Abstract
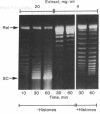
Extracts of Drosophila embryos can mediate the assembly of a chromatinlike structure from histones and DNA under physiological conditions. The histone-DNA complex formed in vitro contains micrococcal nuclease-sensitive sites spaced at 200-base pair intervals. More extensive digestion of the complex by micrococcal nuclease generates 11S particles which cosediment with nucleosome core particles isolated from native chromatin. These particles contain 140-base pair DNA fragments which upon further cleavage with micrococcal nuclease give rise to a pattern of discretely sized DNA fragments characteristic of nucleosome core particles. We have assayed the chromatin assembly process both qualitatively by measuring the induction of supertwists into a relaxed circular DNA (a process requiring a nicking-closing enzyme) and quantitatively by measuring the formation of micrococcal nuclease-resistant DNA fragments from radioactively labeled linear DNA. The amount of chromatin formed depends primarily on the amount of histones, whereas the rate of assembly depends on the amount of extract protein added. The factors in the extract that mediate chromatin assembly appear to interact first with the DNA because preincubation of the DNA with the extract markedly increases the extent of assembly.
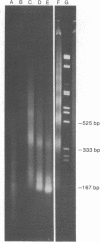
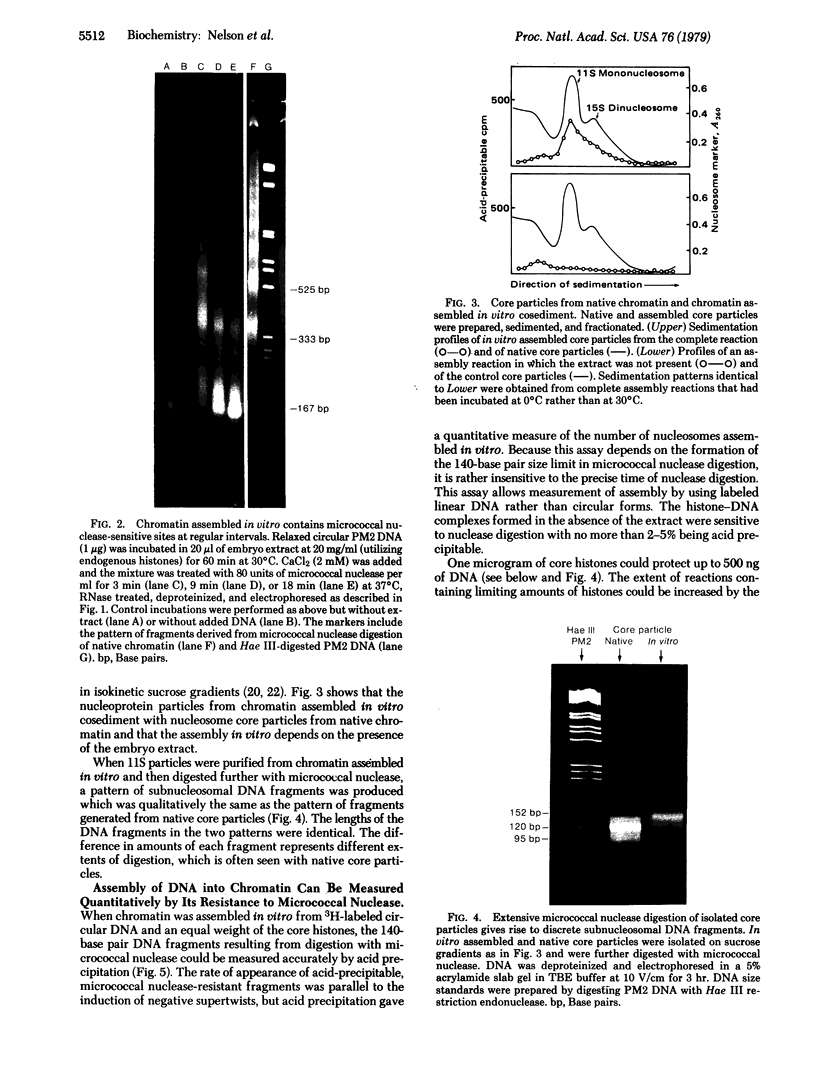
Full text
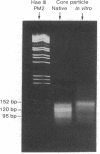
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blumenthal A. B., Kriegstein H. J., Hogness D. S. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Echalier G., Ohanessian A. In vitro culture of Drosophila melanogaster embryonic cells. In Vitro. 1970 Nov-Dec;6(3):162–172. doi: 10.1007/BF02617759. [DOI] [PubMed] [Google Scholar]
- Espejo R. T., Canelo E. S., Sinsheimer R. L. DNA of bacteriophage PM2: a closed circular double-stranded molecule. Proc Natl Acad Sci U S A. 1969 Aug;63(4):1164–1168. doi: 10.1073/pnas.63.4.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Bellard M., Oudet P., Chambon P. Stability of nucleosomes in native and reconstituted chromatins. Nucleic Acids Res. 1976 Nov;3(11):3173–3192. doi: 10.1093/nar/3.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Rouvière-Yaniv J., Yaniv M., Brutlag D. Nicking-closing enzyme assembles nucleosome-like structures in vitro. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3779–3783. doi: 10.1073/pnas.76.8.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. Assembly of new nucleosomal histones and new DNA into chromatin. Proc Natl Acad Sci U S A. 1978 May;75(5):2130–2134. doi: 10.1073/pnas.75.5.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V., Granner D., Chalkley R. Deposition of histone onto the replicating chromosome: newly synthesized histone is not found near the replication fork. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2266–2269. doi: 10.1073/pnas.73.7.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Kingsbury D. T., Helinski D. R. Stimulation by cyclic adenosine monophosphate of plasmid deoxyribonucleic acid replication and catabolite repression of the plasmid deoxyribonucleic acid-protein relaxation complex. J Bacteriol. 1973 May;114(2):577–591. doi: 10.1128/jb.114.2.577-591.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Honda B. M., Mills A. D., Finch J. T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978 Oct 5;275(5679):416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Morris N. R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977 Feb;10(2):237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Leffak I. M., Grainger R., Weintraub H. Conservative assembly and segregation of nucleosomal histones. Cell. 1977 Nov;12(3):837–845. doi: 10.1016/0092-8674(77)90282-3. [DOI] [PubMed] [Google Scholar]
- McEwen C. R. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal Biochem. 1967 Jul;20(1):114–149. doi: 10.1016/0003-2697(67)90271-0. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Panyim S., Jensen R. H., Chalkley R. Proteolytic contamination of calf thymus nucleohistone and its inhibition. Biochim Biophys Acta. 1968 Jun 26;160(2):252–255. doi: 10.1016/0005-2795(68)90095-0. [DOI] [PubMed] [Google Scholar]
- Seale R. L. Studies on the mode of segregation of histone nu bodies during replication in HeLa cells. Cell. 1976 Nov;9(3):423–429. doi: 10.1016/0092-8674(76)90087-8. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Camerini-Otero R. D., Felsenfeld G. Chromatin structure as probed by nucleases and proteases: evidence for the central role of histones H3 and H4. Cell. 1976 Sep;9(1):179–193. doi: 10.1016/0092-8674(76)90063-5. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Butler P. J. The nucleosome core protein. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):119–125. doi: 10.1101/sqb.1978.042.01.013. [DOI] [PubMed] [Google Scholar]