Abstract
Objective
Macrophage migration inhibitory factor (MIF) is a proinflammatory mediator involved in the pathogenesis of rheumatoid arthritis. This study was undertaken to identify the MIF promoter elements responsible for regulating gene expression.
Methods
Luciferase reporter gene assays were used to identify the MIF promoter sequence responsible for basal activity. Bioinformatic analysis was used to predict transcription factor binding sites, and electrophoretic mobility shift assay (EMSA) was used to demonstrate transcription factor binding. Chromatin immunoprecipitation (ChIP) was used to demonstrate transcription factor loading on the MIF promoter.
Results
We identified the minimal promoter sequence required for basal MIF promoter activity that was also capable of conferring glucocorticoid-dependent inhibition in a T lymphocyte model cell line. Deletion studies and EMSA revealed 2 elements in the MIF promoter that were responsible for basal promoter activity. The 5′ element binds CREB/activating transcription factor 1, and the 3′ element is a functional hypoxia-responsive element binding hypoxia-inducible factor 1α. Further studies demonstrated that the cis elements are both required for glucocorticoid-dependent inhibition. ChIP demonstrated glucocorticoid-dependent recruitment of glucocorticoid receptor α to the MIF promoter in lymphocytes within 1 hour of treatment and a concomitant decrease in acetylated histone H3.
Conclusion
Our findings indicate that hypoxia and glucocorticoid signaling converge on a single element regulating MIF; this regulatory unit is a potential interacting node for microenvironment sensing of oxygen tension and glucocorticoid action in foci of inflammation.
Macrophage migration inhibitory factor (MIF) is a proinflammatory mediator that is widely expressed. Currently, the exact role of MIF in the regulation of the immune response is a subject of controversy. Numerous studies have shown that MIF activates or promotes the expression of tumor necrosis factor α (TNFα), interleukin-1 (IL-1), IL-2, IL-6, IL-8, and IL-12 from a variety of cell types, including macrophages and T cells (1-6); other studies have failed to establish a cytokine or cytokine-inducing role of MIF (7-10). However, MIF is capable of overriding the antiinflammatory actions of glucocorticoids, thus impairing glucocorticoid action (1,2,11).
MIF plays an important role in the pathogenesis of inflammatory arthritis. Elevated MIF expression has been reported in rheumatoid arthritis (RA) synovial fluid and serum compared with healthy controls and controls with osteoarthritis (12,13). RA fibroblast-like synoviocyte (FLS)–derived MIF induces monocyte TNF in vitro (12), and MIF up-regulates FLS IL-1 messenger RNA expression (14). Experimental models of arthritis demonstrate a role of MIF, with MIF antagonism capable of delaying the onset and disease frequency of collagen-induced arthritis (CIA) (15), and inhibiting adjuvant-induced arthritis (AIA) in both rats (16) and mice (11) in a dose-dependent manner. MIF−/− mice have a decreased severity of AIA (17,18) and decreased joint inflammation and destruction in a model of passive CIA (19). Increased p53 expression in MIF−/− mice with AIA, together with the observation that MIF inhibits RA synoviocyte p53 expression and subsequent apoptosis in vitro, implicates MIF in the hyperplasia of RA synovium and the formation of pannus (18). Recently, MIF gene expression has been shown to be driven by hypoxia-inducible factor 1α (HIF-1α), under normoxic and hypoxic conditions (20), through an element close to the transcription start site. Therefore, in foci of inflammation, in which oxygen tension is reduced, hypoxia, rather than proinflammatory cytokine action, may be the principal signal for MIF production.
Glucocorticoids exert major effects on the inflammatory process, acting via glucocorticoid receptor α (GRα) to inhibit expression of cytokines, enzymes, and adhesion molecules. Glucocorticoids also inhibit cell proliferation or induce apoptosis, actions which oppose the effects of HIF-1α. Indeed, HIF-1α, acting through MIF expression, has been shown to delay cell senescence and promote cell proliferation (20). Early studies showed an acute, stimulatory effect of glucocorticoids on secretion of MIF (2), but we have found a more complex pattern of glucocorticoid regulation with cell type–specific glucocorticoid inhibition of MIF gene transcription (21). In addition, there is a close interaction between the actions of HIF-1α and activated GRα, with evidence that GRα can potentiate the transactivation functions of HIF-1α (22). Therefore, MIF expression is regulated by both HIF-1α and GRα, 2 important transcription factors involved in cell fate decisions, for which there is previous evidence of functional interaction. However, nothing is currently known about how these signaling cascades converge to regulate MIF expression.
Using the human lymphocyte cell line CEMC7A, we identified an MIF glucocorticoid-responsive module comprising 2 closely linked cis elements. The 5′ element bound activating transcription factor 1 (ATF-1)/CREB, whereas the 3′ element bound HIF-1α and was a functional hypoxia-responsive element (HRE). CEMC7A cells expressed HIF-1α under both normoxic and hypoxic conditions. We demonstrated glucocorticoid-dependent recruitment of GRα to the module and observed the concomitant deacetylation of associated histone H3 at the MIF promoter. A human epithelial cell line, A549, did not show glucocorticoid repression of the MIF promoter, despite the presence of functional GRα, did not express HIF-1α under basal conditions, and did not express proteins capable of binding to the 3′ MIF element. Therefore, we show convergence of hypoxia and glucocorticoid signaling on a short sequence of the MIF gene flanking the transcription start site, with antagonistic activity on MIF gene expression.
MATERIALS AND METHODS
Construction of MIF luciferase constructs
The MIF-173*G plasmid has been described previously (23); this was used as the parental plasmid to make truncations of the MIF promoter. Exonuclease III digestion produced the constructs −482 to +85 MIF Luc, −460 to +85 MIF Luc, −442 to +85 MIF Luc, −71 to +85 MIF Luc, and +3 to +85 MIF Luc. To make constructs containing deletions of putative activator protein 1 (AP-1) sites, site-directed mutagenesis was performed using −71 to +85 MIF Luc and the QuikChange site-directed mutagenesis kit, according to the recommendations of the manufacturer (Stratagene, La Jolla, CA). All plasmid constructs were sequenced to confirm the presence of the predicted changes.
Cell culture
CEMC7A cells (human T lymphoblasts) and A549 cells (human lung epithelial cells) were obtained from the European Collection of Cell Cultures (Porton Down, UK). CEMC7A cells were cultured in RPMI 1640 (Gibco, Grand Island, NY), and A549 cells were cultured in Dulbecco’s modified Eagle’s medium with Glutamax (Gibco). Media were supplemented with 10% fetal bovine serum, and cells were grown at 37°C in 5% CO2. Cells grown under hypoxic conditions were grown at 37°C in 5% CO2, 5% H2, and 90% N2.
Transfections and plasmids
Cells were transfected as described previously (23), with pRLCMV (Promega, Madison, WI) used as a transfection control in all experiments except under hypoxic conditions, where it is responsive. In these cases, total protein was assayed by the Bradford method. After transfection, cells were left untreated or were treated with dexamethasone (DEX). Cells were then harvested and luciferase assays performed using a Dual Luciferase assay kit according to the recommendations of the manufacturer (Promega). Experiments were performed in triplicate on at least 3 occasions. TAT3Luc has been described previously (24).
Western blotting
Cells were treated as indicated in the figure legends. For whole cell lysates, cells were harvested and lysed in radioimmunoprecipitation assay buffer (100 mM Tris [pH 7.4], 150 mM NaCl, 1% Nonidet P40, 2.5% sodium deoxycholate, and 1 mM EDTA) with Complete protease inhibitor (Roche, Indianapolis, IN). Cell lysates and nuclear extracts were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed with one of the following antibodies: anti-GRα (1:1,000 dilution) (GRα-clone 41; BD Biosciences, San Jose, CA) anti-phosphoGR (1:2,000; New England Biolabs, Ipswich, MA), anti–ATF-1 (1:2,000; Santa Cruz Biotechnology, Santa Cruz, CA), anti-CREB (1:2,000; Santa Cruz Biotechnology), anti HIF-1α (1:2,000; Santa Cruz Biotechnology), and anti-tubulin (1:2,000; Sigma, St. Louis, MO). Membranes were probed with the appropriate horseradish peroxidase–conjugated secondary antibody diluted 1:5,000, treated with enhanced chemiluminescent substrate (Amersham, Little Chalfont UK), and then exposed to film.
Preparation of nuclear extract
CEMC7A cells were grown to a density of 1 × 105 cells/ml in 1 liter of RPMI 1640 media, and A549 cells were grown in forty 15-cm2 plates until they were ~80% confluent. Cells were harvested and washed twice in phosphate buffered saline. Nuclear extract was then prepared as described previously (25). Treatments were carried out as indicated in the figure legends.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using radiolabeled double-stranded DNA (dsDNA) oligonucleotides corresponding to the putative AP-1 sites found in the MIF promoter. The oligonucleotides used were as follows: for consensus AP-1, forward 5′-CGCTTGATGACTCAGCCGGAA-3′ and reverse 5′-TTCCGGCTGAGTCATCAAGCG-3′; for mutated AP-1, forward 5′-CGCTTGAACCCGTGGCCGGAA-3′ and reverse 5′-TTCCGGCCACGGGTTCAAGCG-3′; for consensus cAMP response element (CRE), forward 5′-AGAGATTGCCTGACGTCAGAGAGCTAG-3′ and reverse 5′-CTAGC-TCTCTGACGTCAGGCAATCTCT-3′; for MIF-1, forward 5′-GCGGTGGCGTCACAAAAGGCG-3′ and reverse 5′-CGCCTTTTGTGACGCCACCGC-3′; for MIF-2, forward 5′-TGTCCGAGAAGTCAGGCACGT-3′ and reverse 5′-ACGTGCCTGACTTCTCGGACA-3′; and for MIF-3, forward 5′-GCACGTAGCTCAGCGGCGGC-3′ and reverse 5′-GCCGCCGCTGAGCTACGTGC-3′. (The mutated bases are underlined. The consensus-binding sites are boldfaced.)
For competition studies examining the sequence-specific binding requirements of the MIF-3 complex, the following oligonucleotides were used: for M1, forward 5′-GCCCGTAGCTCAGCGGCGGC-3′ and reverse 5′-GCCGCCGCTGAGCTACGGGC-3′; for M2, forward 5′-GCAAGTAGCTCAGCGGCGGC-3′ and reverse 5′-GCCGCCGCTGAGCTACTTGC-3′; for M3, forward 5′-GCACTTAGCTCAGCGGCGGC-3′ and reverse 5′-GCCGCCGCTGAGCTAAGTGC-3′; for M4, forward 5′-GCACGGAGCTCAGCGGCGGC-3′ and reverse 5′-GCCGCCGCTGAGCTCCGTGC-3′; for M5, forward 5′-GCACGTCGCTCAGCGGCGGC-3′ and reverse 5′-GCCGCCGCTGAGCGACGTGC-3′; for M6, forward 5′-GCACGTATCTCAGCGGCGGC-3′ and reverse 5′-GCCGCCGCTGAGATACGTGC-3′; for M7, forward 5′-GCACGTAGATCAGCGGCGGC-3′ and reverse 5′-GCCGCCGCTGATCTACGTGC-3′; for consensus HIF, forward 5′-TCTGTACGTGACCACACTCACCTC-3′ and reverse 5′-GAGGTGAGTGTGGTCACGTACAGA-3′; and for mutated HIF, forward 5′-TCTGTAAAAGACCACACTCACCTC-3′ and reverse 5′-GAGGTGAGTGTGGTCTTTTACAGA-3′. (The mutated bases are underlined. The consensus-binding sites are boldfaced.)
Annealed oligonucleotides were end-labeled with γ32P-ATP (Amersham) using T4 polynucleotide kinase (Promega) and purified using G50 columns (Amersham). Reactions contained 10 μl of buffer C (10 mM Tris [pH 7.8], 50 mM NaCl, 1 mM dithiothreitol [DTT], 1 mM EDTA, and 5% glycerol), 2 μg of bovine serum albumin, 100 ng of poly(dI-dC), 0.1 ng of labeled probe, and 5 μg of nuclear extract. Competition experiments were performed by adding unlabeled oligonucleotides to the reaction mixtures prior to addition of radiolabeled probe. Super-shift experiments were performed by the addition of 2 μg of antibody prior to the addition of radiolabeled probe. Reactions were then incubated at room temperature for 1 hour and loaded onto a 5% 0.5× Tris–borate–EDTA gel run at 150V for ~3 hours. Gels were then dried and autoradiographed.
DNA affinity chromatography
DNA affinity chromatography was performed using biotinylated dsDNA oligonucleotides, which incorporate MIF-3, mutant MIF-3, or the HIF response element consensus sequence. The oligonucleotides used were as follows: for MIF-3, forward 5′-GGGACCACAGTGGTGTCCGAGAAGTCAGGCACGTAGCTCAGCGGCGGCCGCGGCGCGTGCGTCTGTGCCTC-3′ and reverse 5′-GAGGCACAGACGCACGCGCCGCGGCCGCCGCTGAGCTACGTGCCTGACTTCTCGGACACCACTGTGGTCCC-3′; for mutant MIF-3, forward 5′-GGGACCACAGTGGTGTCCGAGAAGTCAGGCAAAAAGCTCAGCGGCGGCCGCGGCGCGTGCGTCTGTGCCT-C-3′ and reverse 5′-GAGGCACAGACGCACGCGCCGCGGCCGCCGCTGAGCTTTTTGCCTGACTTCTCGGACACCACTGTGGTCCC-3′; and for consensus HRE, forward 5′-TCTGTACGTGACCACACTCACCTCTCTGTACGTGACCACACTCACCTCTCTGTACGTGACCACA CTC-ACCTC-3′ and reverse 5′-GAGGTGAGTGTGGTCACGTACAGAGAGGTGAGTGTGGTCACGTACAGAGAGGTGAGTGTGGTCACGTACAGA-3′. (The mutated bases are underlined.)
Double-stranded biotinylated oligonucleotides were attached to streptavidin-coated Dynabeads, according to the recommendations of the manufacturer (Invitrogen, San Diego, CA). Nuclear extract prepared from CEMC7A cells was precleared using Dynabeads and poly(dI-dC) for 1 hour and 30 minutes. A total of 50 μg of nuclear extract was then incubated at 4°C for 1 hour and 45 minutes, with 10 μg of either HRE consensus, MIF-3, or mutated MIF probe coupled to the Dynabeads. After incubation, bound protein–DNA Dynabead complexes were isolated, resuspended in 1× Laemmli sample buffer (50 mM Tris HCl [pH 6.8], 2% SDS, 10% glycerol, and 5 mM DTT), and subjected to SDS-PAGE and immunoblot analysis.
Chromatin immunoprecipitation (ChIP) assay
Cells were grown to a final density of ~1 × 106 cells/sample and treated as indicated in the figure legends, and ChIP assay was performed as described previously (26). For immunoprecipitations, 4 μg of nonspecific rabbit IgG (Upstate Biotechnology, Lake Placid, NY) was used as a control, and 4 μg of anti-GRα M-20 antibody (Santa Cruz Biotechnology) or anti-acetylated H3K9 antibody (Upstate Biotechnology) was used. Real-time quantitative polymerase chain reaction was performed using an ABI 7000 instrument and SYBR Green Master Mix (Sigma), 0.2 μM primers, 1M betaine, and 4 μl of extracted DNA product. The primers used were as follows: for GAPDH, forward 5′-TTGCCATCAATGACCCCTTCA-3′ and reverse 5′-CGCCCCACTTGATTTTGGA-3′; and for MIF-5, forward 5′-GCGGTGACTTCCCCACTC-3′ and reverse 5′-TACGATGAACATCGGCATGA-3′.
Statistical analysis
Analysis of variance followed by Dunnett’s multiple comparison test or Bonferroni post-test was performed using GraphPad Prism software, version 4 (GraphPad Software, San Diego, CA). P values less than 0.05 were considered significant.
RESULTS
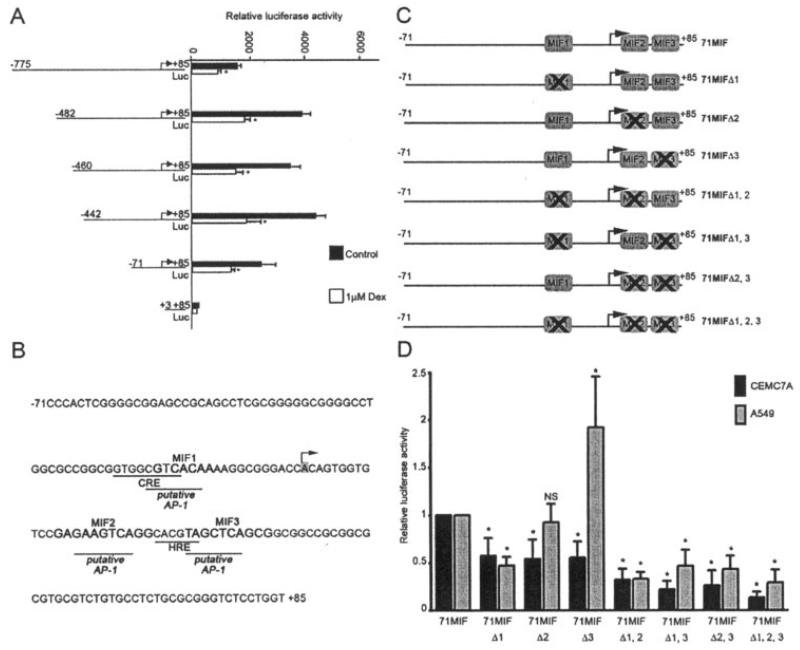
To identify the region of the MIF promoter required for basal promoter activity, as well as conferring glucocorticoid-dependent inhibition of MIF in CEMC7A cells, a series of reporter gene constructs composed of truncated versions of the MIF gene promoter were created by linearization and subsequent exonuclease III digestion of the MIF-173*G plasmid 5′ of the MIF promoter insert. The resulting constructs were transfected into CEMC7A cells and treated with 1 μM glucocorticoid. All of the truncated MIF gene constructs retained basal promoter activity, with the exception of the +3MIF deletant, and all were repressed by glucocorticoids (Figure 1A).
Figure 1.
Basal macrophage migration inhibitory factor (MIF) promoter activity is limited to positions −71 to +85 and putative transcription factor binding sites that regulate MIF promoter activity. A, Relative luciferase activity of MIF promoter constructs in CEMC7A cells. Cells were transfected with the indicated constructs and were left untreated (control) or were treated with 1 μM dexamethasone (DEX) for 20 hours. After incubation, luciferase assays were performed and values were normalized to Renilla. Bars show the mean and SD. * = P < 0.05 versus control. B, Analysis of positions −71 to +85 of the MIF promoter, using AliBaba version 2.1 to predict transcription factor binding sites. Previously identified sites and putative activator protein 1 (AP-1) sites are underlined. Regions of interest were termed MIF-1, MIF-2, and MIF-3, and bases are shown in boldface. The arrow shows the transcription start site. CRE = cAMP response element; HRE = hypoxia-responsive element. C, Reporter gene constructs containing deletions of the MIF-1, MIF-2, and MIF-3 sites. D, Relative luciferase activity of MIF promoter constructs in CEMC7A and A549 cells transfected with the constructs shown in C. Cells were harvested 20 hours after transfection. Luciferase assays were performed, and values were normalized to Renilla. Data represent normalized luciferase activity relative to that observed with the wild-type 71 MIF construct. Bars show the mean and SD. * = P < 0.01 versus wild-type 71 MIF. NS = not significant.
The minimal −71 to +85 MIF gene promoter responsible for basal promoter activity and conferring glucocorticoid-dependent inhibition was analyzed for GR and other transcription factor binding sites (Figure 1B). No GR consensus sites were found, but 3 putative cis regulatory sites termed MIF-1, MIF-2, and MIF-3 were present. These sites lie in close proximity to, or overlap, the previously characterized transcription factor binding sites CRE and HRE.
To determine whether these sites play a functional role in regulating basal MIF promoter activity, a series of reporter gene constructs containing deletions of the MIF-1, MIF-2, and MIF-3 sites were made. The sites were deleted singly and in combination, as shown schematically in Figure 1C, so that any functional interactions between the 3 sites could also be determined. These constructs were then transfected into CEMC7A cells or A549 cells. Deletions of each site, singly or in combination with the other sites, significantly reduced basal promoter activity in CEMC7A cells in all cases. In contrast, in A549 cells deletion of the MIF-1 site reduced activity, deletion of the MIF-2 site had no effect, and deletion of the MIF-3 site resulted in a significant increase in activity (Figure 1D). These data therefore suggest that all 3 sites play a role in regulating basal MIF promoter activity but that MIF-2 and MIF-3 may have different functions depending on cell context.
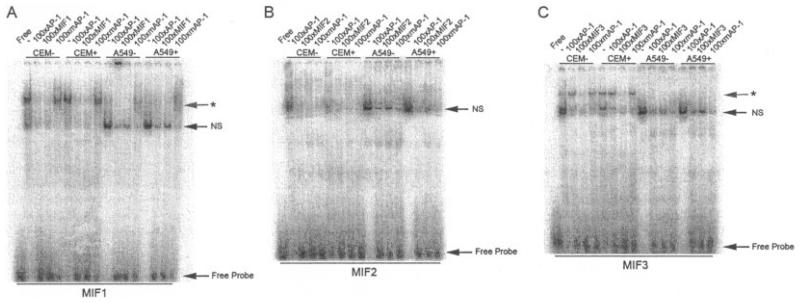
Next, EMSAs were performed using nuclear extracts from CEMC7A and A549 cells in order to determine transcription factor binding to MIF-1, MIF-2, and MIF-3. These assays showed that complexes bound all 3 oligonucleotides, irrespective of cellular glucocorticoid incubation (Figures 2A-C). However, competition experiments performed by adding excess corresponding cold MIF-specific oligonucleotides and mutated sequences (mutated AP-1) showed DNA sequence–specific complex binding only on oligonucleotides MIF-1 and MIF-3 (Figures 2A and C). In the case of MIF-1, the sequence–specific complex was also competed with addition of excess cold consensus AP-1 oligonucleotide, suggesting that this complex is indeed AP-1 or AP-1–like in nature; however, the complex appears to be present at a lower concentration in A549 nuclear extracts. EMSAs performed using MIF-3 (Figure 2C) revealed a sequence-specific complex present only in CEMC7A cells that was not competed by cold consensus AP-1 sequence.
Figure 2.
Sequence-specific complexes bind the MIF-1 and MIF-3 sites in the MIF promoter. A–C, Electrophoretic mobility shift assays of nuclear extracts prepared from untreated CEMC7A cells (CEM−), CEMC7A cells treated with 1 μM DEX for 20 hours (CEM+), untreated A549 cells (A549−), and A549 cells treated with 1 μM DEX for 20 hours (A549+) incubated with radiolabeled oligonucleotides corresponding to the putative transcription factor binding sites found in the MIF gene promoters MIF-1 (A), MIF-2 (B), and MIF-3 (C). Competition experiments were performed by adding unlabeled oligonucleotides as indicated. Asterisks show sequence-specific complexes. Results are representative of at least 3 separate experiments. NS = nonspecific complex (see Figure 1 for other definitions).
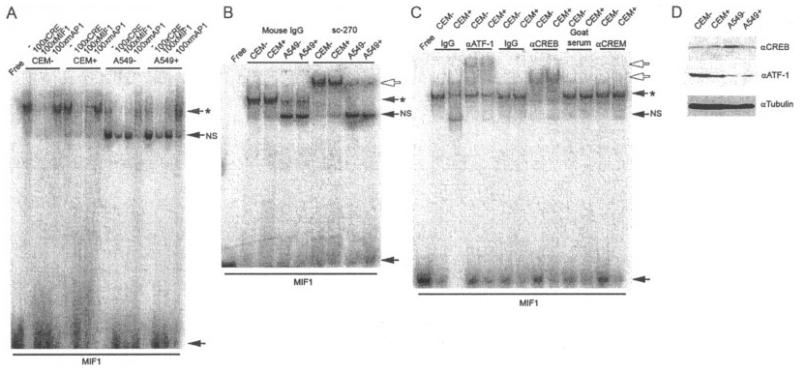
The MIF-1 complex encompasses a previously identified CRE site (27) (Figure 1B) and can be competed by a consensus CRE, in addition to the consensus AP-1 element (Figures 2A and 3A). Since the MIF-1 complex may comprise AP-1, CREB, or CREB-related proteins (components of which are known to heterodimerize [28]), EMSA super-shift analysis was performed. Numerous antibodies specific to components of the AP-1 complex were used, none of which resulted in a shift of the sequence-specific complex (results not shown); however, an antibody that cross-reacted with CREB, ATF-1, and CREM did super shift the sequence-specific complex in both cell lines, irrespective of cellular glucocorticoid incubation (Figure 3B). Further analysis using specific antibodies to these proteins demonstrated that the MIF-1 site can be bound by both ATF-1 and CREB (Figure 3C). Western blotting of cell lysates prepared from untreated and glucocorticoid-treated A549 and CEMC7A cells using antibodies specific to these proteins showed high expression of ATF-1 in CEMC7A cells both in the untreated and glucocorticoid-treated lysates, as compared with A549 cells (Figure 3D), explaining the more abundant MIF-1 complex in CEMC7A cells. CREB was detected in untreated and glucocorticoid-treated cells of both cell lines, with little difference in expression between cell lines, in contrast to the findings for ATF-1 (Figure 3D).
Figure 3.
The MIF-1 complex is composed of activating transcription factor 1 (ATF-1) and CREB. A, Electrophoretic mobility shift assay (EMSA) of nuclear extracts prepared from untreated CEMC7A cells (CEM−), CEMC7A cells treated with 1 μM DEX for 20 hours (CEM+), untreated A549 cells (A549−), and A549 cells treated with 1 μM DEX for 20 hours (A549+) incubated with radiolabeled oligonucleotides corresponding to MIF-1. Competition experiments were performed by adding unlabeled oligonucleotides as indicated. B, EMSA of nuclear extracts from treated and untreated CEMC7A and A549 cells and 2 μg of mouse IgG or sc-270, an antibody that cross-reacts with ATF-1, CREB, or CREM-1. C, EMSA of nuclear extracts from treated and untreated CEMC7A and A549 cells and 2 μg of mouse IgG, antibody to ATF-1 (αATF-1), rabbit IgG, antibody to CREB, goat serum, or antibody to CREM-1. Asterisks show sequence-specific complexes; open arrows show super-shifted complexes; solid arrows show free probe. NS = nonspecific complex. D, Immunoblot of whole cell lysates prepared from treated and untreated CEMC7A and A549 cells performed with anti-CREB, anti–ATF-1, and anti-tubulin antibodies. Results are representative of at least 3 separate experiments. See Figure 1 for other definitions.
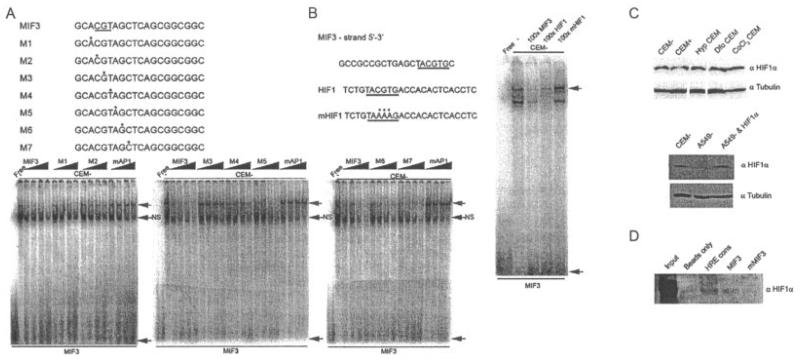
Competition and antibody super-shift studies of AP-1 or CREB-related proteins failed to identify binding of these transcription factors to the MIF-3 element, despite the sequence similarity to consensus-binding sites for these families of transcription factors (Figure 2C and results not shown). Therefore, to define the bases important for sequence-specific binding of this complex, a series of probes containing single-base purine–pyrimidine substitutions were used in competition experiments (Figure 4A). Point mutations of bases in the oligonucleotides M2, M3, and M4 resulted in loss of competition, indicating that a core CGT trinucleotide sequence is required for sequence-specific binding of the MIF-3 complex (Figure 4A), which is 5′ of the putative AP-1 site identified by bioinformatic analysis. This overlaps the previously identified HRE site (27) (Figure 1B). Nuclear extracts prepared from CEMC7A cells grown under normoxic conditions contained abundant HIF-1α, which did not appear to increase when cells were exposed to hypoxia or hypoxic mimetics. In contrast, in A549 cells, no HIF-1α expression was observed under normoxic conditions, indicating that HIF-1α may indeed be binding MIF-3 in CEMC7A cells under normoxic conditions (Figure 4C).
Figure 4.
The MIF-3 complex is composed of hypoxia-inducible factor 1α (HIF-1α). A, Electrophoretic mobility shift assays (EMSAs) of nuclear extracts prepared from untreated CEMC7A cells (CEM−) incubated with radiolabeled oligonucleotides corresponding to MIF-3. Competition experiments using increasing concentrations (5×−100×) of a series of probes (M1–M7) containing purine-pyrimidine substitutions were performed. Point mutations of bases in the oligonucleotides M2, M3, and M4 resulted in loss of competition. B, EMSA of nuclear extracts prepared from untreated CEMC7A cells incubated with radiolabeled oligonucleotide corresponding to MIF-3. Competition experiments were performed by adding unlabeled oligonucleotides as indicated. In A and B, asterisks show mutated bases in oligonucleotides used in the competition experiments. Nucleotides required for binding are underlined. Shaded arrows show sequence-specific complexes; solid arrows show free probe. NS = nonspecific complex. C, Immunoblots of nuclear extracts prepared from untreated CEMC7A cells, CEMC7A cells treated with 1 μM DEX for 20 hours (CEM+), CEMC7A cells grown under hypoxic (Hyp) conditions for 20 hours, CEMC7A cells treated with 100 μM desferrioxamine (Dfo) for 20 hours, CEMC7A cells treated with 100 μM CoCl2 for 20 hours, untreated A549 cells (A549–), and A549 cells transfected with HIF-1α expression vector. D, DNA affinity chromatography using CEMC7A nuclear extract and the HIF response element consensus (HRE cons), MIF-3, or mutated MIF-3 (mMIF-3) sequences. Bound protein–DNA complexes were subjected to immunoblotting. Results are representative of at least 3 separate experiments. See Figure 1 for other definitions.
To determine whether HIF-1α binds to MIF-3, consensus HIF elements were used in competition experiments (Figure 4B), although super-shift studies with HIF-1α–specific antibodies did not confirm that HIF-1α is a component of the MIF-3 complex(results not shown). However, DNA affinity chromatography using CEMC7A nuclear extract and the MIF-3 sequence as bait demonstrated that MIF-3 is an HIF-1α-binding sequence (Figure 4D). The lower abundance of HIF-1α binding to MIF-3 likely reflects the 3 HRE tandem repeats present in the HRE consensus oligonucleotide (Figure 4D). This suggests that the failure of HIF-1α–specific antibodies to super shift the MIF-3 complex in EMSAs is due to failure of antibody binding to the native conformation of the protein bound to DNA. ChIP studies of HIF-1α were unsuccessful, even on control genes, due to antibody limitations.
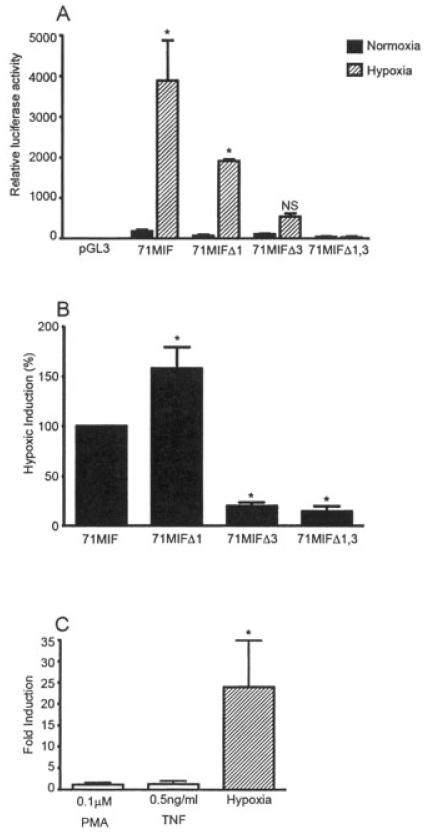
To determine whether the MIF-3 sequence confers hypoxia induction to the MIF promoter, the deletion constructs used previously (Figure 1C) were transfected into CEMC7A cells, and the cells were either left under normoxic conditions or placed in a hypoxic chamber for 20 hours. The wild-type MIF promoter was dramatically induced under hypoxic conditions, as was the construct containing the deletion of MIF-1. However, when either MIF-3 or both MIF-1 and MIF-3 were deleted, basal activity of the promoter was reduced, and hypoxia induction was lost, demonstrating that MIF-3 is indeed an HRE that also functions to regulate the basal promoter activity of the MIF gene under normoxic conditions in CEMC7A cells (Figures 5A-C). Further-more, hypoxia was the most powerful stimulus observed for MIF promoter activation. We found that hypoxia resulted in ~25-fold induction of promoter activity compared with minimal induction by phorbol myristate acetate (PMA; 1.2-fold) or TNFα (1.3-fold) (Figure 5C).
Figure 5.
The MIF-3 site is a functional hypoxia-responsive element. A, Relative luciferase activity in CEMC7A cells transfected with the indicated constructs. Cells were grown under hypoxic conditions for 20 hours or in control cultures (under normoxic conditions). After incubation, luciferase assays were performed, and luciferase values were normalized to protein content. Bars show the mean and SD. * = P < 0.01 versus cells grown under normoxic conditions. NS = not significant. B, Percentage induction of MIF promoter activity in the groups shown in A, where 100% represents hypoxic induction of wild-type 71 MIF, to aid comparison of the hypoxic induction of each of the constructs. Bars show the mean and SD. * = P < 0.01 versus 71 MIF. C, Fold induction of MIF promoter activity over control in CEMC7A cells transfected with the 71 MIF construct and treated with phorbol myristate acetate (PMA) or tumor necrosis factor (TNF) or grown under hypoxic conditions. Cells were treated for 20 hours prior to harvesting, when luciferase assays were performed. Luciferase values were normalized to protein content. Bars show the mean and SD. * = P < 0.01 versus control. See Figure 1 for other definitions.
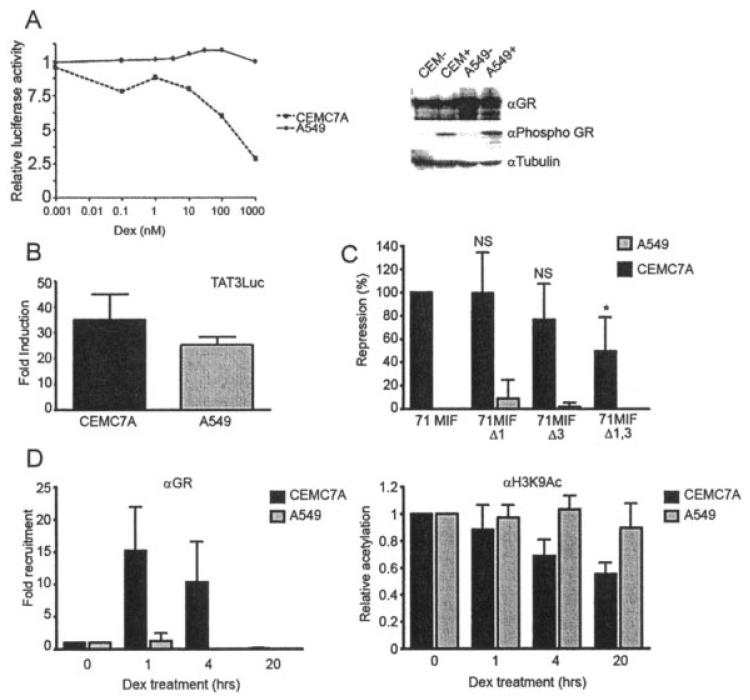
We next wanted to examine the role of the MIF-1 and MIF-3 elements in glucocorticoid repression of the MIF promoter in CEMC7A cells. As demonstrated previously (21), glucocorticoid treatment inhibited MIF promoter activity in CEMC7A cells in a dose-dependent manner, with ~50% inhibition occurring with 0.1 μM DEX. This inhibition was not observed in A549 cells, despite expression of GRα in A549 cells and appropriate induction of phosphoSer211 GRα in response to glucocorticoid treatment (Figure 6A). Importantly, this regulation of the MIF promoter was specific, since the cytomegalovirus–Renilla construct was not regulated by glucocorticoid treatment over this time course (data not shown). In fact, although both cell lines express GRα, there is clearly higher GRα expression in A549 cells, with commensurate greater induction of phosphoSer211 GRα after glucocorticoid treatment (Figure 6A). Also, glucocorticoid activates the GRα-responsive TAT3 promoter in both A549 cells and CEMC7A cells, demonstrating functional endogenous GRα in both cell types (Figure 6B) (29-31).
Figure 6.
Cell type–specific glucocorticoid-dependent repression of the MIF promoter acts via the MIF-1 and MIF-3 basal elements. A, Relative luciferase activity in A549 and CEMC7A cells transfected with the construct MIF-173*G and treated with the indicated concentrations of DEX for 20 hours (left). After incubation, luciferase assays were performed, and luciferase values were normalized to Renilla. An immunoblot of untreated CEMC7A cells (CEM−), CEMC7A cells treated with 1 μM DEX for 1 hour (CEM+), untreated A549 cells (A549−), and A549 cells treated with 1 μM DEX for 1 hour (A549+) is shown (right). B, Fold induction over control of the TAT3 promoter in CEMC7A and A549 cells treated with 1 μM DEX for 20 hours. After incubation, luciferase assays were performed, and luciferase values were normalized to Renilla. Bars show the mean and SD. C, Percentage repression of MIF promoter activity in CEMC7A and A549 cells transfected with the indicated constructs and treated with 1 μM DEX for 20 hours, where 100% represents DEX repression of 71 MIF. After incubation, luciferase assays were performed, and luciferase values were normalized to Renilla. Bars show the mean and SD. * = P < 0.01 versus 71 MIF. NS = not significant. D, Fold recruitment of glucocorticoid receptor α (GRα) to the MIF promoter (left) and relative acetylation of the MIF promoter (right) in CEMC7A and A549 cells treated with 1 μM DEX for the indicated time periods. Chromatin immunoprecipitation was performed using anti-GR or anti–acetylated H3K9 antibodies (αH3K9Ac). Results were normalized to those obtained using rabbit nonspecific IgG and DNA input. Bars show the mean and SD. See Figure 1 for other definitions.
Deletion of either MIF-1 or MIF-3 alone did not abolish the repressive effect of glucocorticoid on MIF promoter activity in CEMC7A cells; however, when both sites were deleted, there was a statistically significant reduction in the repressive effect of glucocorticoid treatment in this cell line (Figure 6C). In contrast, in A549 cells none of the constructs were repressed by glucocorticoid exposure (Figure 6C). We therefore sought evidence of recruitment of GRα to the endogenous MIF gene proximal promoter using ChIP. In CEMC7A cells, GRα was recruited within 1 hour of glucocorticoid treatment; by 4 hours, GRα abundance declined, and by 20 hours, there was very little GRα present (Figure 6D). In contrast, no recruitment of GRα to the MIF promoter was seen in A549 cells, demonstrating cell type–specific recruitment of GRα to the MIF promoter in CEMC7A cells. To determine whether the recruited GRα resulted in transcriptional repression, we also measured histone H3 acetylation of the proximal MIF promoter. These studies showed a progressive decline in acetyl H3 in CEMC7A cells following glucocorticoid exposure. Consistent with the lack of GRα recruitment, and the lack of promoter repression, there was no observed change in acetyl H3 associated with the MIF promoter in A549 cells (Figure 6D).
DISCUSSION
MIF is a key immunomodulatory molecule, capable of stimulating cyclooxygenase 2 and matrix metalloproteinase activity as well as counteracting the antiinflammatory effects of glucocorticoids (1,2,14,32). MIF has also been shown to be expressed at high levels in FLS, synovial fluid, and serum from RA patients, leading to the belief that MIF modulates the inflammatory response in the early stages of the disease (12). In addition, MIF has been shown to be regulated by hypoxia, to be proangiogenic in cancers, and to limit cell senescence (20,27,33,34). This is important because sites of inflammation have lower oxygen tension, and the attendant HIF-1α stabilization drives glycolysis as the principle source of cellular ATP. Activated GRα has been shown to potentiate HIF-1α transactivation of target genes, including glucose transporter 3 (22). While there is clear evidence of GRα regulation of MIF gene expression, this effect is dependent on cell context (21), and the mechanism of action is undefined.
Limited analysis of regulatory elements present in the MIF promoter has been performed previously; a CRE-binding protein CREB, which was present at positions −48 to −41 in the murine MIF promoter, was shown to be essential for corticotropin-releasing factor–induced MIF transcription (35). This corresponds to position −12 in the human MIF gene promoter (36). Later work confirmed that this site was indeed a CREB site in the human MIF promoter (27). In addition, an HRE capable of binding HIF-1α centered on position +25 in the 5′-untranslated region of the human gene has been reported to mediate MIF induction under hypoxic conditions (27). This sequence is partially conserved in the murine gene, centered on position +6. The murine gene element has been shown to be essential for hypoxic induction of the MIF promoter (20).
In this study, we identified the minimal MIF promoter sequence that is required for basal gene expression and discovered within this sequence the 2 regulatory elements that control MIF expression both in the basal state and in activated/repressed states. The first, MIF-1, encompasses the previously described CREB binding element (27,35), although bioinformatic analysis suggested greater homology with the AP-1 consensus sequence. The second, MIF-3, overlaps with the putative HIF-1α binding site (27). We have determined the exact location of these binding sites in the MIF promoter region, and shown that these sites function to regulate basal MIF gene expression and the response to hypoxia and glucocorticoids.
The results of reporter gene assays suggest that the MIF-1 site serves to potentiate basal gene expression of MIF in both CEMC7A cells and A549 cells. EMSA demonstrated that the MIF-1 complex was more abundant in CEMC7A cells than in A549 cells and appeared to consist of CREB and/or ATF-1. It is possible that this site binds heterodimers of CREB/ATF-1 as well as homodimers of CREB, since super-shift analysis using ATF-1–specific antibodies of the complex showed incomplete shifting, while a super-shift analysis using a CREB-specific antibody was complete.
We have demonstrated that MIF-3 regulates basal gene expression, contains a functional HRE, and binds a complex including HIF-1α even under normoxic conditions. Therefore, MIF-3 is important for both basal and hypoxic activation of the MIF promoter. Although HIF-1α is regulated by stabilization of the protein under hypoxic conditions, it can also be stabilized under normoxic conditions by growth factors and cytokines (37). Moreover, deletion of HIF-1α has been shown to inhibit basal production of MIF, even under normoxic conditions in mouse embryonic fibroblasts, indicating regulation of MIF by HIF under physiologic conditions in nontransformed cells (20). Our findings indicate greatly enhanced MIF promoter activation in hypoxia, which is mediated via the MIF-3 element, despite negligible changes in HIF-1α protein abundance. These findings suggest further posttranslational modification of HIF-1α, resulting in an enhanced transactivation, possibly via the recruitment of histone acetyltransferases (HATs) such as CREB binding protein (CBP)/p300 (38). Indeed, it would appear that hypoxia is a far more potent activator of transcription from the minimal MIF promoter than either of the proinflammatory mediators PMA and TNFα in CEMC7A cells.
Since GRα regulation of MIF may play a critical role in antiinflammation, GRα regulation of the minimal MIF promoter, via MIF-1 and MIF-3, was sought. Both MIF-1 and MIF-3 play a role in conferring GRα inhibition; EMSA studies did not show any differences in complex generation, suggesting a tethering mechanism of GRα action (39). ChIP was therefore used to confirm recruitment of GRα to the MIF gene promoter. We propose that GRα can be recruited to MIF-1 via ATF-1/CREB and to MIF-3 via HIF-1α. Indeed, an interaction between GRα and CREB has previously been demonstrated (40), and there is evidence of functional crosstalk between GRα and HIF-1α (22). The close proximity of these 2 elements makes distinguishing GRα binding between them in vivo by ChIP impossible, but clearly, both contribute to the observed glucocorticoid effect.
The failure of glucocorticoid treatment to repress the MIF gene in A549 cells is not due to lack of functional GRα, but likely reflects altered transcription factor loading, and so failure to recruit GRα (39). We have previously shown loading of GRα on the IL-8 promoter in response to glucocorticoids in A549 cells, suggesting that the failure to detect GRα recruitment to the proximal MIF promoter is a genuine, gene-specific effect (31). Recruitment of GRα to the MIF promoter is then proposed to assemble transcription comodulators and histone deacetylases (HDAs), such as HDA-2, and inhibit the recruitment of HATs such as CBP/p300, resulting in inhibition of gene transcription, as shown by ChIP for modified histone H3 (41,42).
These data have implications for inflammatory arthritis. The inflamed joint is known to be hypoxic (43); we propose that this results in augmented MIF expression, a consequence of which is an increase in the expression of mediators of inflammation, hyperplasia of the synovium, and the inhibition of the antiinflammatory effects of endogenous glucocorticoid. MIF is also known to lead to increased expression of HIF-1α (44), which in turn, further drives MIF expression (20). This feed-forward loop promotes local inflammation, glucocorticoid resistance, and joint destruction. This HIF-MIF circuit can be interrupted by activated GRα, acting through the 2 MIF regulatory elements MIF-1 and MIF-3. Thus, microenvironmental sensing via HIF-1α and glucocorticoid action can converge to regulate the expression of MIF.
ACKNOWLEDGMENTS
We would like to thank Kaye Williams and James Lynch for allowing the use of the hypoxic chambers and for informative discussions regarding hypoxia.
Supported by the Arthritis Research Campaign (ARC grants 16404 and 17900), the Manchester Academic Health Sciences Centre, and the NIHR Manchester Biomedical Research Centre. Dr. Beaulieu is recipient of a Frederik Craven Moore award.
REFERENCES
- 1.Bacher M, Metz CN, Calandra T, Mayer K, Chesney J, Lohoff M, et al. An essential regulatory role for macrophage migration inhibitory factor in T-cell activation. Proc Natl Acad Sci U S A. 1996;93:7849–54. doi: 10.1073/pnas.93.15.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calandra T, Bernhagen J, Metz CN, Spiegel LA, Bacher M, Donnelly T, et al. MIF as a glucocorticoid-induced modulator of cytokine production. Nature. 1995;377:68–71. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- 3.Senter PD, Al-Abed Y, Metz CN, Benigni F, Mitchell RA, Chesney J, et al. Inhibition of macrophage migration inhibitory factor (MIF) tautomerase and biological activities by acetamino-phen metabolites. Proc Natl Acad Sci U S A. 2002;99:144–9. doi: 10.1073/pnas.011569399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Jong YP, Abadia-Molina AC, Satoskar AR, Clarke K, Rietdijk ST, Faubion WA, et al. Development of chronic colitis is dependent on the cytokine MIF. Nat Immunol. 2001;2:1061–6. doi: 10.1038/ni720. [DOI] [PubMed] [Google Scholar]
- 5.Santos LL, Lacey D, Yang Y, Leech M, Morand EF. Activation of synovial cell p38 MAP kinase by macrophage migration inhibitory factor. J Rheumatol. 2004;31:1038–43. [PubMed] [Google Scholar]
- 6.Onodera S, Nishihira J, Koyama Y, Majima T, Aoki Y, Ichiyama H, et al. Macrophage migration inhibitory factor up-regulates the expression of interleukin-8 messenger RNA in synovial fibroblasts of rheumatoid arthritis patients: common transcriptional regulatory mechanism between interleukin-8 and interleukin-1β. Arthritis Rheum. 2004;50:1437–47. doi: 10.1002/art.20190. [DOI] [PubMed] [Google Scholar]
- 7.Sugie K, Tomura T, Takakura K, Kawano T, Taniguchi M, Grey HM, et al. Target cells for an immunosuppressive cytokine, glycosylation-inhibiting factor. Int Immunol. 1999;11:1149–56. doi: 10.1093/intimm/11.7.1149. [DOI] [PubMed] [Google Scholar]
- 8.Tomura T, Watarai H, Honma N, Sato M, Iwamatsu A, Kato Y, et al. Immunosuppressive activities of recombinant glycosylation-inhibiting factor mutants. J Immunol. 1999;162:195–202. [PubMed] [Google Scholar]
- 9.Watarai H, Nozawa R, Tokunaga A, Yuyama N, Tomas M, Hinohara A, et al. Posttranslational modification of the glycosylation inhibiting factor (GIF) gene product generates bioactive GIF. Proc Natl Acad Sci U S A. 2000;97:13251–6. doi: 10.1073/pnas.230445397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kudrin A, Scott M, Martin S, Chung CW, Donn R, McMaster A, et al. Human macrophage migration inhibitory factor: a proven immunomodulatory cytokine? J Biol Chem. 2006;281:29641–51. doi: 10.1074/jbc.M601103200. [DOI] [PubMed] [Google Scholar]
- 11.Santos L, Hall P, Metz C, Bucala R, Morand EF. Role of macrophage migration inhibitory factor (MIF) in murine antigen-induced arthritis: interaction with glucocorticoids. Clin Exp Immunol. 2001;123:309–14. doi: 10.1046/j.1365-2249.2001.01423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leech M, Metz C, Hall P, Hutchinson P, Gianis K, Smith M, et al. Macrophage migration inhibitory factor in rheumatoid arthritis: evidence of proinflammatory function and regulation by glucocorticoids. Arthritis Rheum. 1999;42:1601–8. doi: 10.1002/1529-0131(199908)42:8<1601::AID-ANR6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 13.Onodera S, Tanji H, Suzuki K, Kaneda K, Mizue Y, Sagawa A, et al. High expression of macrophage migration inhibitory factor in the synovial tissues of rheumatoid joints. Cytokine. 1999;11:163–7. doi: 10.1006/cyto.1998.0402. [DOI] [PubMed] [Google Scholar]
- 14.Onodera S, Kaneda K, Mizue Y, Koyama Y, Fujinaga M, Nishihira J. Macrophage migration inhibitory factor up-regulates expression of matrix metalloproteinases in synovial fibroblasts of rheumatoid arthritis. J Biol Chem. 2000;275:444–50. doi: 10.1074/jbc.275.1.444. [DOI] [PubMed] [Google Scholar]
- 15.Mikulowska A, Metz CN, Bucala R, Holmdahl R. Macrophage migration inhibitory factor is involved in the pathogenesis of collagen type II-induced arthritis in mice. J Immunol. 1997;158:5514–7. [PubMed] [Google Scholar]
- 16.Leech M, Metz C, Santos L, Peng T, Holdsworth SR, Bucala R, et al. Involvement of macrophage migration inhibitory factor in the evolution of rat adjuvant arthritis. Arthritis Rheum. 1998;41:910–7. doi: 10.1002/1529-0131(199805)41:5<910::AID-ART19>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 17.Santos LL, Dacumos A, Yamana J, Sharma L, Morand EF. Reduced arthritis in MIF deficient mice is associated with reduced T cell activation: down-regulation of ERK MAP kinase phosphorylation. Clin Exp Immunol. 2008;152:372–80. doi: 10.1111/j.1365-2249.2008.03639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leech M, Lacey D, Xue JR, Santos L, Hutchinson P, Wolvetang E, et al. Regulation of p53 by macrophage migration inhibitory factor in inflammatory arthritis. Arthritis Rheum. 2003;48:1881–9. doi: 10.1002/art.11165. [DOI] [PubMed] [Google Scholar]
- 19.Ichiyama H, Onodera S, Nishihira J, Ishibashi T, Nakayama T, Minami A, et al. Inhibition of joint inflammation and destruction induced by anti-type II collagen antibody/lipopolysaccharide (LPS)-induced arthritis in mice due to deletion of macrophage migration inhibitory factor (MIF) Cytokine. 2004;26:187–94. doi: 10.1016/j.cyto.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Welford SM, Bedogni B, Gradin K, Poellinger L, Broome PM, Giaccia AJ. HIF1α delays premature senescence through the activation of MIF. Genes Dev. 2006;20:3366–71. doi: 10.1101/gad.1471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alourfi Z, Donn RP, Stevens A, Berry A, McMaster A, Ray DW. Glucocorticoids suppress macrophage migration inhibitory factor (MIF) expression in a cell-type-specific manner. J Mol Endocrinol. 2005;34:583–95. doi: 10.1677/jme.1.01647. [DOI] [PubMed] [Google Scholar]
- 22.Kodama T, Shimizu N, Yoshikawa N, Makino Y, Ouchida R, Okamoto K, et al. Role of the glucocorticoid receptor for regulation of hypoxia-dependent gene expression. J Biol Chem. 2003;278:33384–91. doi: 10.1074/jbc.M302581200. [DOI] [PubMed] [Google Scholar]
- 23.Donn R, Alourfi Z, Zeggini E, Lamb R, Jury F, Lunt M, et al. A functional promoter haplotype of macrophage migration inhibitory factor is linked and associated with juvenile idiopathic arthritis. Arthritis Rheum. 2004;50:1604–10. doi: 10.1002/art.20178. [DOI] [PubMed] [Google Scholar]
- 24.Waters CE, Stevens A, White A, Ray DW. Analysis of co-factor function in a glucocorticoid-resistant small cell carcinoma cell line. J Endocrinol. 2004;183:375–83. doi: 10.1677/joe.1.05804. [DOI] [PubMed] [Google Scholar]
- 25.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elsby LM, O’Donnell AJ, Green LM, Sharrocks AD, Roberts SG. Assembly of transcription factor IIB at a promoter in vivo requires contact with RNA polymerase II. EMBO Rep. 2006;7:898–903. doi: 10.1038/sj.embor.7400767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baugh JA, Gantier M, Li L, Byrne A, Buckley A, Donnelly SC. Dual regulation of macrophage migration inhibitory factor (MIF) expression in hypoxia by CREB and HIF-1. Biochem Biophys Res Commun. 2006;347:895–903. doi: 10.1016/j.bbrc.2006.06.148. [DOI] [PubMed] [Google Scholar]
- 28.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci U S A. 1991;88:3720–4. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthews L, Berry A, Ohanian V, Ohanian J, Garside H, Ray D. Caveolin mediates rapid glucocorticoid effects and couples glucocorticoid action to the antiproliferative program. Mol Endocrinol. 2008;22:1320–30. doi: 10.1210/me.2007-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NFκB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14:2314–29. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garside H, Stevens A, Farrow S, Normand C, Houle B, Berry A, et al. Glucocorticoid ligands specify different interactions with NF-κB by allosteric effects on the glucocorticoid receptor DNA binding domain. J Biol Chem. 2004;279:50050–9. doi: 10.1074/jbc.M407309200. [DOI] [PubMed] [Google Scholar]
- 32.Sampey AV, Hall PH, Mitchell RA, Metz CN, Morand EF. Regulation of synoviocyte phospholipase A2 and cyclooxygenase 2 by macrophage migration inhibitory factor. Arthritis Rheum. 2001;44:1273–80. doi: 10.1002/1529-0131(200106)44:6<1273::AID-ART219>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 33.Koong AC, Denko NC, Hudson KM, Schindler C, Swiersz L, Koch C, et al. Candidate genes for the hypoxic tumor phenotype. Cancer Res. 2000;60:883–7. [PubMed] [Google Scholar]
- 34.Bacher M, Schrader J, Thompson N, Kuschela K, Gemsa D, Waeber G, et al. Up-regulation of macrophage migration inhibitory factor gene and protein expression in glial tumor cells during hypoxic and hypoglycemic stress indicates a critical role for angiogenesis in glioblastoma multiforme. Am J Pathol. 2003;162:11–7. doi: 10.1016/S0002-9440(10)63793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waeber G, Thompson N, Chautard T, Steinmann M, Nicod P, Pralong FP, et al. Transcriptional activation of the macrophage migration-inhibitory factor gene by the corticotropin-releasing factor is mediated by the cyclic adenosine 3′,5′-monophosphate responsive element-binding protein CREB in pituitary cells. Mol Endocrinol. 1998;12:698–705. doi: 10.1210/mend.12.5.0109. [DOI] [PubMed] [Google Scholar]
- 36.Paralkar V, Wistow G. Cloning the human gene for macrophage migration inhibitory factor (MIF) Genomics. 1994;19:48–51. doi: 10.1006/geno.1994.1011. [DOI] [PubMed] [Google Scholar]
- 37.Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1: interaction between H-ras and hypoxia. J Biol Chem. 2001;276:9519–25. doi: 10.1074/jbc.M010144200. [DOI] [PubMed] [Google Scholar]
- 38.Ema M, Hirota K, Mimura J, Abe H, Yodoi J, Sogawa K, et al. Molecular mechanisms of transcription activation by HLF and HIF1α in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18:1905–14. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMaster A, Ray DW. Drug insight: selective agonists and antagonists of the glucocorticoid receptor. Nat Clin Pract Endocrinol Metab. 2008;4:91–101. doi: 10.1038/ncpendmet0745. [DOI] [PubMed] [Google Scholar]
- 40.Imai E, Miner JN, Mitchell JA, Yamamoto KR, Granner DK. Glucocorticoid receptor-cAMP response element-binding protein interaction and the response of the phosphoenolpyruvate carboxykinase gene to glucocorticoids. J Biol Chem. 1993;268:5353–6. [PubMed] [Google Scholar]
- 41.Ito K, Barnes PJ, Adcock IM. Glucocorticoid receptor recruitment of histone deacetylase 2 inhibits interleukin-1β-induced histone H4 acetylation on lysines 8 and 12. Mol Cell Biol. 2000;20:6891–903. doi: 10.1128/mcb.20.18.6891-6903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fryer CJ, Kinyamu HK, Rogatsky I, Garabedian MJ, Archer TK. Selective activation of the glucocorticoid receptor by steroid antagonists in human breast cancer and osteosarcoma cells. J Biol Chem. 2000;275:17771–7. doi: 10.1074/jbc.M908729199. [DOI] [PubMed] [Google Scholar]
- 43.Taylor PC, Sivakumar B. Hypoxia and angiogenesis in rheumatoid arthritis. Curr Opin Rheumatol. 2005;17:293–8. doi: 10.1097/01.bor.0000155361.83990.5b. [DOI] [PubMed] [Google Scholar]
- 44.Oda S, Oda T, Nishi K, Takabuchi S, Wakamatsu T, Tanaka T, et al. Macrophage migration inhibitory factor activates hypoxia-inducible factor in a p53-dependent manner. PLoS ONE. 2008;3:e2215. doi: 10.1371/journal.pone.0002215. [DOI] [PMC free article] [PubMed] [Google Scholar]