +O, +C) | Yes | |
| Passage 3 | |||||||
| Donor BM3164 | Quantum | 2.00E+07 | 7.91E+07 | 18.2 | Yes | Yes (+A, +O, +C) | Yes |
| 2.00E+07 | 6.15E+07 | 17.8 | |||||
| 2.00E+07 | 7.18E+07 | 18.1 | |||||
| TCPS | 1.71E+06 | 1.16E+07 | 23.4 | Yes | Yes (+A, +O, +C) | Yes | |
| Donor BM3346 | Quantum | 2.00E+07 | 2.07E+08 | 21.3 | Yes | Yes (+A, +O, +C) | Yes |
| 2.00E+07 | 2.04E+08 | 21.2 | |||||
| 2.00E+07 | 2.04E+08 | 21.2 | |||||
| TCPS | 1.71E+06 | 3.28E+07 | 26.2 | Yes | Yes (+A, +O, +C) | Yes | |
| Donor BM3437 | Quantum | 2.00E+07 | 1.36E+08 | 20.0 | Yes | Yes (+A, +O, +C) | Yes |
| 2.00E+07 | 1.56E+08 | 20.2 | |||||
| 2.00E+07 | 1.42E+08 | 20.1 | |||||
| TCPS | 1.71E+06 | 2.62E+07 | 25.8 | Yes | Yes (+A, +O, +C) | Yes | |
| Passage 4 | |||||||
| Donor BM3164 | Quantum | 2.00E+07 | 6.20E+07 | 19.7 | Yes | Yes (+A, +O, +C) | Yes |
| 2.00E+07 | 5.88E+07 | 19.6 | |||||
| 2.00E+07 | 5.60E+07 | 19.5 | |||||
| TCPS | 1.71E+06 | 1.10E+07 | 26.1 | Yes | Yes (+A, +O, +C) | Yes | |
| Donor BM3346 | Quantum | 2.00E+07 | 1.97E+08 | 24.6 | Yes | Yes (+A, +O, +C) | Yes |
| 2.00E+07 | 1.91E+08 | 24.5 | |||||
| 2.00E+07 | 2.22E+08 | 24.7 | |||||
| TCPS | 1.71E+06 | 1.08E+07 | 28.9 | Yes | Yes (+A, +O, +C) | Yes | |
| Donor BM3437 | Quantum | 2.00E+07 | 1.81E+08 | 23.4 | Yes | Yes (+A, +O, +C) | Yes |
| 2.00E+07 | 2.00E+08 | 23.5 | |||||
| 2.00E+07 | 1.72E+08 | 23.3 | |||||
| TCPS | 1.71E+06 | 1.36E+07 | 28.8 | Yes | Yes (+A, +O, +C) | Yes | |
Cumulative doublings are summarized for Quantum and corresponding control TCPS. The “yes” or “no” designations indicate that the ISCT criteria for hMSC morphology, tri-lineage differentiation and cell surface biomarker expression were reached during specified passages. Detailed morphology, surface biomarker expression, and tri-lineage differentiation results are presented in Figures 2, 3 and 4.
Morphology
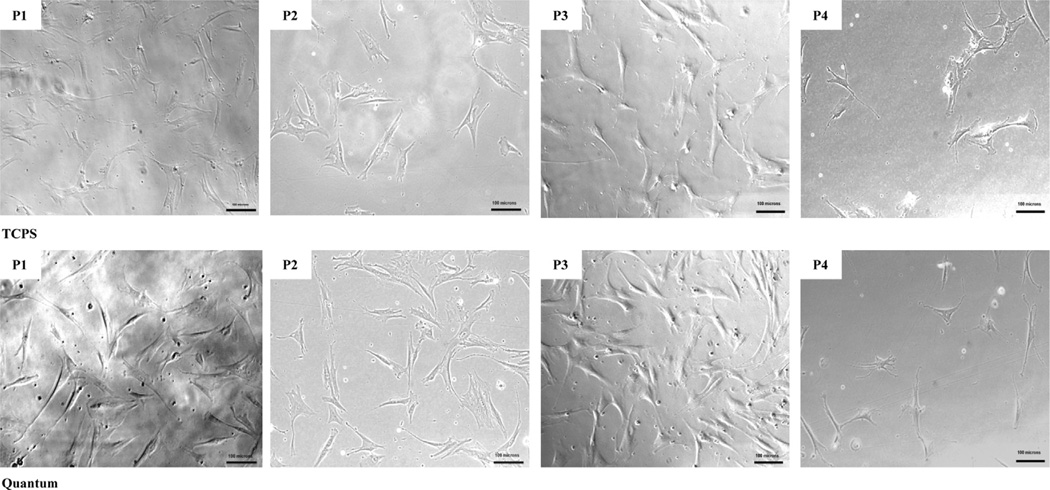
In Figure 2, phase-contrast images of hMSC lineages from the Quantum and flask (TCPS) were recorded 1 day after harvest to document cell attachment to a plastic surface and morphology. The results from all three qualified donors suggest that bone marrow-derived adherent cells expanded in the Quantum hollow fiber bioreactor exhibited a spindle-fibroblastic shape with minimal debris and performed in a similar manner to cells expanded in TCPS over passages 1–4, which is consistent with the ISCT qualifying criteria for MSCs. In our study, the morphology of the cells is in accordance with the type 1 stem cell fibroblast-like progenitor cells previously described by Mets et al. (23) and the fibroblastoid cell sub-classifications of BM-hMSCs previously outlined by Montesinos et al. (24) (i.e., lamellipodia-lamellipodia, lamellipodia-filopodia, filopodia-filopodia and triangular).
Figure 2.
Representative day 1 hMSC morphology images across 4 passages. (Magnification ×100, scale bars =100 µm.)
hMSC immunophenotyping
Specific membrane surface antigen expression MSCs derived from the three bone donor aspirates were expanded in the Quantum and tissue culture flasks from three donors and analyzed by immunophenotyping using flow cytometry stains and compared based on the ISCT criteria for defining multi-potent stromal cells through 4 passages as represented in Figure 3.
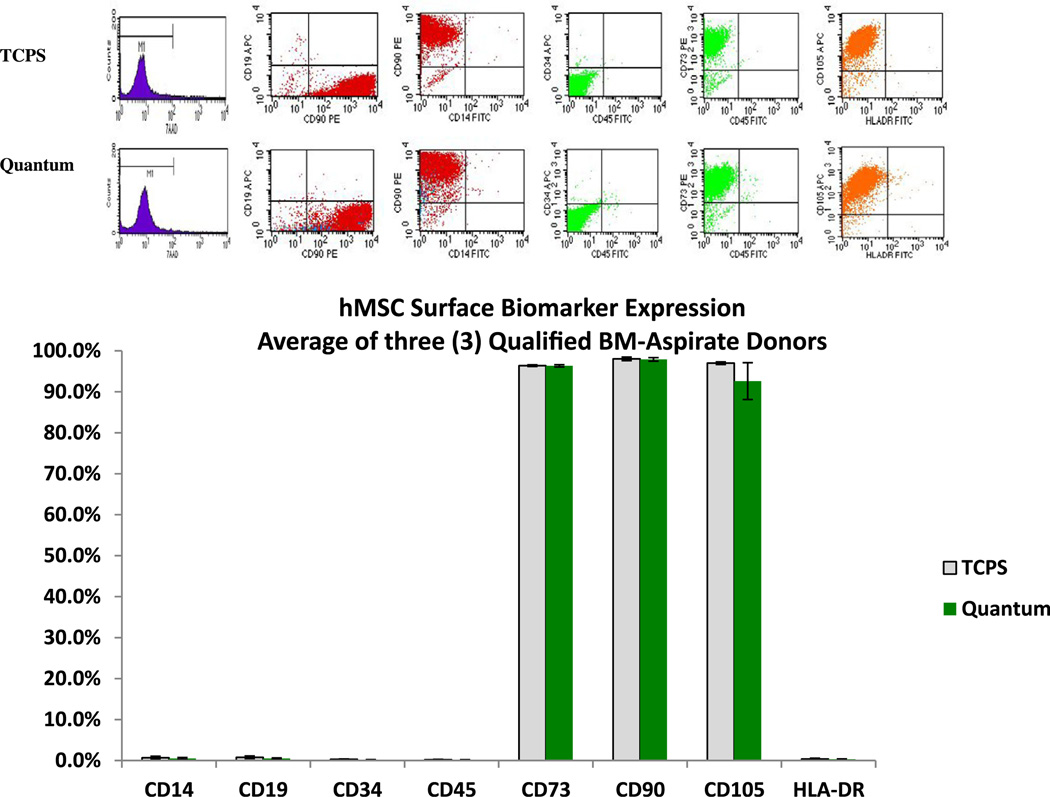
Figure 3.
Representative flow cytometry dot-plots and analysis comparing hMSC viability (7-AAD) and surface biomarker expression profiles of cells selected and expanded in Quantum and TCPS.
Surface biomarker expression of BM-hMSCs was profiled over 4 passages (n = 12). For hMSCs expanded in the Quantum, the negative panel values are CD14+ (0.5%; standard error ± 0.2%), CD19+ (0.5% ± 0.1%), CD34+ (0.2% ± 0.0%), CD45 (0.2% ± 0.0%) and HLA-DR (0.3% ± 0.1%), and the positive panel values are CD73+ (96.4% ± 0.3%), CD90+ (97.9% ± 0.4%) and CD105+ (92.6% ± 4.5%). For hMSCs expanded in TCPS, negative panel values are CD14+ (0.7% ± 0.4%), CD19+ (0.8% ± 0.3%), CD34+ (0.3% ± 0.1%), CD45 (0.2% ± 0.1%) and HLA-DR (0.4% ± 0.2%), and positive panel values are CD73+ (96.4% ± 0.2%), CD90+ (98.0% ± 0.4%) and CD105+ (97.0% ± 0.3%). In addition, cell viability was assessed by 7-AAD staining. All mean values had corresponding P values ≥0.193.
The results of the flow analysis demonstrate that there is no significant difference between the expression of hMSC negative markers (CD14+, CD19+, CD34+, CD45+, HLA-DR+) and hMSC positive markers (CD73+, CD90+, CD105+) with respect to growth conditions in either Quantum or TCPS. Also, there was no significant difference in cell viability between the two methods of expansion. The Quantum method generated an average hMSC viability of 97.6% (±0.3%), and the flask (TCPS) method generated an average hMSC viability of 97.5% (±0.4%) over passages 1–4 (P = 0.902).
hMSC multi-potency (tri-lineage differentiation)
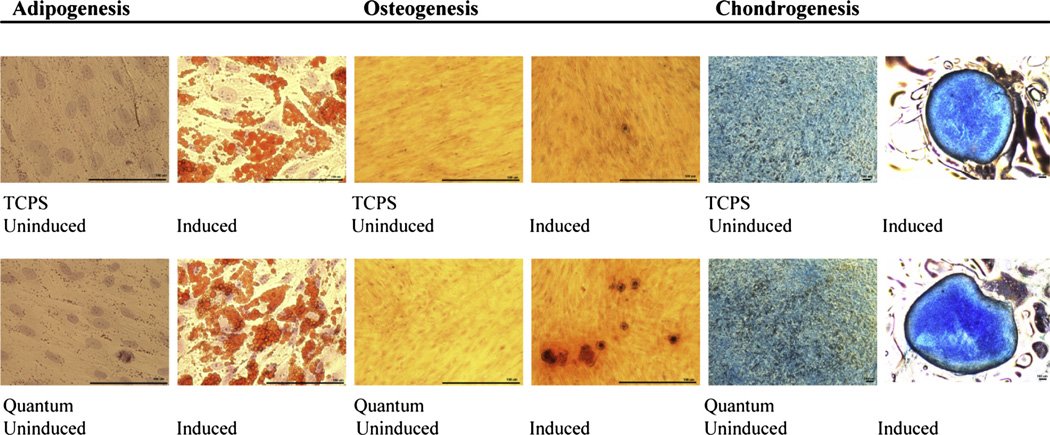
hMSCs derived from all three bone marrow donors and their corresponding Quantum and TCPS harvests were seeded into six-well plates and grown under un-induced and induced conditions to determine their response to multi-lineage differentiation. Images were captured using a phase-contrast microscope equipped with a charge-coupled device camera at 40×/400× magnification and demonstrated tri-lineage differentiation from hMSC lineages of all three donors (Figure 4).
Figure 4.
Representative images of hMSC tri-lineage differentiation from bone marrow-derived cells cultured in un-induced and induced ex vivo conditions. Fixed and stained adipocytes display oil red O-stained lipid-rich vesicles, osteocytes display alizarin red-stained calcium matrix formations and chondrocytes display alcian blue-stained glycosaminoglycan matrix. (Scale bars = 100 µm.)
In the chondrogenesis assay, un-induced cells did not maintain their micro-mass colonization, whereas the induced cells did maintain their micro-mass colony appearance. The lack of hMSC osteogenesis differentiation at passage 1 in cells from donor BM3164 may have been due to the use of FBS-supplemented media. In their review, Jung et al. (25) suggested that FBS-supplemented media or media without basic fibroblast growth factor can contribute to enlargement of hMSCs, which are considered more mature and may have a diminished potential for multipotency. The 1-day shorter culture time during passage 1 expansion also may have contributed to reduced differentiation.
SKY
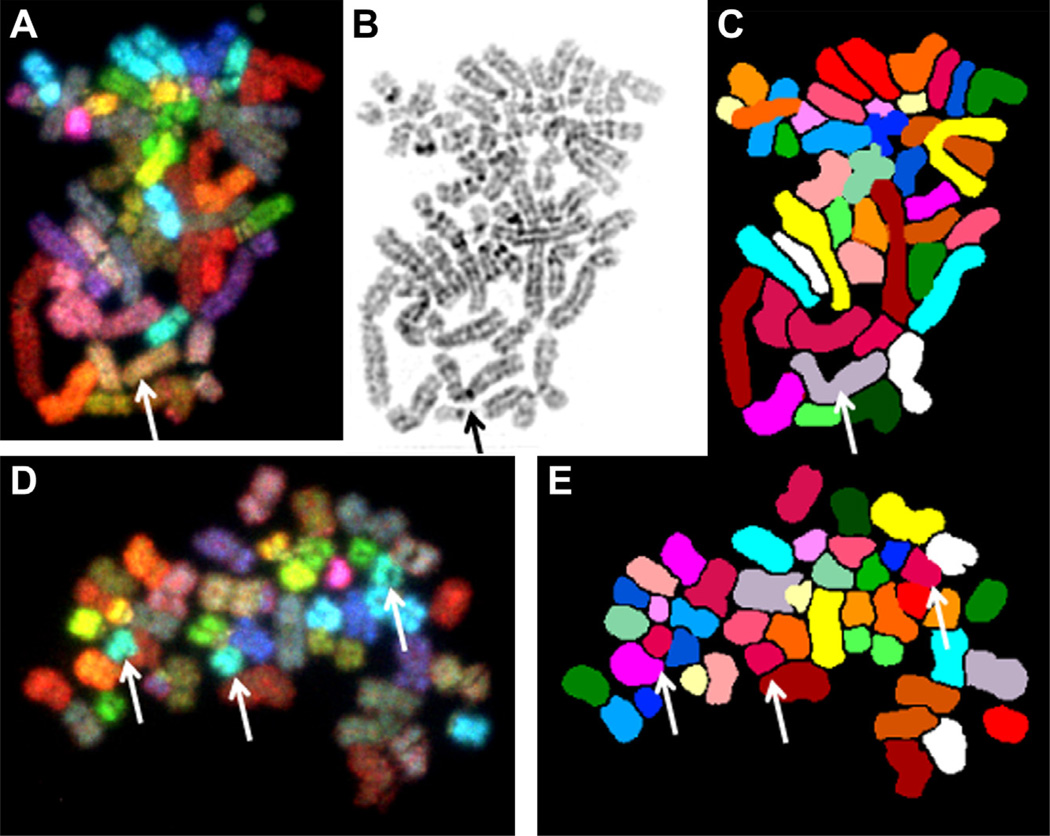
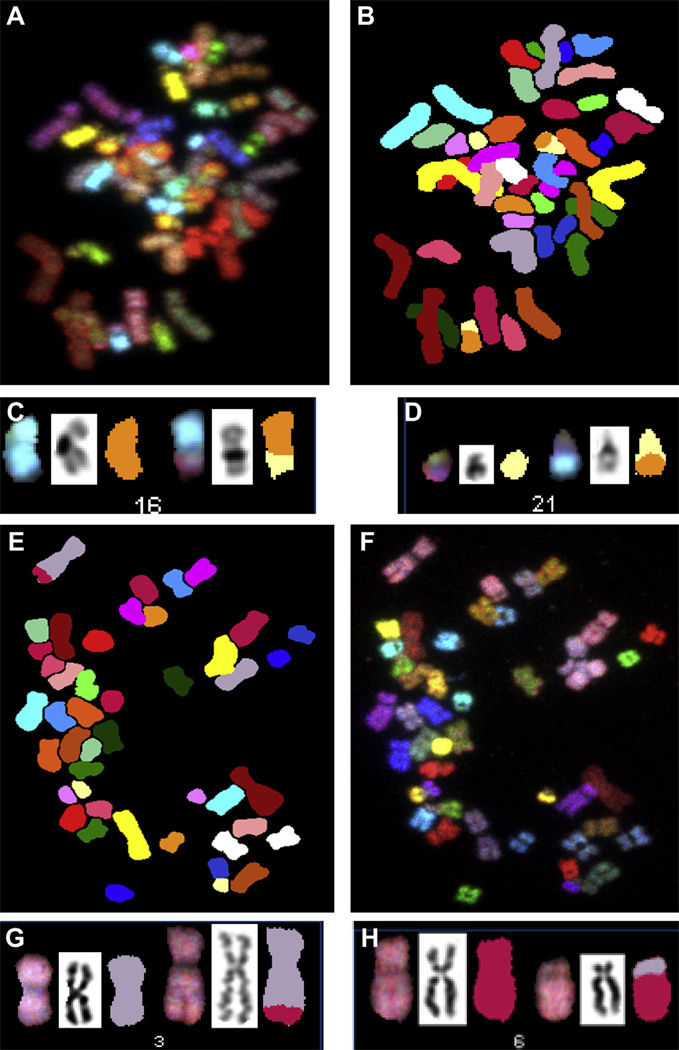
Donors BM3164 and BM3437 showed male karyotype 46,XY; donor 5 (BM3346) showed female karyotype 46,XX. Table II summarizes the results of complete spectral karyotyping analyses in the specimens cultured in the TCPS and Quantum Systems. Overall, 89% (range, 84–93%) and 92% (range, 86–97%) of cells had normal karyotype in the TCPS and Quantum specimens, respectively, and there was no significant difference between the two groups (Yates χ2 = 2.031; P = 0.362). A cell was considered carrying a normal karyotype when presenting 22 autosomes and two sex chromosomes (XX or XY) without any structural abnormality or achromatic lesion. The most common abnormalities were numerical changes, represented by gains or loss of a single chromosome. A metaphase spread from donor BM3164 Quantum with chromosomal loss (45,XY,–3) is illustrated in Figure 5A–C, and a metaphase cell from donor BM3164 TCPS with chromosomal gain (47,XY,+ 18) is shown in Figure 5D,E. Structural chromosomal abnormalities were rare (2% in TCPS and 3% in Quantum) and were represented by chromosomal break [46,XY,chrb(10)(p11.2) in Quantum] and reciprocal translocations [46,XY,t(3;6)(q29;p21.1) and 46,XY,t(1;13)(p32;q21) in TCPS, 46,XY,t(16,21) (q13;q21), 46,XY,t(1;15)(p33;q26) and 46,XY, t(3;13)(p15;q31),t(6;7)(p23;q32) in Quantum]. Reciprocal translocations are illustrated in Figure 6A–D for donor BM3164 Quantum and Figure 6E–H for donor 5 TCPS. None of these abnormalities were considered clonal because they all were seen in a single cell per specimen (Table III).
Table II.
Results of bone marrow-derived human mesenchymal stromal cell spectral karyotyping analysis from tissue culture polystyrene substrates and Quantum lineages.
| TCPS |
Quantum |
|||||||
|---|---|---|---|---|---|---|---|---|
| Specimen | Number of metaphases |
Normal karyotype |
Numerical abnormality |
Structural abnormality |
Number of metaphases |
Normal karyotype |
Numerical abnormality |
Structural abnormality |
| 2 | 40 | 93% | 8% | 0% | 37 | 86% | 8% | 5% |
| 5 | 44 | 84% | 14% | 2% | 38 | 97% | 3% | 0% |
| 6 | 42 | 90% | 7% | 2% | 43 | 93% | 2% | 5% |
| All | 126 | 89% | 10% | 2% | 118 | 92% | 4% | 3% |
Figure 5.
SKY images of specimen BM3164 Quantum showing a cell with hypodiploid male karyotype 45,XY,–3. Spectral imaging (A), inverted DAPI image (B) and classified image (C). A single copy of chromosome 3 is indicated by the arrow. A cell from donor 2 TPCS showing the hyperdiploid karyotype 47,XY,+ 18 in spectral imaging (D) and classified image (E). Arrows identify the three copies of chromosome 18.
Figure 6.
SKY images of specimen BM3164 Quantum showing a cell with karyotype 46,XY,t(16;21)(q13;q21). Spectral imaging (A), classified image (B) and spectral, inverted DAPI and classified images (C and D) for chromosomes 16 and 21. SKY images of specimen donor 5 TCPS showing a cell with karyotype 46,XY,t(3;6)(q29;p21.1). Spectral imaging (E), classified image (F) and spectral, inverted DAPI and classified images (G and H) for chromosomes 3 and 6.
Table III.
Abnormalities of bone marrow-derived human mesenchymal stromal cell spectral karyotyping analysis from tissue culture polystyrene substrates and Quantum lineages.
| Abnormalities in TCPS |
Abnormalities in Quantum |
|||
|---|---|---|---|---|
| Donor | Numerical | Structural | Numerical | Structural |
| 2 | 45,XY,−21 | 45,XY,−14 | 46,XY,t(16;21) | |
| 47,XY,+Y | 45,XY,−3 | 46,XY,chrb(10)(p11.2) | ||
| 47,XY,+18 | 47,XY,+3 | |||
| 5 | 45,XX,−2 | 46,XX,t(3;6)(q29;p21.1) | 92,XXXX | |
| 45,XX,−12 | ||||
| 45,XX,−21 | ||||
| 47,XX,+2 | ||||
| 47,XX,+7 | ||||
| 93,XXXX,+16 | ||||
| 6 | 46, XY,−2,+15 | 45,XY,t(1;13)(p32;q21) | 47,XY,+10 | 46,XY,t(1;15)(p33;q26) |
| 45, XY,–21 | 46,XY,t(3;13(p15;q31),t(6;7)(p23;q32) | |||
| 45, XY,–12 | ||||
All chromosome numbers deviating from euploidy were scored as aneuploid. The aneuploidy rate was defined as the number of aneuploid cells divided by the total cells analyzed. As a result, the aneuploidy rates for the TCPS hMSC lineage were 0.08, 0.11 and 0.07 and the aneuploidy rates for the Quantum hMSC lineage were 0.08, 0.00 and 0.02 for cells derived from donors BM3164(2), BM3346(5) and BM3437(6) at passage 2 (Table IV).
Table IV.
Aneuploidy rate of bone marrow-derived human mesenchymal stromal cell spectral karyotyping analysis from tissue culture polystyrene substrates and Quantum lineages.
| Donor | Treatment group |
Total cells analyzed |
Aneuploid cells |
Aneuploidy rate |
|---|---|---|---|---|
| 2 | TCPS | 37 | 3 | 0.08 |
| 2 | Quantum | 40 | 3 | 0.08 |
| 5 | TCPS | 44 | 5 | 0.11 |
| 5 | Quantum | 38 | 0 | 0 |
| 6 | TCPS | 42 | 3 | 0.07 |
| 6 | Quantum | 43 | 1 | 0.02 |
Micronucleus formation
Formation of MN is the response of cells during mitosis to both structural and numerical chromosome alterations associated with clastogenic or aneugenic activity. The measurement of MN by flow cytometry offers the benefit of higher throughput than manual methods. Differentially staining cells in the in vitro micronucleus (MNvit) assay overcomes issues associated with separating MN from cellular debris.
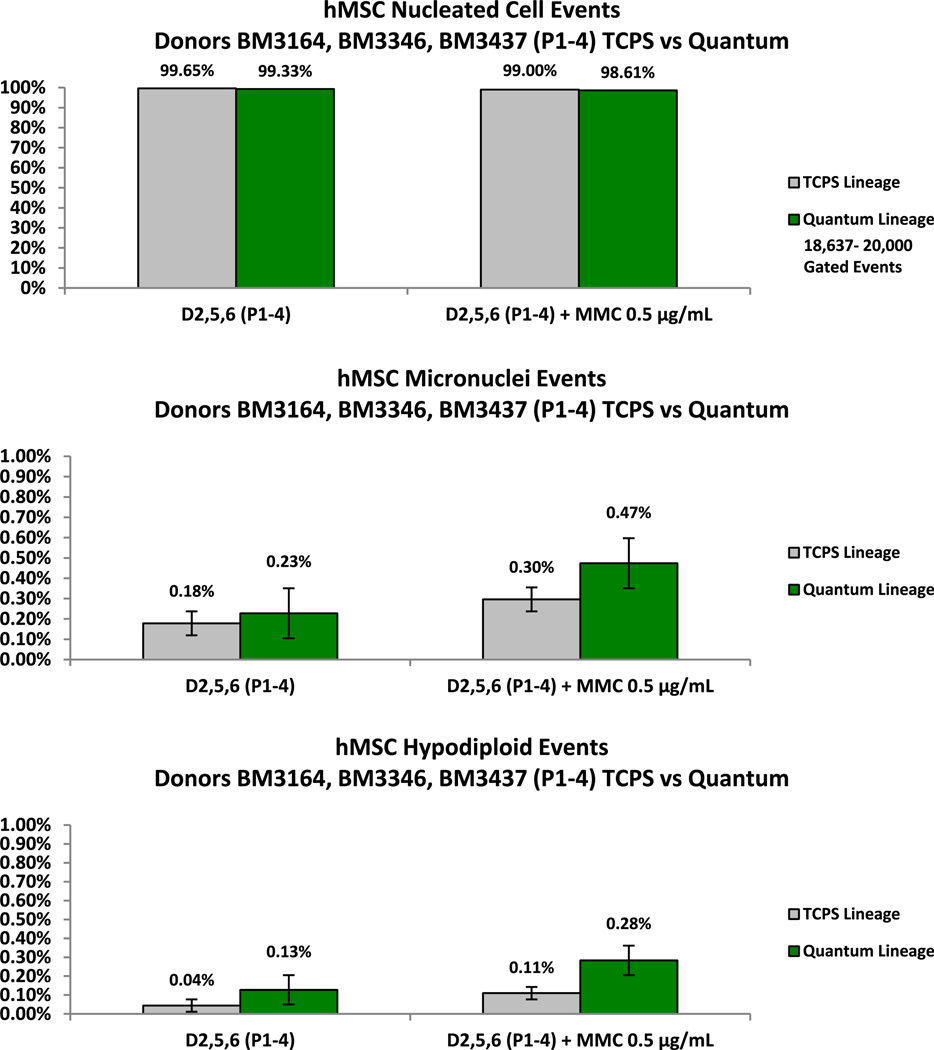
Flow cytometry software was set to collect 20,000 gated events in each sample of 5 × 105 cells. Nucleated, MN and hypodiploid events were acquired simultaneously via the multi-gating strategy in the assay template. In Figure 7, the frequency of nucleated cells, MN events and hypodiploid events was 99.65%, 0.18% and 0.04% for hMSCs expanded in the TCPS (n = 12). The frequency of nucleated cells, MN events and hypodiploid events was 99.33%, 0.23%, and 0.13% for hMSCs expanded in the Quantum (n = 12). In this study of three human bone marrow aspirate lineages, there were no significant differences in the percentage of nucleated, MN or hypodiploid cells between the BM-hMSC flask or Quantum lineages over 5 passages (15.8–28.9 cell doublings) as measured by the MNvit assay. As expected, both hMSC lineages responded to MN induction by treatment with the DNA alkylating agent MMC after exposure over 1.5–2.0 doublings.
Figure 7.
Comparison of average nucleated events from TCPS-derived BM-hMSCs and Quantum-derived BM-hMSCs over 5 passages (n = 12; P = 0.273 with 95% CI). Comparison of average MN events from un-induced TCPS-derived BM-hMSCs and Quantum-derived BM-hMSCs over 5 passages (n = 12; P = 0.546 with 95% CI). The response of both BM-hMSC lineages to MN induction by MMC is also shown. Comparison of average hypodiploid events from un-induced TCPS-derived BM-hMSCs and Quantum-derived BM-hMSCs over 5 passages (n = 12; P = 0.295 with 95% CI).
hMSC xenograft studies in athymic mouse model
Previous xenograft studies by Ra et al. (26) with non-transformed hMSCs in severe combined immunodeficiency (SCID) mice suggest a lack of spontaneous tumor formation 90 days after subcutaneous injection. Wang et al. (27) isolated a putative endothelial CD133+ sub-population (CD73−, CD90−, CD105−; non-hMSC phenotype) in ex vivo culture from bone marrow. This bone marrow-derived CD133+ sub-population displayed elevated telomerase activity, aneuploidy and chromosome translocations, which formed malignant sarcomas in sub-lethally irradiated non-obese diabetic/SCID mice that were implanted either intraperitoneally or intravenously. In other tumorigenicity studies by Li et al. (28), hMSCs immortalized with H-RAS or hTERT gene constructs gave rise to osteosarcomas or teratomas in SCID mice after long-term culture and orthotopic injection. Although the most common site of osteosarcoma is the metaphysis or growth zone of bone, osteosarcomas can occur in soft tissue (28). Rygaard et al. (13) conducted immune surveillance studies of athymic mice and observed that spontaneous lymphomas can occur at day 156, although the average is typically closer to 1 year.
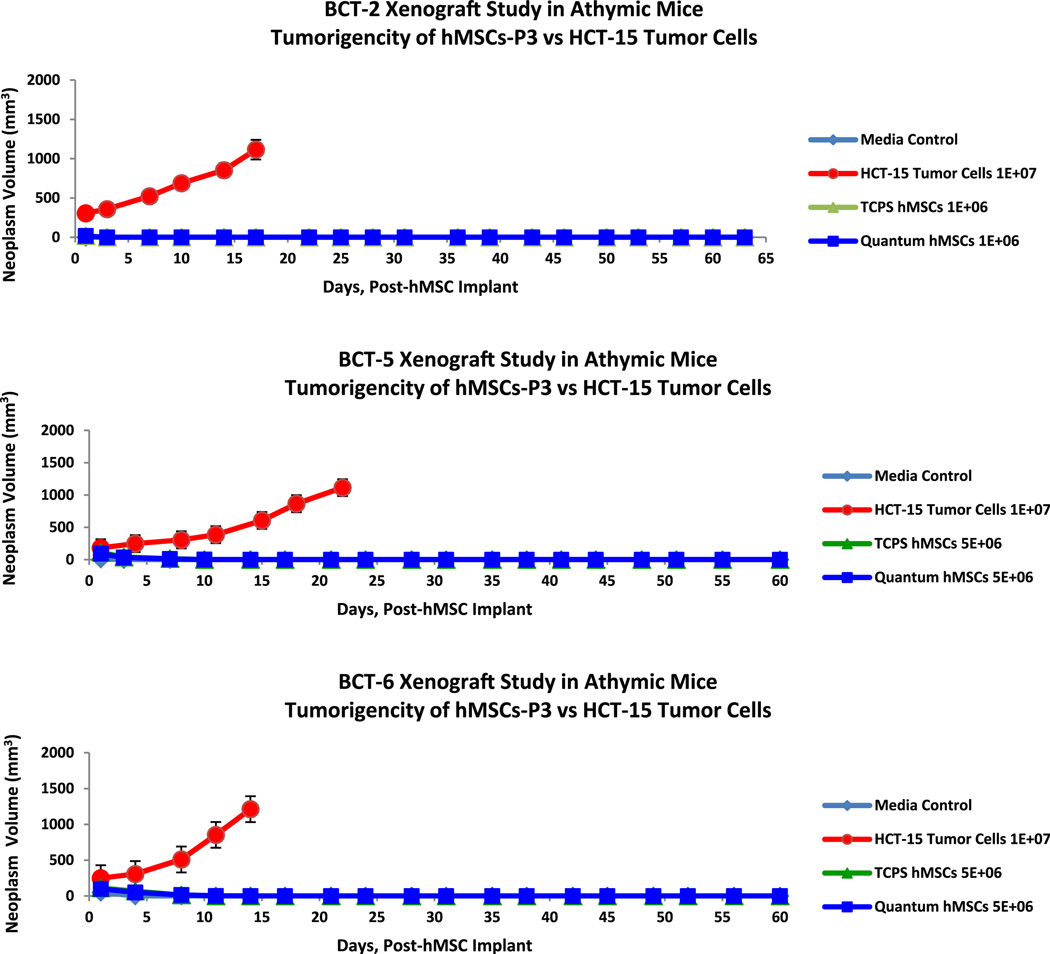
To reduce the background incidence of spontaneous lymphoma in the host animal, the xenograft experiments in this study were conducted with athymic (nu/nu) mice that were subcutaneously implanted with BM-hMSCs (passage 2, 15.8–22.0 doublings) at 1–5 × 106 and monitored for tumorigenesis for 60 days post-injection (120 days of age). As represented in Figure 8, all three experiments (BCT-2, BCT-5 and BCT-6) showed no detectable tumor volume associated with malignant neoplasm after the initial cell implantation of 0.2 mL was assimilated by the host animal in the media control, hMSC flask and hMSC Quantum groups. Only the mice in the positive control arm (group B) of each study that were implanted with HCT-15 human colorectal adenocarcinoma cells developed malignant neoplasm (i.e., tumor volume ≥ 150–200 mm3).
Figure 8.
Xenograft experiments BCT-2, BCT-5 and BCT-6. Lack of malignant neoplasm development (volume) was shown in both the TCPS BM-hMSC and the Quantum BM-hMSC lineages versus HCT-15 colorectal adenoma cells in the athymic (nu/nu) mouse model (n = 8/arm).
Tumor-bearing and non-tumor-bearing host animals were removed from the study when animal health care endpoints were met per Institutional Animal Care & Use Committee protocol. At the conclusion of the xenograft experiments, tumor or dermal tissue samples from the area of cell implantation were snap-frozen in liquid nitrogen, prepared for hematoxylin and eosin staining and histologically examined by the Department of Pathology, University of Colorado Hospital.
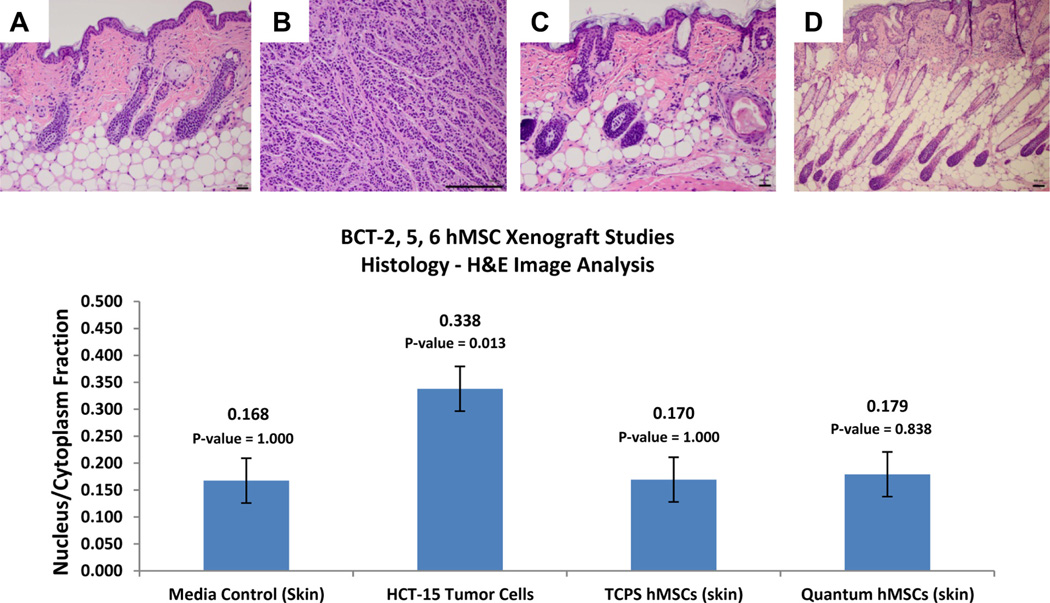
Under the ex vivo expansion conditions, no measurable malignant neoplasm was detected in the cell culture base media, TCPS hMSC or Quantum hMSC groups in the three xenograft studies (Figure 5). Only the experimental group implanted with HCT-15 tumor cells showed a measurable malignant neoplasm volume (BCT-2 = 305–1115 mm3; BCT-5 = 183–1114 mm3; BCT-6 = 249–1213 mm3) over the course of the study (Figure 8). In addition, all tissue samples from the media control groups as well as the TCPS hMSC and Quantum hMSC groups of the xenograft studies were histologically confirmed negative for malignancy, and only the experimental groups engrafted with HCT-15 tumor cells were confirmed positive for malignancy in the athymic mouse model as presented in Figure 9.
Figure 9.
Xenograft hematoxylin and eosin histology. (A–D) Representative sections of host-animal tumor and injection site dermal tissue for BCT-2, BCT-5 and BCT-6 experiments. Groups A (negative control; α-MEM-based cell culture media), C (TCPS-derived BM-hMSCs) and D (Quantum-derived BM-hMSCs) are negative for malignant neoplasm in the three experiments. Only group B (containing HCT-15 tumor cells) arms were positive for malignant neoplasm. Image analysis of hematoxylin and eosin histology sections from all three xenograft studies is also shown. Nucleus/cytoplasm ratios were computed from LABCAT digital processing software. These data indicate that there is no significant difference between the nucleus/cytoplasm ratios of host animal tissue implanted with BM-hMSCs expanded in either TCPS or the Quantum. Student t test P values for HCT-15 tumor cells, TCPS BM-hMSCs and Quantum BM-hMSCs all were comparisons with the media control group results. (Scale bars = 100 µm.)
Discussion
Quantum and TCPS BM-hMSC selection and expansion
The selection and expansion of hMSCs in both the Quantum and the flask lineages generated cells with normal spindle-shaped and fibroblast morphology, while displaying adherence to plastic (TCPS) surfaces. Additionally, the cells expressed a surface biomarker profile (CD73+, CD90+, CD105+ >95% and CD14+, CD19+, CD34+, CD45+, HLA-DR ≤2%) and were induced to tri-lineage differentiation during passages 0–5 (11.4–28.9 cell doublings), which is consistent with ISCT criteria for MSCs (5).
SKY Analysis
SKY is a genome scanning technique with advantages and limitations. The technique is most useful in identifying chromosomal material lacking banding information (rings, markers, double minutes), defining subtle rearrangements and identifying translocations unresolved by G-banding. Limitations of the technique are defined by the length of the hybridized metaphase chromosomes, the dye combinations involved in the abnormality and the quality of the hybridization.
The SKY analysis was done using the SKYView application (ASI), and the results are presented with three image types: the spectral image, the inverted DAPI image and the classified image (Figure 3). On starting analysis, the spectral image and the inverted DAPI counterpart are displayed. After contours have been defined for each chromosome using the spectral and DAPI images, the SKY karyotype can be established, rendering the classified image. The classified image is a pseudo-colored image with distinct colors for easy interpretation of the spectral content for each chromosome. The conversion of the emission spectra to the display colors was achieved by assigning blue, green and red colors to specific sections of the emission spectrum. Each of the images offers a piece of information, which altogether reveals the true content of the genomic material. For accurate interpretation of the content of each chromosome, close evaluation of all three images (spectral, inverted DAPI and classified) was performed.
The evaluation of hMSCs for biopharmaceutical development is strengthened by the use of SKY to characterize the genetic stability for tissue engraftment and regeneration. Genomic stability appears to depend on the duration of cell culture or specific mechanisms of genetic transformation (28–30). For example, neoplastic transformation has been reported in long-term hMSC cultures or in H-Ras or hTERT transfected MSCs (27,28,30).
Wang et al. (27) reported aneuploidy, translocations by gray banding, and high levels of telomerase in a CD133+ bone marrow-derived cell clone that was initially morphologically distinct at passage 3 compared with normal hMSCs. In other studies by Li et al. (28), hMSCs transfected with Tag and Ras genes appeared normal, but became more complex after transformation and similar to the karyotype of osteosarcoma cells. Ra et al. (26) showed that adipose-derived MSCs at passages 4, 7, 10 and 12 retained their normal karyotype during repeated ex vivo expansion. Previous studies by Miura et al. (31) with murine MSCs cultured continuously in vitro for >1 year (>400 cell doublings) reported fibrosarcomas when transplanted into immunocompromised mice. These murine MSCs exhibited clonality for chromosome 2 copy gain at P27, deleted, fragmented chromosome 14 at P55 and increased c-myc/telomerase activity.
In our experiments, the quality of the hybridization was excellent, and some limitations were minimized. However, owing to chromatin condensation, abnormalities affecting <3 Mb nucleotide pairs would likely be undetectable (on average, 1 pixel = 3 Mb pairs), and deletions and paracentric inversions would be undetectable. The SKY analyses from all three donor specimens indicate that there is no clonality of chromosomal aberrations from either flask-derived or Quantum-derived hMSCs and that most cells in both lineages display normal metaphase chromosome structure and distributions based on International System for Human Cytogenetic Nomenclature (ISCN) guidelines (14).
Automated micronucleus analysis
Previous multi-laboratory studies of TK6 lymphoblasts and lymphocytes with the in vitro Micro Flow assay (MNvit) suggest that this flow cytometry is a more robust method by which to enumerate MN induction (15). The advantages of this differential staining assay include rapid event acquisition, while separating MN from apoptotic and necrotic cells.
In these experiments, we report that the MNvit flow cytometry assay, along with other assays, has been used to evaluate hMSCs for aneugenic and clastogenic events. Evaluations by Gisselsson et al. (32) showed a correlation between the increased frequency of MN formation and unstable chromosomes from soft tissue tumors. Previously, Ebert et al. (33) showed, by manual counting, that the MN frequency in primary bone marrow stromal cells ranges from 1.7–8.0% when cultivated on glass slides. The results of our experiments show that the average frequency of MN formation in hMSCs is not significantly different for cells expanded on TCPS (0.18%) or in the Quantum (0.23%) across 4 serial passages (P = 0.546, 95% CI).
hMSC xenograft studies
The lack of tumorigenesis from BM-hMSCs from three donors over a dose range of 4 × 107 to 2 × 108/ kg body weight in athymic mice is consistent with stem cell genetic stability and the results of earlier xenograft studies with adipose-derived hMSCs at a dose range of 2 × 106 to 2 × 108 adipose-derived hMSCs/kg body weight (26). From an interspecies perspective, hMSC dose ranges for human studies have been reported to be 5 × 105 to 9 × 106/kg body weight (34,35). Our xenograft studies in immunocompromised mice evaluated the effects of hMSC engraftment at doses 22-fold to 80-fold greater than somatic stem cell dosages designed for human clinical studies.
In conclusion, based on the results of our experimental findings, these studies demonstrate that the Quantum supports the genetic stability of human BM-hMSC grown with ex vivo selection and expansion protocols. Under these conditions, BM-hMSCs have been shown to:
Meet the ISCT criteria for MSC phenotype and tri-lineage differentiation
Exhibit normal metaphase karyotype by SKY
Display normal nuclei formation by automated MN analysis
Lack the ability to transform into malignant neoplasm in the athymic mouse model
In addition, these data suggest a linkage between MN analysis by flow cytometry and the assessment of neoplasm in immunocompromised mouse models. The results indicate that the examination of MN formation can be used as a proxy for xenograft studies to affirm hMSC genetic stability of hMSCs.
Acknowledgments
The authors thank Michelle Wallace and the vivarium staff in the Office of Laboratory Animal Resources at the University of Colorado Denver-AMC for their support during the xenograft studies and to David Davis, BS, HT (ASCP), QIHC, Donna Tucker, HT (ASCP), QIHC, in the Histology Laboratory at the University of Colorado Hospital, School of Medicine for the tissue specimen preparation, and Kim Nguyen and Robert Schuyler at Terumo BCT for their support.
Footnotes
Disclosure of interest: Mark Jones, Brian Nan-kervis, Rebecca Peters, Boah Vang, Michelle Brecheisen, Thomas Startz and Nathan Frank are associates of Terumo BCT, Inc, Lakewood, CO, USA. Dr. Marileila Varella-Garcia and Margaret Skokan are on staff in the Division of Medical Oncology, School of Medicine at the University of Colorado–Denver, Anschutz Medical Campus, Aurora, CO, USA. Dr. Jeffrey Schowinsky is on staff in the Department of Pathology at the University of Colorado Hospital, Anschutz Medical Campus, Aurora, CO, USA. The studies at the University of Colorado–Denver AMC and Hospital were funded under contract by Terumo BCT, Inc. Steven Bryce is an associate of Litron Laboratories, Inc, Rochester, NY, USA.
References
- 1.Friedenstein AJ, Piatetzky-Shaprio II, Petrakova KV. Osteogenesis in transplants of bone marrow cells. J Embryol Exp Morph. 1966;16:381–390. [PubMed] [Google Scholar]
- 2.Friedenstein AJ, Petrakova KV, Kurolesova KI, Frolova GP. Heterotopic transplants of bone marrow. Transplantation. 1968;6:230–247. [PubMed] [Google Scholar]
- 3.Latsinik NV, Luria EA, Friedenstein AJ, Samoylina NL, Chertkov JL. Colony-forming cells in organ cultures of embryonal liver. J Cell Physiol. 1970;75:163–166. doi: 10.1002/jcp.1040750205. [DOI] [PubMed] [Google Scholar]
- 4.Pittenger MF, Martin BJ. Mesenchymal stem cells and their potential as cardiac therapeutics. Circ Res. 2004;95:9–20. doi: 10.1161/01.RES.0000135902.99383.6f. [DOI] [PubMed] [Google Scholar]
- 5.Dominici M, Le Blanc K, Slaper-Cortenbach I, Marini FC, Krause DS, Deans RJ, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–331. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Huso DL, Harrington J, Kellner J, Jeong DK, Turney J, McNiece IK. Outgrowth of a transformed cell population derived from normal human BM mesenchymal stem cell culture. Cytotherapy. 2005;7:509–519. doi: 10.1080/14653240500363216. [DOI] [PubMed] [Google Scholar]
- 7.Varella-Garcia M, Chen L, Powell RL, Hirsch FR, Kennedy TC, Keith R, et al. Spectral karyotyping detects chromosome damage in bronchial cells of smokers and patients with cancer. Am J Respir Crit Care Med. 2007;176:505–512. doi: 10.1164/rccm.200609-1329OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mateuca R, Lombaert N, Aka PV, Decordier I, Kirsch-Volders M. Chromosomal changes: induction, detection methods and applicability in human biomonitoring. Biochimie. 2006;88:1515–1531. doi: 10.1016/j.biochi.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Fenech M, Kirsch-Volders M, Natarajan AT, Surrallas J, Crott JW, Parry J, et al. Molecular mechanisms of micro-nucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011;26:125–132. doi: 10.1093/mutage/geq052. [DOI] [PubMed] [Google Scholar]
- 10.Savage JRK. [Accessed online June 8, 2012];Micronuclei: pitfalls and problems. Atlas of Genetics and Cytogenetics in Oncology and Haematology. 2000 < http://AtlasGeneticsOncology.org/Deep/MicronucleiID20016.html>;
- 11.Bryce S, Bemis JC, Avlasevich SL, Dertinger SD. In vitro micronucleus assay scored by flow cytometry provides a comprehensive evaluation of cytogenetic damage and cytotoxicity. Mutat Res. 2007;630:78–91. doi: 10.1016/j.mrgentox.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rygaard J, Povlsen CO. Heterotransplantation of a human malignant tumour to “nude” mice. Acta Pathol Microbiol Scand. 1969;77:758–760. doi: 10.1111/j.1699-0463.1969.tb04520.x. [DOI] [PubMed] [Google Scholar]
- 13.Rygaard J, Povlsen CO. The nude mouse vs. the hypothesis of immunological surveillance. Transplant Rev. 1976;28:43–61. doi: 10.1111/j.1600-065x.1976.tb00192.x. [DOI] [PubMed] [Google Scholar]
- 14.Shaffer LG, Slovak ML, Campbell LJ. International Standing Committee on Human Cytogenetic Nomenclature; An International System for Human Cytogenetic Nomenclature. ISCN; Basel. 2009. [Google Scholar]
- 15.Bryce SM, Avlasevich SL, Bemis JC, Lukamowicz M, Elhajouji A, Van Goethem F, et al. Interlaboratory evaluation of a flow cytometric, high content in vitro micronucleus assay. Mutat Res. 2008;650:181–195. doi: 10.1016/j.mrgentox.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicolette J, Diehl M, Sohnders P, Bryce S, Blomme E. In vitro micronucleus screening of pharmaceutical candidates by flow cytometry in Chinese hamster V79 cells. Environ Mol Mutagen. 2011;52:355–362. doi: 10.1002/em.20631. [DOI] [PubMed] [Google Scholar]
- 17.Avlasevich S, Bryce S, De Boeck M, Elhajouji A, Van Goethem F, Lynch A, et al. Flow cytometric analysis of micronuclei in mammalian cell cultures: past, present, and future. Mutagenesis. 2011;26:147–152. doi: 10.1093/mutage/geq058. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto Y, Shudo K. Chemical modification of DNA with MutaCarcinogens. III. Reductive alkylation of DNA with mitomycin C. Environ Health Perspect. 1985;62:219–222. doi: 10.1289/ehp.8562219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Euhus DM, Hudd C, LaRegina M, Johnson F. Tumor measurement in the nude mouse. J Surg Oncol. 1986;21:229–234. doi: 10.1002/jso.2930310402. [DOI] [PubMed] [Google Scholar]
- 21.Demma M, Maxwell E, Ramos R, Liang L, Li C, Hesk D, et al. SCH529074, a small molecule activator of mutant p53, which binds p53 DNA binding domain (DBD), restores growth-suppressive function to mutant p53 and interrupts HDM2-mediated ubiquitination of wild type p53. J Biol Chem. 2010;285:10198–10212. doi: 10.1074/jbc.M109.083469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojewski M, Fekete N, Baila S, Nguyen K, Fürst D, Antwiler D, et al. GMP-compliant isolation and expansion of bone marrow-derived MSCs in the closed, automated device Quantum Cell Expansion System [published online ahead of print October 25, 2012] [Accessed on March 6, 2013];Cell Transplant. 2012 doi: 10.3727/096368912X657990. http://dx.doi.org/10.3727/096368912X657990. [DOI] [PubMed] [Google Scholar]
- 23.Mets T, Verdonk G. Variation in the stromal cell population of human bone marrow during aging. Mech Ageing Dev. 1981;15:41–49. doi: 10.1016/0047-6374(81)90006-3. [DOI] [PubMed] [Google Scholar]
- 24.Montesinos JJ, Flores-Figueroa E, Castillo-Medina S, Flores-Guzmán P, Hernández-Estevez E, Fajardo-Orduna G, et al. Human mesenchymal stromal cells from adult and neonatal sources: comparative analysis of their morphology, immunophenotype, differentiation patterns and neural protein expression. Cytotherapy. 2009;11:163–176. doi: 10.1080/14653240802582075. [DOI] [PubMed] [Google Scholar]
- 25.Jung S, Panchalingam K, Rosenberg L, Behie L. Ex vivo expansion of human mesenchymal stem cells in defined serum-free media. [Accessed online December 11, 2012];Stem Cells Int. 2012 2012:1–21. doi: 10.1155/2012/123030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ra JC, Shin IS, Kim SH, Kang SK, Kang BC, Lee HY, et al. Safety of intravenous infusion of human adipose tissuederived mesenchymal stem cells in animals and humans. Stem Cells Dev. 2011;20:1297–1308. doi: 10.1089/scd.2010.0466. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Huso DL, Harrington J, Kellner J, Jeong DK, Turney J, et al. Outgrowth of a transformed cell population derived from normal human BM mesenchymal stem cell culture. Cytotherapy. 2005;7:509–519. doi: 10.1080/14653240500363216. [DOI] [PubMed] [Google Scholar]
- 28.Li N, Yang R, Zhang W, Dorfman H, Rao P, Gorlick R. Genetically transforming human mesenchymal stem cells to sarcomas. [Accessed on May 29, 2012];Cancer. 2009 Oct 15;115:4795–4806. doi: 10.1002/cncr.24519. http://dx.doi.org/10.1002/cncr.24519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loring JF, Rao MS. Establishing standards for the characterization of human embryonic stem cell lines. Stem Cells. 2006;24:145–150. doi: 10.1634/stemcells.2005-0432. [DOI] [PubMed] [Google Scholar]
- 30.Serkinci N, Guldberg P, Burns JS, Abdallah B, Schrødder H, Jensen T, et al. Adult human mesenchymal stem cell as a target for neoplastic transformation. Oncogene. 2004;23:5095–5098. doi: 10.1038/sj.onc.1207651. [DOI] [PubMed] [Google Scholar]
- 31.Miura M, Miura Y, Padilla-Nash HM, Molinolo AA, Fu B, Patel V, et al. Accumulated chromosomal instability in murine bone marrow mesenchymal stem cells leads to malignant transformation. Stem Cells. 2006;24:1095–1103. doi: 10.1634/stemcells.2005-0403. [DOI] [PubMed] [Google Scholar]
- 32.Gisselsson D, Björk J, Höglund M, Mertens F, Dal Cin P, Akerman M, et al. Abnormal nuclear shape in solid tumors reflects mitotic instability. Am J Pathol. 2001;158:199–206. doi: 10.1016/S0002-9440(10)63958-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebert R, Ulmer M, Zeck S, Messner-Weigl J, Schneider D, Stopper H, et al. Selenium supplementation restores the antioxidative capacity and prevents cell damage in bone marrow cells in vitro. Stem Cells. 2006;24:1226–1235. doi: 10.1634/stemcells.2005-0117. [DOI] [PubMed] [Google Scholar]
- 34.Giordano A, Galderisi U, Marino IR. From the laboratory bench to the patient’s bedside: an update on clinical trials with mesenchymal stem cells. J Cell Physiol. 2007;211:27–35. doi: 10.1002/jcp.20959. [DOI] [PubMed] [Google Scholar]
- 35.Lund TC, Kobs A, Blazar BR, Tolar J. Mesenchymal stromal cells from donors varying widely in age are of equal cellular fitness after in vitro expansion under hypoxic conditions. Cytotherapy. 2010;12:971–981. doi: 10.3109/14653249.2010.509394. [DOI] [PMC free article] [PubMed] [Google Scholar]