Abstract
Integrin receptors provide a dynamic tightly-regulated link between the extracellular matrix (or cellular counter-receptors) and intracellular cytoskeletal and signalling networks, enabling cells to sense and respond to their chemical and physical environment. Talins and kindlins, two families of FERM–domain proteins, bind the cytoplasmic tail of integrins, recruit cytoskeletal and signalling proteins involved in mechano-transduction, and synergise to activate integrin binding to extracellular ligands. New data reveal the domain structure of full-length talin, provide insights into talin-mediated integrin activation, and show that RIAM recruits talin to the plasma membrane while vinculin stabilises talin in cell–matrix junctions. How Kindlins’ act is less well defined, but disease-causing mutations show that kindlins are also essential for integrin activation, adhesion, cell spreading and signalling.
Cell proliferation, cell migration, tissue morphogenesis and homeostasis all depend on cell–cell and cell–extracellular matrix (ECM) interactions, and the integrin family of adhesion receptors play essential roles in these processes. Integrins are heterodimeric type I transmembrane proteins comprised of α and β subunits, both of which have an extracellular ligand-binding region and a generally short cytoplasmic tail that binds multiple cytoskeletal and adaptor proteins that regulate the affinity of integrin for ligands (integrin activation; Box 1). Ligand binding and subsequent integrin clustering lead to the recruitment of additional actin-binding, scaffolding and signalling proteins, resulting in the transmission of mechanical and chemical signals into the cell. Here, we focus on two structurally and functionally related protein families, the talins and kindlins, which bind β-integrin cytoplasmic tails and are essential for integrin activation and signalling.
BOX 1. Integrin structure and activation.
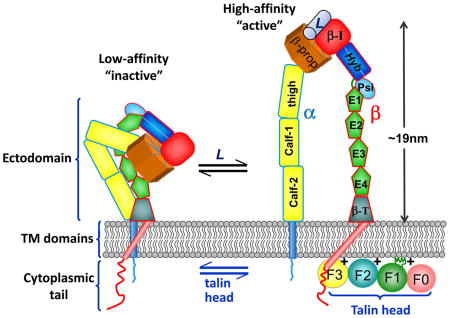
Integrin adhesion receptors are obligate αβ heterodimers. In mammals the 18 α subunits and 8 β subunits combine to form 24 different heterodimeric receptors, each with different ligand-binding specificities and a different tissue distribution149. The subunits have large ectodomains that are constructed from various modular units, including Calf and Thigh domains, and the crystal structures of various integrin ectodomains have been solved150. Each α and β subunit has a single trans-membrane (TM) helix and, usually, a short unstructured cytoplasmic tail21. Integrins exist in equilibrium between a bent, low-affinity state and an upright, high-affinity state. They can be activated by the binding of extracellular ligands (L) to integrin ectodomains (known as ‘outside-in’ activation) and by the binding of the talin head (an atypical FERM domain with F0, F1, F2 and F3 domains26) to β-integrin tails and acidic membrane phospholipids (known as ‘inside-out’ activation)20–22. Evidence for a ‘bent’ to ‘upright’ change in the integrin heterodimer on activation includes an electron-microscopy study of αIIbβ3 integrin embedded in membrane nanodiscs, in which talin was observed to cause extension of the integrin conformation107.
Talin was first identified in membrane ruffles and cell–ECM junctions (focal adhesions (FAs)) 30 years ago1, and the discovery that it bound two other FA proteins, vinculin and integrins, attracted much interest. Sequencing revealed a highly-conserved, 2541 amino acid protein composed of an N-terminal FERM domain (known as the talin head) linked to a long C-terminal region, known as the talin rod. Initial evidence that talin is important for FA assembly came from antibody microinjection, gene down-regulation and gene deletion experiments (reviewed in 2), while the first mechanistic insights into how talin functions came from the landmark discovery that the talin head binds directly to the cytoplasmic tails of β-integrin subunits3, and performs the key final step in integrin activation4. This, combined with structural studies on the integrin–talin head interaction5,6, began to explain how talin activates integrins from within the cell (inside-out activation)7. Talin is conserved throughout metazoans, and studies in D. melanogaster8 and C. elegans9 confirm its importance in integrin function. Vertebrates have two talin genes encoding closely related proteins with distinct but overlapping functions2, and talin2 upregulation in talin1 knockout mouse embryo fibroblasts compensates for loss of talin110. However, talin2 knockdown in these cells reveals that talins are essential for linking activated integrins to cytoskeletal actin, for FA assembly and the exertion of force on the ECM10. Note that endothelial cells only express talin111,12, making them ideal for talin1 structure–function studies.
Given the focus on talin in integrin activation, data showing that a lack of kindlin-3 results in severe integrin activation defects in platelets was unexpected13. A role for kindlin in integrin function first emerged when C. elegans kindlin (UNC-112) was shown to be essential for the organisation of integrins at muscle attachment sites14. In 2003, mutations in a human UNC-112 homolog were shown to cause Kindler syndrome15,16, a rare autosomal recessive disease characterized by skin blisters, photosensitivity, mucosal errosion and gastro-intestinal ulcers. Expression of this gene (which was named kindlin-1) is largely restricted to epithelia, and kindlin-1-deficient cells exhibit defects in spreading, polarity, migration, survival and ECM organization17. Sequence similarity between kindlin and the talin FERM domain, and the ability of both proteins to bind integrin β tails and to localize to FAs18, heightened the interest in kindlins. Humans have three closely related kindlins: kindlin-1, kindlin-2 and kindlin-3, and all three mammalian kindlins have now been implicated in integrin activation. Mutations in kindlin-3 cause leukocyte adhesion deficiency type III (LAD-III) - reviewed in17,19. While the detailed mechanisms by which kindlins exert their effects on integrin activation remain uncertain, it is clear that kindlins cooperate with talin during integrin activation, and that a direct interaction of kindlin with the integrin β tail is required, but apparently not sufficient, for integrin activation.
Here we focus on recent advances in talin structure, the mechanisms by which the Rap1A-effector RIAM binds and recruits talin to dynamic nascent adhesions at the leading edge of cells, and the way that force-induced conformational changes in the talin rod lead to a switch from talin–RIAM to talin–vinculin complexes, stabilising FAs. We also discuss new information on kindlin domain organization and the possible mechanisms underlying cooperation between kindlins and talin in integrin activation. We refer readers to more in depth reviews on the structural aspects of talin-mediated integrin activation20–22, and vinculin structure and function23,24.
STRUCTURAL INSIGHTS INTO TALIN FUNCTION
Understanding the mode(s) of action of talin at the cellular and organismal levels requires information about talin structure, but so far, efforts to crystallize talin have failed, probably because of its inherent flexibility25. However, completion of the structures of all 18 domains in full-length talin represents a significant advance26–35, which has allowed a model of full-length talin to be built35 (FIG 1A,B). The structures of individual domains, together with those of domains in complex with binding partners, show that talin has evolved to regulate integrin activity, to couple integrins to cytoskeletal actin and to act as a mechanosensitive protein in which ligand binding to the talin rod is regulated by force.
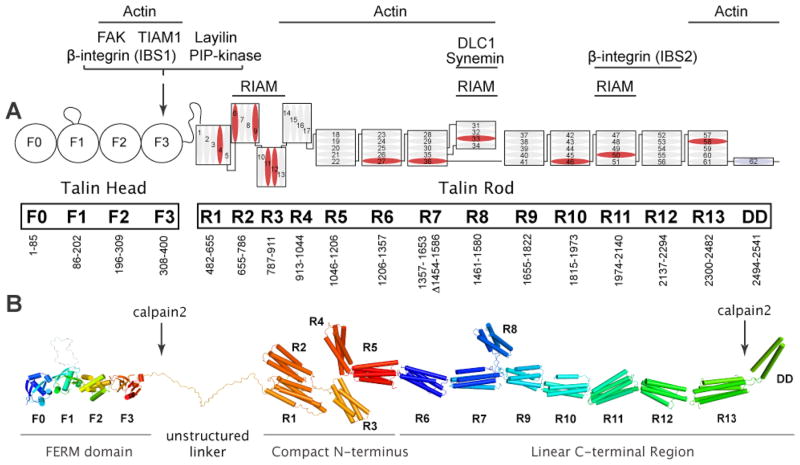
FIG 1. Domain organisation and structural model of full-length talin.
A: The domain organisation of talin135. The N-terminal talin head, which is comprised of an atypical talin FERM domain containing F0, F1, F2, and F3 domains, is joined by an unstructured linker of ~80 residues to the flexible talin rod. The rod is made up of 62 α-helices (numbered blue cylinders) that are organised into thirteen 4- or 5-helix bundles (R1–R13), with a single helical dimerisation domain (DD) at the C-terminus. Domain boundaries and the interaction sites for talin-binding proteins are indicated (IBS; integrin binding site). Helices that bind vinculin are in blue. Talin2 is predicted to have the same domain structure. B: Structural model of talin assembled from the crystal and NMR structures of the various domains. The position of the calpain2 cleavage sites are indicated. The R1 and R2 domains interact via an extensive hydrophobic interface34, and a long common helix joins R11 and R1230 (not shown). Otherwise, helical bundles are joined by short linkers (not shown since their structures were not determined). Because the N- and C-termini of the three 4-helix bundles (R2R3R4) are positioned at the same end of the bundle, this region will be more compact than the long succession of 5-helix bundles linked via their N- and C-termini.
Structural model of full-length talin
The N-terminal talin head is an atypical FERM domain with an F0 domain in addition to the three domains (F1, F2, F3) that are characteristic of other FERM domains32 (Fig 1). Moreover, the crystal structure shows that it adopts an extended rather than the clover-leaf structure characteristic of other FERM domains26. The F3 phosphotyrosine binding (PTB)-like domain binds β-integrin tails6,36,37, the cytoplasmic tail of the hyaluronan receptor layilin38 and the type 1 PIP-kinase γ-isoform39; structures of these complexes reveal similar modes of talin binding. The F3 domain of talin also reportedly binds focal adhesion kinase (FAK)40, a tyrosine kinase that is phosphorylated in response to integrin ligation, and the Rac1 exchange factor TIAM141, although as yet there are no structural details on binding mechanisms.
The talin head is linked by a large unstructured region (with the potential to span 20nm when fully extended42) to the talin rod, 62 amphipathic α-helices organised into thirteen 4 or 5-helix bundles (R1–R13)35; the single helix at the C-terminal end of the rod serves as the dimerisation domain (DD)27. Crystal structures of the R1R234, R7R828 and R11R1230 double domains have been determined, while the structures of other domains have been solved by NMR (FIG 1B). The rod contains multiple vinculin-binding sites (VBSs)31, and at least two actin-binding sites2, the best characterised of which is in the C-terminal R13 domain and requires talin dimerisation to bind actin27. The rod also contains a second integrin-binding site, IBS230,43, and several binding sites for the Rap1A-GTPase effector RIAM35 that is involved in recruiting talin to membranes to activate integrins44–46. The tumour suppressor DLC1 (a RhoGAP)47 and synemin (which links intermediate filament proteins to integrin-containing adherens junctions in striated muscle48) both bind the R8 domain that is inserted into a loop in the R7 domain (FIG 1). The availability of high-resolution structures for all talin domains should facilitate the detailed mapping of interaction surfaces and therefore studies on talin function.
Mechanism of talin-mediated integrin activation
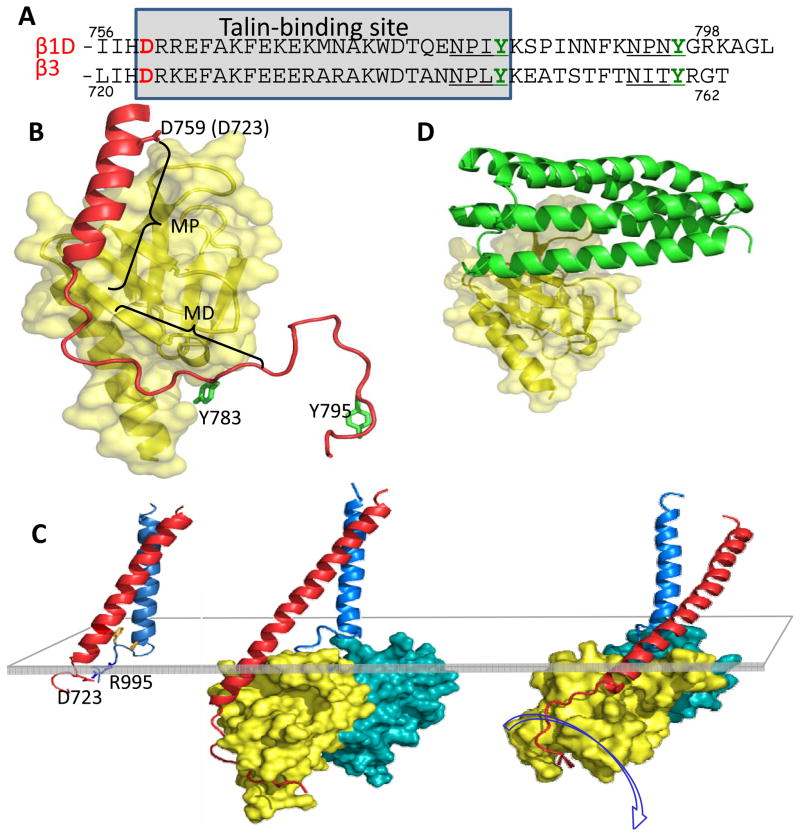
Biochemical3 and crystallographic studies5 established that a complex is formed between the talin F3 domain and the first NPxY motif in integrin β tails (FIG 2A), and that this interaction is required for the talin-mediated inside-out activation of integrins in vitro4 and in vivo49. More recent studies of complexes formed with larger β tail fragments and talin subdomains6,37 (FIG 2B), and structures of integrin transmembrane (TM) segments50, have extended our knowledge of integrin–talin–membrane complexes and have led to plausible models for talin-mediated integrin activation20–22 (FIG 2C and Box 1). In brief, the close association of the TM helices of the integrin subunits results in a low-affinity receptor. Structures of the αIIbβ3 TM complex show that, in this state, the αIIb TM helix is roughly perpendicular to the membrane while the β3 TM helix is tilted50,51 (FIG 2C). The regions of the αIIb and β3 TM helices nearer the extracellular face of these integrin subunits pack closely together, but contact at their intracellular ends involves an unusual folding back of the α-chain that promotes electrostatic interactions between αIIb (R995) and β3 (D723)50 (Fig 2C). Recent evidence indicates that a conserved β3 TM lysine helps orient the TM helix tilt by placing its positive charge at the membrane–water interface52. During physiological activation, talin F3 binds to the first NPxY motif in the β tail; this interaction stabilizes additional interactions between F3 and the membrane-proximal region of the β integrin tail, extending the β TM helix (FIG 2B,C)6. Talin F3 also forms a salt bridge with the conserved membrane-proximal Asp in β integrin (D723 in β3)37, which likely disrupts the inhibitory interaction of the β integrin tail with the conserved R995 residue in αIIb, thereby contributing to integrin activation. These findings are consistent with multiscale molecular dynamics simulations where binding of talin F2F3 to a membrane-embedded αIIbβ3 dimer fragment causes tilting and reorientation of the β TM helix, and dissociation of the α and β TM contacts53 (Fig. 2C and BOX 1). Experiments with fluorophores attached to regions of the β TM segment at the membrane–water interfaces also show that talin induces changes in β TM segment positioning in the membrane 54.
FIG 2. Mechanism of talin-mediated integrin activation.
A: The sequences of two representative integrin β-tails, those of integrin β1D and integrin β3, are shown. The two NPxY-like motifs are underlined, and the conserved Asp723 (numbered using the β3 sequence) that forms a salt bridge with R995 in the αIIb tail, and leads to a low affinity state, is highlighted in red. Talin binds to the indicated region of β-tails via its F3 domain. B: The complex between talin2 F3 (yellow) and the β1D tail (red)37 using β1D numbering to indicate key residues. The membrane-proximal (MP) and membrane-distal (MD) regions of the complex are indicated. The first NPxY-like region in the β1D tail is indicated by Y783. The second NPxY-like region in the β1D tail was not seen in the X-ray structure, but is modelled here (indicated by Y795) to show that it is very exposed, and has the potential to bind kindlins. C: The NMR structure of the transmembrane segments of the αIIbβ3 integrin is shown on the left50. This NMR structure and the structure of the talin2 F2F3 domains bound to the β1D-integrin tail37 were used to form the composite structure in the centre. The structure on the right was obtained after 100ns of molecular dynamics simulation in the presence of a membrane bilayer. Formation of favorable electrostatic interactions between talin and the membrane causes rotation of the talin–integirn β tail complex (centre and right structures), increasing the tilt of the β TM helix. This leads to separation of the α and β TM regions53, and hence to integrin activation. The translucent rectangle indicates the position of the cytoplasmic face of the membrane bilayer. D: The structure of the autoinhibitory complex between the talin1 F3 domain (yellow) and the R9 domain of the talin1 rod (green)65. The talin1 F3 domain is shown in approximately the same orientation as in B. Note how binding of the talin rod to the talin F3 domain occludes the F3 binding site for the membrane-proximal portion of the integrin β tail, effectively preventing integrin binding and activation.
Although interactions between talin F3 and β-integrin tails are required for integrin activation they are not sufficient, and interactions between the talin head and the membrane play a key role. The talin head has a series of basic residues along its membrane-proximal surface26 that bind acidic phospholipids, and mutations in basic residues in F237 and in an inserted loop in F132 markedly impair membrane binding and integrin activation. Moreover, acidic phospholipids have been shown to increase greatly the affinity of β3-tails for the talin head55, suggesting that acidic plasma membrane phospholipids such as PIP2 play a key role in orientating the talin head such that it can engage and activate integrins26 (BOX 1).
Whether the above model applies to activation of most integrins is controversial, although there is general agreement that the activity of leukocyte56 and platelet integrins57–59 are regulated by talin, and talin-mediated activation of integrins containing β1, β2 and β3 subunits has been reported in a range of cell types4,60. Integrin activation is generally assessed by measuring the binding of soluble ligands or reporter antibodies that selectively or preferentially bind the active integrin61. While over-expressing the talin head is sufficient for integrin activation, full-length talin also activates integrins, and mutagenesis confirms that this relies on interactions between the talin head and the integrin β tail44,61,62. Nonetheless, full-length talin is a less potent integrin activator because it can adopt an auto-inhibited conformation62–65. The interaction of full-length talins with integrins is also implicated in integrin clustering8,66,67, and as many integrin ligands are multivalent, this could greatly increase adhesion through avidity modulation68. Moreover, clustered talin-bound integrins serve as a link to the actin cytoskeleton and acts as a signalling hub.
REGULATION OF TALIN FUNCTION
Since talin plays a key role in integrin activation, its function is tightly regulated. Mechanisms involved include changes in talin conformation and localisation, which are induced by signalling pathways and mechanical force, the action of competitor proteins and limited proteolysis of talin by calpain2.
Talin conformations and auto-inhibition
Biochemical data and electron microscopy25,69 show that talin exists in both globular and extended conformations (~60 nm long), and a structural explanation for this has recently begun to emerge. Specifically, the talin F3 domain binds the R9 domain in the talin rod62,64,65, which sterically inhibits binding of the F3 domain to the membrane-proximal region of β-integrin tails (compare FIG 2B and D). Further insights into the structure of auto-inhibited talin are provided by a 3D model of full-length talin derived from EM reconstruction studies, and the shapes of individual domains and inter-domain angles determined by small angle X-ray scattering (SAXS)63. The model indicates that talin can form a compact dimer (12.5nm × 11nm × 9.5nm) in which the two talin rods form a donut-shaped structure, with the talin heads packed in the centre of the donut. The model recapitulates the high affinity interaction between the F3 head and R9 rod domains, plus several weaker inter-subunit interactions that have been detected by NMR.
The concept of auto-inhibited talin is consistent with observations that much of talin is cytosolic70, and that it translocates to the membrane in response to activation of the Rap1A GTPase, a key regulator of integrin activity45,46. The positively-charged residues on F3 that interact with the negatively-charged face on R9 also interact with acidic membrane phospholipids (Box 1). PIP2 has been shown to relieve the F3–R9 interaction62, and could therefore activate talin. Note: the interactions that stabilise auto-inhibited talin62 appear much weaker than those in auto-inhibited vinculin71.
As predicted by the structural data, mutations that compromise the talin F3–R9 interaction accelerate the rate of FA assembly11. However, cell fractionation studies show that, on its own, disrupting the F3–R9 interaction is not sufficient to drive talin from the cytosol to the plasma membrane; additional interactions between the F2F3 FERM domains and R1R2 rod domains mask membrane-targeting sequences in the FERM domain70. Interestingly, R1R2 contains 3 VBSs (FIG 1A), and expression of the talin-binding domain of vinculin (Vd1)24 in cells was sufficient to drive talin to the plasma membrane. Thus, vinculin could play a significant role in talin activation, a conclusion supported by the observation that Vd1 stabilises FAs and locks integrins into the activated state72,73. Moreover, vinculin Vd1 increases binding of αIIbβ3 integrin to recombinant talin in vitro and in cells74.
Rap1A–RIAM in talin-mediated integrin activation
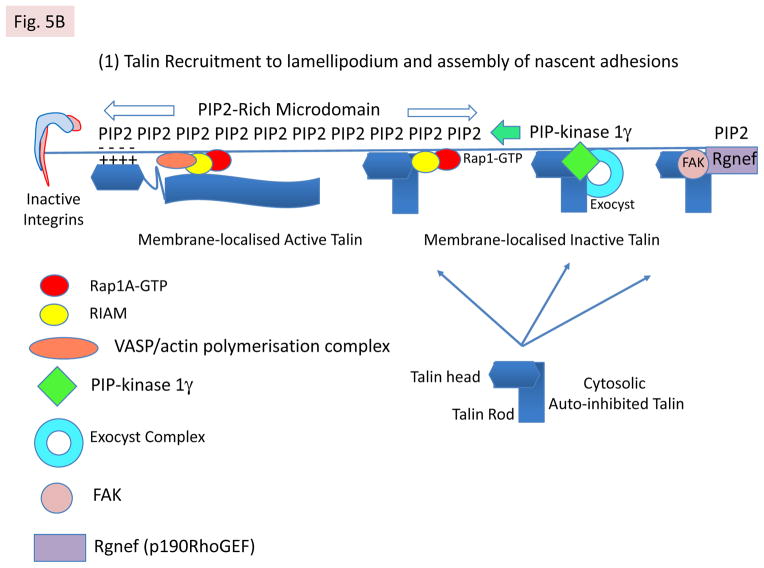
Elegant reconstitution studies of αIIbβ3 integrin activation in Chinese hamster ovary cells have defined a pathway in which activated Rap1A and its effector RIAM recruit talin to the cytoplasmic domain of integrins, which leads to integrin activation44–46. A short amphipathic helix (residues 6–30) in the N-terminal region of RIAM binds talin, and a protein containing this helix fused to the membrane-targeting sequence of Rap1A recruits talin to the plasma membrane, and supports integrin activation45. Talin also binds the N-terminal helix of lammellipodin45, another member of the MRL family of proteins75. RIAM and lamellipodin contain adjacent RA and PH domains, and the structure of the RIAM RA–PH double domain suggests that they act as a proximity detector for activated Rap1A and PIP276; that is, RIAM and therefore talin is only recruited to Rap1-GTP embedded in PIP2 rich microdomains. Interestingly the N-terminal region of RIAM inhibits its recruitment to membranes, raising the possibility that talin might contribute to RIAM activation. This leaves open the question as to whether RIAM activates talin.
The talin–PIPkinase type 1γ interaction
Cells lacking PIP-kinase type 1γ, the isoform of PIP-kinase that binds talin, show reduced initial rates of attachment to ECM, impaired recruitment of both talin and vinculin (but not kindlin-2 or paxillin) to FAs, and reduced ability of β1 integrins to exert force on ECM77. A similar phenotype was obtained with wild-type cells expressing a K274E mutation in F2 of the the talin FERM domain that inhibits PIP2 binding and dramatically reduces integrin activation37. The authors conclude that the major role of PIP-kinase type 1γ in FAs is the local synthesis of PIP2 that is required to orientate the talin head such that it can activate integrins77. However, PIP2 also likely plays a role in the RIAM-mediated recruitment of talin to the membrane76 and talin activation62. Moreover, PIP-kinase type 1γ and talin associate with the exocyst complex to deliver integrins to the leading edge78. Thus, PIP-kinase type 1γ can modulate talin function and cell adhesion in several ways. Interestingly, ubiquitination of PIP-kinase type 1γ has recently been shown to regulate FA turnover79.
FAK-mediated recruitment of talin to nascent adhesions
Evidence that FAK also plays a key role in recruiting talin to nascent adhesions has recently emerged40, challenging the conventional model of FA assembly in which integrin–talin–actin complexes precede the recruitment of other FA proteins. Although talin and paxillin colocalised in FAs formed 15 min after plating mouse embryo fibroblasts on fibronectin, talin was not detectable in early paxillin-positive FAs formed by FAK null cells even though it was present in mature FAs. Moreover, talin co-localised with FAK in early FAs in cells expressing a β1-integrin tail mutation (Y783A) that abrogates talin binding, suggesting that talin can be recruited to FAs independently of integrin binding. What recruits FAK to these early adhesions has not been established although this might involve p190RhoGEF (also known as Rgnef), which binds FAK80. Recent developments in single-protein tracking and super-resolution imaging81 may help clarify the kinetics of FAK and talin recruitment during adhesion initiation and maturation. Interestingly, talin-deficient adhesions in FAK null cells stain with the 9EG7 antibody that detects activated β1 integrin, and mammary epithelial cells depleted of talin1 also contain activated β1 integrins and assemble FAs, although these are deficient in vinculin, paxillin and ILK82. Whether integrin activation, and also the early stages of cell spreading10, in talin knockdown cells is driven by ECM binding, via other tail-binding proteins, or by residual talin below the limits of detection, is unclear.
Inhibitors of talin–integrin interactions
Several mechanisms have the potential to modulate the talin–integrin interaction and therefore integrin activation (FIG 3), even if talin itself remains activated and membrane-bound7,83. Phosphorylation of the membrane-proximal NPxY motif in integrin β tails by Src family kinase (SFK) directly inhibits binding of the talin F3 PTB-like domain84,85, and so impairs integrin activation. However, this tail phosphorylation also promotes the binding of other PTB domain proteins, such as the scaffold protein DOK184–86; competition of these proteins with talin is likely to further suppress integrin activation (FIG 3). Other proteins that bind β integrin tails can also interfere with talin binding, the best characterised of which is filamin. Structural studies reveal that filamin binds β-integrin tails at a site overlapping that for talin, and hence filamin competes with talin for β integrin binding87. Filamin also likely competes with kindlin for β integrin tail binding (FIG 3). Consistent with this, loss of filamin expression enhances integrin activation87, and expression of the filamin-binding protein migfilin, which occupies the integrin-binding site on filamin88,89, enhances integrin activation90. However, migfilin knockout mice either exhibit no phenotype91 or the effects are restricted to bone remodeling by osteoblast progenitors92, raising questions about the wider relevance of migfilin in regulating integrin activation. Filamin binding to β2-integrins is also inhibited by 14-3-3 proteins that bind to β2 tails that are phosphorylated on Thr75893.
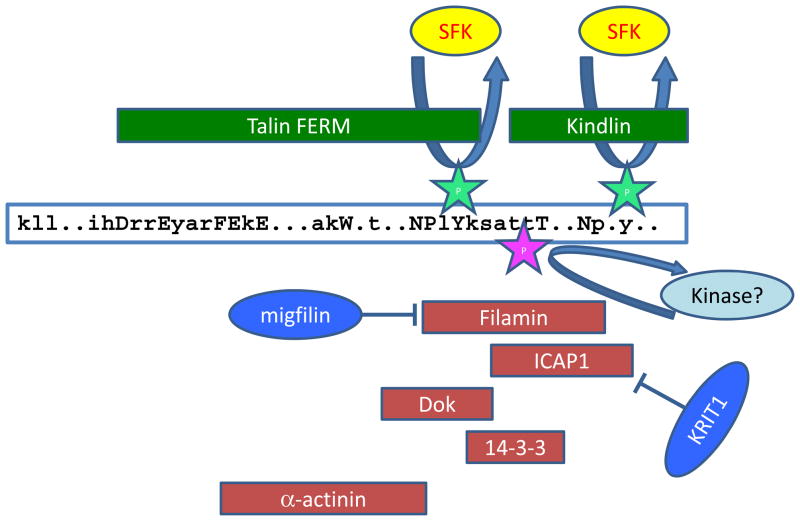
FIG 3. Regulators of talin and kindlin binding to integrins.
A schematic of the integrin β tail showing conserved residues: uppercase for near-invariant residues, lowercase for conserved residues with all other residues marked by a dot. Binding sites for integrin activators (green) and inhibitors of integrin activation (red), based on structural and biophysical studies87,93,95,96,113, are indicated by the box that encompasses the protein in question. Talin and kindlin can bind the integrin β tail simultaneously106, but binding sites for the proteins that inhibit their binding, and thus inhibit integrin activation, overlap, suggesting that only one can bind at a time. Src-family kinase (SFK)-mediated tyrosine phosphorylation of the membrane-proximal or membrane-distal NPxY motif can inhibit talin and kindlin binding, respectively, and enhance binding of the inhibitor Dok184,85,143. Threonine phosphorylation at residues between the NPxY-motifs has the potential to activate or inhibit integrin activation — it suppresses binding of the integrin inhibitor filamin87,93 and generates a binding site for 14-3-3 poteins that inhibit integrin activation93. α-actinin can both positively and negatively regulate integrin–talin interactions, depending on the β tail in question96. Binding of other proteins, such as migfilin or KRIT189,90,95, to the integrin inhibitors filamin and ICAP1, respectively, prevents these inhibitors from binding to integrins and hence favours integrin activation.
Another well-characterized integrin inhibitor is integrin cytoplasmic domain-associated protein 1 (ICAP1)94 (FIG 3). The PTB-domain in ICAP1 binds the membrane-distal kindlin-binding NPxY motif in β1 integrins, but also inhibits talin binding to integrin83. The crystal structure of the integrin β1–ICAP1 complex has now been solved95 and talin-binding residues are not involved in the interface, suggesting that the inhibitory effect of ICAP1 is not via direct competition. Nonetheless, ICAP1 expression impairs talin-mediated β1 activation, and ICAP1 mutants defective in integrin binding do not inhibit this activation. Furthermore, KRIT1 binding to ICAP1 displaces ICAP1 from integrins, facilitating talin-mediated integrin activation95.
Intriguingly, a recent study implicates the actin-bundling protein α-actinin in both positive and negative regulation of integrin–talin interactions96. The reported α-actinin binding site on integrin β tails96 overlaps the talin-binding site (Fig 3), suggesting that α-actinin and talin may compete for binding, and this is now reported for β3 integrins96. Consistent with this, α-actinin binding suppresses αIIbβ3 activation in platelets97. However, α-actinin apparently enhances talin binding to β1 integrins96. The data highlight the need to consider α-actinin as a modulator of talin binding and adhesion site maturation, although the different effects on β1 and β3 integrins remain to be explained.
Integrin–talin interactions may also be inhibited by proteins that bind the talin F3-PTB domain. For example, over-expression of the type 1 PIP-kinase γ-isoform, that binds the talin F3 domain, suppresses integrin activation98, even though talin dimers have the potential to engage two different F3-domain ligands simultaneously. In addition, inhibitors of integrin activation that bind the α tail have been identified83, one of which, SHARPIN, is reported to inhibit talin and kindlin binding to the β integrin tail through an unknown mechanism. Thus, a variety of protein–protein interactions can modulate talin–integrin interactions and hence control integrin activation.
CO-OPERATION BETWEEN TALIN AND KINDLIN
Over the past 5 years it has become evident that the kindlins, another family of integrin β-tail-binding, FERM-domain-containing proteins, also play key roles in integrin activation and signalling17,19,99,100. Kindlins directly bind integrin β tails, and kindlin or integrin mutations that inhibit this binding impair talin-mediated integrin activation, strongly suggesting that a direct kindlin—integrin interaction is required for maximal integrin activation. However, in most cell-culture systems, kindlin over-expression does not activate integrins (in some cases it can even suppresses activation101), but when co-expressed with talin head, kindlins can strongly potentiate αIIbβ3 activation101–103. Surprisingly, even when co-expressed with talin head, over-expressed kindlin does not activate α5β1 integrins101. Indeed, kindlin co-expression can suppress talin head-mediated β1 activation. The basis for this suppression, and for the differential behaviour of β1 and β3 integrins, requires further study. Nonetheless, β1 integrin activation is kindlin-dependent as kindlin knockout or knockdown impairs β1 activation103–105. Consistent with a role for kindlins as co-activators of integrins, rather than as direct activators, NMR data indicate that kindlin-2, unlike the talin head, is unable to unclasp the inhibitory αIIb-β3 tail interaction106. Furthermore, in engineered cell systems, mutations that block talin binding to β integrin tails block both talin- and kindlin-driven integrin activation, while mutations that inhibit kindlin binding still permit talin-mediated activation, although they block the kindlin enhancement effect107. Thus, kindlins apparently modulate talin-mediated integrin activation; the major question is how.
Kindlins are structurally related to the talin head
Much less structural information is available for kindlins than for talins, partly because kindlins are difficult to express. Kindlins are highly conserved and revised sequence alignments show that they have a similar domain organisation to the atypical talin FERM domain108 (FIG 4a). Like talins, kindlins have an F0 domain plus a large unstructured insert in F1. However, unlike talins, kindlin F2 domains contain an inserted PH domain. Structures of kindlin F0108,109 and PH domains110–112 have been solved and recent SAXS studies indicate that intact kindlin-3 is relatively elongated in solution113, like the talin head26 (FIG 4b). Consistent with the similarities to the talin FERM domain, pull-down assays demonstrate that kindlins directly bind integrin β-tails and that point mutations in the kindlin F3 domain inhibit this binding13,101,102. Kindlin binds the second NPxY motif in integrin β tails and mutations at this site, or at conserved threonine residues preceding it, inhibit binding. These results have recently been confirmed using various biophysical techniques106,113. In summary, while we still lack high-resolution structures of intact kindlins, the available evidence points to a high degree of similarity to the talin head with the exceptions that kindlins contain an inserted PH domain and bind to the membrane-distal NPxY motif (rather that the membrane-proximal NPxY motif) in integrin β tails (Fig 3).
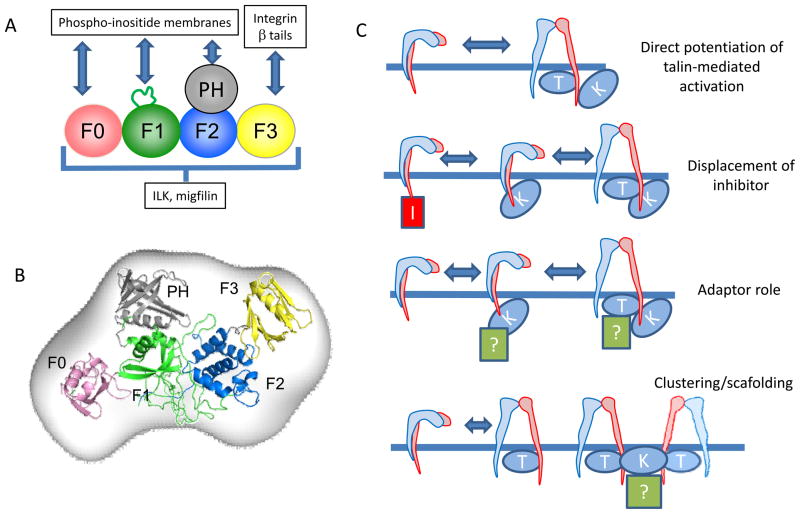
FIG 4. Kindlin: an integrin co-activator.
A: A schematic representation of kindlin domain organization, showing where kindlin interaction partners bind. Kindlin is predicted to fold as an atypical FERM domain composed of 5 subdomains (F0–F3 plus a PH domain). Similar to the talin head (Fig 1)26, kindlin is thought to form an extended structure113. β Integrin tails bind to the F3 subdomain while phospho-inositide membrane-binding sites have been identified in the kindlin F0 and PH domains, and in the large unstructured loop in F1 109,110,112,116. Binding sites for ILK and migfilin have yet to be definitively mapped. B: Possible orientation of kindlin domains based on x-ray scattering113, the kindlin-1 PH domain crystal structure112, and NMR structures of the kindlin-1 F0 domain108 and the talin FERM domain26. Domains are coloured as in part A. C: Models for cooperation between talin and kindlin during integrin activation. Binding of kindlin to the β integrin tail may directly potentiate talin-mediated integrin activation (top panel), perhaps by binding both the integrin β tail and the membrane to cause optimal exposure of the talin-binding site in the β tail. Alternatively, kindlin binding to the integrin may displace inhibitors, facilitating talin binding and activation (upper middle panel). In addition, kindlin may recruit other activating or adaptor proteins that cooperate with talin to activate integrins (lower middle panel). Finally, Kindlin may directly or indirectly (via another kindlin binding protein, labelled with a question mark) induce clustering of talin-activated integrins to increase avidity (bottom panel). Note: The Z-band alternatively spliced PDZ-motif containing protein (Zasp) also co-operates with talin to activate α5β1 integrin151, although It is unknown whether kindlins have any role in this process.
Kindlins’ roles in talin-mediated integrin activation
As described earlier, the molecular basis of talin-mediated integrin activation has been worked out in considerable detail and, in purified systems, talin binding is sufficient to activate membrane-embedded αIIbβ3 integrins107. However, kindlin knockout, knockdown and over-expression, along with integrin mutants defective in kindlin binding, all implicate kindlins in integrin activation. While we still lack a definitive understanding of how kindlins activate integrins, a number of models have been proposed22,100,114 (Fig 4c). In vitro, the talin head, kindlin and integrin β tails form a ternary complex106,113, and this may be essential for integrin activation, although the sequential binding of these components, or their binding to adjacent integrins114, have not been excluded. Recent data suggest that the binding of kindlin to integrins neither enhances talin—integrin binding, nor increases talin targeting to the membrane106,115, and no direct talin—kindlin interaction has been reported. This has led to suggestions that kindlin influences events occurring after talin recruitment to the integrin115. Consistent with this, in neutrophils, talin1 is required to generate the intermediate-affinity state of αLβ2 integrin that is responsible for the initial slow rolling of neutrophils, while both talin1 and kindlin-3 are required for high-affinity αLβ2 and neutrophil arrest56. Kindlins might enhance talin-mediated integrin tail separation by binding to the second NPxY-like motif in the β integrin tail and anionic membrane phospholipids. Indeed, the kindlin F0 domain, F1 loop and the PH domain can each bind anionic phospholipids (FIG 4a), and such binding is required for kindlin to fully coactivate αIIbβ3 integrin109,110,112,116. This hypothesis could be tested in purified reconstituted systems such as the integrin nanodiscs already used to show talin-mediated integrin activation107. If kindlin potentiates talin-mediated integrin activation in vitro, then it would strongly suggest that the ternary complex is the key to integrin activation. Alternatively, in cells, kindlin binding to the integrin tail may recruit additional activator or signalling proteins, or displace inhibitory proteins that modify talin’s ability to activate integrins. In this regard it is noteworthy that the structurally-defined binding sites for the inhibitors ICAP195 and filamin87 in integrin β tails overlap with the kindlin-binding site (FIG 3), suggesting that kindlin binding will displace these inhibitors. Similarly, the binding of 14-3-3 protein to phosphorylated β2 tails is likely to inhibit kindlin binding93. Kindlins might also influence integrin clustering and so enhance binding of multivalent ligands via increased avidity, rather than through conformational changes that lead to increased affinity for monovalent ligand. Available data suggests kindlins are monomeric so clustering might be mediated by kindlin-binding proteins such as migfilin and ILK, both of which have been implicated in promoting integrin activation90,117. Thus, kindlins’ adaptor functions may be critical for enhancing integrin activation and/or clustering.
Integrin and kindlin isoform specificity
All three kindlins bind integrins and impact their activation, but loss of kindlin-1, kindlin-2 or kindlin-3 results in markedly different phenotypes that are only partly due to differential expression17,114. Of the eight different human integrin β subunits, kindlins are currently known to bind β1, β2, β3 and β699,114,118. Kindlin-2 preferentially binds β1 and β3 integrins, binds more weakly to β2 and exhibits very little binding to β6106,118. Kindlin-1 binds strongly to both β1 and β6 integrins118. Kindlin-3 binds β1, β2 and β3 integrins and, while differential integrin binding has not been reported for kindlin-3, it has been suggested to preferentially regulate β1 integrin-dependent processes119. Differential integrin binding may have consequences in cells such as keratinocytes, which express both kindlin-1 and kindlin-2118. In addition to differences in integrin binding, different kindlins also have different subcellular localization and function119,120. Furthermore, despite all three kindlins binding to β1 and β3 integrin tails, kindlin-1 and kindlin-2, but not kindlin-3, co-activate αIIbβ3 integrins in CHO cells13,101. Likewise, while loss of kindlin-1 or kindlin-2 impairs β1 integrin activation, co-expressing talin head and kindlin does not activate β1 integrins in CHO cells101. The bases for these integrin- and kindlin-specific effects are unknown.
TALIN – BEYOND INTEGRIN ACTIVATION
While talins’ roles in integrin activation have been the focus of attention over the last 15 years, talins also play vital roles linking activated integrins to the actin cytoskeleton10, sensing and reinforcing the response to mechanical force121 and regulating adhesion formation and turnover.
Role of talin in mechano-transduction
The talin rod is largely made up of 5-helix bundles in which the N-termini and C-termini are positioned at opposite ends; this means that these bundles form a linear chain35 (FIG 1). However, three 4-helix bundles (R2–R4) interrupt the succession of 5-helix bundles, and since their N-termini and C-termini are at the same end of the bundle, they must adopt a more compact structure. Thus, the conformation of the talin rod will change in response to force exerted on integrin–talin–actin complexes, and this will likely impact on its interaction with certain ligands.
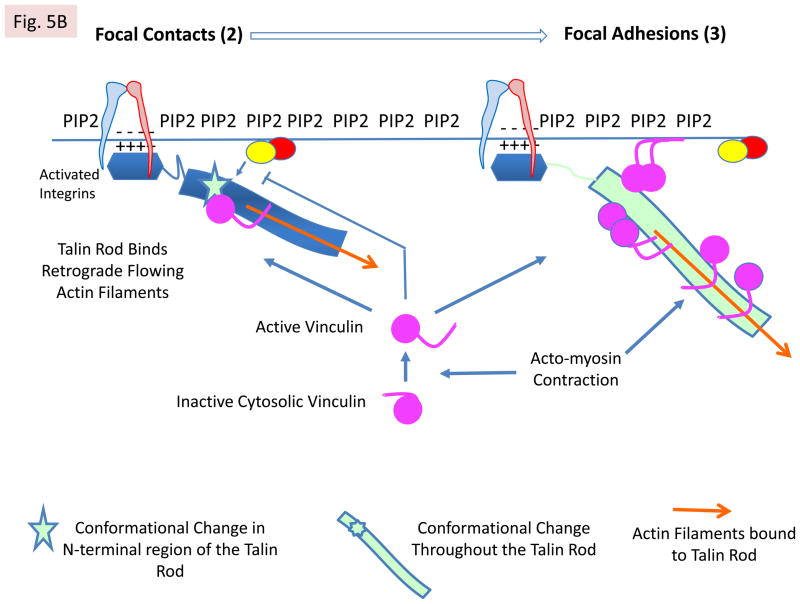
The multiple VBSs in the talin rod (FIG 1A) are each defined by hydrophobic residues on one face of an amphipathic helix31. These are normally buried within the core of the helical bundles, and vinculin binding therefore requires domain unfolding2. Elegant single-molecule experiments show that VBSs can be activated by force122, and the recruitment of vinculin to FAs is myosin-II dependent123. Moreover, talin undergoes repeated cycles of actomyosin-dependent stretching in the direction of actin flow124. In this context, the R2R3 4-helix bundles contained within the compact N-terminal region of the rod are of particular interest. Unusually, each contains two VBSs (FIG 1A), and while vinculin binding to R2 is inhibited because R2 is stabilised by extensive contacts with R134, R3 is destabilised by a unique cluster of threonine residues buried within its hydrophobic core35. Therefore, R3 is likely to be amongst the first of the rod domains to bind vinculin.
An additional feature of R2 and R3 is that they each bind the RIAM N-terminal peptide (residues 6–30) albeit with low affinity35. However, it is now apparent that the N-terminal region of RIAM (residues 1–127) contains two talin-binding sites that bind synergistically to R2R335. Significantly, the talin-binding domain of vinculin, Vd1, displaces RIAM 1–127 from R2R335 and inhibits binding of the RIAM 6–30 peptide to full-length recombinant talin74, suggesting that RIAM and vinculin binding to talin is mutually exclusive. Indeed, RIAM is localised preferentially at the membrane and in nascent adhesions where it supports cell protrusion74, while vinculin is abundant in mature FAs35,74. This suggests a model (FIG 5) in which RIAM binding to talin R2R3 initially recruits talin to the plasma membrane in a Rap1-GTP dependent manner. Here, talin activates integrins and triggers the assembly of dynamic nascent adhesions. RIAM also recruits VASP and the actin polymerisation machinery required to drive membrane protrusion. As talin binds F-actin flowing away from the leading edge, force-induced conformational changes in talin R2R3 disrupt high-affinity RIAM binding, exposing the VBSs. Vinculin recruitment stabilises FAs by promoting talin binding to integrins (this is Rap1-GTP independent74), therefore maintaining integrins in the activated state72,74. Moreover, vinculin can potentially cross-link talin to actin or acidic membrane phospholipids24 (FIG 5). Vinculin also regulates the recruitment and release of other core FA proteins in a force-dependent manner73.
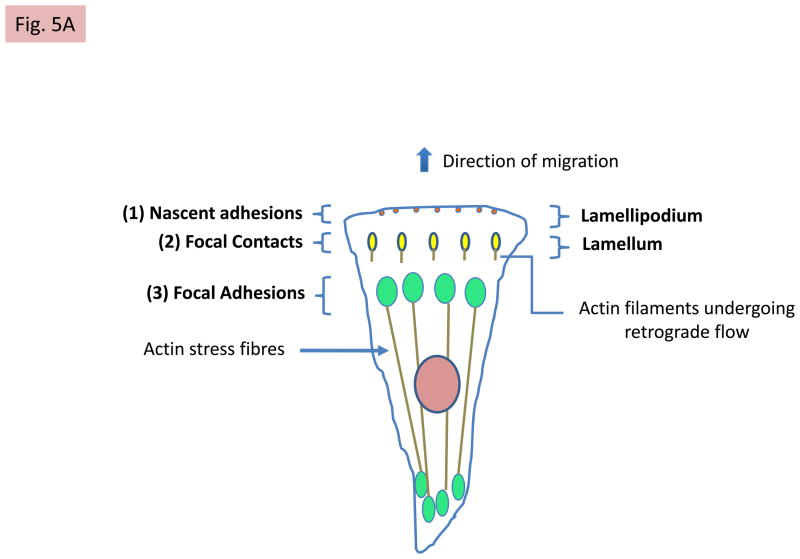
FIG 5. Talin changes partners during adhesion assembly and maturation.
A: Mechanisms of talin recruitment to the lamellipodium. The model envisages that auto-inhibited talin in the cytosol can be recruited to PIP2-rich microdomains in the plasma membrane via several mechanisms. First, by binding RIAM, which is complexed to membrane localised Rap1A-GTP44,45,74, via the N-terminal region of the talin rod35. Second, by exocyst complexes78 that contain integrins and PIP-kinase type1γ, which creates the PIP2-rich microdomains. Third, by FAK40 complexed to Rngef80, a p190RRhoGEF with a PH domain that binds phosphoinositides. Both PIP2 and RIAM binding may contribute to talin activation. Positively charged residues on one surface of the talin head are then envisaged to interact with PIP226 (see Box 1). Note that only one subunit of talin is shown for simplicity. B: Integrin activation following talin activation. Ba PIP2 binding to the talin head increases its affinity for β-integrin tails55, leading to integrin binding and activation. This triggers the assembly of nascent adhesions. RIAM also drives membrane protrusion74, probably by binding VASP, which recruits the actin polymerisation machinery. Bb The talin rod then binds F-actin undergoing retrograde flow, and the force exerted on talin124 is envisaged to alter the conformation of the N-terminal part of the talin rod. This disrupts the RIAM binding site and increases vinculin binding35, which reinforces the connection of talin to F-actin and causes dynamic focal contacts to form. The vinculin Vd1 domain binds talin, while its C-terminal tail binds F-actin; it may also cross-link talin to PIP2 in the membrane24. Bc Further increases in force exerted on talin by acto-myosin contraction induce more extensive conformational changes in both the talin rod and the unstructured linker between the head and rod. This enhances vinculin binding and promotes the maturation of dynamic focal contacts into more stable focal adhesions that are associated with actin stress fibres. Note that vinculin exists in an auto-inhibited cytoplasmic form and the mechansims by which it is activated have yet to be defined24, although force exerted by acto-myosin contraction is involved73. A schematic diagram showing the relative positions of nascent adhesions, focal contacts and focal adhesions in a migrating cell, and linking them to the relevant state of integrin activation, is shown.
Several regions of talin bind F-actin2, and may therefore play a role in initiating force-induced conformational changes in talin. The best characterised F-actin binding site is in the C-terminal R13 rod domain27, and mutations that compromise this site markedly reduce the ability of talin to rescue cell spreading and FA assembly in talin1 knockdown endothelial cells11. Interestingly, helix 1 in the R13 5-helix bundle negatively regulates actin binding27,125, and mutations that relieve this inhibition increase FA size and stability11. This suggests that force exerted on the R13 bundle may increase its affinity for F-actin, strengthening the talin–actin connection. However, the C-terminal actin-binding site in vinculin is also essential for force transduction, and the repolarisation of cells in response to stretch73. In summary, talin is a mechanosensitive protein that likely changes conformation in response to force; this is predicted to regulate its affinity for RIAM, vinculin and F-actin, and therefore its role during adhesion assembly versus FA maturation.
Role of the integrin-binding site IBS2 in the talin rod
In addition to the well characterized integrin-binding site in the talin F3 FERM domain (IBS1), the talin rod contains an integrin-binding site (IBS2)43 that spans two 5-helix bundles, R11 and R1230. Studies using D. melanogaster have established that, despite some redundancy, IBS1 maintains the link between integrins and the ECM, presumably by activating integrins, while IBS2 stabilises the link between integrins and the intracellular adhesion complex that includes paxillin, PINCH and FAK126. Interactions between β1 integrins and an IBS2-containing talin rod polypeptide have been detected by FRET, although surprisingly, no interaction was detected between β1 integrins and the talin head127. This suggests that IBS2 forms a relatively stable association with integrins although it does not activate integrins43; in constrast the IBS1–integrin interaction is more dynamic. Factors that determine which of the two IBSs in talin engage β integrins remain to be defined. Super-resolution fluorescence microscopy of FAs shows that the talin head is close to integrin tails while the C-terminus of the rod co-localises with actin ~40nm from the membrane128, an orientation supported by single molecule analysis of talin in cells124. This would suggest that IBS2, which is close to the C-terminal end of talin, is not engaged in FAs.
A role for C-terminal talin polypeptides?
Although talin-depleted mammary epithelial cells still spread on ECM proteins, FAK signalling is compromised, p21 levels are elevated and proliferation is inhibited; this phenotype is rescued by expressing membrane-localized FAK, but not a kinase dead mutant82. FAK signalling and proliferation was also rescued by expression of just the C-terminal region of talin (residues 1974–2541; FIG 1), suggesting that this part of the talin rod plays a role in assembling FAK signalling complexes, although FAK has not been shown to bind the talin rod. Intriguingly, a similar C-terminal talin polypeptide is generated in cells by calpain2-mediated proteolysis of talin between residues 1902–1903, coupled to arginylation of the liberated 70 kDa talin polypeptide129. Moreover, the talin2 gene has at least two internal promoters that encode C-terminal talin2 rod polypeptides130. These results raise the possibility that C-terminal polypeptides generated from both talin1 and talin2 may have physiological relevance.
Talin, FA dynamics and turnover
Studies using single protein tracking and super-resolution microscopy have recently begun to provide remarkable new insights into the dynamics of individual molecules within FAs. Integrin immobilisation in FAs requires simultaneous binding to both matrix and talin, while integrins outside FAs are in free diffusion81. Freely diffusing talin in the cytosol is recruited directly to FAs where it becomes immobilised, implying that talin is activated within FAs, and immediately engages and activates integrins. Intriguingly, integrins within FAs go through periods of immobilisation and slow free diffusion, suggesting that they cycle between active and less active states.
Turnover of FAs is essential for cell migration, and calpain2-mediated cleavage of talin between the head and rod131 and at a second site that removes the C-terminal dimerisation domain, promotes FA turnover42. Interestingly, a FAK mutant (FAKE1015A) that is unable to bind talin fails to support calpain2-mediated talin cleavage and FA turnover40. The possibility that the talin head liberated by calpain2 cleavage is physiologically important is raised by the discovery that its levels appear to be tightly regulated. Thus, the talin head undergoes Smurf1-mediated ubiquitinylation, which leads to its degradation, promoting FA turnover. Conversely, Ser425 phosphorylation by cdk5 stabilises the talin head and FAs132. Thus, turnover of the liberated talin head might support cycling between active and inactive integrins during FA remodelling, while the liberated talin rod might maintain FAK signalling and therefore cell-cycle progression82.
Talin, kindlin and integrin recycling
The Rab GTPases play key roles in integrin recycling133. However, the possibility that talin might also contribute was raised by early observations that talin depletion in Hela cells results in aberrant α5β1 processing134, and its delayed export from a secretory compartment135. Moreover, talin1 knockout ES cells show a reduced steady state level of β1 integrins136, and recent studies show that talin1 (but not talin2) protects β1-integrins from proteasomal degradation, and that this is important in epithelial morphogenesis137. New evidence points to a role for a talin–PIP-kinase type 1γ–exocyst complex in promoting trafficking of integrins to the leading edge, the establishment of cell polarity and directional cell migration78. Interestingly, talin1 also binds directly to the Rac1-GEF TIAM141, which in turn associates with the PAR3 component of the PAR polarity complex. The talin1–TIAM1 interaction is important in transient integrin-mediated Rac1 activation, which occurs via outside-in signalling, and the subsequent lamellipodia protrusion and cell spreading. TIAM1 also co-localises with talin in a sub-population of larger FAs in the front of migrating cells and regulates their turnover. It now emerges that talin also plays a role in α5β1 internalisation, while kindlin-2 binding to the membrane-distal NPxY motif in β1 tails appears to prevent lysosomal degradation and promote recycling of internalised, activated α5β1 integrin138. However, two recent papers suggest that kindlins dissociate from internalised integrins and that the FERM domain-containing protein sorting nexin-17 interacts with the kindlin binding site on integrin β tails in early endosomes and drives recycling139,140. In conclusion, both talin and kindlins can influence integrin function at several levels.
OUTSTANDING ISSUES
Mechanisms involved in talin-mediated integrin activation are now well understood, but how kindlins and talins cooperate during integrin activation remains a major question. Nonetheless, progress in characterising kindlin structures and interactions, combined with the reconstitution of integrin activation in purified systems107, advanced imaging techniques such as FRET, super-resolution microscopy and fluorescence correlation spectroscopy81,127,128,141, all hold promise for addressing this question.
A second major question relates to the regulation of talin and kindlin functions. Again, more is known for talin, although the mechanisms remain ill defined. While Rap1A–RIAM recruits talin to the plasma membrane44–46 (FIG 5), it is unclear whether the RIAM binding sites in auto-inhibited talin are exposed and whether RIAM activates talin. Interestingly, the PIP-kinase type 1γ binding site in auto-inhibited talin is exposed65, so perhaps it targets talin to the plasma membrane (for example, as part of the integrin-containing exocyst complex78) (FIG 5), and synthesises the PIP2 that activates talin62 (FIG 5). However, if PIP-kinase type 1γ occupies talin F3, how does talin bind integrins? In principle, talin dimers can bind both molecules, but could this support integrin clustering? The affinity of talin for integrins is greatly increased by PIP255, so once talin has been delivered to the plasma membrane, PIP2 could shift the binding equilibrium in favour of talin–integrin complexes. Indeed, talin appears to be activated within FAs81, and PIP2 localised in FAs appear to be important in integrin adhesion and force coupling77. The roles of FAK40 and vinculin70,73 in recruiting talin to FAs, and in the regulation of talin-mediated integrin activation, also require further clarification (FIG 5).
Once integrin–talin complexes engage F-actin, force-induced conformational changes in the talin rod may displace RIAM and promote the vinculin binding required for adhesion maturation35,74 (FIG 5). It will be important to establish first, whether the 11 VBSs in the talin rod are differentially activated in response to increasing force, second, whether force relaxation allows the helical bundles in the talin rod to refold displacing vinculin and third, whether vinculin stablises FAs by cross-linking talin to actin or PIP2 or both? Both in vitro molecular tweezer approaches and in vivo FRET tension sensors may help to address these questions. In the case of kindlin, similar mechanisms are likely to regulate its activation, namely membrane binding, competition with other partners, phosphorylation and calpain cleavage109,110,113,116,142–144, and possibly even conformational rearrangements145, but these studies are at an early stage.
Ideas about the regulation and function of talins and kindlins obtained from in vitro or cell culture approaches need testing in whole organisms. While progress has been made using talin-knockout in D. melanogaster8 and conditional talin1 knockout mice (Table 1), studies in vertebrates are complicated by talin2, the function of which remains obscure130,146, although the fact that talin2 expression is tightly regulated by multiple pathways146 suggests an important function. Likewise, potential redundancy between kindlins can make interpretation of mouse and D. melanogaster kindlin knockout phenotypes difficult, and it will be important to assess the isoform-specific interactions and functions of the different kindlins. Although many questions remain unanswered, our understanding of the ‘integrin, talin, kindlin story’ has made remarkable progress in the 30 years since talin was first discovered. It will be fascinating to see how this impacts on our understanding of human diseases, and several recent reports suggest a role for talin-mediated integrin activation in metastasis147,148
TABLE 1.
Summary of Tln1 and Tln2 gene deletion experiments
| Genetic modification | Phenotypic effects | References |
|---|---|---|
| Tln1 | ||
| Tln1 knockout | Embryonic Lethal E8.5 due to gastrulation defects. | 152 |
| Tln1fl/fl; Tie2-Cre (Endothelial cells) | Lethal by E10.5 due to haemorrhage. Defects in angiogenesis and endothelial cell spreading in vivo. Development of heart and other tissues apparently unaffected. Thus, talin1 is not essential for the later stages of development in most tissues. | 12 |
| Tln1fl/fl; CreER Tamoxifen injected into pregnant mothers at E8.5, E9.5 or E10.5 | Angiogenesis defects and bleeding within 48hrs; death after 72hrs. Development of heart and other tissues apparently unaffected. | 12 |
| Tln1fl/fl; PF4-Cre (Megakaryocytes) | Defects in membrane tethering to the cytoskeleton; membrane blebbing. Phenocopied by knockout of the PIP-kinase type1γ isoform. | 153 |
| Tln1fl/fl; PF4-Cre (Platelets) | Spontaneous haemorrhaging. Prolonged bleeding times. Defective αIIbβ3 and α2β1 activation and platelet adhesion. | 57,58 |
| Tln1 L325R knockin (Platelets) | Defective agonist-induced integrin activation and fibrin clot retraction. | 59 |
| Tln1fl/fl;/CD4-Cre (T-lymphocytes) | Talin1 is required to stabilise the immune synapse and to support T-cell stopping and contact-dependent cell proliferation. | 154 |
| Tln1fl/fl;/CD19-Cre (B-lymphocytes) | Talin1 is required for integrin-dependent B-cell homing to lymph nodes, but is not required for follicular B-cell maturation in the spleen. | 155 |
| Tln1fl/fl;/Mx1-Cre (Leukocytes) | Dendritic cell (DC) migration in 3D is not integrin or talin1 dependent. However, DC extravasation from the blood, which involves adhesion to the endothelium, is integrin and talin dependent. | 156 |
| Tln1fl/fl;/Human skeletal α-actin-Cre (Skeletal muscle) | Myotendinous junction stability defect. Mild muscular dystrophy and reduced ability of muscle to generate force. | 157 |
| Tln1fl/fl;/Cathepsin K-Cre (Osteoclasts) | Impaired M-CSF-stimulated integrin activation, reduced adhesion and migration on ECM. Arrested osteoclast maturation into mature resorptive cells leading to increased bone mass. | 158 |
| Tln1fl/fl;/α-myosin heavy chain-Cre (Heart) | Talin2 is the predominant isoform in cardiomyocytes and talin1 is not required for heart development or basal function. Deletion of talin1 attenuates the hypertrophic response of heart to stress. | 159 |
| Tln2 | ||
| Tln2 gene traps | No true talin2 null alleles and no phenotype. | 130 |
| Tln2 knockout Complete gene deletion | Mice are viable and fertile although they are difficult to breed. Mildly dystrophic phenotype slightly more severe than muscle-specific Tln1 knockout. | 146 |
| Tln2 exon1 deletion | Ablates talin2 expression in skeletal muscle but substantial expression of talin2 in other tissues. Myotendinous junction defects. Mildly dystrophic phenotype slightly more severe than muscle-specific Tln1 knockout. | 160 |
| Tln1 and Tln2 | ||
| Tln1fl/fl;/Human skeletal α-actin-Cre; Tln2 exon1 deletion | Deletion of Tln1 and Tln2 from skeletal muscle results in defects in myoblast fusion and sarcomere assembly. Myoblasts still express active β1-integrins, but show defects in coupling integrins to cytoskeletal actin. | 160 |
The system or cell type in which the gene was modified is shown in brackets.
Tln1fl/fl indicates that both copies of the Tln1 gene contain a pair of LoxP. Crossing these mice with mice expressing Cre-recombinase from a tissue-specific promoter allows for deletion of the Tln1 gene in selected tissues. Cre-ER is a ubiquitously expressed form of Cre-recombinase that can be activated by injecting tamoxifen into the animal.
The phenotypes of mice lacking the various integrin subunits, and integrin activators (including kindlins) and inhibitors, is summarised elsewhere161.
Acknowledgments
Work in our laboratories was supported by the National Institutes of Health, the Medical Research Council, the Wellcome Trust, Cancer Research UK and the Medical Research Council. We thank members of the Calderwood lab for their input during preparation of this review, Ben Goult for help in preparing Figure 1, and Antreas Kalli for contributions to Figure 2c. We apologize to colleagues whose work was not cited or fully discussed due to space limitations.
Glossary
- Leukocyte adhesion deficiency type III
LAD-III is a rare genetic disease characterized by severe bacterial infections and bleeding disorders. It is caused by mutations in the gene encoding kindlin-3 in hematopoietic cells
- FERM domains
found in various cytoskeletal-associated proteins, including band 4.1, ezrin, radixin and moesin, the proteins after which this domain was named. One role of these domains is to localize proteins to the plasma membrane. FERM domains typically contain three sub-domains, F1, F2 and F3, normally arranged in a cloverleaf formation
- Multiscale molecular dynamics (MD) simulations
these numerically solve Newton’s equations for interacting particles and follow their trajectories. MD has become a powerful way of fine-tuning protein structure and dynamics. Major limitations are the relatively small size of the systems and the short timescales of the trajectories that can be studied. Multiscale simulations that combine simplified representations of molecules (coarse grain) with all atom (atomistic) representations circumvent some limitations
- Negative headgroup
Lipid headgroups come in various forms with various charged properties. For example, phosphatidyl serine and phosphatidylinositol 4,5-bisphosphate (PIP2) have a net negative charge that promotes the binding of some proteins, for example talin, to the membrane
- RA domain
The Ras association domain (RA) is found in many effector proteins for the Ras family of small GTPases. The domain adopts a ubiquitin-like fold and supports binding of proteins containing an RA domain to ‘active’ GTP-loaded Ras family proteins
- PH domain
The pleckstrin homology (PH) domain is found in a wide range of intracellular signalling proteins. It often binds to membranes via interactions with PIP2
- PTB domain
The phosphotyrosine binding (PTB) domain is found in a wide range of intracellular signalling proteins. It commonly binds NPxY motifs; the Y may, or may not, require phosphorylation to support binding
- KRIT1
K Ras Interaction Trapped-1, the product of the CCM1 gene, is a multi-domain adaptor protein important for cell–cell and cell matrix adhesion. Loss-of-function mutations in CCM1 cause predisposition to cerebral cavernous malformations, neurovascular anomalies that increase the risk of haemorrhagic stroke
- Exocyst complex
An octameric protein complex involved in vesicle trafficking. It targets post-Golgi vesicles to the plasma membrane prior to vesicle fusion
- Nanodiscs
A nanodisc is a synthetic model membrane system made from lipids and a scaffold protein. They are useful for the study of incorporated membrane proteins, such as integrins, because they are relatively small, monodisperse and homogenous, yet provide a native-like environment
- SAXS
small angle X-ray scattering is a technique that gives low-resolution information about the shape of objects. It depends on analysis of the angular intensity distribution of X-rays scattered by molecules in solution
Biographies
David Calderwood is an Associate Professor at Yale University School of Medicine, where his group studies integrin activation, signaling and the link to the cytoskeleton. He earned his Ph.D. in Martin Humphries’ laboratory at the University of Manchester, U.K., investigating integrin–ligand interactions. In his postdoctoral work with Mark Ginsberg at The Scripps Research Institute, LA Jolla, CA, USA, he identified talin as a key regulator of integrin activation.
Iain Campbell is Emeritus Professor of Structural Biology at the University of Oxford. Trained as a physicist at St Andrews University, he had three main stages in his research career: first developing NMR methods for studying biological systems, second determining the structure of numerous recurring protein module structures, such as Epidermal Growth Factor and, lastly, focusing on structures and interactions associated with focal adhesions.
David Critchley is Emeritus Professor of Biochemistry at the University of Leicester, where he led a multidisciplinary team studying the structure and function of focal adhesion proteins, including talin, vinculin and α-actinin.
Contributor Information
David A Calderwood, Email: david.calderwood@yale.edu, Departments of Pharmacology and of Cell Biology, Yale University School of Medicine, New Haven, CT, USA.
Iain D Campbell, Email: Iain.campbell@bioch.ox.ac.uk, Department of Biochemistry, University of Oxford, S. Parks Rd., Oxford, OX1 3QU, UK.
David R Critchley, Email: drc@le.ac.uk, Department of Biochemistry, University of Leicester, Leicester LE1 7RH.
References
- 1.Burridge K, Connell L. A New Protein of Adhesion plaques and ruffling membranes. J Cell Biol. 1983;97:359–367. doi: 10.1083/jcb.97.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys. 2009;38:235–54. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 3.Calderwood DA, et al. The talin head domain binds to integrin b subunit cytoplasmic tails and regulates integrin activation. J Biol Chem. 1999;274:28071–28704. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 4.Tadokoro S, et al. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 2003;302:103–6. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Alvarez B, et al. Structural determinants of integrin recognition by talin. Mol Cell. 2003;11:49–58. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- 6.Wegener KL, et al. Structural basis of integrin activation by talin. Cell. 2007;128:171–82. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 7.Calderwood DA. Integrin activation. J Cell Sci. 2004;117:657–66. doi: 10.1242/jcs.01014. [DOI] [PubMed] [Google Scholar]
- 8.Brown NH, et al. Talin is essential for integrin function in Drosophila. Dev Cell. 2002;3:569–579. doi: 10.1016/s1534-5807(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 9.Cram EJ, Clark SG, Schwarzbauer JE. Talin loss-of-function uncovers roles in cell contractility and migration in C. elegans. J Cell Sci. 2003;116:3871–8. doi: 10.1242/jcs.00705. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, et al. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat Cell Biol. 2008;10:1062–8. doi: 10.1038/ncb1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopp PM, et al. Studies on the morphology and spreading of human endothelial cells define key inter- and intramolecular interactions for talin1. Eur J Cell Biol. 2010;89:661–73. doi: 10.1016/j.ejcb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monkley SJ, et al. Endothelial cell talin1 is essential for embryonic angiogenesis. Dev Biol. 2011;349:494–502. doi: 10.1016/j.ydbio.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moser M, Nieswandt B, Ussar S, Pozgajova M, Fassler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med. 2008;14:325–30. doi: 10.1038/nm1722. First demonstration that loss of kindlin-3 results in severe defects in platelet integrin activation, revealing the importance of kindlins in integrin activation. [DOI] [PubMed] [Google Scholar]
- 14.Rogalski TM, Mullen GP, Gilbert MM, Williams BD, Moerman DG. The UNC-112 gene in Caenorhabditis elegans encodes a novel component of cell-matrix adhesion structures required for integrin localization in the muscle cell membrane. J Cell Biol. 2000;150:253–64. doi: 10.1083/jcb.150.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegel DH, et al. Loss of kindlin-1, a human homolog of the Caenorhabditis elegans actin-extracellular-matrix linker protein UNC-112, causes Kindler syndrome. Am J Hum Genet. 2003;73:174–87. doi: 10.1086/376609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jobard F, et al. Identification of mutations in a new gene encoding a FERM family protein with a pleckstrin homology domain in Kindler syndrome. Human molecular genetics. 2003;12:925–35. doi: 10.1093/hmg/ddg097. [DOI] [PubMed] [Google Scholar]
- 17.Karakose E, Schiller HB, Fassler R. The kindlins at a glance. Journal of cell science. 2010;123:2353–6. doi: 10.1242/jcs.064600. [DOI] [PubMed] [Google Scholar]
- 18.Kloeker S, et al. The Kindler syndrome protein is regulated by transforming growth factor-beta and involved in integrin-mediated adhesion. J Biol Chem. 2004;279:6824–33. doi: 10.1074/jbc.M307978200. [DOI] [PubMed] [Google Scholar]
- 19.Plow EF, Qin J, Byzova T. Kindling the flame of integrin activation and function with kindlins. Curr Opin Hematol. 2009;16:323–8. doi: 10.1097/MOH.0b013e32832ea389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye F, Kim C, Ginsberg MH. Molecular mechanism of inside-out integrin regulation. Journal of thrombosis and haemostasis : JTH. 2011;9 (Suppl 1):20–5. doi: 10.1111/j.1538-7836.2011.04355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anthis NJ, Campbell ID. The tail of integrin activation. Trends Biochem Sci. 2011;36:191–198. doi: 10.1016/j.tibs.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: the end game. Nat Rev Mol Cell Biol. 2010;11:288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng X, Nelson ES, Maiers JL, DeMali KA. New insights into vinculin function and regulation. Int Rev Cell Mol Biol. 2011;287:191–231. doi: 10.1016/B978-0-12-386043-9.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–60. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Winkler J, Lunsdorf H, Jockusch BM. Energy-filtered electron microscopy reveals that talin is a highly flexible protein composed of a series of globular domains. Eur J Biochem. 1997;243:430–436. doi: 10.1111/j.1432-1033.1997.0430a.x. [DOI] [PubMed] [Google Scholar]
- 26.Elliott PR, et al. The Structure of the talin head reveals a novel extended conformation of the FERM domain. Structure (London, England : 1993) 2010;18:1289–99. doi: 10.1016/j.str.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gingras AR, et al. The structure of the C-terminal actin-binding domain of talin. EMBO J. 2008;27:458–69. doi: 10.1038/sj.emboj.7601965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gingras AR, et al. Central region of talin has a unique fold that binds vinculin and actin. J Biol Chem. 2010;285:29577–87. doi: 10.1074/jbc.M109.095455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gingras AR, et al. Structural and dynamic characterization of a vinculin binding site in the talin rod. Biochemistry. 2006;45:1805–17. doi: 10.1021/bi052136l. [DOI] [PubMed] [Google Scholar]
- 30.Gingras AR, et al. Structural determinants of integrin binding to the talin rod. J Biol Chem. 2009;284:8866–76. doi: 10.1074/jbc.M805937200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gingras AR, et al. Mapping and Consensus Sequence Identification for Multiple Vinculin Binding Sites within the Talin Rod. J Biol Chem. 2005;280:37217–24. doi: 10.1074/jbc.M508060200. [DOI] [PubMed] [Google Scholar]
- 32.Goult BT, et al. Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation. EMBO J. 2010;29:1069–80. doi: 10.1038/emboj.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goult BT, et al. The domain structure of talin: residues 1815–1973 form a five-helix bundle containing a cryptic vinculin-binding site. FEBS Lett. 2010;584:2237–41. doi: 10.1016/j.febslet.2010.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papagrigoriou E, et al. Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. EMBO J. 2004;23:2942–2951. doi: 10.1038/sj.emboj.7600285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goult BT, et al. RIAM and Vinculin Binding to Talin Are Mutually Exclusive and Regulate Adhesion Assembly and Turnover. J Biol Chem. 2013;288:8238–49. doi: 10.1074/jbc.M112.438119. Presents a structural model for full-length talin and provides a structural basis for a switch from talin–RIAM to talin–vinculin complexes during adhesion assembly and maturation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anthis NJ, Wegener KL, Critchley DR, Campbell ID. Structural diversity in integrin/talin interactions. Structure. 2010;18:1654–66. doi: 10.1016/j.str.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anthis NJ, et al. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 2009;28:3623–32. doi: 10.1038/emboj.2009.287. The first structure of a full-length β integrin tail bound to the talin F2F3 FERM domains. This, combined with biophysical studies and integrin activation assays, reveals that talin F3 binding to the membrane-proximal helix of the β tail disrupts an inhibitory α tail–β tail interaction, and identifies an important positively charged membrane binding surface on talin F2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wegener KL, et al. Structural Basis for the Interaction between the Cytoplasmic Domain of the Hyaluronate Receptor Layilin and the Talin F3 Subdomain. J Mol Biol. 2008;382:112–126. doi: 10.1016/j.jmb.2008.06.087. [DOI] [PubMed] [Google Scholar]
- 39.de Pereda JM, et al. Structural basis for phosphatidylinositol phosphate kinase type Igamma binding to talin at focal adhesions. J Biol Chem. 2005;280:8381–6. doi: 10.1074/jbc.M413180200. [DOI] [PubMed] [Google Scholar]
- 40.Lawson C, et al. FAK promotes recruitment of talin to nascent adhesions to control cell motility. J Cell Biol. 2012;196:223–32. doi: 10.1083/jcb.201108078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S, et al. Tiam1 interaction with the PAR complex promotes talin-mediated Rac1 activation during polarized cell migration. J Cell Biol. 2012;199:331–45. doi: 10.1083/jcb.201202041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bate N, et al. Talin contains a C-terminal calpain2 cleavage site important in focal adhesion dynamics. PLoS One. 2012;7:e34461. doi: 10.1371/journal.pone.0034461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodius S, et al. The talin rod IBS2 alpha-helix interacts with the beta3 integrin cytoplasmic tail membrane-proximal helix by establishing charge complementary salt bridges. J Biol Chem. 2008;283:24212–23. doi: 10.1074/jbc.M709704200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han J, et al. Reconstructing and Deconstructing Agonist-Induced Activation of Integrin alphaIIbbeta3. Curr Biol. 2006;16:1796–806. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 45.Lee HS, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J Biol Chem. 2009;284:5119–27. doi: 10.1074/jbc.M807117200. Provides the first insights into how the Rap1A effector RIAM binds to talin and recruits it to the membrane to activate integrins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe N, et al. Mechanisms and consequences of agonist-induced talin recruitment to platelet integrin alphaIIbbeta3. J Cell Biol. 2008;181:1211–22. doi: 10.1083/jcb.200803094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li G, et al. Full activity of the deleted in liver cancer 1 (DLC1) tumor suppressor depends on an LD-like motif that binds talin and focal adhesion kinase (FAK) Proc Natl Acad Sci U S A. 2011;108:17129–34. doi: 10.1073/pnas.1112122108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun N, Critchley DR, Paulin D, Li Z, Robson RM. Identification of a repeated domain within mammalian alpha-synemin that interacts directly with talin. Exp Cell Res. 2008;314:1839–1849. doi: 10.1016/j.yexcr.2008.01.034. [DOI] [PubMed] [Google Scholar]
- 49.Petrich BG. Talin-dependent integrin signalling in vivo. Thromb Haemost. 2009;101:1020–4. [PubMed] [Google Scholar]
- 50.Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28:1351–61. doi: 10.1038/emboj.2009.63. An NMR structure that provides key information about the complex formed by the membrane spanning helices in the inactive integrin state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu J, et al. The structure of a receptor with two associating transmembrane domains on the cell surface: integrin alphaIIbbeta3. Mol Cell. 2009;34:234–49. doi: 10.1016/j.molcel.2009.02.022. A stucture of the membrane spanning region of an intact integrin, determined by disulfide crosslinking and molecular modelling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim C, et al. Basic amino-acid side chains regulate transmembrane integrin signalling. Nature. 2011;481:209–13. doi: 10.1038/nature10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalli AC, Campbell ID, Sansom MSP. Multiscale simulations suggest a mechanism for integrin inside-out activation. Proc Natl Acad Sci U S A. 2011;108:11890–5. doi: 10.1073/pnas.1104505108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim C, Ye F, Hu X, Ginsberg MH. Talin activates integrins by altering the topology of the beta transmembrane domain. J Cell Biol. 2012;197:605–11. doi: 10.1083/jcb.201112141. Demonstrates, using environmentally sensitive fluorophores, that talin binding to the integrin β transmembrane domain (TMD) alters the membrane embedding of the β TMD (see also ref 52) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moore DT, et al. Affinity of talin-1 for the beta3-integrin cytosolic domain is modulated by its phospholipid bilayer environment. Proc Natl Acad Sci U S A. 2012;109:793–8. doi: 10.1073/pnas.1117220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lefort CT, et al. Distinct roles for talin-1 and kindlin-3 in LFA-1 extension and affinity regulation. Blood. 2012;119:4275–82. doi: 10.1182/blood-2011-08-373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nieswandt B, et al. Loss of talin1 in platelets abrogates integrin activation, platelet aggregation, and thrombus formation in vitro and in vivo. J Exp Med. 2007;204:3113–8. doi: 10.1084/jem.20071827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petrich BG, et al. Talin is required for integrin-mediated platelet function in hemostasis and thrombosis. J Exp Med. 2007;204:3103–11. doi: 10.1084/jem.20071800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haling JR, Monkley SJ, Critchley DR, Petrich BG. Talin-dependent integrin activation is required for fibrin clot retraction by platelets. Blood. 2011;117:1719–22. doi: 10.1182/blood-2010-09-305433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–5. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 61.Bouaouina M, Harburger DS, Calderwood DA. Talin and signaling through integrins. Methods Mol Biol. 2012;757:325–47. doi: 10.1007/978-1-61779-166-6_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goksoy E, et al. Structural basis for the autoinhibition of talin in regulating integrin activation. Mol Cell. 2008;31:124–33. doi: 10.1016/j.molcel.2008.06.011. The first biochemical and structural data to show how binding of the talin head to the rod occludes the integrin binding site in the head, and how talin auto-inhibition might be relieved by PIP2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goult B, et al. Structural studies on full-length talin1 reveal a compact auto-inhibited dimer: Implications for talin activation. J Struct Biol. 2013 May 30; doi: 10.1016/j.jsb.2013.05.014. pii: S1047-8477(13)00146-9 Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goult BT, et al. The structure of an interdomain complex that regulates talin activity. J Biol Chem. 2009;284:15097–106. doi: 10.1074/jbc.M900078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song X, et al. A novel membrane-dependent on/off switch mechanism of talin FERM domain at sites of cell adhesion. Cell Res. 2012;22:1533–1545. doi: 10.1038/cr.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saltel F, et al. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. J Cell Biol. 2009;187:715–31. doi: 10.1083/jcb.200908134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Helsten TL, et al. Differences in regulation of Drosophila and vertebrate integrin affinity by talin. Mol Biol Cell. 2008;19:3589–98. doi: 10.1091/mbc.E08-01-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bunch TA. Integrin alphaIIbbeta3 activation in Chinese hamster ovary cells and platelets increases clustering rather than affinity. J Biol Chem. 2010;285:1841–9. doi: 10.1074/jbc.M109.057349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Molony L, Mccaslin D, Abernethy J, Paschal B, Burridge K. Properties of talin from chicken gizzard smooth-muscle. J Biol Chem. 1987;262:7790–7795. [PubMed] [Google Scholar]
- 70.Banno A, et al. Subcellular localization of talin is regulated by inter-domain interactions. J Biol Chem. 2012;287:13799–13812. doi: 10.1074/jbc.M112.341214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bakolitsa C, et al. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–6. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- 72.Humphries JD, et al. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J Cell Biol. 2007;179:1043–57. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carisey A, et al. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr Biol. 2013;23:271–81. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee HS, Anekal P, Lim CJ, Liu CC, Ginsberg MH. Two Modes of Integrin Activation Form a Binary Molecular Switch in Adhesion Maturation. Mol Biol Cell. 2013;24:1354–1362. doi: 10.1091/mbc.E12-09-0695. Provides evidence that vinculin displaces RIAM from talin, and that this drives the transition from dynamic RIAM-positive nascent adhesions that support membrane protrusion to more stable vinculin-rich force-bearing FAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Colo GP, Lafuente EM, Teixido J. The MRL proteins: Adapting cell adhesion, migration and growth. Eur J Cell Biol. 2012;91:861–8. doi: 10.1016/j.ejcb.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 76.Wynne JP, et al. Rap1-interacting adapter molecule (RIAM) associates with the plasma membrane via a proximity detector. J Cell Biol. 2012;199:317–29. doi: 10.1083/jcb.201201157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Legate KR, et al. Integrin adhesion and force coupling are independently regulated by localized PtdIns(4,5)2 synthesis. EMBO J. 2011;30:4539–53. doi: 10.1038/emboj.2011.332. Provides definitive evidence that PIP2 generated by the talin-binding isoform of PIP-kinase type 1γ in focal adhesions is essential for formation of initial integrin-mediated cell attachment, and for the subsequent linkage of integrins to cytoskeletal actin and the exertion of force on matrix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thapa N, et al. Phosphoinositide signaling regulates the exocyst complex and polarized integrin trafficking in directionally migrating cells. Developmental cell. 2012;22:116–30. doi: 10.1016/j.devcel.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li X, et al. Ubiquitination of PIPKIgamma90 by HECTD1 regulates focal adhesion dynamics and cell migration. J Cell Sci. 2013 doi: 10.1242/jcs.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller NL, Lawson C, Chen XL, Lim ST, Schlaepfer DD. Rgnef (p190RhoGEF) knockout inhibits RhoA activity, focal adhesion establishment, and cell motility downstream of integrins. PLoS One. 2012;7:e37830. doi: 10.1371/journal.pone.0037830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rossier O, et al. Integrins beta(1) and beta(3) exhibit distinct dynamic nanoscale organizations inside focal adhesions. Nat Cell Biol. 2012;14:1057–67. doi: 10.1038/ncb2588. [DOI] [PubMed] [Google Scholar]
- 82.Wang P, Ballestrem C, Streuli CH. The C-terminus of talin links integrins to cell cycle progression. J Cell Biol. 2011;195:499–513. doi: 10.1083/jcb.201104128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pouwels J, Nevo J, Pellinen T, Ylanne J, Ivaska J. Negative regulators of integrin activity. J Cell Sci. 2012;125:3271–80. doi: 10.1242/jcs.093641. [DOI] [PubMed] [Google Scholar]
- 84.Anthis NJ, et al. Beta integrin tyrosine phosphorylation is a conserved mechanism for regulating talin-induced integrin activation. J Biol Chem. 2009;284:36700–10. doi: 10.1074/jbc.M109.061275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oxley CL, et al. An integrin phosphorylation switch: the effect of beta3 integrin tail phosphorylation on Dok1 and talin binding. J Biol Chem. 2008;283:5420–6. doi: 10.1074/jbc.M709435200. [DOI] [PubMed] [Google Scholar]
- 86.Calderwood DA, et al. Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc Natl Acad Sci U S A. 2003;100:2272–7. doi: 10.1073/pnas.262791999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kiema T, et al. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21:337–47. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 88.Ithychanda SS, et al. Migfilin, a molecular switch in regulation of integrin activation. J Biol Chem. 2009;284:4713–22. doi: 10.1074/jbc.M807719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lad Y, et al. Structural basis of the migfilin-filamin interaction and competition with integrin beta tails. J Biol Chem. 2008;283:35154–63. doi: 10.1074/jbc.M802592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Das M, Ithychanda SS, Qin J, Plow EF. Migfilin and filamin as regulators of integrin activation in endothelial cells and neutrophils. PloS one. 2011;6:e26355. doi: 10.1371/journal.pone.0026355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moik DV, Janbandhu VC, Fassler R. Loss of migfilin expression has no overt consequences on murine development and homeostasis. J Cell Sci. 2011;124:414–21. doi: 10.1242/jcs.075960. [DOI] [PubMed] [Google Scholar]
- 92.Xiao G, et al. Critical role of filamin-binding LIM protein 1 (FBLP-1)/migfilin in regulation of bone remodeling. J Biol Chem. 2012;287:21450–60. doi: 10.1074/jbc.M111.331249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takala H, et al. Beta2 integrin phosphorylation on Thr758 acts as a molecular switch to regulate 14-3-3 and filamin binding. Blood. 2008;112:1853–62. doi: 10.1182/blood-2007-12-127795. [DOI] [PubMed] [Google Scholar]
- 94.Millon-Fremillon A, et al. Cell adaptive response to extracellular matrix density is controlled by ICAP-1-dependent beta1-integrin affinity. J Cell Biol. 2008;180:427–41. doi: 10.1083/jcb.200707142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu W, Draheim KM, Zhang R, Calderwood DA, Boggon TJ. Mechanism for KRIT1 Release of ICAP1-Mediated Suppression of Integrin Activation. Mol Cell. 2013;49:719–729. doi: 10.1016/j.molcel.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Roca-Cusachs P, et al. Integrin-dependent force transmission to the extracellular matrix by alpha-actinin triggers adhesion maturation. Proc Natl Acad Sci USA. 2013;110:E1361–70. doi: 10.1073/pnas.1220723110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tadokoro S, et al. A potential role for alpha-actinin in inside-out alphaIIbbeta3 signaling. Blood. 2011;117:250–8. doi: 10.1182/blood-2009-10-246751. [DOI] [PubMed] [Google Scholar]
- 98.Calderwood DA, Tai V, Di Paolo G, De Camilli P, Ginsberg MH. Competition for talin results in trans-dominant inhibition of integrin activation. J Biol Chem. 2004;279:28889–95. doi: 10.1074/jbc.M402161200. [DOI] [PubMed] [Google Scholar]
- 99.Bouaouina M, Calderwood DA. Kindlins. Current biology : CB. 2011;21:R99–101. doi: 10.1016/j.cub.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 100.Ye F, Petrich BG. Kindlin: helper, co-activator, or booster of talin in integrin activation? Curr Opin Hematol. 2011;18:356–60. doi: 10.1097/MOH.0b013e3283497f09. [DOI] [PubMed] [Google Scholar]
- 101.Harburger DS, Bouaouina M, Calderwood DA. Kindlin-1 and -2 directly bind the C-terminal region of beta integrin cytoplasmic tails and exert integrin-specific activation effects. J Biol Chem. 2009;284:11485–97. doi: 10.1074/jbc.M809233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ma YQ, Qin J, Wu C, Plow EF. Kindlin-2 (Mig-2): a co-activator of beta3 integrins. J Cell Biol. 2008;181:439–46. doi: 10.1083/jcb.200710196. Early evidence for cooperativity between the talin head and kindlin-2 in integrin activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Montanez E, et al. Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 2008;22:1325–30. doi: 10.1101/gad.469408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Malinin NL, et al. A point mutation in KINDLIN3 ablates activation of three integrin subfamilies in humans. Nat Med. 2009;15:313–8. doi: 10.1038/nm.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qu H, et al. Kindlin-2 regulates podocyte adhesion and fibronectin matrix deposition through interactions with phosphoinositides and integrins. J Cell Sci. 2011;124:879–91. doi: 10.1242/jcs.076976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bledzka K, et al. Spatial coordination of kindlin-2 with talin head domain in interaction with integrin beta cytoplasmic tails. J Biol Chem. 2012;287:24585–94. doi: 10.1074/jbc.M111.336743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ye F, et al. Recreation of the terminal events in physiological integrin activation. J Cell Biol. 2010;188:157–73. doi: 10.1083/jcb.200908045. Establishes that purified talin components, in an in vitro nanodisc system, are sufficient to activate isolated membrane-embedded integrins, even in the absence of kindlin. Also provides support for a model where integrins become more ‘upright’ when activated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Goult BT, et al. The Structure of the N-Terminus of Kindlin-1: A Domain Important for alphaIIbbeta3 Integrin Activation. J Mol Biol. 2009;394:944–56. doi: 10.1016/j.jmb.2009.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Perera HD, et al. Membrane binding of the N-terminal ubiquitin-like domain of kindlin-2 is crucial for its regulation of integrin activation. Structure. 2011;19:1664–71. doi: 10.1016/j.str.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu J, et al. Structural basis of phosphoinositide binding to kindlin-2 protein pleckstrin homology domain in regulating integrin activation. J Biol Chem. 2011;286:43334–42. doi: 10.1074/jbc.M111.295352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu Y, Zhu Y, Ye S, Zhang R. Crystal structure of kindlin-2 PH domain reveals a conformational transition for its membrane anchoring and regulation of integrin activation. Protein & cell. 2012;3:434–40. doi: 10.1007/s13238-012-2046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yates LA, et al. Structural and functional characterisation of the kindlin-1 pleckstrin homology domain. J Biol Chem. 2012;287:43246–61. doi: 10.1074/jbc.M112.422089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yates LA, Fuzery AK, Bonet R, Campbell ID, Gilbert RJC. Biophysical Analysis of Kindlin-3 Reveals an Elongated Conformation and Maps Integrin Binding to the Membrane-distal beta-Subunit NPXY Motif. J Biol Chem. 2012;287:37715–31. doi: 10.1074/jbc.M112.415208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moser M, Legate KR, Zent R, Fassler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–9. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 115.Kahner BN, et al. Kindlins, integrin activation and the regulation of talin recruitment to alphaIIbbeta3. PLoS One. 2012;7:e34056. doi: 10.1371/journal.pone.0034056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bouaouina M, et al. A conserved lipid-binding loop in the kindlin FERM F1 domain is required for kindlin-mediated alphaIIbbeta3 integrin coactivation. J Biol Chem. 2012;287:6979–90. doi: 10.1074/jbc.M111.330845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Honda S, et al. Integrin-linked kinase associated with integrin activation. Blood. 2009;113:5304–13. doi: 10.1182/blood-2008-07-169136. [DOI] [PubMed] [Google Scholar]
- 118.Bandyopadhyay A, Rothschild G, Kim S, Calderwood DA, Raghavan S. Functional differences between kindlin-1 and kindlin-2 in keratinocytes. J Cell Sci. 2012;125:2172–84. doi: 10.1242/jcs.096214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bialkowska K, et al. The integrin co-activator Kindlin-3 is expressed and functional in a non-hematopoietic cell, the endothelial cell. J Biol Chem. 2010;285:18640–9. doi: 10.1074/jbc.M109.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ussar S, et al. Loss of Kindlin-1 causes skin atrophy and lethal neonatal intestinal epithelial dysfunction. PLoS Genet. 2008;4:e1000289. doi: 10.1371/journal.pgen.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roca-Cusachs P, Gauthier NC, Del Rio A, Sheetz MP. Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v)beta(3) and talin enable mechanotransduction. Proc Natl Acad Sci USA. 2009;106:16245–50. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.del Rio A, et al. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–41. doi: 10.1126/science.1162912. Use of magnetic tweezers, total internal reflection fluorescence, and atomic force microscopy showed that physiologically relevant forces cause stretching of talin and result in exposure of cryptic vinculin-binding sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J Cell Biol. 2010;188:877–90. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Margadant F, et al. Mechanotransduction in vivo by repeated talin stretch-relaxation events depends upon vinculin. PLoS Biol. 2011;9:e1001223. doi: 10.1371/journal.pbio.1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Smith SJ, McCann RO. A C-Terminal Dimerization Motif Is Required for Focal Adhesion Targeting of Talin1 and the Interaction of the Talin1 I/LWEQ Module with F-Actin. Biochemistry. 2007;46:10886–98. doi: 10.1021/bi700637a. [DOI] [PubMed] [Google Scholar]
- 126.Ellis SJ, Pines M, Fairchild MJ, Tanentzapf G. In vivo functional analysis reveals specific roles for the integrin-binding sites of talin. J Cell Sci. 2011;124:1844–56. doi: 10.1242/jcs.083337. [DOI] [PubMed] [Google Scholar]
- 127.Parsons M, Messent AJ, Humphries JD, Deakin NO, Humphries MJ. Quantification of integrin receptor agonism by fluorescence lifetime imaging. J Cell Sci. 2008;121:265–71. doi: 10.1242/jcs.018440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–4. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang F, Saha S, Kashina A. Arginylation-dependent regulation of a proteolytic product of talin is essential for cell-cell adhesion. J Cell Biol. 2012;197:819–36. doi: 10.1083/jcb.201112129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Debrand E, et al. Talin 2 is a large and complex gene encoding multiple transcripts and protein isoforms. FEBS J. 2009;276:1610–28. doi: 10.1111/j.1742-4658.2009.06893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Franco SJ, et al. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat Cell Biol. 2004;6:977–83. doi: 10.1038/ncb1175. [DOI] [PubMed] [Google Scholar]
- 132.Huang C, et al. Talin phosphorylation by Cdk5 regulates Smurf1-mediated talin head ubiquitylation and cell migration. Nat Cell Biol. 2009;11:624–30. doi: 10.1038/ncb1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bridgewater RE, Norman JC, Caswell PT. Integrin trafficking at a glance. J Cell Sci. 2012;125:3695–701. doi: 10.1242/jcs.095810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Albiges-Rizo C, Frachet P, Block MR. Down regulation of talin alters cell adhesion and the processing of the a5b1 integrin. J Cell Sci. 1995;108:3317–3329. doi: 10.1242/jcs.108.10.3317. [DOI] [PubMed] [Google Scholar]
- 135.Martel V, et al. Talin controls the exit of the integrin a5b1 from an early compartment of the secretory pathway. J Cell Sci. 2000;113:1951–1961. doi: 10.1242/jcs.113.11.1951. [DOI] [PubMed] [Google Scholar]
- 136.Priddle H, et al. Disruption of the talin gene compromises focal adhesion assembly in undifferentiated but not differentiated ES cells. J Cell Biol. 1998;142:1121–1133. doi: 10.1083/jcb.142.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu J, et al. Talin1 regulates integrin turnover to promote embryonic epithelial morphogenesis. Mol Cell Biol. 2011;31:3366–77. doi: 10.1128/MCB.01403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Margadant C, Kreft M, de Groot DJ, Norman JC, Sonnenberg A. Distinct Roles of Talin and Kindlin in Regulating Integrin alpha5beta1 Function and Trafficking. Curr Biol. 2012;22:1554–63. doi: 10.1016/j.cub.2012.06.060. [DOI] [PubMed] [Google Scholar]
- 139.Bottcher RT, et al. Sorting nexin 17 prevents lysosomal degradation of beta1 integrins by binding to the beta1-integrin tail. Nat Cell Biol. 2012;14:584–92. doi: 10.1038/ncb2501. [DOI] [PubMed] [Google Scholar]
- 140.Steinberg F, Heesom KJ, Bass MD, Cullen PJ. SNX17 protects integrins from degradation by sorting between lysosomal and recycling pathways. J Cell Biol. 2012;197:219–30. doi: 10.1083/jcb.201111121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Choi CK, Zareno J, Digman MA, Gratton E, Horwitz AR. Cross-correlated fluctuation analysis reveals phosphorylation-regulated paxillin-FAK complexes in nascent adhesions. Biophys J. 2011;100:583–92. doi: 10.1016/j.bpj.2010.12.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhao Y, et al. Regulation of cell adhesion and migration by Kindlin-3 cleavage by calpain. J Biol Chem. 2012;287:40012–20. doi: 10.1074/jbc.M112.380469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bledzka K, et al. Tyrosine phosphorylation of integrin beta3 regulates kindlin-2 binding and integrin activation. J Biol Chem. 2010;285:30370–4. doi: 10.1074/jbc.C110.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu Y, Zhu Y, Ye S, Zhang R. Crystal structure of kindlin-2 PH domain reveals a conformational transition for its membrane anchoring and regulation of integrin activation. Protein Cell. 2012;3:434–40. doi: 10.1007/s13238-012-2046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qadota H, Moerman DG, Benian GM. A molecular mechanism for the requirement of PAT-4 (integrin-linked kinase (ILK)) for the localization of UNC-112 (Kindlin) to integrin adhesion sites. J Biol Chem. 2012;287:28537–51. doi: 10.1074/jbc.M112.354852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Debrand E, et al. Mice carrying a complete deletion of the talin2 coding sequence are viable and fertile. Biochem Biophys Res Commun. 2012;426:190–5. doi: 10.1016/j.bbrc.2012.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Desiniotis A, Kyprianou N. Significance of talin in cancer progression and metastasis. Int Rev Cell Mol Biol. 2011;289:117–47. doi: 10.1016/B978-0-12-386039-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kato H, et al. The primacy of beta1 integrin activation in the metastatic cascade. PLoS One. 2012;7:e46576. doi: 10.1371/journal.pone.0046576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 150.Dong X, et al. alpha(V)beta(3) integrin crystal structures and their functional implications. Biochemistry. 2012;51:8814–28. doi: 10.1021/bi300734n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bouaouina M, et al. Zasp regulates integrin activation. J Cell Sci. 2012 doi: 10.1242/jcs.103291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Monkley SJ, et al. Disruption of the talin gene arrests mouse development at the gastrulation stage. Dev Dynamics. 2000;219:560–574. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1079>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 153.Wang Y, et al. Loss of PIP5KIgamma, unlike other PIP5KI isoforms, impairs the integrity of the membrane cytoskeleton in murine megakaryocytes. J Clin Invest. 2008;118:812–9. doi: 10.1172/JCI34239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wernimont SA, et al. Contact-Dependent T Cell Activation and T Cell Stopping Require Talin1. J Immunol. 2011;187:6256–6267. doi: 10.4049/jimmunol.1102028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Manevich-Mendelson E, et al. Talin1 is required for integrin-dependent B lymphocyte homing to lymph nodes and the bone marrow but not for follicular B-cell maturation in the spleen. Blood. 2010;116:5907–18. doi: 10.1182/blood-2010-06-293506. [DOI] [PubMed] [Google Scholar]
- 156.Lammermann T, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–5. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 157.Conti FJ, et al. Progressive myopathy and defects in the maintenance of myotendinous junctions in mice that lack talin 1 in skeletal muscle. Development. 2008;135:2043–53. doi: 10.1242/dev.015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zou W, et al. Talin1 and Rap1 are Critical for Osteoclast Function. Mol Cell Biol. 2012;33:830–844. doi: 10.1128/MCB.00790-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Manso A, et al. Talin1 has unique expression versus talin 2 in the heart and modifies the hypertrophic response to pressure overload. J Biol Chem. 2013;288:4252–4264. doi: 10.1074/jbc.M112.427484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Conti FJ, Monkley SJ, Wood MR, Critchley DR, Muller U. Talin 1 and 2 are required for myoblast fusion, sarcomere assembly and the maintenance of myotendinous junctions. Development. 2009;136:3597–606. doi: 10.1242/dev.035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bouvard D, Pouwels J, De Franceschi N, Ivaska J. Integrin inactivators: balancing cellular fiunctions in vitro and in vivo. Nature Rev Mol Cell Biol. 2013;14:432–44. doi: 10.1038/nrm3599. [DOI] [PubMed] [Google Scholar]