Abstract
Fatty acid synthase is up-regulated in a variety of cancers, including prostate cancer. Up-regulation of fatty acid synthase not only increases production of fatty acids in tumors but also contributes to the transformed phenotype by conferring growth and survival advantages. In addition, increased fatty acid synthase expression in prostate cancer correlates with poor prognosis, although the mechanism(s) by which this occurs are not completely understood. Because fatty acid synthase is expressed at low levels in normal cells, it is currently a major target for anticancer drug design. Fatty acid synthase is normally found in the cytosol; however, we have discovered that it also localizes to the nucleus in a subset of prostate cancer cells. Analysis of the fatty acid synthase protein sequence indicated the presence of a nuclear localization signal, and subcellular fractionation of LNCaP prostate cancer cells, as well as immunofluorescent confocal microscopy of patient prostate tumor tissue and LNCaPs confirmed nuclear localization of this protein. Finally, immunohistochemical analysis of prostate cancer tissue indicated that nuclear localization of fatty acid synthase correlates with Gleason grade, implicating a potentially novel role in prostate cancer progression. Possible clinical implications include improving the accuracy of prostate biopsies in the diagnosis of low- versus intermediate-risk prostate cancer and the uncovering of novel metabolic pathways for the therapeutic targeting of androgen-independent prostate cancer.
Fatty acid synthase (FASN) is a large, multifunctional enzyme that is responsible for the de novo synthesis of long chain fatty acids. Because it is typically unnecessary for FASN to produce fatty acids in cells because of sufficient intake of fatty acids in the diet, FASN is expressed at low levels in most normal tissues. However, FASN expression has been found to be up-regulated in many cancers1, and in several cancers, including prostate cancer, it correlates with poor prognosis.2–4 In immortalized human prostate epithelial cells and in the LNCaP human prostate cancer cell line, FASN expression increases cell proliferation and growth in soft agar and results in androgen receptor-dependent formation of invasive adenocarcinoma.5 That same study also reported that transgenic expression of FASN in mouse prostate epithelial cells led to prostatic intraepithelial neoplasia and protected against castration- and chemotherapeutic-induced apoptosis, whereas siRNA knockdown of FASN in LNCaP cells resulted in apoptosis.5
A number of theories have been proposed to explain FASN up-regulation in cancers, which ultimately results in a metabolic shift toward producing large amounts of fatty acids. One such theory is related to the Warburg effect, a phenomenon that is observed in most cancers. This involves an increase in the use of the glycolytic pathway for energy production, which also leads to an increase in the substrates used in de novo fatty acid synthesis.6,7 It has also been postulated that fatty acid production is necessary to supply the structural components of the cell membrane for actively proliferating tumor cells.7 Alternatively, in the hypoxic environment of tumors, FASN may provide a means for balancing redox through its ability to consume reducing molecules such as NADPH.5–7 Regardless of the exact function(s) of FASN in carcinogenesis, it clearly contributes to the transformed phenotype by conferring growth and survival advantages.
Although FASN is an attractive therapeutic target in the treatment of cancer because of its low level of expression in normal tissues and concomitant overexpression in many cancers, small molecule inhibitors designed against this oncogenic protein have experienced limitations in their effectiveness because of poor bioavailability, lack of specificity, and significant side effects.7–10 To improve on therapeutics to target FASN, it is important to understand the precise mechanisms by which FASN promotes carcinogenesis.
The normal location of FASN in the cytoplasm is consistent with its known role in fatty acid synthesis. However, when we were examining immunohistochemical images of FASN expression in prostate cancer specimens, we observed that FASN appeared to be localized to the nucleus in a subset of the tumor cells. Through examination of an online database (The Human Protein Atlas, http://www.proteinatlas.org, last accessed March 15, 2013) that contains patient prostate cancer sections subjected to multiple antibodies raised against FASN, nuclear localization of FASN in a subset of tumor cells was again observed. We sought to confirm the nuclear localization of FASN in prostate cancer and to determine whether it could be used as a clinical marker for this disease. To our knowledge, this is the first observation of FASN localizing to the nucleus in any cell type.
Materials and Methods
Immunoblot Analysis
LNCaP whole-cell extracts (30 μm) and nuclear extracts isolated with hypotonic buffer were run on an 8% SDS-PAGE gel, transferred to polyvinylidene difluoride membrane, blocked in 5% bovine serum albumin, incubated with a monoclonal anti-FASN antibody (dilution 1:2500; BD Biosciences, San Jose, CA,), washed in Tris-buffered saline and Tween 20, incubated with anti-mouse antibody (Sigma-Aldrich, St. Louis, MO), washed, incubated with electrochimiluminescence reagent, and exposed to film. Treatments with 1 nmol/L testosterone and/or 17β-estradiol (Sigma-Aldrich) were performed, and quantitation was accomplished with ImageJ version 1.47v (NIH, Bethesda, MD).
Immunofluorescence and Confocal Microscopy
Frozen sections of Gleason 7 prostate cancer specimens [acquired from the University of Pittsburgh Health Sciences Tissue Bank and approved by the University of Pittsburgh Institutional Review Board (protocol 0506140)] or subconfluent LNCaP cells were used. Slides were fixed in 2% paraformaldehyde, incubated in 0.1% Triton X-100, rehydrated, blocked in 2% bovine serum albumin, washed in 0.5% bovine serum albumin, incubated with monoclonal anti-FASN clone 3F2-1F3 (dilution 1:250; Sigma-Aldrich) in 0.5% bovine serum albumin, washed, incubated with Alexa Fluor 488 anti-mouse (3.5 μg/mL; Invitrogen, Eugene, OR), washed, incubated with Draq5 nuclear stain (dilution 1:1000; eBioscience, Inc., San Diego, CA), and covered with a coverslip. Confocal microscopy was performed on a Leica (Wetzlar, Germany) TCS SP confocal microscope at ×63 magnification in the University of Pittsburgh Cell and Tissue Imaging Facility.
Immunohistochemistry
Formalin-fixed, paraffin-embedded prostate cancer tissue of various Gleason grades from 45 patients who had undergone radical prostatectomy were obtained from the University of Pittsburgh Health Sciences Tissue Bank and approved by the University of Pittsburgh Institutional Review Board (protocol PR011110523). Two samples of benign prostate glands were also obtained from deceased tissue donors. Slides were heated in a 60°C oven, deparaffinized, placed in boiling 0.01 mol/L citrate buffer (PolyScientific, Bay Shore, NJ) for antigen retrieval, and incubated in 3% H2O2 to block endogenous peroxidase activity. The M.O.M. kit (Vector Laboratories Inc., Burlingame, CA) was used for the application of antibody according to the manufacturer's instructions. Slides were incubated with primary anti-FASN (dilution 1:250; Sigma-Aldrich), flooded with NovaRED solution (Vector Laboratories Inc., Burlingame, CA) as per the manufacturer's instructions, counterstained with hematoxylin, dehydrated, applied coverslip with Permount mounting medium (Thermo Fisher Scientific, Waltham, MA), and air-dried overnight.
Correlation of Nuclear FASN with Gleason Grade
Final pathology of each specimen was confirmed in a blinded review with a genitourinary pathologist (A.V.P.). Each slide was digitally scanned as a whole slide image with the use of ScanScope XT (Aperio, Vista, CA) and viewed with Aperio ImageScope version 11.2.0.780. Each specimen was examined for the region with the most representative pathology and the most consistently highest staining intensity, which was then used for the analysis. Of the 49 original prostatectomy samples, 4 were excluded from being analyzed because of either significant staining artifact or small and poorly preserved cancer regions.
The immunohistochemical Nuclear Image Analysis algorithm version 9.1 (Aperio) was used to quantify the percentage of mild, moderate, and strongly staining nuclei. The default values for the parameters nuclear stain and positive stain provided by Aperio for the detection of the brown staining were used. In addition, the default values (range, 0 to 255) for the weak (210 to 189), moderate (188 to 163), and strong (162 or less) staining intensity thresholds were not changed. These are graded on an inverse scale of light transmission, with the value of 255 representing no staining, or high light transmission, and 0 representing the darkest staining possible. The algorithm parameters of curvature threshold (2), minimum nuclear size in square micrometers (30 μm2), maximum nuclear size in square micrometers (500 μm2), minimum roundness (0.4), minimum compactness (0.3), and minimum elongation (0.2) were modified during an initial validation process to produce the most consistent and accurate results across the pathological spectrum of the samples.
The scoring output by the image analysis software was reported as a percentage of nuclei staining at each threshold. This was converted into a composite nuclear staining score to account for the varying staining intensities and to allow appropriate comparison between specimens. The conversion required multiplying the weak percentage by 1, the moderate percentage by 2, and the strong percentage by 3, and then summing these amounts for a maximum score of 300.
The output results of the algorithm were confirmed in a second blinded review by a genitourinary pathologist (A.V.P.). Statistical analysis was performed with STATA version 12.1 (StataCorp LP, College Station, TX). A two-sample Wilcoxon rank sum (Mann–Whitney) test was performed to detect a difference in the median percentages of strongly staining nuclei between different cancer grades.
Results
Presence of an NLS in FASN and Prediction of Nuclear Localization
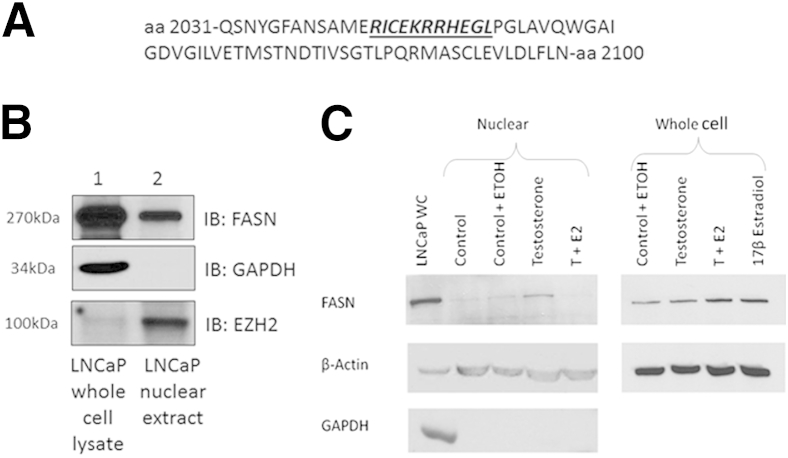
After observing nuclear localization of FASN in a subset of prostate cancer cells via immunohistochemistry, we wanted to examine whether FASN contained a nuclear localization signal (NLS). We used the online program cNLS Mapper to determine the presence of an NLS in the FASN protein sequence.11 This program found a monopartite NLS, beginning at amino acid 2043 (Figure 1A), which would allow for nuclear localization of FASN.
Figure 1.
The online program cNLS Mapper indicates the presence of an NLS within the FASN protein sequence (A), immunoblot analysis of LNCaP nuclear lysates indicates that FASN is localized to the nucleus in LNCaP cells (B), and overall expression of FASN is stimulated by testosterone and/or 17β-estradiol (C), but nuclear expression is abolished in the presence of 17β-estradiol. A: The FASN amino acid sequence is shown with the NLS in bold italics and underlined. B: Immunoblot analysis of FASN expression in LNCaP whole cell and nuclear extracts. Levels of nuclear FASN in LNCaPs can vary according to the passage. C: LNCaP cells were treated with 1 nmol/L testosterone, 1 nmol/L 17-β estradiol, or vehicle alone for 4.5 hours. Whole-cell or nuclear extracts were run and anti-FASN, an antibody detecting beta actin, and an antibody detecting the cytoplasmic marker GAPDH are shown. E2, 17-β estradiol; ETOH, ethanol; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; T, testosterone; WC, whole-cell extract.
In addition, we used the FASN protein sequence as input into the cell localization prediction tool WoLF PSORT.12 This program predicts cytoplasmic and/or nuclear localization of proteins on the basis of sorting signals, amino acid composition, and functional motifs.12 This program also predicted both nuclear and cytoplasmic localization of FASN in cells.
Evidence for Nuclear Localization of FASN in LNCaP Prostate Cancer Cells
Next, we examined whether cultured prostate cancer cells expressed nuclear FASN. Nuclear and whole-cell lysates were prepared from the LNCaP prostate cancer cell line and analyzed for FASN expression. Immunoblot analysis found that the 270-kDa FASN is expressed in LNCaP whole-cell lysates as well as nuclear extracts (Figure 1B). The nuclear extracts were determined to be free of cytoplasmic contamination because of the absence of the cytoplasmic protein glyceraldehyde-3-phosphate dehydrogenase. The LNCaP nuclear extracts were also enriched for the nuclear marker EZH2 compared with the whole-cell lysates. Nuclear and whole-cell lysates from the DU145, PC3, and BPH1 prostate cell lines were also examined for nuclear FASN via immunoblot analysis; however, these prostate cell lines do not express nuclear FASN (data not shown) (Supplemental Figure S1).
We then wanted to determine what regulated nuclear localization of FASN. Hormones, growth factors, and cytokines are known to regulate FASN expression.13 To determine whether hormone treatment could induce nuclear localization of FASN, LNCaP cells were treated with 1 nmol/L testosterone (or the testosterone analogue R1881; data not shown), 1 nmol/L testosterone plus 1 nmol/L 17β-estradiol, 17β-estradiol alone, or vehicle (ethanol) alone for 4.5 hours. Cells were harvested, nuclear and whole-cell lysates were prepared as described above, and immunoblot analysis for FASN was performed (Figure 1C). Treatment with testosterone increased overall expression of FASN protein by 54%, and nuclear localization of FASN was increased similarly by 59%. However, the addition of 17β-estradiol almost completely abolished nuclear localization of FASN, whereas treatment with both testosterone and 17β-estradiol or with 17β-estradiol alone increased overall expression of FASN protein by 140% and 119% over vehicle alone, respectively. LNCaP cells were also exposed to hypoxic conditions (1% O2), which induces a cytokine response, or treated with epidermal growth factor; however, these treatments/conditions did not induce nuclear localization of FASN (data not shown). These data suggest that 17β-estradiol is able to regulate nuclear localization of FASN protein.
Confirmation of Nuclear Localization of FASN in Prostate Cancer Cells
To further confirm nuclear localization of FASN in prostate cancer cells, we performed immunofluorescence on prostate cancer specimens obtained from patients. Confocal microscopic analysis of the immunofluorescence on patient prostate tumor tissues showed in a three-dimensional manner that FASN localizes to the nucleus in a subset of cancer cells (Figure 2). In addition, immunofluorescence for FASN was performed on cultured LNCaP prostate cancer cells, and confocal analysis of these cells also showed localization of FASN to the nucleus (Supplemental Figure S2).
Figure 2.
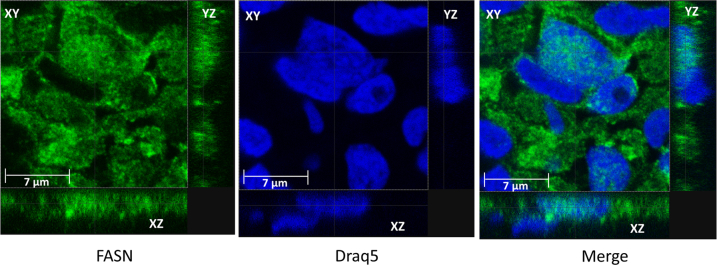
Confocal microscopy confirms nuclear localization of FASN in a subset of prostate cancer cells. FASN (green), Draq5 nuclear stain (blue), and the merge of the two stainings.
Correlation of Nuclear FASN with Gleason Grade
Finally, we wanted to determine whether nuclear FASN expression correlated with Gleason grade via immunohistochemical analysis. Forty-one cancer specimens were stained, and four patient samples were used as negative controls with secondary antibody staining only. There were two Gleason score (GS) 3 + 3 = 6 negative controls and two GS 3 + 4 = 7 negative controls. The 41 tumor specimens were composed of 6 specimens GS = 6, 26 specimens GS = 7, and 9 specimens GS ≥ 8, with 2 additional specimens from benign tissue donors. High-grade prostatic intraepithelial neoplasia (HGPIN) regions (n = 21) were found throughout the already used cancer specimens and were also analyzed (Table 1).
Table 1.
Results of Immunohistochemical Nuclear Staining Analysis
| Pathology | Median composite nuclear score | 95% CI | No. of specimens |
|---|---|---|---|
| Benign prostate epithelium | 0.43 | 0.11–0.74 | 2 |
| High-grade prostate intraepithelial neoplasia | 3.63 | 2.41–6.58 | 21 |
| Prostate cancer | 41 | ||
| Gleason pattern 3 + 3 = 6 | 3.39 | 0.03–4.96 | 6 |
| Gleason pattern = 7 (combined) | 22.73 | 15.09–26.22 | 26 |
| 3 + 4 = 7 | 20.19 | 14.09–26.03 | 22 |
| 4 + 3 = 7 | 26.36 | 14.28–47.93 | 4 |
| Gleason pattern 4 + 4 = 8 or higher | 43.59 | 4.02–60.13 | 9 |
| Negative controls (combined) | 0 | 0–3.2 | 4 |
| Gleason pattern 3 + 3 = 6 | 0 | 0 | 2 |
| Gleason pattern 3 + 4 = 7 | 0 | 0–3.2 | 2 |
Composite nuclear score combines the percentage of nuclei that have mild, moderate, and strong intensity staining for a maximum score of 300.
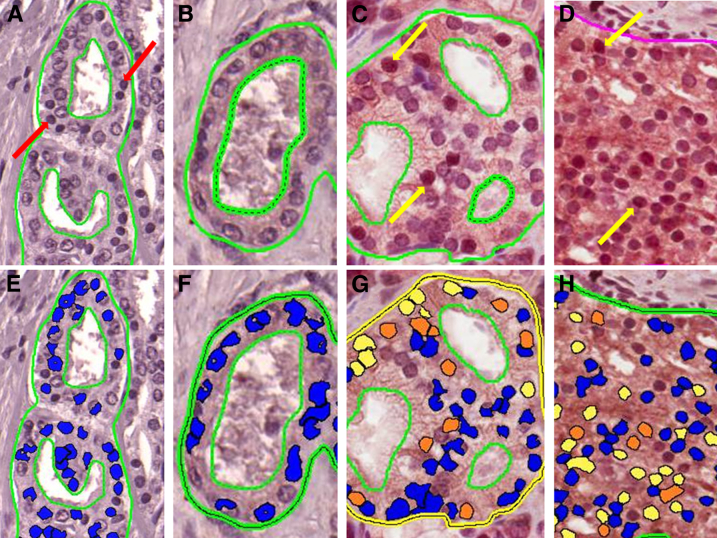
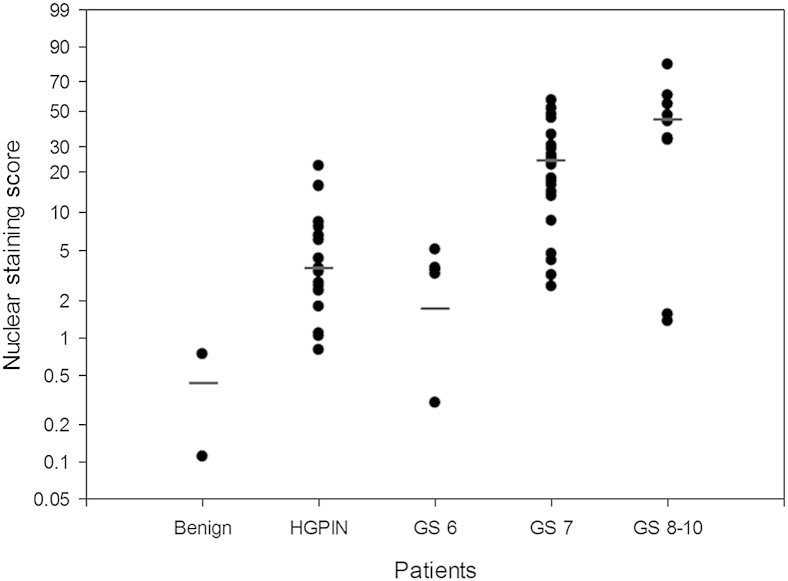
Representative analysis outputs by the nuclear staining algorithm for the negative controls and various Gleason patterns are shown in Figure 3. The median composite nuclear scores for GS = 6 and GS = 7 negative controls were 0 (0 to 0) and 1.6 (0 to 3.2), respectively. The median composite nuclear score was 0.43 (95% CI, 0.11–0.74) for the benign glands, 3.63 (95% CI, 2.41–6.58) for the HGPIN regions, 3.39 (95% CI, 0.03–4.96) for GS = 6, 22.73 (95% CI, 15.09–26.22) for GS = 7, and 43.59 (95% CI, 4.02–60.13) for GS ≥ 8 (Figure 4). The skewness and kurtosis test for normality, as well as the Shapiro–Wilk normality test, confirmed a nonparametric distribution of the results.
Figure 3.
Immunohistochemical staining and automated analysis output. Gleason score 3 + 4 = 7 negative control with only secondary antibody staining (A) and the resulting scoring output (E) from the Nuclear Image Analysis algorithm version 9.1 (Aperio). The lines drawn on each section are examples of areas chosen to be anaylzed by the Aperio software. Dashed lines represent areas to be removed from the analysis. A: Red arrows indicate dark nuclei that are still scored as negative because of lack of brown staining. C and D: Yellow arrows indicate specific nuclear localization of FASN. E–H: Blue output coloring from the algorithm indicates no nuclear staining, whereas yellow, orange, and red indicate mild, moderate, and strong nuclear staining, respectively. Gleason pattern 3 + 3 = 6 prostate cancer stained with anti-FASN antibody (B) and scoring output (F), Gleason pattern 3 + 4 = 7 prostate cancer stained with anti-FASN antibody (C) and scoring output (G), and Gleason pattern 4 + 4 = 8 prostate cancer stained with anti-FASN antibody (D) and scoring output (H).
Figure 4.
Median and composite nuclear score by pathological group. GS, Gleason score; HGPIN, high-grade prostatic intraepithelial neoplasia.
A significant increase existed in nuclear FASN staining between GS = 6 and cancers that were GS ≥ 7 (P = 0.0008), between only GS = 6 compared with GS = 7 cancers (P = 0.0005), and between GS = 6 and GS ≥ 8 cancers (P = 0.0251). No difference was found between HGPIN and GS = 6 cancers (P = 0.255). Significant increases in FASN nuclear staining were found between HGPIN and GS = 7 (P < 0.0001) and between HGPIN and GS ≥ 8 (P = 0.0062). The increase between GS = 7 and GS 9 to 10 cancers was also significant (P = 0.004). Finally, as an extension of the Wilcoxon rank sum test, we confirmed a significant trend for the composite nuclear scores across each Gleason score group (P < 0.001).
Discussion
This is the first report of FASN localization to the nucleus. Because this phenomenon occurs almost exclusively in tumor cells and increases in frequency along with disease grade implies that mislocalization of this protein to the nucleus is involved in prostate carcinogenesis and progression and may be a novel marker of more aggressive disease. Our observation was supported by bioinformatic analysis of the FASN protein sequence, which showed the presence of a previously unreported NLS sequence, and nuclear fractionation of the LNCaP cell line which indicated that at least a portion of the FASN expressed in these cells is localized to the nucleus. The DU145, PC3, and BPH1 prostate cell lines were also examined for nuclear FASN expression via nuclear fractionation and immunoblot analysis; however, these cell lines did not express nuclear FASN. LNCaP cells are arguably more representative of true prostate cancer than the other lines (eg, they express the androgen receptor, prostate-specific antigen, prostate-specific membrane antigen, and prostatic acid phosphatase). In addition, it is known that molecular changes occur in cells when they are taken from patients and grown in culture, so perhaps nuclear FASN is lost in some cell lines when grown in culture. We then verified nuclear localization of FASN in a subset of prostate cancer cells via confocal microscopy on prostate cancer patient tissue specimens and in cultured LNCaP prostate cancer cells. Importantly, we demonstrated that 17β-estradiol abolished nuclear localization of FASN, giving insight into regulation of nuclear FASN. Hormones are known to regulate FASN. Most notably, testosterone induces FASN expression. The regulation of FASN by 17β-estradiol is likely complex, and we plan to investigate this further in the future. Finally, we used digital imaging and automated nuclear analysis and quantitation to indicate a significant relationship between the frequency and intensity of nuclear FASN expression and Gleason grade; we found minimal to no nuclear staining in benign prostate epithelial cells, HGPIN, and low-grade prostate tumor cells (Gleason score = 6). These findings are consistent with a previous report that found a significant increase in the expression of FASN mRNA in cancers with GS = 7 and above compared with low-grade disease.14
Previous studies do exist with evidence to support the concept of FASN nuclear localization. In a large-scale report published in 2006, Olsen et al15 looked at the subcellular localization and phosphorylation status of proteins in HeLa cells in the presence or absence of epidermal growth factor treatment. The investigators found that treatment with epidermal growth factor induced nuclear accumulation of FASN. In a separate study examining binding partners of nuclear versus cytoplasmic histone deacetylase 5, Greco et al16 showed that FASN interacts with the predominantly nuclear form of histone deacetylase 5. Moreover, another large-scale study showed that FASN interacts with Sirtuin 7, a primarily nuclear protein.17 These findings, along with what we have shown in the present study, provide evidence to support the idea that FASN can localize to the nucleus in specific cells under certain conditions. There are two reports of perinuclear localization of FASN, one as part of the pathogenesis of dengue virus and the second in oral squamous cell carcinoma cells18,19; however, this is different from the nuclear localization that we observe here.
The potential function of nuclear FASN is largely unknown. As reviewed by Rossi et al,20 knockdown of FASN results in inhibition of DNA synthesis, implicating a potential nuclear role for FASN. Alternatively, the crystal structure of mammalian FASN indicated that this large protein contains a conserved methyltransferase domain.21 It is unknown whether this methyltransferase domain is functional, but, if it is, nuclear FASN may be contributing to the altered epigenetic landscape in prostate cancer via DNA methylation or methylation of other nuclear proteins such as histones. Furthermore, one of the enzymatic domains of FASN that is used during fatty acid synthesis is an acetyl-transferase. Therefore, another potential function for nuclear FASN may be through modification of chromatin via acetylation.
The immunohistochemical analysis of nuclear FASN expression in the different Gleason grade prostate cancers used automated analysis technology. There are an increasing number of reports that use the many different options for automated analysis, including studies specifically using the Aperio image analysis software to investigate nuclear proteins.22,23 The benefits of an automated software package include decreased interobserver variability and increased reproducibility, also known as precision, because the parameters do not change between specimens or interpreters.24 Although these algorithms still require validation with another form of measurement to confirm their accuracy, usually from correlation with a pathologist's findings, the removal of some of the biases inherent in visual interpretation increases precision and should theoretically improve the overall application of the results. In fact, a recent report found that, by using image analysis tools, the quantifying of nuclear proteins in breast cancer was improved and allowed for lower thresholds to be used to predict tumor responsiveness to certain hormone therapies.23 Because the degree of nuclear staining can be difficult to interpret, especially in the context of concurrent cytoplasmic staining, we consider the use of our automated algorithm as a major strength to our findings.
Two areas exist in which nuclear localization of FASN could have novel effects on the management of prostate cancer. The first is in improving the accuracy of prostate needle biopsies in the diagnosis of low- versus intermediate-risk prostate cancer. The second area in which this discovery may prove beneficial is in the treatment of castration-resistant or androgen-independent prostate cancer. The normal regulatory control of FASN expression is lost in more advanced cancers and is often no longer manipulated by androgens. In addition, in human prostate cancer specimens, as well as in vitro and in vivo models, FASN expression was highest in androgen-independent cells and in fact represented the progression from androgen-dependent to androgen-independent cancers.25 This same study also confirmed that inhibition of FASN led to death of these cells. It would be most useful to discover the role of nuclear localization of FASN in this progression, and whether there are any opportunities to prevent it from occurring. In addition, because no cure for androgen-independent prostate cancer is available, a therapy that targets FASN directly, especially one that is specifically targeted to the nucleus, could be quite effective and potentially decrease side effects from nonspecific inhibition of FASN in the cytoplasm of normal cells.
In conclusion, we report here the novel observation that FASN can localize to the nucleus in a subset of prostate cancer cells. Although overall FASN expression is already known to correlate with poor prognosis in prostate cancer, we show that nuclear localization of FASN correlates with Gleason grade. Because it is still unclear exactly how FASN promotes tumorigenesis, it is important to investigate the function of nuclear FASN, because the nuclear form of this protein may promote carcinogenesis. Nuclear FASN expression reaches significant levels in intermediate- and high-grade cancers, potentially implicating nuclear FASN as an important factor in the progression of prostate cancer from clinically indolent to clinically significant. Future investigations are required to examine how nuclear FASN is regulated, what its function is within the nucleus, and how these factors may be used to develop more specific therapeutic agents to target aggressive prostate cancer.
Acknowledgments
We thank Dr. Donna Stolz (University of Pittsburgh Cell and Tissue Imaging Facility) for help with the confocal microscopy, the University of Pittsburgh Health Sciences Tissue Bank and Cancer Center for frozen and formalin-fixed, paraffin-embedded prostate cancer specimens, Dr. Milon Amin for assistance with reviewing the pathological specimens, and Malini Srinivasan and Jon Duboy for assistance with scanning slides and using the Aperio software package.
Footnotes
Supported by U.S. Department of Defense grants PC110816 (A.A.M. and D.S.O'K.) and W81XWH-09-1-0171 (Y.J.D.), NIH grants R01CA138444 (D.S.O'K., D.J.B., K.M.S., A.V.P., and J.L.C.) and P30CA047904 (A.V.P. and P.H.B.), and King Saud University graduate student fellowship (A.M.B.).
The U.S. Army Medical Research Acquisition Activity (Fort Detrick, MD) is the awarding and administering acquisition office. The content of this publication does not necessarily reflect the position or policy of the government, and no official endorsement should be inferred. The conclusions do not necessarily reflect the opinions of the U.S. Government.
A.A.M. and K.J.R. contributed equally to this work.
Disclosures: None declared
Supplemental Data
Immunoblot analysis of nuclear and whole-cell lysates from DU145 and PC3 cells indicates that FASN is not present in the nucleus of these prostate cancer cell lines.
Confocal microscopy confirms nuclear localization of FASN in a subset of cultured LNCaP prostate cancer cells. Green staining represents FASN, blue is the Draq5 nuclear stain, and the image is a merge of both stains.
References
- 1.Flavin R., Peluso S., Nguyen P.L., Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010;6:551–562. doi: 10.2217/fon.10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shurbaji M.S., Kuhajda F.P., Pasternack G.R., Thurmond T.S. Expression of oncogenic antigen 519 (OA-519) in prostate cancer is a potential prognostic indicator. Am J Clin Pathol. 1992;97:686–691. doi: 10.1093/ajcp/97.5.686. [DOI] [PubMed] [Google Scholar]
- 3.Shurbaji M.S., Kalbfleisch J.H., Thurmond T.S. Immunohistochemical detection of a fatty acid synthase (OA-519) as a predictor of progression of prostate cancer. Hum Pathol. 1996;27:917–921. doi: 10.1016/s0046-8177(96)90218-x. [DOI] [PubMed] [Google Scholar]
- 4.Epstein J.I., Carmichael M., Partin A.W. OA-519 (fatty acid synthase) as an independent predictor of pathologic state in adenocarcinoma of the prostate. Urology. 1995;45:81–86. doi: 10.1016/s0090-4295(95)96904-7. [DOI] [PubMed] [Google Scholar]
- 5.Migita T., Ruiz S., Fornari A., Fiorentino M., Priolo C., Zadra G., Inazuka F., Grisanzio C., Palescandolo E., Shin E., Fiore C., Xie W., Kung A.L., Febbo P.G., Subramanian A., Mucci L., Ma J., Signoretti S., Stampfer M., Hahn W.C., Finn S., Loda M. Fatty acid synthase: a metabolic enzyme and candidate oncogene in prostate cancer. J Natl Cancer Inst. 2009;101:519–532. doi: 10.1093/jnci/djp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menendez J.A., Lupu R. Oncogenic properties of the endogenous fatty acid metabolism: molecular pathology of fatty acid synthase in cancer cells. Curr Opin Clin Nutr Metab Care. 2006;9:346–357. doi: 10.1097/01.mco.0000232893.21050.15. [DOI] [PubMed] [Google Scholar]
- 7.Flavin R., Zadra G., Loda M. Metabolic alterations and targeted therapies in prostate cancer. J Pathol. 2011;223:283–294. doi: 10.1002/path.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilvo M., Denkert C., Lehtinen L., Muller B., Brockmoller S., Seppanen-Laakso T., Budczies J., Bucher E., Yetukuri L., Castillo S., Berg E., Nygren H., Sysi-Aho M., Griffin J.L., Fiehn O., Loibl S., Richter-Ehrenstein C., Radke C., Hyotylainen T., Kallioniemi O., Iljin K., Oresic M. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011;71:3236–3245. doi: 10.1158/0008-5472.CAN-10-3894. [DOI] [PubMed] [Google Scholar]
- 9.Puig T., Aguilar H., Cufi S., Oliveras G., Turrado C., Ortega-Gutierrez S., Benhamu B., Lopez-Rodriguez M.L., Urruticoechea A., Colomer R. A novel inhibitor of fatty acid synthase shows activity against HER2+ breast cancer xenografts and is active in anti-HER2 drug-resistant cell lines. Breast Cancer Res. 2011;13:R131. doi: 10.1186/bcr3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen A.M., Eisenberg B.L., Kuemmerle N.B., Flanagan A.J., Morganelli P.M., Lombardo P.S., Swinnen J.V., Kinlaw W.B. Fatty acid synthesis is a therapeutic target in human liposarcoma. Int J Oncol. 2010;36:1309–1314. doi: 10.3892/ijo_00000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosugi S., Hasebe M., Tomita M., Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menendez J.A., Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 14.Swinnen J.V., Vanderhoydonc F., Elgamal A.A., Eelen M., Vercaeren I., Joniau S., Van Poppel H., Baert L., Goosens K., Heyns W., Verhoeven G. Selective activation of the fatty acid synthesis pathway in human prostate cancer. Int J Cancer. 2000;88:176–179. doi: 10.1002/1097-0215(20001015)88:2<176::aid-ijc5>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 15.Olsen J.V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 16.Greco T.M., Yu F., Guise A.J., Cristea I.M. Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.004317. M110.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai Y.C., Greco T.M., Boonmee A., Miteva Y., Cristea I.M. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics. 2012;11:60–76. doi: 10.1074/mcp.A111.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agostini M., Silva S.D., Zecchin K.G., Coletta R.D., Jorge J., Loda M., Graner E. Fatty acid synthase is required for the proliferation of human oral squamous carcinoma cells. Oral Oncol. 2004;40:728–735. doi: 10.1016/j.oraloncology.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Heaton N.S., Perera R., Berger K.L., Khadka S., Lacount D.J., Kuhn R.J., Randall G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossi S., Graner E., Febbo P., Weinstein L., Bhattacharya N., Onody T., Bubley G., Balk S., Loda M. Fatty acid synthase expression defines distinct molecular signatures in prostate cancer. Mol Cancer Res. 2003;1:707–715. [PubMed] [Google Scholar]
- 21.Maier T., Leibundgut M., Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 22.Laurinavicius A., Laurinaviciene A., Ostapenko V., Dasevicius D., Jarmalaite S., Lazutka J. Immunohistochemistry profiles of breast ductal carcinoma: factor analysis of digital image analysis data. Diagn Pathol. 2012;7:27. doi: 10.1186/1746-1596-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rexhepaj E., Brennan D.J., Holloway P., Kay E.W., McCann A.H., Landberg G., Duffy M.J., Jirstrom K., Gallagher W.M. Novel image analysis approach for quantifying expression of nuclear proteins assessed by immunohistochemistry: application to measurement of oestrogen and progesterone receptor levels in breast cancer. Breast Cancer Res. 2008;10:R89. doi: 10.1186/bcr2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurinavicius A., Laurinaviciene A., Dasevicius D., Elie N., Plancoulaine B., Bor C., Herlin P. Digital image analysis in pathology: benefits and obligation. Anal Cell Pathol (Amst) 2012;35:75–78. doi: 10.3233/ACP-2011-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pizer E.S., Pflug B.R., Bova G.S., Han W.F., Udan M.S., Nelson J.B. Increased fatty acid synthase as a therapeutic target in androgen-independent prostate cancer progression. Prostate. 2001;47:102–110. doi: 10.1002/pros.1052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunoblot analysis of nuclear and whole-cell lysates from DU145 and PC3 cells indicates that FASN is not present in the nucleus of these prostate cancer cell lines.
Confocal microscopy confirms nuclear localization of FASN in a subset of cultured LNCaP prostate cancer cells. Green staining represents FASN, blue is the Draq5 nuclear stain, and the image is a merge of both stains.