Abstract
A number of single nucleotide polymorphisms have been associated with disease predisposition in chronic lymphocytic leukemia. A single nucleotide polymorphism in the MDM2 promotor region, MDM2SNP309, was shown to soothe the p53 pathway. In the current study, we aimed to clarify the effect of the MDM2SNP309 on chronic lymphocytic leukemia characteristics and outcome. We performed a meta-analysis of data from 2598 individual patients from 10 different cohorts. Patients’ data and genetic analysis for MDM2SNP309 genotype, immunoglobulin heavy chain variable region mutation status and fluorescence in situ hybridization results were collected. There were no differences in overall survival based on the polymorphism (log rank test, stratified by study cohort; P=0.76; GG genotype: cohort-adjusted median overall survival of 151 months; TG: 153 months; TT: 149 months). In a multivariable Cox proportional hazards regression analysis, advanced age, male sex and unmutated immunoglobulin heavy chain variable region genes were associated with inferior survival, but not the MDM2 genotype. The MDM2SNP309 is unlikely to influence disease characteristics and prognosis in chronic lymphocytic leukemia. Studies investigating the impact of individual single nucleotide polymorphisms on prognosis are often controversial. This may be due to selection bias and small sample size. A meta-analysis based on individual patient data provides a reasonable strategy for prognostic factor analyses in the case of small individual studies. Individual patient data-based meta-analysis can, therefore, be a powerful tool to assess genetic risk factors in the absence of large studies.
Introduction
A number of single nucleotide polymorphisms (SNP) of different genes have been associated with disease predisposition.1–4 While the potential effects of gene polymorphisms are widely acknowledged, most studies investigating the impact of individual SNPs on prognosis in chronic lymphocytic leukemia (CLL) have been controversial.5–11 None of the allelic variants described so far contribute to relevant current clinical risk models in CLL.12,13
Sample sizes of studies to detect associations between SNP markers and clinical end points should be reasonably large, taking into account the very large number of candidate SNPs. In the absence of large studies, individual patient data (IPD)-based meta-analyses can provide a reliable assessment of possibly prognostic SNP markers. However, few IPD-based meta-analyses on SNP data have been performed so far.
A number of years ago, a single nucleotide polymorphism in the MDM2 promotor region (IVS1+309) was discovered and shown to soothe the p53 pathway by influencing MDM2 transcript and protein levels.14,15 Patients with the Li-Fraumeni syndrome develop cancers a decade earlier, when carrying the G/G allele of MDM2SNP309. These important findings have led to the investigation of the role of the MDM2 polymorphism in a variety of cancers with conflicting results. A combined meta-analysis of breast, colorectal and lung cancers found no effect of the SNP on predisposition in colorectal and breast cancer, but showed a small but significant predisposition to lung cancer.16 Most positive findings on the MDM2SNP309 genotype were related to disease onset rather than outcome.16 The exceptionally well documented functional consequences of the MDM2SNP309 and the important prognostic role of p53,17,18 as well as the particularly striking effects of the SNP in CLL in one study, has led to the study of the MDM2SNP309 in many centers, providing conflicting results.5,6,9–11,19
In the current study, we aimed to clarify the prognostic value of MDM2SNP309, as well as the correlation between this SNP and the characteristics of CLL, by using an IPD-based meta-analysis.20 To this end, we have collected data from 10 European centers contributing pre-determined clinical and genetic information on their patient cohorts. The combined data set is the largest cohort analyzed for the prognostic impact of genetic risk groups in CLL and thus constitutes a powerful data set to approach prognostic models in this disease.
Methods
Cohorts
This meta-analysis includes individual patient data from 10 cohorts (n=2598) (Figure 1). The cohorts have, in part, been published previously.5–9,11 Two data sets were unavailable either because of regulatory issues21 or due to the decision of the investigator.22,23 Tables 1 and 2 show the characteristics of the meta-analysis data set. Details are provided in the Online Supplementary Appendix.
Figure 1.
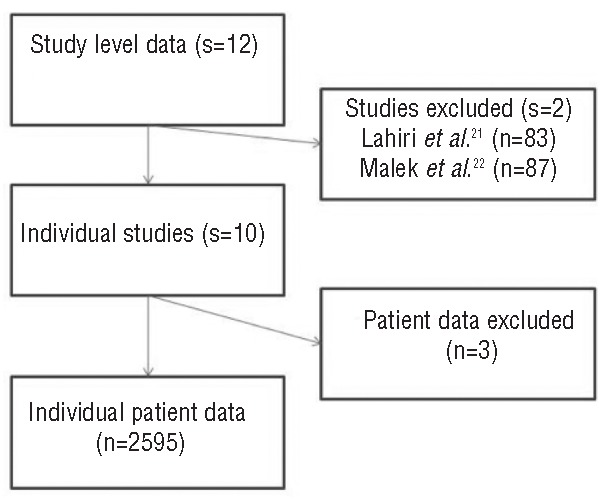
Summary of individual studies as reported.
Table 1.
Comparison of clinical and biological characteristics between the ten study cohorts.
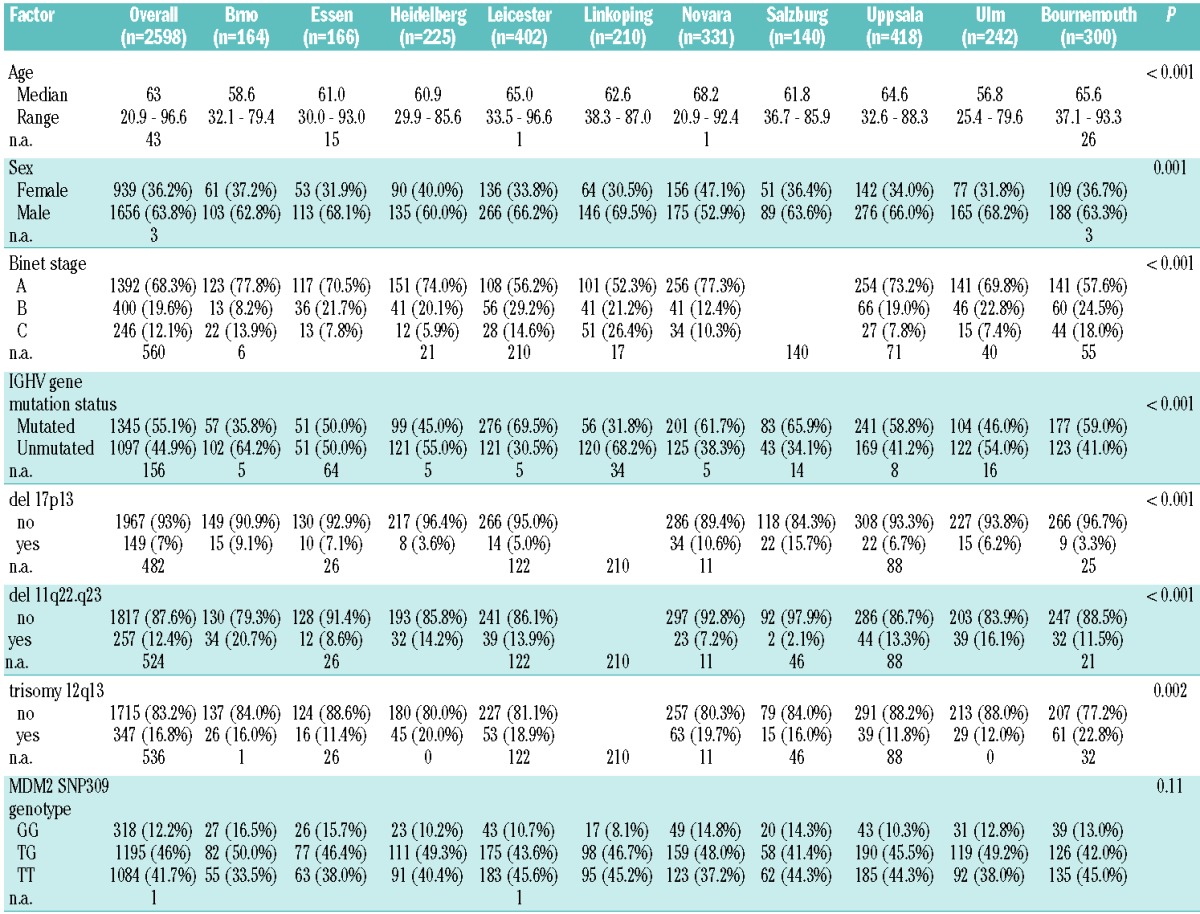
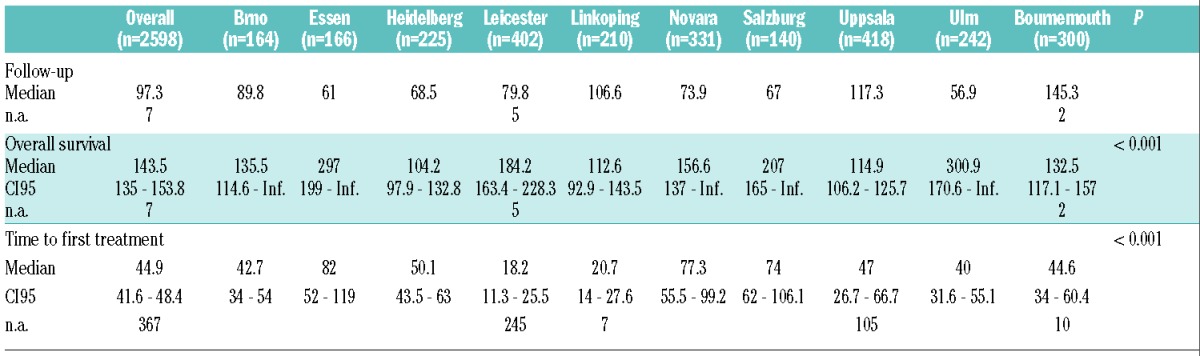
Table 2.
Comparison of time-to-event characteristics between the ten study cohorts.
Statistical analysis
Meta-analyses are applied to combine the results of related studies to produce an overall effect estimate. The large majority of meta-analyses use summary statistics supplied by the trial investigators or extracted from trial reports. Trial-level results are then combined to produce an overall effect estimate. A more powerful and less biased approach collects individual patient data (IPD) directly from the researchers responsible for each trial. Use of IPD has several advantages over the aggregate data approach, including standardization of statistical analyses, the assessment of potential causes of heterogeneity, and the investigation of interactions and non-linear effects. IPD-based meta-analysis is especially useful when time-to-event data are of interest. For survival analysis, the approach uses individual survival times and takes account of censoring. This provides a more powerful and flexible approach compared to the use of aggregate data since power is lost if time is ignored.
In our study, we apply an approach based on a single proportional hazards regression model stratified by trial. In order to incorporate random effects, a mixed-effect regression model is used. The primary end point of the IPD-based meta-analysis study presented here was overall survival (OS), and the second was time to first treatment (TFT). OS was defined as the time from diagnosis to death (event) or last follow up. TFT was defined as the time interval between the date of diagnosis and date of first CLL-specific treatment or death. TFT was censored at last treatment-free follow-up date. The log rank test, stratified by center, was used to compare survival curves with respect to MDM2SNP309 genotypes. For multivariable survival analysis, the proportional hazards regression model of Cox was applied including patients’ sex, age, IGVH gene mutation status and FISH results. For the MDM2SNP309, an additive genetic model was used with values 0, 1, and 2, indicating the number of copies of the variant G allele for each patient. A stratified approach was used, allowing each center to have a different base-line hazard. We applied mixed effects Cox proportional hazards models with a random center effect to consider between-center heterogeneity.24 Univariable survival distributions according to genotype are provided as direct adjusted survival curves, adjusted for cohort.25 Forest plots were used to present results for individual centers and the combined analysis. Cochran’s Q was applied to test heterogeneity, whereas the I2 index was used to quantify the degree of heterogeneity between cohorts. Bias and systematic heterogeneity was checked by contour-enhanced funnel plots, where the standard error is plotted against the log hazard ratio estimate.26 The contour enhanced funnel plots also indicate regions of statistical significance at levels of 0.01, 0.05 and 0.1, testing the null hypotheses that log hazard ratios are equal to zero.
Results
A total of 2598 patients were included in this study, gathered from 10 different single-center series from Europe. The comparison of the base-line characteristics of different cohorts is shown in Table 1, while time-to-event data (survival time, time to first treatment) are shown in Table 2. Median overall survival from diagnosis ranged from a median of 104 months to 301 months. Not surprisingly, there was a difference in outcome of the cohorts with respect to overall survival and time to first treatment reflecting the patient cohort differences and the respective referral patterns. As expected, the most important known genetic risk factors (17p-, 11q-, unmutated IGHV genes) showed hierarchical impact on OS (data not shown).
MDM2SNP309 genotype and disease characteristics
There was no significant difference in genotype distribution among cohorts (P=0.11). Furthermore, the MDM2SNP309 was not associated with particular disease characteristics, such as age of onset, 17p-, 11q-, +12q, or IGHV gene mutation status (Table 3). To assess the potential influence of the genotype on the patients’ prognosis, we analyzed TFT and OS using an additive genetic model. Results of the analyses of OS in individual cohorts and the summarizing IPD meta-analysis are illustrated by the forest plot shown in Figure 2. The funnel plot of individual studies was roughly symmetrical, indicating no evidence of publication bias in this study (Figure 3). Results for TFT were comparable. To account for heterogeneity, meta-analysis was carried out stratified for study cohorts. Stratified log rank tests showed no significant differences based on the polymorphism with respect to OS and TFT (P=0.76 and P=0.58, respectively). Specifically, the GG genotype group had a cohort-adjusted median OS of 151 months compared to 153 months for the TG genotype group and 149 months for the TT genotype group (Figure 4). Cohort-adjusted median TFT was 43 months for the GG genotype group, and 42 months for the TG and the TT genotype group.
Table 3.
Association of the MDM2SNP309 genotype and patients’ characteristics.
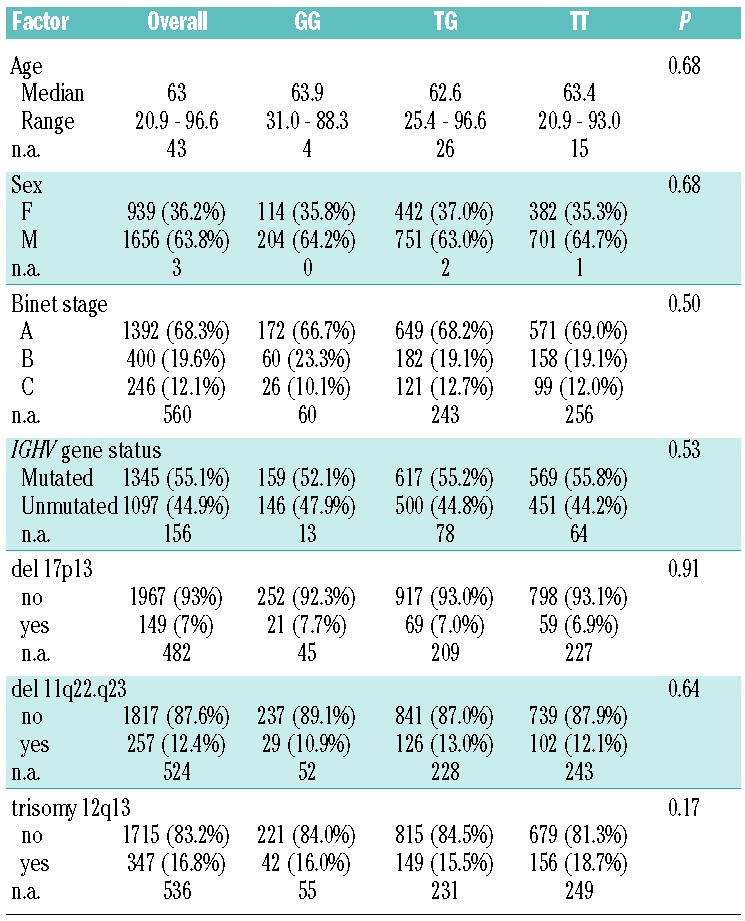
Figure 2.
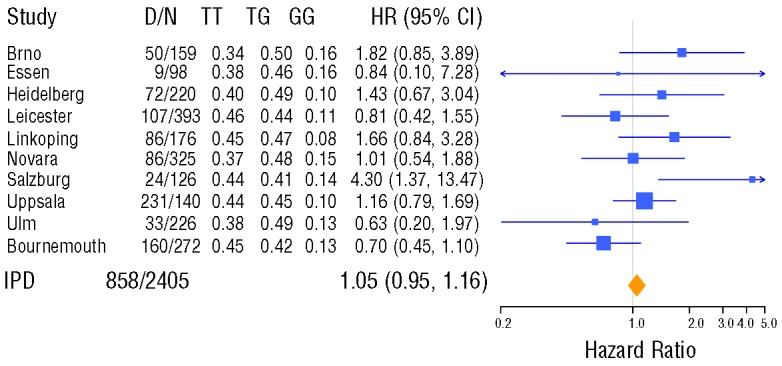
Forest plot of the IPD-based meta-analysis for overall survival. The plot shows the individual results of univariable Cox regression models for each study cohort together with the meta-analysis result with respect to the MDM2SNP309 genotype using an additive genetic model. The estimated hazard ratios, HR, together with their 95% confidence intervals are shown numerically and graphically. In addition the numbers of events, D, and observations, N, as well as the proportions of the three genotypes are provided. The results of individual studies are shown as squares centered on each cohort’s point estimate of the hazard ratio (HR). Size of a square represents the precision of the estimate (the inverse of the squared standard error). 95% confidence intervals are represented by horizontal lines. The overall HR estimate from the IPD meta-analysis and its confidence interval are shown at the bottom, represented by a diamond. Testing heterogeneity resulted in Q(df=9) = 14.9, P = 0.10, with I2 =39%.
Figure 3.
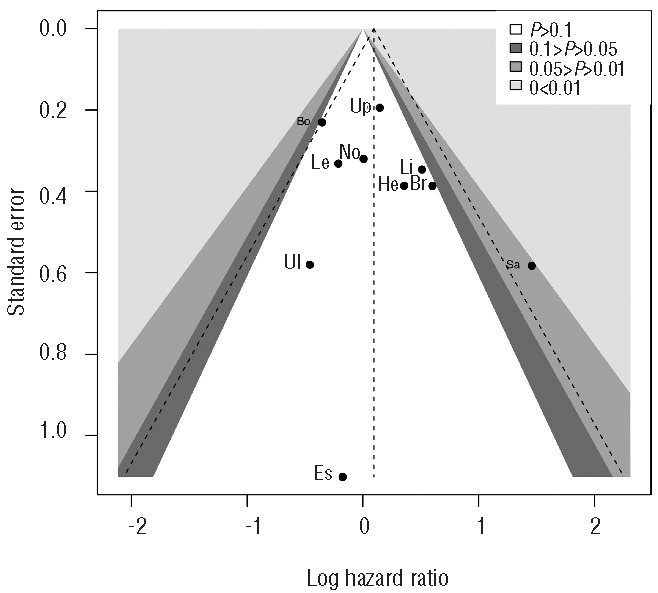
Contour-enhanced funnel plot of the OS analysis. The black dots indicate the cohort-specific estimates of the log hazard ratio and their standard errors; the vertical dashed line shows the summary estimate. Cohorts are indicated by the first two letters of their names. The shaded regions reveal the P value in each study relating to the test of the null hypothesis that the log (hazard ratio) is zero. The funnel plot of the OS analyses with respect to MDM2SNP309 genotypes is roughly symmetrical, indicating no evidence of publication bias in this study.
Figure 4.
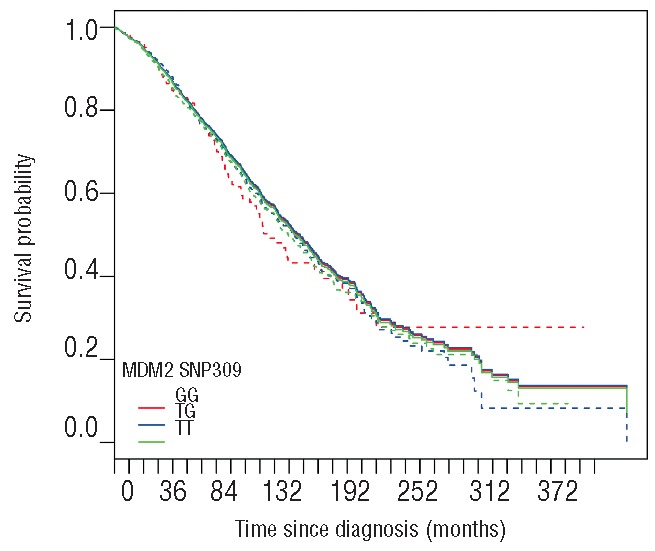
Estimated distributions of overall survival. The solid lines show the direct adjusted estimates of survival curves corresponding to MDM2 genotype. Adjustment was made with respect to the different cohorts of the study. Dashed lines represent the unadjusted Kaplan-Meier estimates.
Because of the known interaction of MDM2 with the p53 pathway, we also assessed the impact of the SNP in the group with and the group without 17p deletion. In the subset of patients without 17p deletion, the survival distribution in the sets defined by the three genotypes were comparable (stratified log rank test: P=0.58). Accordingly, no statistically significant impact of the genotype could be demonstrated in the subset with 17p deletion (stratified log rank test: P=0.56). Similar results were obtained for TFT.
Multivariable meta-analysis
We performed a cohort-stratified Cox proportional hazards regression analysis to assess the impact of the MDM2 genotype on overall and treatment-free survival considering known prognostic factors. The first model included MDM2 genotype, sex, age and IGHV gene mutation status as well as the stratification according to cohorts. As shown in Table 4, advanced age, male sex and unmutated IGHV genes were significantly associated with inferior survival but not the MDM2 genotype (P=0.31). Results for TFT were comparable.
Table 4.
Cox regression models for overall survival, stratified by study center. Stratified Cox proportional hazards model including covariates age, sex, IGHV mutation status and MDM2SNP309 (10 centers) and extended model, additionally including cytogenetic aberrations (17p-,11q- and +12q; 9 centers).
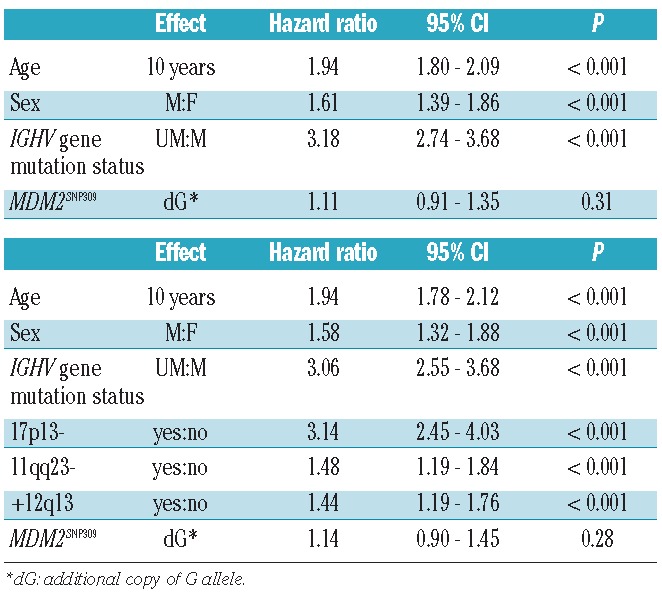
In a separate model, we included cytogenetic aberrations (17p-, 11q-, +12q). In this model, sex, age, IGHV gene mutation status, and cytogenetic aberrations were associated with increased risk of death. Again, the MDM2SNP309 genotype was not significantly associated with overall survival (P=0.28; Table 4). Considering treatment-free survival, age (P=0.10) and the MDM2SNP309 genotype (P=0.85) were the only factors that were not statistically significant.
Discussion
The current study used individual patient data to assess the role of a particular host genetic factor in CLL. In contrast to other large studies, we focused on the prognostic impact of the MDM2 polymorphism. The size of the cohort allowed us to perform subgroup analysis with sufficient statistical power to account for the biological heterogeneity of CLL.
In response to cellular stress, the p53 protein is stabilized and regulates the activity of key effectors of cellular processes, such as DNA repair, cell-cycle arrest, senescence, and apoptosis. These p53-mediated responses are crucial both in reducing cancer frequency and in mediating the response of commonly used cancer therapies. Not surprisingly, p53 loss or mutation leads to very poor outcome in hematologic malignancies, and the impact is particularly striking in CLL. There is a large body of evidence suggesting that the p53 pathway harbors functional inherited single-nucleotide polymorphisms (SNPs) that affect p53 signaling in cells, resulting in differences in cancer risk and clinical outcome in humans (reviewed by Grochola et al.27). Recent elegant work with mice carrying the MDM2SNP309G allele showed that MDM2SNP309G/G cells exhibit elevated mdm2 levels, reduced p53 levels, and decreased apoptosis with the MDM2SNP309G allele potentiating the tumor phenotype, and altered the tumor spectrum in mice inheriting a p53 hot-spot mutation.28
Although the number of studies on the prognostic impact of individual SNPs in cancer, including CLL, is rapidly increasing, the validation of the results obtained in these studies is often disappointing. Meta-analysis has become an essential part of the process of SNP evaluation. In CLL, IPD-based meta-analysis have so far been limited to clinical trial analysis focusing on clinical questions such as early versus late treatment,29 immunoglobulin prophylaxis30 or response to treatment.31,32 This is, therefore, the first IPD based meta-analysis in which the clinical and prognostic significance of a biomarker such as MDM2 gene polymorphism is evaluated. To that purpose, we have compiled a unique data set of 2598 patients from 10 centers using IPD-based meta-analysis to analyze the association between MDM2SNP309 polymorphism, genetic and clinical features and clinical outcome in CLL. We observed a strong heterogeneity in the cohorts studied, likely to be due to referral patterns and health care differences across countries. The incidence of the MDM2SNP309 genotype in different studies argues for comparability of the different techniques used to detect it.
In contrast to single center studies, we did not find a correlation between MDM2SNP309, disease characteristics and outcome, including patients’ age at diagnosis, genetic lesions, IGHV mutations, TFT or overall survival. On the other hand, the current study provides a unique dataset to estimate the impact of genetic aberrations and IGHV mutational status for outcome of patients with CLL. There are additional smaller studies that we have not been able to include in the analysis for various reasons;21–23,33 based on the limited number of cases involved the omission is unlikely to affect the results.
In our analysis, we have used an additive genetic model considering the number of copies of the variant G allele and stratification by study cohort. We have also analyzed the data using mixed effects Cox models with random cohort effect providing comparable results (data not shown). One shortcoming of our study is the lack of data on TP53 mutations, which are now accepted as markers of poor prognosis in CLL but which were not widely available when the series analyzed here were investigated. The analysis within the group of patients with 17p deletion did not show an impact of the MDM2SNP309 suggesting that, in CLL, interaction with p53 status is unlikely. While no sample exchange and validation experiments were performed, the incidence of the SNP allele distribution was similar across centers suggesting that technology reproducibility was not a confounding factor. While comprehensive data on the mRNA or Protein expression of mdm2 in CLL are missing, a larger study detected no impact of the MDM2SNP309 on mRNA levels.11 Based on our data, further meta-analyses are encouraged; this will help classify and develop the hierarchy of relevant prognostic factors in this disease. This is particularly important as, with the identification of novel mutations (e.g. SF3B1, NOTCH1, BIRC3, ATM, MYD88), the number of prognostic factors has increased, necessitating large cohorts to avoid over-fitted models.
Footnotes
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Di Bernardo MC, Crowther-Swanepoel D, Broderick P, Webb E, Sellick G, Wild R, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40(10):1204–10 [DOI] [PubMed] [Google Scholar]
- 2.Crowther-Swanepoel D, Broderick P, Di Bernardo MC, Dobbins SE, Torres M, Mansouri M, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 2010;42(2):132–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Bernardo MC, Broderick P, Catovsky D, Houlston R. Common genetic variation contributes significantly to the risk of developing chronic lymphocytic leukemia. Haematologica. 2013;98(3):e23–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slager SL, Skibola CF, Di Bernardo MC, Conde L, Broderick P, McDonnell SK, et al. Common variation at 6p21.31 (BAK1) influences the risk of chronic lymphocytic leukemia. Blood. 2012;120(4):843–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gryshchenko I, Hofbauer S, Stoecher M, Daniel PT, Steurer M, Gaiger A, et al. MDM2 SNP309 is associated with poor outcome in B-cell chronic lymphocytic leukemia. J Clin Oncol. 2008;26(14):2252–7 [DOI] [PubMed] [Google Scholar]
- 6.Kaderi MA, Mansouri M, Zainuddin N, Cahill N, Gunnarsson R, Jansson M, et al. Lack of association between the MDM2 promoter polymorphism SNP309 and clinical outcome in chronic lymphocytic leukemia. Leuk Res. 2010;34(3):335–9 [DOI] [PubMed] [Google Scholar]
- 7.Majid A, Richards T, Dusanjh P, Kennedy DBJ, Miall F, Gesk S, et al. TP53 codon 72 polymorphism in patients with chronic lymphocytic leukaemia: identification of a subgroup with mutated IGHV genes and poor clinical outcome. Br J Haematol. 2011; 153(4):533–5 [DOI] [PubMed] [Google Scholar]
- 8.Rasi S, Forconi F, Bruscaggin A, Sozzi E, Gaidano G, Rossi D. Impact of the host genetic background on prognosis of chronic lymphocytic leukemia. Blood. 2010;115(5):1106–7 [DOI] [PubMed] [Google Scholar]
- 9.Willander K, Ungerback J, Karlsson K, Fredrikson M, Soderkvist P, Linderholm M. MDM2 SNP309 promoter polymorphism, an independent prognostic factor in chronic lymphocytic leukemia. Eur J Haematol. 2010;85(3):251–6 [DOI] [PubMed] [Google Scholar]
- 10.Zenz T, Benner A, Stilgenbauer S. MDM2 promotor polymorphism and disease characteristics in CLL. Leuk Res. 2010;34(5):578–9 [DOI] [PubMed] [Google Scholar]
- 11.Zenz T, Habe S, Benner A, Kienle D, Dohner H, Stilgenbauer S. The MDM2 - 309 T/G promoter single nucleotide polymorphism does not alter disease characteristics in chronic lymphocytic leukemia. Haematologica. 2008; 93(7):1111–3 [DOI] [PubMed] [Google Scholar]
- 12.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008; 111(12):5446–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10(1):37–50 [DOI] [PubMed] [Google Scholar]
- 14.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119(5):591–602 [DOI] [PubMed] [Google Scholar]
- 15.Bond GL, Hu W, Levine A. A single nucleotide polymorphism in the MDM2 gene: from a molecular and cellular explanation to clinical effect. Cancer Res. 2005; 65(13):5481–4 [DOI] [PubMed] [Google Scholar]
- 16.Wilkening S, Bermejo JL, Hemminki K. MDM2 SNP309 and cancer risk: a combined analysis. Carcinogenesis. 2007; 28(11):2262–7 [DOI] [PubMed] [Google Scholar]
- 17.Zenz T, Mohr J, Edelmann J, Sarno A, Hoth P, Heuberger M, et al. Treatment resistance in chronic lymphocytic leukemia: the role of the p53 pathway. Leuk Lymphoma 2009; 50(3):510–3 [DOI] [PubMed] [Google Scholar]
- 18.Zenz T, Mertens D, Dohner H, Stilgenbauer S. Importance of genetics in chronic lymphocytic leukemia. Blood Rev. 2011;25(3):131–7 [DOI] [PubMed] [Google Scholar]
- 19.Asslaber D, Pinon JD, Seyfried I, Desch P, Stocher M, Tinhofer I, et al. microRNA-34a expression correlates with MDM2 SNP309 polymorphism and treatment-free survival in chronic lymphocytic leukemia. Blood. 2010;115(21):4191–7 [DOI] [PubMed] [Google Scholar]
- 20.Stewart LA, Tierney JF. To IPD or not to IPD¿ Eval Health Prof. 2002;25(1):76–97 [DOI] [PubMed] [Google Scholar]
- 21.Lahiri O, Harris S, Packham G, Howell M. p53 pathway gene single nucleotide polymorphisms and chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2007; 179(1):36–44 [DOI] [PubMed] [Google Scholar]
- 22.Malek SN. MDM2-SNP 309 allele status does not affect sensitivity to MDM2 inhibitors in CLL. Blood. 2008;112(5):2169 [Google Scholar]
- 23.Saddler C, Ouillette P, Kujawski L, Shangary S, Talpaz M, Kaminski M, et al. Comprehensive biomarker and genomic analysis identifies p53 status as the major determinant of response to MDM2 inhibitors in chronic lymphocytic leukemia. Blood. 2008;111(3):1584–93 [DOI] [PubMed] [Google Scholar]
- 24.Therneau TM, Grambsch PM, Pankratz VS. Penalized survival models and frailty. J Comp Graph Stat. 2003;12(1):156–75 [Google Scholar]
- 25.Therneau TM, Grambsch PM. Modeling survival data: Extending the Cox model. 1st ed New York: Springer; 2000. p. 261–87 [Google Scholar]
- 26.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Contour-enhanced meta-analysis funnel plots help distinguish publication bias from other causes of asymmetry. J Clin Epidemiol. 2008;61(10):991–6 [DOI] [PubMed] [Google Scholar]
- 27.Grochola LF, Zeron-Medina J, Meriaux S, Bond GL. Single-nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb Perspect Biol. 2010;2(5):a001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Post SM, Quintas-Cardama A, Pant V, Iwakuma T, Hamir A, Jackson JG, et al. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell. 2010;18(3):220–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mhaskar AR, Quinn G, Vadaparampil S, Djulbegovic B, Gwede CK, Kumar A. Timing of first-line cancer treatments - early versus late - a systematic review of phase III randomized trials. Cancer Treat Rev. 2010;36(8):621–8 [DOI] [PubMed] [Google Scholar]
- 30.Raanani P, Gafter-Gvili A, Paul M, Ben-Bassat I, Leibovici L, Shpilberg O. Immunoglobulin prophylaxis in chronic lymphocytic leukemia and multiple myeloma: systematic review and meta-analysis. Leuk Lymphoma. 2009;50(5):764–72 [DOI] [PubMed] [Google Scholar]
- 31.Chemotherapeutic options in chronic lymphocytic leukemia: a meta-analysis of the randomized trials. CLL Trialists’ Collaborative Group. J Natl Cancer Inst. 1999;91(10):861–8 [DOI] [PubMed] [Google Scholar]
- 32.Steurer M, Pall G, Richards S, Schwarzer G, Bohlius J, Greil R. Purine antagonists for chronic lymphocytic leukaemia. Cochrane Database Syst Rev. 2006;3:CD004270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong HJ, Fang C, Fan L, Zhu DX, Wang DM, Zhu HY, et al. MDM2 promoter SNP309 is associated with an increased susceptibility to chronic lymphocytic leukemia and correlates with MDM2 mRNA expression in Chinese patients with CLL. Int J Cancer. 2012;130(9):2054–61 [DOI] [PubMed] [Google Scholar]


