Abstract
Phosphopantetheinyltransferase (PPTase) enzymes fulfil essential roles in primary and secondary metabolism in prokaryotes, archaea and eukaryotes. PPTase enzymes catalyse the essential modification of the carrier protein domain of fatty acid synthases, polyketide synthases (PKSs) and non-ribosomal peptide synthetases (NRPSs). In bacteria and fungi, NRPS and PKS enzymes are often responsible for the biosynthesis of secondary metabolites with clinically relevant properties; these secondary metabolites include a variety of antimicrobial peptides. We have previously shown that in the Gram-negative bacterium Serratia marcescens Db10, the PPTase enzyme PswP is essential for the biosynthesis of an NRPS-PKS dependent antibiotic called althiomycin. In this work we utilize bioinformatic analyses to classify PswP as belonging to the F/KES subfamily of Sfp type PPTases and to putatively identify additional NRPS substrates of PswP, in addition to the althiomycin NRPS-PKS, in Ser. marcescens Db10. We show that PswP is required for the production of three diffusible metabolites by this organism, each possessing antimicrobial activity against Staphylococcus aureus. Genetic analyses identify the three metabolites as althiomycin, serrawettin W2 and an as-yet-uncharacterized siderophore, which may be related to enterobactin. Our results highlight the use of an individual PPTase enzyme in multiple biosynthetic pathways, each contributing to the ability of Ser. marcescens to inhibit competitor bacteria by the production of antimicrobial secondary metabolites.
Introduction
Phosphopantetheinyltransferase (PPTase) enzymes have essential roles in the biosynthetic pathways of primary and secondary metabolism in prokaryotes, archaea and eukaryotes (Beld et al., 2014; Lambalot et al., 1996). Fatty acid synthases (FASs), polyketide synthases (PKSs) and non-ribosomal peptide synthetases (NRPSs) are all dependent upon the catalytic action of PPTase enzymes to function (Beld et al., 2014). FASs, PKSs and NRPSs are generally large, multimodular proteins. Each module within the protein incorporates one acyl or amino acid unit into the growing metabolite and comprises a minimum of three essential domains, including the carrier protein domain (Fischbach & Walsh, 2006). PPTase enzymes catalyse the transfer of a 4′-phosphopantetheine (PPT) prosthetic group from coenzyme A to a conserved serine residue in the carrier protein domain of FASs, PKSs and NRPSs (Beld et al., 2014; Elovson & Vagelos, 1968; Lambalot et al., 1996). The 4′-phosphopantetheine prosthetic group of the carrier protein domain fulfils two main functions: the thiol-terminated 4′-phosphopantetheine arm serves as the point of attachment for the FAS, PKS or NRPS intermediate to the biosynthetic machinery and the flexibility of this arm allows the biosynthetic intermediate access to the catalytic reaction centres within the FAS, PKS or NRPS module (Beld et al., 2014).
PPTase enzymes can be divided into three families: the Sfp family whose prototype is Sfp of Bacillus subtilis (Lambalot et al., 1996; Reuter et al., 1999); the AcpS family whose prototype is AcpS of Escherichia coli (Elovson & Vagelos, 1968; Lambalot & Walsh, 1995); and a third family whose members are integrated as domains within the FAS or PKS enzyme that they modify (Beld et al., 2014; Copp & Neilan, 2006; Lambalot et al., 1996; Walsh et al., 1997). The Sfp family of PPTases have broad substrate specificity and are generally required for the activation of PKS and NRPS enzymes of secondary metabolism (Mootz et al., 2001; Quadri et al., 1998); these enzymes are approximately 230 residues in length and exist as monomers (Reuter et al., 1999). Based on sequence comparisons, members of the Sfp family were further divided into the F/KES and W/KEA subfamilies (Copp & Neilan, 2006; Lambalot et al., 1996). F/KES subfamily members were shown to largely be associated with NRPS enzymes whereas W/KEA subfamily members were more often associated with PKS enzymes (Copp & Neilan, 2006). Members of the AcpS family of PPTases are generally around 120 residues in length and are thought to exist as trimers (Chirgadze et al., 2000; Parris et al., 2000). The AcpS family of PPTases show a higher degree of specificity towards the carrier protein domain that they modify and are generally required for the modification of FAS enzymes from primary metabolism (Lambalot et al., 1996; Mootz et al., 2001; Parris et al., 2000). Despite sharing limited overall sequence similarity, the AcpS and Sfp family members share homology at the structural level; an AcpS dimer resembles one Sfp monomer (Lambalot et al., 1996; Reuter et al., 1999). As previously described, there exists a third family of PPTases that are translationally fused to the FAS or PKS enzyme that they modify (Copp & Neilan, 2006; Fichtlscherer et al., 2000; Lambalot et al., 1996). Examples of these integrated PPTases are few and, to date, only two bacterial integrated PPTases have been experimentally validated (Murugan & Liang, 2008; Weissman et al., 2004).
Serratia marcescens is a Gram-negative bacterium belonging to the family Enterobacteriaceae. Members of this genus are known to produce a variety of secondary metabolites (Fineran et al., 2005; Kadouri & Shanks, 2013; Thomson et al., 2000). Many strains, including Ser. marcescens ATCC 274, produce the secondary metabolite prodigiosin that gives the bacterium a distinctive red colour (Williamson et al., 2006). Ser. marcescens Db10 is a model insect pathogen but lacks the genes to produce prodigiosin and so is an example of a non-pigmented strain of Serratia (Flyg et al., 1980). However, we recently described the production of the antibiotic secondary metabolite, althiomycin, by Ser. marcescens Db10 (Gerc et al., 2012). Althiomycin is the product of the six-gene alb operon, which encodes the following: a hybrid NRPS-PKS enzyme, appropriate tailoring enzymes and an export/resistance protein. The PPTase encoded by SMA2452, which is not genetically linked with the alb operon, was shown to be required for althiomycin biosynthesis in this organism (Gerc et al., 2012). PswP, in Ser. marcescens ATCC 274, is the PPTase required for the production of prodigiosin and the surfactant serrawettin W1 (Sunaga et al., 2004). Ser. marcescens Db10 does not produce serrawettin W1 dependent on the NRPS SwrW (Li et al., 2005), instead, it produces serrawettin W2 dependent on the NRPS SwrA (Matsuyama et al., 1992). Serrawettins W1 and W2 are both cyclic lipopeptides but have quite distinct chemical structures (Matsuyama et al., 1992; Wasserman et al. 1962). The PPTase SMA2452 and PswP from Ser. marcescens ATCC 274 show 96 % identity at the protein level; therefore, SMA2452 will henceforth be referred to as PswP.
In this study, we provide evidence that PswP is required for the biosynthesis of two further secondary metabolites in Ser. marcescens Db10 and, in addition to althiomycin, these metabolites have an antimicrobial effect against Staphylococcus aureus. The production of one of these secondary metabolites, serrawettin W2, is known to be dependent on the action of PswP (Pradel et al., 2007). We further show that the second PswP-dependent metabolite is a siderophore, likely enterobactin or a related molecule. Together, althiomycin, serrawettin W2 and the siderophore comprise a repertoire of secondary metabolites that have antimicrobial activity and are dependent on PswP for their biosynthesis.
Methods
Strains and culture media.
Strains used in this study are detailed in Table 1. E. coli was routinely cultured in Luria–Bertani (LB) medium (1 %, w/v, Bacto tryptone, 0.5 %, w/v, Bacto yeast extract, 1 %, w/v, NaCl), Ser. marcescens in low salt Luria–Bertani (LB) medium (1 %, w/v, Bacto tryptone, 0.5 %, w/v, Bacto yeast extract, 0.5 %, w/v, NaCl) and Sta. aureus in tryptic soya broth (TSB) (0.5 %, w/v, NaCl, 0.5 %, w/v, soytone, 1.5 %, w/v, tryptone). Chrome azurol S (CAS) agar plates were prepared as described by Schwyn & Neilands (1987). Growth media were solidified through addition of agar to 1.5 % (w/v). When required, media were supplemented with FeCl3, to a final concentration of 50 μM, or with antibiotics: 100 µg ampicillin ml−1; 1000 µg kanamycin ml−1 and 100 µg streptomycin ml−1.
Table 1. Bacterial strains and plasmids used in this study.
| Strain/plasmid | Description | Source or reference |
| Strains | ||
| Db10 | Serratia marcescens wild-type | Flyg et al. (1980) |
| Staphylococcus aureus 113 | ATCC 35556 | Professor T. Palmer (University of Dundee) |
| Staphylococcus aureus | Newman spa : : tet sbi : : kan | Sibbald et al. (2010) |
| JESM267 | Db10 (swrA : : Tn5) | Pradel et al. (2007) |
| SAN5 | Db10 (Δalb4-5) in-frame | Gerc et al. (2012) |
| SAN112 | Db10 (ΔpswP : : cml) | Gerc et al. (2012) |
| SAN124 | Db10 (Δalb4-5, swrA : : Tn5) | This study |
| SAN176 | Db10 (ΔentB) in-frame | This study |
| SAN180 | Db10 (Δalb4-5, ΔentB) | This study |
| SAN181 | Db10 (Δalb4-5, swrA : : Tn5, ΔentB) | This study |
| Escherichia coli MC1061 | F′lacIQ lacZM15 Tn10 (TetR), cloning host | Perego & Hoch (1988) |
| E. coli CC118λpir | Cloning host and donor strain for pKNG101-derived marker exchange plasmids (λpir) | Herrero et al. (1990) |
| E. coli HH26 pNJ5000 | Mobilizing strain for conjugal transfer | Grinter (1983) |
| Plasmids | ||
| pBluescript KS(+) | High copy cloning vector (AmpR) | Stratagene |
| pKNG101 | Suicide vector for marker exchange (SmR, sacBR, mobRK2, ori R6K) | Kaniga et al. (1991) |
| pSUPROM | Vector for constitutive expression of cloned genes under the control of the E. coli tat promoter (KanR) | Jack et al. (2004) |
| pNW572 | pKNG101-derived marker exchange plasmid for the generation of chromosomal Δalb4-5 | Gerc et al. (2012) |
| pSAN69 | pKNG101-derived marker exchange plasmid for the generation of chromosomal ΔentB | This study |
| pSAN46 | pswP coding sequence in pSUPROM | Gerc et al. (2012) |
AmpR, ampicillin resistance; KanR , kanamycin resistance; SmR, streptomycin ; TetR, tetracycline resistance.
Construction of strains and plasmids.
A Ser. marcescens chromosomal mutant with an in-frame deletion in entB was constructed by marker (allelic) exchange using the suicide vector pKNG101 (Kaniga et al., 1991), as described previously (Coulthurst et al., 2006). Briefly, the upstream and downstream flanking regions of the gene SMA4415 (entB) were cloned into pBluescript KS(+) so as to generate a non-polar, in-frame deletion of the gene. This deletion allele was then cloned into pKNG101 and the resulting marker exchange plasmid introduced into Ser. marcescens by conjugation. Selection on streptomycin-containing agar and then on high sucrose agar allowed isolation of mutants in which the deletion allele had replaced the wild-type copy. The plasmids used in this study are detailed in Table 1. The upstream and downstream regions of entB were amplified using the primers AG157 (TATAAAGCTTTTTTGGAATGGCCATCGA) and AG158 (TATAGGGCCCACATCCTGACGACGCAAG) (upstream), and AG159 (TATAAAGCTTGAAGAGAAAGCCTGATTTTAATAATG) and AG160 (TATATCTAGATATTTGCCGGTGTCGGTC) (downstream). The resulting product was cloned into pBluescript KS(+) using restriction sites engineered into the primers (underlined in the primer sequences). Following marker exchange, the integrity of the disrupted region was confirmed by DNA sequencing. The Δalb4-5 ΔentB double mutant (SAN180) was constructed by introduction of the entB deletion into SAN5 (Δalb4-5), as described above. The swrA : : Tn5 mutation was introduced into the single Δalb4-5 mutant and the double Δalb4-5 ΔentB mutant by phage ϕIF3-mediated transduction as described previously (Petty et al., 2006), generating strains SAN124 and SAN181, respectively. Growth curve measurement was also performed on each mutant strain, this showed that the growth rate was unaffected compared with wild-type Ser. marcescens Db10 (data not shown).
Antimicrobial activity assays.
Sta. aureus was grown in liquid culture in a volume of 5 ml in a universal tube at 37 °C with rotation and 100 µl culture was spread onto an agar plate. Strains of Ser. marcescens being tested for antimicrobial production were grown in liquid culture in a volume of 5 ml in a universal tube at 30 °C with rotation and 10 µl culture was then spotted on the Sta. aureus lawn. The plates were incubated (15 h) at 30 °C prior to photography.
CAS assay.
Ser. marcescens strains were grown to stationary phase at 30 °C and 10 µl of culture was spotted on to the CAS agar plate. The plates were incubated (15 h) at 30 °C prior to photography. For complementation analysis, plasmids were maintained by the addition of kanamycin to both the culture medium and to the CAS agar plates.
Bioinformatic analysis.
The genome of Ser. marcescens strain Db11 was sequenced by the Pathogen Sequencing Unit, the Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK. Ser. marcescens strain Db11 is a spontaneous Sm-resistant mutant of Ser. marcescens strain Db10 and hence the Db11 genome information can be used for Db10. To identify NRPS or PKS secondary metabolite gene clusters encoded within the Ser. marcescens Db11 genome, the bioinformatics tool antiSMASH was employed (Blin et al., 2013). The genome was uploaded as a nucleotide sequence and the bacterial, archaeal and plant plastid translational code, with a minimal gene length of 50, was used for gene finding by glimmer. Within antiSMASH, the following parameters were selected: smCOG analysis for functional prediction and phylogenetic analysis of genes, gene cluster blast analysis, subcluster blast analysis, whole genome pfam analysis and secondary metabolite detection on all possible ORFs. To investigate the likely function of NRPS or PKS products identified by antiSMASH, further manual analysis using blastp (Altschul et al., 1997) was performed.
Results and Discussion
The PswP PPTase of Ser. marcescens Db10 is required for the biosynthesis of more than one metabolite with antimicrobial activity
During a prior study investigating the mechanism of althiomycin biosynthesis in Ser. marcescens Db10, it was noted that while Sta. aureus was sensitive to althiomycin, disruption of althiomycin biosynthesis did not completely abolish the antimicrobial effect of Ser. marcescens Db10 against Sta. aureus (Gerc et al., 2012). Aiming to investigate this further, we discovered that deletion of the gene encoding the PPTase protein PswP (which is required for althiomycin biosynthesis) was sufficient to negate the antimicrobial activity of Ser. marcescens Db10 against Sta. aureus (Fig. 1a). To confirm that the loss of antimicrobial activity associated with deletion of pswP was specific to deletion of this gene, the PswP mutant phenotype was complemented by expression of PswP in trans. Expression of PswP in trans restored the antimicrobial activity of Ser. marcescens Db10 against Sta. aureus (Fig. 1b). PPTase enzymes are known to modify the carrier protein domain of FAS, PKS and NRPS enzymes (Lambalot et al., 1996). We hypothesized that, in addition to being required for althiomycin biosynthesis, PswP was required for the biosynthesis of one or more FAS, PKS or NRPS product(s) with antimicrobial activity against Sta. aureus. Following this initial observation, we aimed to determine the identity of this metabolite(s).
Fig. 1.
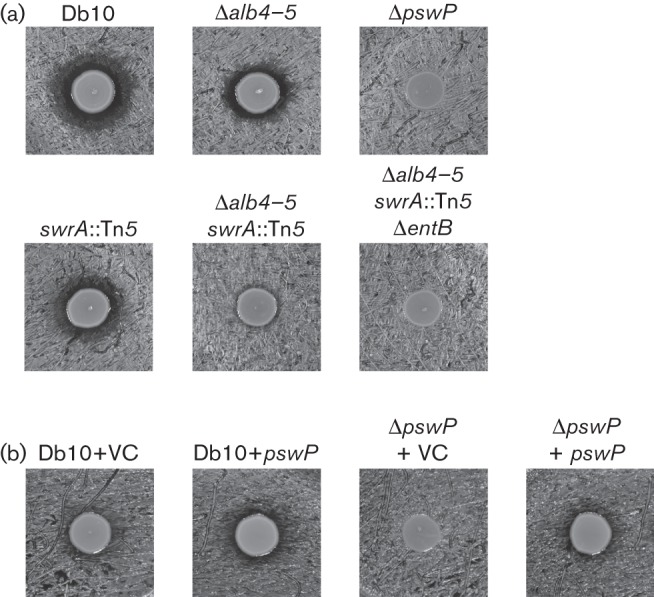
The PPTase PswP is required for the biosynthesis of three secondary metabolites with antimicrobial activity against Sta. aureus. (a) Antimicrobial activity assays using Sta. aureus 113 as the indicator lawn. The producer strains are indicated above: Db10 (wild-type Ser. marcescens Db10), Δalb4-5 (SAN5), ΔpswP (SAN112), swrA : : Tn5 (JESM267), Δalb4-5 swrA : : Tn5 (SAN124) and Δalb4-5 swrA : : Tn5 ΔentB (SAN181). (b) Antimicrobial activity assays show complementation of the pswP deletion by expression of the pswP gene in trans, using kanamycin-resistant Sta. aureus Newman spa : : tet sbi : : kan as the indicator lawn. The producer strains are indicated above: Db10+VC (Ser. marcescens Db10 pSUPROM, vector control), Db10+pswP (Ser. marcescens Db10 pSAN46), ΔpswP+VC (SAN112 pSUPROM) and ΔpswP+pswP (SAN112 pSAN46).
Bioinformatic analysis of PswP and potential target proteins
PswP belongs to the Sfp class of PPTases and is one of three PPTases encoded by Ser. marcescens Db10 (Gerc et al., 2012). There exists a second Sfp-type PPTase, SMA4147, which was previously identified during a search for the PPTase required for the biosynthesis of althiomycin; however, no biological function for SMA4147 has been ascribed (Gerc et al., 2012). SMA3052 represents a third PPTase encoded by Ser. marcescens Db10 that shows homology to AcpS family PPTases (data not shown) and is therefore likely involved in primary metabolism (Lambalot et al., 1996). Therefore, further bioinformatic analysis of the PPTases encoded by Ser. marcescens Db10 focused solely on PswP. Based on the presence of the conserved residues within motifs 1A, 1, 2 and 3, we assigned PswP to the F/KES subfamily of Sfp-type PPTases (Fig. 2).
Fig. 2.

Identification of conserved motifs of the F/KES subfamily of Sfp-type PPTases within PswP of Ser. marcescens Db10. Conserved PPT motifs 1A, 1, 2 and 3 are denoted with a dashed boxed. Residues previously identified as being highly conserved across the F/KES subfamily are indicated below the amino acid sequence; residues of PswP that match the consensus shown are shaded black. Analysis is based on the motif alignment analysis performed by Copp & Neilan (2006).
In Ser. marcescens Db10, the pswP gene is not embedded within or adjacent to an NRPS or PKS biosynthetic gene cluster. We were therefore unable to predict additional PKS or NRPS biosynthetic pathways that might require the activity of PswP based on its genomic context. The bioinformatics tool, antiSMASH, allows the identification and the preliminary analysis of secondary metabolite gene clusters in bacterial and fungal genomes to be performed in an unbiased manner (Blin et al., 2013). Therefore, antiSMASH was employed to identify all genomic regions containing putative PKS, NRPS and NRPS-PKS gene clusters in the Ser. marcescens Db10 genome, on the basis that one or more of these might direct the biosynthesis of the secondary metabolite(s) active against Sta. aureus. The results of this search are summarized in Table 2. Five genomic regions containing NRPS or NRPS-PKS gene clusters, whose products could potentially be modified by the action of PswP, were identified. Upon closer inspection of the NRPS and NRPS-PKS gene clusters, the althiomycin biosynthetic machinery and the machinery required for the biosynthesis of the surfactant, serrawettin W2, were identified in clusters three and four (Gerc et al., 2012; Pradel et al., 2007). Published data for the three remaining gene clusters was not available; therefore, a bioinformatic approach was adopted. In an attempt to deduce the likely product of the three remaining NRPS biosynthetic gene clusters, the protein sequences encoded by the NRPS genes(s) identified by antiSMASH and the products of their flanking genes were subjected to database searching using blastp (Altschul et al., 1997). Analysis of the NRPSs in gene cluster one revealed that this locus likely encodes proteins involved in the biosynthesis of a microcin (Table 2). SMA1574A shows 41 % identity with microcin N of E. coli 2424 (Corsini et al., 2010) and the NRPS proteins, SMA1572 and SMA1571, are likely involved in the biosynthesis of a modifying group added to the microcin peptide encoded by this gene cluster (Rebuffat, 2012). In contrast, gene clusters two and five appear to direct the synthesis of multiple siderophores (Table 2). The NRPS encoded within gene cluster two shares homology with chrysobactin synthetase component F (Persmark et al., 1989). Within gene cluster five is a locus containing an NRPS and associated genes conserved with the enterobactin biosynthesis and utilization genes from E. coli (see below) (Raymond et al., 2003).
Table 2. Characterization of NRPS- or PKS-encoding genes identified by antiSMASH (Blin et al., 2013).
| Gene cluster | Genomic region predicted by antiSMASH | Enzyme predicted by antiSMASH | Gene(s) encoding NRPS (or NRPS-PKS) | Previously reported product | Putative product | Comments |
| 1 | SMA1552–1589 | NRPS | SMA1571-1572 | – | Unknown | * |
| 2 | SMA1705–1744 | NRPS | SMA1729 | – | Siderophore | † |
| 3 | SMA2274-1309 | NRPS-PKS | SMA2289-2290 | Althiomycin | na | ‡ |
| 4 | SMA3659–3702 | NRPS | SMA3680 | Serrawettin W2 | na | § |
| 5 | SMA4386–4438 | NRPS | SMA4411 | – | Siderophore | || |
| SMA4402-4404, SMA4406 | – | Unknown | ¶ |
na, not applicable.
Predicted NRPS encoded within a cluster of genes (SMA1575–1568) showing similarity with genes involved in the biosynthesis and secretion of microcins; may synthesize a modifying group added to the microcin.
Predicted NRPS similar to chrysobactin synthetase (64 % identity over the whole length) and flanked by genes encoding proteins with similarity to proteins mediating siderophore export, uptake and iron release.
Althiomycin is a broad-spectrum antibiotic. NRPS-PKS is encoded within the six-gene alb operon (SMA2288–2293), which also encodes tailoring and export functions (Gerc et al., 2012).
Serrawettin W2 is a biosurfactant (Pradel et al., 2007).
||Predicted NRPS is similar to enterobactin synthetase EntF and within cluster of genes (SMA4408–4415) similar to the enterobactin biosynthesis, export and uptake cluster of E. coli (Fig. 3).
Encoded NRPS proteins share similarity with siderophore synthetase enzymes from other organisms but number and nature of product(s) unclear.
The PswP-dependent surfactant serrawettin W2 has antimicrobial activity against Sta. aureus
Biosurfactants are amphipathic molecules whose primary function is to reduce surface tension to allow bacterial spreading across surfaces (Matsuyama et al., 2011). Biosurfactants have been shown to possess several properties, including antimicrobial activity (Singh & Cameotra, 2004). Of particular note, serrawettin W1 produced by Ser. marcescens ATCC 274 has been shown to have antimicrobial properties against meticillin-resistant Sta. aureus (MRSA) (Kadouri & Shanks, 2013). In Ser. marcescens Db10, PswP was previously shown to be required for the biosynthesis of serrawettin W2 (Pradel et al., 2007). Therefore, to determine whether serrawettin W2 also possessed antimicrobial activity against Sta. aureus, a serrawettin W2 single mutant was obtained (Pradel et al., 2007) and used to construct a double mutant (Δalb4-5 swrA : : Tn5) that was unable to produce althiomycin or serrawettin W2. Both mutant strains were tested for antimicrobial activity against Sta. aureus (Fig. 1a). Compared with Ser. marcescens Db10, the swrA : : Tn5 mutant showed a slight reduction in antimicrobial activity against Sta. aureus. The reduction in size of the antibiosis halo observed with the double mutant (Δalb4-5 swrA : : Tn5) was much more pronounced compared with either single mutant; however, a small antibiosis halo could still be observed (Fig. 1a). Therefore, we concluded that serrawettin W2 does indeed possess antimicrobial activity against Sta. aureus, but that Ser. marcescens Db10 must produce another antimicrobial, in addition to serrawettin W2 and althiomycin, which is dependent on the presence of PswP for biosynthesis.
The SMA4408-4415 gene cluster encodes a siderophore that is dependent on PswP for biosynthesis and has antimicrobial activity against Sta. aureus
In light of the results presented above, we reasoned that one or more of the three remaining NRPS gene clusters identified by antiSMASH should be responsible for the antimicrobial effect against Sta. aureus observed in the absence of althiomycin and serrawettin W2. Two of the three remaining NRPS gene clusters, identified by antiSMASH, encoded proteins with similarity to siderophore biosynthetic proteins (Table 2, Fig. 3). We therefore hypothesized that either a siderophore produced by Ser. marcescens Db10 was directly toxic toward Sta. aureus, or that siderophore-dependent removal of iron from the environment prevented growth of Sta. aureus. In order to investigate these possibilities, FeCl3 was added to the growth media and the antimicrobial bioassay repeated. The small antibiosis halo observed in the absence of additional iron with the double Δalb4-5 swrA : : Tn5 mutant disappeared upon the addition of FeCl3 (Fig. 4). From these results two scenarios are possible: 1) addition of excess iron negates the iron depletion effect of a siderophore(s), or 2) the addition of iron negatively affects the production of siderophores by Ser. marcescens Db10, as observed in E. coli and many other Gram-negative bacteria (Crosa & Walsh, 2002). In either case, these findings indicated that siderophore production by Ser. marcescens Db10 was likely to inhibit Sta. aureus growth.
Fig. 3.
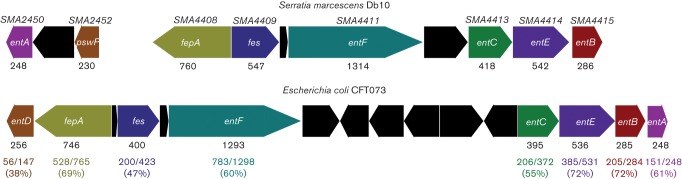
Comparison of the SMA2450–2452 and SMA4408–4415 gene clusters of Ser. marcescens Db10 with the enterobactin biosynthetic gene cluster of E. coli CFT073 (c0668–c0683). Genes are drawn approximately to scale and protein length is indicated below the encoding gene (as the number of amino acids). Homologous genes are indicated by the same colour, the level of identity between homologous proteins is shown below the corresponding genes as an absolute value and as a percentage.
Fig. 4.
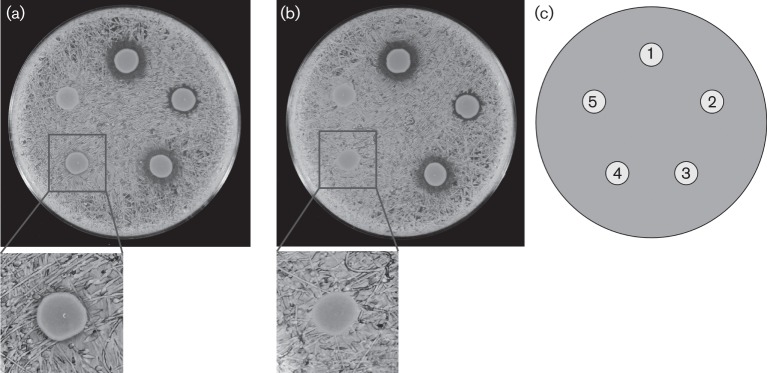
An althiomycin and serrawettin W2 mutant of Ser. marcescens Db10 is unable to kill Sta. aureus in the presence of additional iron. Antimicrobial activity assays in the absence (a) or presence (b) of 50 μM FeCl3, using Sta. aureus as the indicator lawn, with a schematic of the location of each producer strain shown (c). The producing strains were: 1, Ser. marcescens Db10 (wild-type); 2, SAN5 (Δalb4-5); 3, JESM267 (swrA : : Tn5); 4, SAN124 (Δalb4-5 swrA : : Tn5); 5, SAN112 (ΔpswP). The insets show a magnified image of producer strain 4 SAN124 in both (a) and (b).
As mentioned above, a set of genes (SMA4408–4415) within cluster five show homology and synteny with the enterobactin gene cluster of E. coli (Fig. 3). It therefore seemed likely that these genes are involved in the synthesis of enterobactin or a closely related molecule. However, it should be noted that the only close homologue of entA, which is located immediately upstream of entB in E. coli, was identified as SMA2450 in Ser. marcescens Db11. In contrast with E. coli, the SMA2450 gene is located at a distant locus from the other enterobactin-like genes; however, it is closely genetically linked with pswP (SMA2452) in Ser. marcescens. Given both the linkage between entA and pswP in Ser. marcescens and that the SMA4408–4415 genomic region showed the most convincing similarity with a well-characterized cluster of genes involved in siderophore biosynthesis and utilization, we tested whether the product of these genes was responsible for the PswP- and iron-dependent growth inhibition observed in the Δalb4-5 swrA : : Tn5 double mutant. The product of SMA4415 shares significant homology with entB of the enterobactin biosynthetic gene cluster (Fig. 3). Since EntB is essential for enterobactin biosynthesis in E. coli (Crosa & Walsh, 2002), we reasoned that SMA4415 would be essential for biosynthesis of this siderophore in Ser. marcescens Db10. An in-frame deletion in entB was therefore constructed in Ser. marcescens Db10, and also in the althiomycin and serrawettin W2 mutant background to give a triple althiomycin, serrawettin W2 and ΔentB mutant (Δalb4-5 swrA : : Tn5 ΔentB). To compare siderophore production by wild-type Ser. marcescens Db10 with that of the single ΔentB mutant, the triple Δalb4-5 swrA : : Tn5 ΔentB mutant and the ΔpswP mutant, the relevant strains were grown on CAS indicator plates. CAS/hexadecyltrimethylammonium bromide complexed with ferric iron present in these plates serves as an indicator of siderophore biosynthesis; in the presence of an iron chelator this indicator turns from a blue to an orange colour (Schwyn & Neilands, 1987). In wild-type Ser. marcescens Db10, production of a diffusible siderophore was clearly observed (Fig. 5a). In contrast, no siderophore production was observed in either the ΔentB or PswP PPTase (ΔpswP) single mutants (Fig. 5a). Furthermore, no siderophore production was observed in the althiomycin, serrawettin W2 and ΔentB triple mutant (Δalb4-5 swrA : : Tn5 ΔentB) (Fig. 5a). The loss of siderophore production was specific to the deletion of entB or pswP, with siderophore production being unaffected in strains unable to produce althiomycin and/or serrawettin (Fig. 5a) These results clearly show that both entB and pswP are essential for the biosynthesis of a siderophore in Ser. marcescens Db10. In addition, this assay confirmed that construction of the pswP (SMA2452) PPTase mutant did not affect siderophore biosynthesis through downstream polar effects on the expression of SMA2450, which is predicted to encode an EntA homologue (Fig. 3), since the loss of siderophore biosynthesis observed with the pswP mutant could be complemented by the expression of pswP in trans (Fig. 5b). Having shown that entB was required for biosynthesis of a siderophore in Ser. marcescens Db10, the triple althiomycin, serrawettin W2 and siderophore mutant was tested for antimicrobial activity against Sta. aureus. No antibiosis halo was observed with the Δalb4-5 swrA : : Tn5 ΔentB triple mutant (Fig. 1a), thus providing a phenocopy of the PswP PPTase mutant and identifying the entB-dependent siderophore as the third PswP-dependent antimicrobial secondary metabolite produced by Ser. marcescens Db10.
Fig. 5.
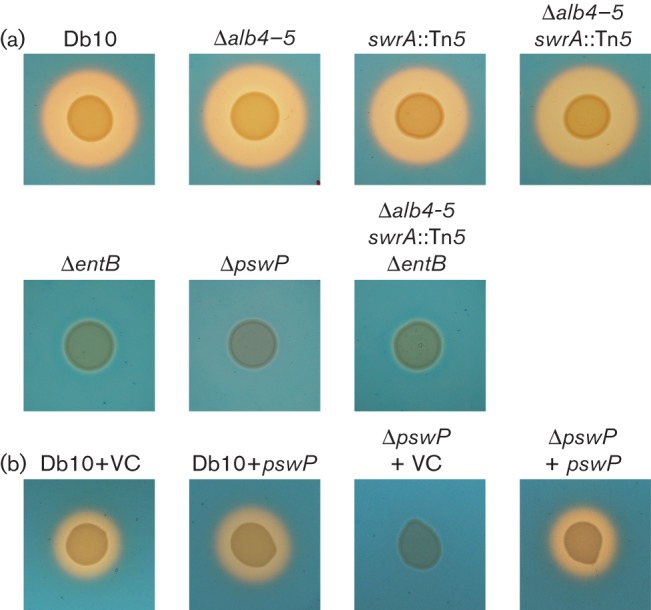
The PPTase PswP and SMA4415 (EntB) are required for the biosynthesis of a siderophore in Ser. marcescens Db10. CAS assay for the detection of siderophore biosynthesis, as indicated by the presence of an orange halo. (a) The producer strains are indicated above: Db10 (wild-type Ser. marcescens Db10), Δalb4-5 (SAN5), swrA : : Tn5 (JESM267), Δalb4-5 swrA : : Tn5 (SAN124), ΔentB (SAN176), ΔpswP (SAN112) and Δalb4-5 swrA : : Tn5 ΔentB (SAN181). (b) Complementation of the pswP deletion by expression of the pswP gene in trans. The producer strains are indicated above: Db10+VC (Ser. marcescens Db10 pSUPROM, vector control), Db10+pswP (Ser. marcescens Db10 pSAN46), ΔpswP+VC (SAN112 pSUPROM) and ΔpswP+pswP (SAN112 pSAN46).
Concluding remarks
In this study, we have shown that Ser. marcescens Db10 produces three diffusible molecules with antimicrobial activity against the clinically relevant pathogen Sta. aureus. Production of all three requires PswP, consistent with their biosynthesis being dependent on NRPS enzymes. Genetic evidence has revealed these three metabolites to be: the antibiotic althiomycin, the product of Alb1-6 (Gerc et al., 2012); the biosurfactant serrawettin W2, the product of SwrA (Pradel et al., 2007); and a siderophore whose production is associated with NRPS-containing cluster of genes, SMA4408–4415. Importantly, this is the first time, to our knowledge, that the latter two molecules have been shown to act against Sta. aureus. Furthermore, production of this siderophore and its association with PswP and the SMA4408–4415 gene cluster has not been reported previously in Ser. marcescens. Whilst we speculate that this molecule is likely to be closely related, or even identical, to enterobactin produced by E. coli, its chemical structure remains to be confirmed. Whilst we speculate that the ability to use this siderophore to scavenge Fe3+ is likely to be important for Ser. marcescens to survive within the iron-limited environment of the host during infection (Miethke & Marahiel, 2007), this remains to be investigated. In addition, our data highlight how an individual bacterial PPTase enzyme can play an essential role in the biosynthesis of multiple secondary metabolites with disparate physiological roles. PswP provides another example, in addition to others reported previously (Beld et al., 2014), illustrating that PPTase enzymes are not necessarily pathway-specific or genetically linked with their substrates but may act on multiple NRPS enzymes. Finally, our data also illustrate how several distinct secondary metabolites can contribute to the inhibition of growth of competitor bacteria, a scenario that is likely to occur at polymicrobial infection sites and in varied environmental niches. In particular, we note the potential utility of iron restriction to inhibit the growth of clinically significant pathogens such as Sta. aureus.
Note added in proof
The siderophore product dependent on the enterobactin-like gene cluster SMA4408-4415 is likely to be identical or related to the serratiochelin molecules recently reported to be produced by Serratia sp. V5 using a ‘shuffled’ combination of enterobactin-like and vibriobactin-like genes (Seyedsayamdost et al., 2012).
Acknowledgements
The authors wish to thank Lijiang Song for helpful discussions. Jonathan J. Ewbank and Jan Maarten van Dijl are gratefully acknowledged for providing strains JESM267 and Sta. aureus Newman spa : : tet sbi : : kan, respectively. A. J. G is funded by a PhD studentship supported by the Wellcome Trust (093711/B/10/Z) (www.wellcome.ac.uk/) and S. J. C is supported by a Royal Society of Edinburgh/Scottish Government Personal Research Fellowship (www.royalsoced.org.uk/).
Abbreviations:
- CAS
chrome azurol S
- FAS
fatty acid synthase
- NRPS
non-ribosomal peptide synthetase
- PKS
polyketide synthase
- PPTase
phosphopantetheinyltransferase
References
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beld J., Sonnenschein E. C., Vickery C. R., Noel J. P., Burkart M. D. (2014). The phosphopantetheinyl transferases: catalysis of a post-translational modification crucial for life. Nat Prod Rep 31, 61–108. 10.1039/c3np70054b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K., Medema M. H., Kazempour D., Fischbach M. A., Breitling R., Takano E., Weber T. (2013). antiSMASH 2.0–a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 41 (Web Server issue), W204-W212. 10.1093/nar/gkt449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgadze N. Y., Briggs S. L., McAllister K. A., Fischl A. S., Zhao G. (2000). Crystal structure of Streptococcus pneumoniae acyl carrier protein synthase: an essential enzyme in bacterial fatty acid biosynthesis. EMBO J 19, 5281–5287. 10.1093/emboj/19.20.5281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp J. N., Neilan B. A. (2006). The phosphopantetheinyl transferase superfamily: phylogenetic analysis and functional implications in cyanobacteria. Appl Environ Microbiol 72, 2298–2305. 10.1128/AEM.72.4.2298-2305.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsini G., Karahanian E., Tello M., Fernandez K., Rivero D., Saavedra J. M., Ferrer A. (2010). Purification and characterization of the antimicrobial peptide microcin N. FEMS Microbiol Lett 312, 119–125. 10.1111/j.1574-6968.2010.02106.x [DOI] [PubMed] [Google Scholar]
- Coulthurst S. J., Williamson N. R., Harris A. K., Spring D. R., Salmond G. P. (2006). Metabolic and regulatory engineering of Serratia marcescens: mimicking phage-mediated horizontal acquisition of antibiotic biosynthesis and quorum-sensing capacities. Microbiology 152, 1899–1911. 10.1099/mic.0.28803-0 [DOI] [PubMed] [Google Scholar]
- Crosa J. H., Walsh C. T. (2002). Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev 66, 223–249. 10.1128/MMBR.66.2.223-249.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elovson J., Vagelos P. R. (1968). Acyl carrier protein. X. Acyl carrier protein synthetase. J Biol Chem 243, 3603–3611. [PubMed] [Google Scholar]
- Fichtlscherer F., Wellein C., Mittag M., Schweizer E. (2000). A novel function of yeast fatty acid synthase. Subunit alpha is capable of self-pantetheinylation. Eur J Biochem 267, 2666–2671. 10.1046/j.1432-1327.2000.01282.x [DOI] [PubMed] [Google Scholar]
- Fineran P. C., Slater H., Everson L., Hughes K., Salmond G. P. (2005). Biosynthesis of tripyrrole and beta-lactam secondary metabolites in Serratia: integration of quorum sensing with multiple new regulatory components in the control of prodigiosin and carbapenem antibiotic production. Mol Microbiol 56, 1495–1517. 10.1111/j.1365-2958.2005.04660.x [DOI] [PubMed] [Google Scholar]
- Fischbach M. A., Walsh C. T. (2006). Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem Rev 106, 3468–3496. 10.1021/cr0503097 [DOI] [PubMed] [Google Scholar]
- Flyg C., Kenne K., Boman H. G. (1980). Insect pathogenic properties of Serratia marcescens: phage-resistant mutants with a decreased resistance to Cecropia immunity and a decreased virulence to Drosophila. J Gen Microbiol 120, 173–181. [DOI] [PubMed] [Google Scholar]
- Gerc A. J., Song L., Challis G. L., Stanley-Wall N. R., Coulthurst S. J. (2012). The insect pathogen Serratia marcescens Db10 uses a hybrid non-ribosomal peptide synthetase-polyketide synthase to produce the antibiotic althiomycin. PLoS ONE 7, e44673. 10.1371/journal.pone.0044673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinter N. J. (1983). A broad-host-range cloning vector transposable to various replicons. Gene 21, 133–143. 10.1016/0378-1119(83)90155-5 [DOI] [PubMed] [Google Scholar]
- Herrero M., de Lorenzo V., Timmis K. N. (1990). Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol 172, 6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack R. L., Buchanan G., Dubini A., Hatzixanthis K., Palmer T., Sargent F. (2004). Coordinating assembly and export of complex bacterial proteins. EMBO J 23, 3962–3972. 10.1038/sj.emboj.7600409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadouri D. E., Shanks R. M. (2013). Identification of a methicillin-resistant Staphylococcus aureus inhibitory compound isolated from Serratia marcescens. Res Microbiol 164, 821–826. 10.1016/j.resmic.2013.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniga K., Delor I., Cornelis G. R. (1991). A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109, 137–141. 10.1016/0378-1119(91)90599-7 [DOI] [PubMed] [Google Scholar]
- Lambalot R. H., Walsh C. T. (1995). Cloning, overproduction, and characterization of the Escherichia coli holo-acyl carrier protein synthase. J Biol Chem 270, 24658–24661. 10.1074/jbc.270.42.24658 [DOI] [PubMed] [Google Scholar]
- Lambalot R. H., Gehring A. M., Flugel R. S., Zuber P., LaCelle M., Marahiel M. A., Reid R., Khosla C., Walsh C. T. (1996). A new enzyme superfamily - the phosphopantetheinyl transferases. Chem Biol 3, 923–936. 10.1016/S1074-5521(96)90181-7 [DOI] [PubMed] [Google Scholar]
- Li H., Tanikawa T., Sato Y., Nakagawa Y., Matsuyama T. (2005). Serratia marcescens gene required for surfactant serrawettin W1 production encodes putative aminolipid synthetase belonging to nonribosomal peptide synthetase family. Microbiol Immunol 49, 303–310. 10.1111/j.1348-0421.2005.tb03734.x [DOI] [PubMed] [Google Scholar]
- Matsuyama T., Kaneda K., Nakagawa Y., Isa K., Hara-Hotta H., Yano I. (1992). A novel extracellular cyclic lipopeptide which promotes flagellum-dependent and -independent spreading growth of Serratia marcescens. J Bacteriol 174, 1769–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama T., Tanikawa T., Nakagawa T. (2011). Serrawettins and other surfactants produced by Serratia. In Biosurfactants (Microbiology Monographs vol. 20), pp. 93–120. Edited by Soberón-Chávez G. Heidelberg: Springer; 10.1007/978-3-642-14490-5_4 [DOI] [Google Scholar]
- Miethke M., Marahiel M. A. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71, 413–451. 10.1128/MMBR.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz H. D., Finking R., Marahiel M. A. (2001). 4′-phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J Biol Chem 276, 37289–37298. 10.1074/jbc.M103556200 [DOI] [PubMed] [Google Scholar]
- Murugan E., Liang Z. X. (2008). Evidence for a novel phosphopantetheinyl transferase domain in the polyketide synthase for enediyne biosynthesis. FEBS Lett 582, 1097–1103. 10.1016/j.febslet.2008.02.061 [DOI] [PubMed] [Google Scholar]
- Parris K. D., Lin L., Tam A., Mathew R., Hixon J., Stahl M., Fritz C. C., Seehra J., Somers W. S. (2000). Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites. Structure 8, 883–895. 10.1016/S0969-2126(00)00178-7 [DOI] [PubMed] [Google Scholar]
- Perego M., Hoch J. A. (1988). Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production and sporulation in Bacillus subtilis. J Bacteriol 170, 2560–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persmark M., Expert D., Neilands J. B. (1989). Isolation, characterization, and synthesis of chrysobactin, a compound with siderophore activity from Erwinia chrysanthemi. J Biol Chem 264, 3187–3193. [PubMed] [Google Scholar]
- Petty N. K., Foulds I. J., Pradel E., Ewbank J. J., Salmond G. P. (2006). A generalized transducing phage (phiIF3) for the genomically sequenced Serratia marcescens strain Db11: a tool for functional genomics of an opportunistic human pathogen. Microbiology 152, 1701–1708. 10.1099/mic.0.28712-0 [DOI] [PubMed] [Google Scholar]
- Pradel E., Zhang Y., Pujol N., Matsuyama T., Bargmann C. I., Ewbank J. J. (2007). Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci U S A 104, 2295–2300. 10.1073/pnas.0610281104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadri L. E., Weinreb P. H., Lei M., Nakano M. M., Zuber P., Walsh C. T. (1998). Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37, 1585–1595. 10.1021/bi9719861 [DOI] [PubMed] [Google Scholar]
- Raymond K. N., Dertz E. A., Kim S. S. (2003). Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci U S A 100, 3584–3588. 10.1073/pnas.0630018100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebuffat S. (2012). Microcins in action: amazing defence strategies of Enterobacteria. Biochem Soc Trans 40, 1456–1462. 10.1042/BST20120183 [DOI] [PubMed] [Google Scholar]
- Reuter K., Mofid M. R., Marahiel M. A., Ficner R. (1999). Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4′-phosphopantetheinyl transferase superfamily. EMBO J 18, 6823–6831. 10.1093/emboj/18.23.6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwyn B., Neilands J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160, 47–56. 10.1016/0003-2697(87)90612-9 [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost M. R., Cleto S., Carr G., Vlamakis H., João Vieira M., Kolter R., Clardy J. (2012). Mixing and Matching Siderophore Clusters: Structure and Biosynthesis of Serratiochelins from Serratia sp. V4. J Am Chem Soc 134, 13550–13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbald M. J., Winter T., van der Kooi-Pol M. M., Buist G., Tsompanidou E., Bosma T., Schäfer T., Ohlsen K., Hecker M. & other authors (2010). Synthetic effects of secG and secY2 mutations on exoproteome biogenesis in Staphylococcus aureus. J Bacteriol 192, 3788–3800. 10.1128/JB.01452-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Cameotra S. S. (2004). Potential applications of microbial surfactants in biomedical sciences. Trends Biotechnol 22, 142–146. 10.1016/j.tibtech.2004.01.010 [DOI] [PubMed] [Google Scholar]
- Sunaga S., Li H., Sato Y., Nakagawa Y., Matsuyama T. (2004). Identification and characterization of the pswP gene required for the parallel production of prodigiosin and serrawettin W1 in Serratia marcescens. Microbiol Immunol 48, 723–728. 10.1111/j.1348-0421.2004.tb03597.x [DOI] [PubMed] [Google Scholar]
- Thomson N. R., Crow M. A., McGowan S. J., Cox A., Salmond G. P. (2000). Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Mol Microbiol 36, 539–556. 10.1046/j.1365-2958.2000.01872.x [DOI] [PubMed] [Google Scholar]
- Walsh C. T., Gehring A. M., Weinreb P. H., Quadri L. E., Flugel R. S. (1997). Post-translational modification of polyketide and nonribosomal peptide synthases. Curr Opin Chem Biol 1, 309–315. 10.1016/S1367-5931(97)80067-1 [DOI] [PubMed] [Google Scholar]
- Wasserman H. H., Keggi J. J., McKeon J. E. (1962). The structure of serratamolide1–3. J Am Chem Soc 84, 2978–2982. 10.1021/ja00874a028 [DOI] [Google Scholar]
- Weissman K. J., Hong H., Oliynyk M., Siskos A. P., Leadlay P. F. (2004). Identification of a phosphopantetheinyl transferase for erythromycin biosynthesis in Saccharopolyspora erythraea. ChemBioChem 5, 116–125. 10.1002/cbic.200300775 [DOI] [PubMed] [Google Scholar]
- Williamson N. R., Simonsen H. T., Harris A. K., Leeper F. J., Salmond G. P. (2006). Disruption of the copper efflux pump (CopA) of Serratia marcescens ATCC 274 pleiotropically affects copper sensitivity and production of the tripyrrole secondary metabolite, prodigiosin. J Ind Microbiol Biotechnol 33, 151–158. 10.1007/s10295-005-0040-9 [DOI] [PubMed] [Google Scholar]


