Abstract
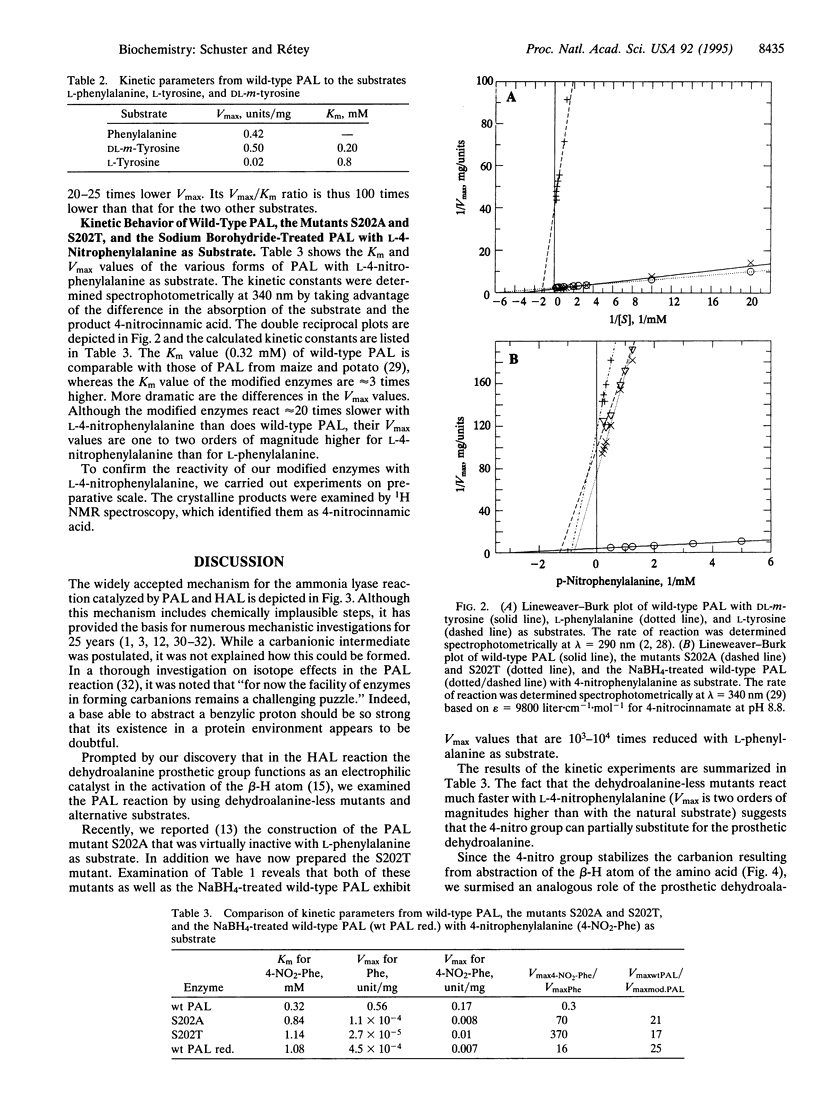
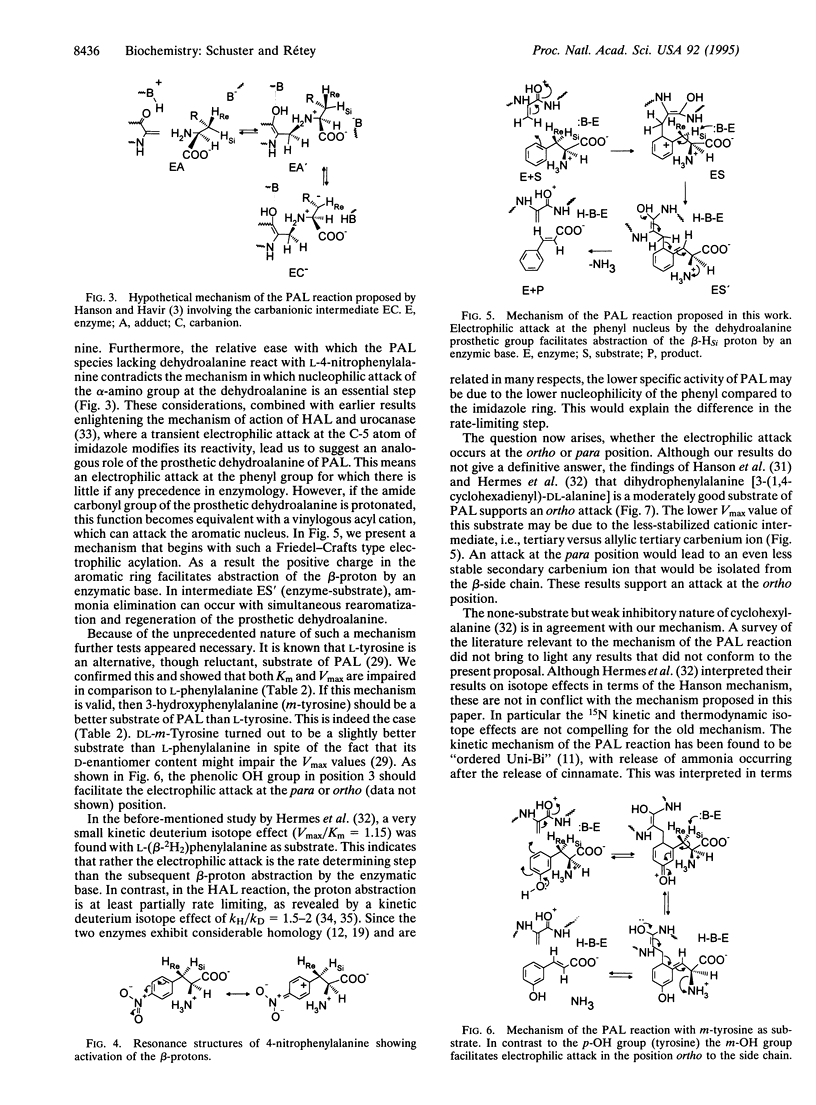
Phenylalanine ammonia-lyase (EC 4.3.1.5) from parsley is posttranslationally modified by dehydrating its Ser-202 to the catalytically essential dehydroalanine prosthetic group. The codon of Ser-202 was changed to those of alanine and threonine by site-directed mutagenesis. These mutants and the recombinant wild-type enzyme, after treatment with sodium borohydride, were virtually inactive with L-phenylalanine as substrate but catalyzed the deamination of L-4-nitrophenylalanine, which is also a substrate for the wild-type enzyme. Although the mutants reacted about 20 times slower with L-4-nitrophenylalanine than the wild-type enzyme, their Vmax for L-4-nitrophenylalanine was two orders of magnitude higher than for L-phenylalanine. In contrast to L-tyrosine, which was a poor substrate, DL-3-hydroxyphenylalanine (DL-m-tyrosine) was converted by phenylalanine ammonia-lyase at a rate comparable to that of L-phenylalanine. These results suggest a mechanism in which the crucial step is an electrophilic attack of the prosthetic group at position 2 or 6 of the phenyl group. In the resulting carbenium ion, the beta-HSi atom is activated in a similar way as it is in the nitro analogue. Subsequent elimination of ammonia, concomitant with restoration of both the aromatic ring and the prosthetic group, completes the catalytic cycle.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Consevage M. W., Phillips A. T. Sequence analysis of the hutH gene encoding histidine ammonia-lyase in Pseudomonas putida. J Bacteriol. 1990 May;172(5):2224–2229. doi: 10.1128/jb.172.5.2224-2229.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givot I. L., Smith T. A., Abeles R. H. Studies on the mechanism of action and the structure of the electrophilic center of histidine ammonia lyase. J Biol Chem. 1969 Dec 10;244(23):6341–6353. [PubMed] [Google Scholar]
- Hanson K. R., Havir E. A. L-Phenylalanine ammonia-lyase (maize, potato, and Rhodotorula glutinis) Explaining the kinetic effects of substrate modification by linear free-energy relationships. Arch Biochem Biophys. 1977 Apr 15;180(1):102–113. doi: 10.1016/0003-9861(77)90013-3. [DOI] [PubMed] [Google Scholar]
- Hanson K. R., Havir E. A. L-phenylalanine ammonia-lyase. IV. Evidence that the prosthetic group contains a dehydroalanyl residue and mechanism of action. Arch Biochem Biophys. 1970 Nov;141(1):1–17. doi: 10.1016/0003-9861(70)90100-1. [DOI] [PubMed] [Google Scholar]
- Hanson K. R., Havir E. A., Ressler C. Phenylalanine ammonia-lyase: enzymic conversion of 3-(1,4-cyclohexadienyl)-L-alanine to trans-3-(1,4-cyclohexadienyl)acrylic acid. Biochemistry. 1979 Apr 17;18(8):1431–1438. doi: 10.1021/bi00575a007. [DOI] [PubMed] [Google Scholar]
- Havir E. A., Hanson K. R. L-phenylalanine ammonia-lyase (maize, potato, and Rhodotorula glutinis). Studies of the prosthetic group with nitromethane. Biochemistry. 1975 Apr 22;14(8):1620–1626. doi: 10.1021/bi00679a012. [DOI] [PubMed] [Google Scholar]
- Havir E. A. Phenylalanine ammonia-lyase: purification and characterization from soybean cell suspension cultures. Arch Biochem Biophys. 1981 Oct 15;211(2):556–563. doi: 10.1016/0003-9861(81)90490-2. [DOI] [PubMed] [Google Scholar]
- Hermes J. D., Weiss P. M., Cleland W. W. Use of nitrogen-15 and deuterium isotope effects to determine the chemical mechanism of phenylalanine ammonia-lyase. Biochemistry. 1985 Jun 4;24(12):2959–2967. doi: 10.1021/bi00333a023. [DOI] [PubMed] [Google Scholar]
- Hernandez D., Phillips A. T. Ser-143 is an essential active site residue in histidine ammonia-lyase of Pseudomonas putida. Biochem Biophys Res Commun. 1994 Jun 30;201(3):1433–1438. doi: 10.1006/bbrc.1994.1863. [DOI] [PubMed] [Google Scholar]
- Kalghatgi K. K., Subba Rao P. V. Microbial L-phenylalanine ammonia-lyase. Purification, subunit structure and kinetic properties of the enzyme from Rhizoctonia solani. Biochem J. 1975 Jul;149(1):65–72. doi: 10.1042/bj1490065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee C. B., Kirk K. L., Cohen L. A. 4-Nitro-L-histidine as a substrate for histidine ammonia-lyase: the role of beta-hydrogen acidity in the rate-limiting step. Biochem Biophys Res Commun. 1979 Mar 15;87(1):343–348. doi: 10.1016/0006-291x(79)91685-1. [DOI] [PubMed] [Google Scholar]
- Klee C. B., Kirk K. L., Cohen L. A., McPhie P. Histidine ammonia-lyase. The use of 4-fluorohistidine in identification of the rate-determining step. J Biol Chem. 1975 Jul 10;250(13):5033–5040. [PubMed] [Google Scholar]
- Klepp J., Fallert-Müller A., Grimm K., Hull W. E., Rétey J. Mechanism of action of urocanase. Specific 13C-labelling of the prosthetic NAD+ and revision of the structure of its adduct with imidazolylpropionate. Eur J Biochem. 1990 Sep 24;192(3):669–676. doi: 10.1111/j.1432-1033.1990.tb19274.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langer M., Lieber A., Rétey J. Histidine ammonia-lyase mutant S143C is posttranslationally converted into fully active wild-type enzyme. Evidence for serine 143 to be the precursor of active site dehydroalanine. Biochemistry. 1994 Nov 29;33(47):14034–14038. doi: 10.1021/bi00251a011. [DOI] [PubMed] [Google Scholar]
- Langer M., Reck G., Reed J., Rétey J. Identification of serine-143 as the most likely precursor of dehydroalanine in the active site of histidine ammonia-lyase. A study of the overexpressed enzyme by site-directed mutagenesis. Biochemistry. 1994 May 31;33(21):6462–6467. doi: 10.1021/bi00187a011. [DOI] [PubMed] [Google Scholar]
- Ma P. X., Schloo B., Mooney D., Langer R. Development of biomechanical properties and morphogenesis of in vitro tissue engineered cartilage. J Biomed Mater Res. 1995 Dec;29(12):1587–1595. doi: 10.1002/jbm.820291215. [DOI] [PubMed] [Google Scholar]
- Schnell N., Entian K. D., Schneider U., Götz F., Zähner H., Kellner R., Jung G. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature. 1988 May 19;333(6170):276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- Schulz W., Eiben H. G., Hahlbrock K. Expression in Escherichia coli of catalytically active phenylalanine ammonia-lyase from parsley. FEBS Lett. 1989 Dec 4;258(2):335–338. doi: 10.1016/0014-5793(89)81687-4. [DOI] [PubMed] [Google Scholar]
- Schuster B., Rétey J. Serine-202 is the putative precursor of the active site dehydroalanine of phenylalanine ammonia lyase. Site-directed mutagenesis studies on the enzyme from parsley (Petroselinum crispum L.). FEBS Lett. 1994 Aug 1;349(2):252–254. doi: 10.1016/0014-5793(94)00681-4. [DOI] [PubMed] [Google Scholar]
- Skaugen M., Nissen-Meyer J., Jung G., Stevanovic S., Sletten K., Inger C., Abildgaard M., Nes I. F. In vivo conversion of L-serine to D-alanine in a ribosomally synthesized polypeptide. J Biol Chem. 1994 Nov 4;269(44):27183–27185. [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Symington J., Green M., Brackmann K. Immunoautoradiographic detection of proteins after electrophoretic transfer from gels to diazo-paper: analysis of adenovirus encoded proteins. Proc Natl Acad Sci U S A. 1981 Jan;78(1):177–181. doi: 10.1073/pnas.78.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Nakata Y., Li H. O., Zhang M., Gao H., Fujita A., Sakatsume O., Ohta T., Yokoyama K. The optimization of preparations of competent cells for transformation of E. coli. Nucleic Acids Res. 1994 Jul 25;22(14):2857–2858. doi: 10.1093/nar/22.14.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. Dehydroalanine in histidine ammonia lyase. J Biol Chem. 1969 Dec 10;244(23):6550–6552. [PubMed] [Google Scholar]
- Zimmermann S., Hahlbrock K. Light-induced changes of enzyme activities in parsley cell suspension cultures. Purification and some properties of phenylalanine ammonia-lyase (E.C.4.3.1.5). Arch Biochem Biophys. 1975 Jan;166(1):54–62. doi: 10.1016/0003-9861(75)90364-1. [DOI] [PubMed] [Google Scholar]




