Abstract
Staphylococcus aureus is a versatile pathogen of humans and a continued public health concern due to the rise and spread of multidrug-resistant strains. As part of an ongoing investigation into the pathogenic mechanisms of this organism we previously demonstrated that an intracellular N-terminal processing protease is required for S. aureus virulence. Following on from this, here we examine the role of CtpA, the lone C-terminal processing protease of S. aureus. CtpA, a member of the S41 family, is a serine protease whose homologues in Gram-negative bacteria have been implicated in a range of biological functions, including pathogenesis. We demonstrate that S. aureus CtpA is localized to the bacterial cell wall and expression of the ctpA gene is maximal upon exposure to conditions encountered during infection. Disruption of the ctpA gene leads to decreased heat tolerance and increased sensitivity when exposed to components of the host immune system. Finally we demonstrate that the ctpA− mutant strain is attenuated for virulence in a murine model of infection. Our results represent the first characterization of a C-terminal processing protease in a pathogenic Gram-positive bacterium and show that it plays a critical role during infection.
Introduction
Carboxy terminal peptidases (CTPs) represent an unusual and poorly understood class of serine proteases. Classified as the S41 family in the MEROPS database, CTPs are involved in C-terminal proteolytic cleavage of proteins (Rawlings et al., 2010). CTPs are found in a broad range of organisms including eukaryotes, prokaryotes and archaea; however, despite their abundance in nature very few have been studied in detail. Perhaps the best-characterized S41 family member is the C-terminal processing protease (CtpA) found in the chloroplasts of plants, algae and cyanobacteria (Shestakov et al., 1994). Here, CtpA cleaves the C terminus of the precursor D1 protein, a critical component of the photosystem II reaction centre. Removal of the C-terminal peptide results in activation of D1, which is essential for photosynthesis.
In bacteria a limited number of CTPs have been studied, primarily in Gram-negative organisms, with diverse roles and targets reported. Typically, CTPs in Gram-negative bacteria are located in the periplasm (Hara et al., 1991; Hoge et al., 2011). The Escherichia coli CTP (called Prc or Tsp) was originally identified as a periplasmic protease responsible for C-terminal processing of penicillin-binding protein 3 (PBP-3) (Hara et al., 1991). A prc− mutant strain demonstrates altered cell morphology and increased sensitivity to thermal and osmotic stress. As a result, the role of Prc is thought to be in maintaining cell-wall integrity. Prc/Tsp has also been shown to play a role in degrading proteins with non-polar C termini (Silber et al., 1992). Other known targets of Gram-negative CTPs include the outer-membrane protein P13 in Borrelia burgdorferi (Noppa et al., 2001). C-terminal processing of P13 by CtpA stabilizes the protein and directs it to the outer membrane where it may be important during infection (Kumru et al., 2011). Although CTP mutants in Gram-negative bacteria display a variety of phenotypes, certain commonalities exist. Altered cell morphology, differences in osmotic/thermal stress resistance and altered susceptibility to antibiotics are common phenotypes of mutants from different species (and may result from alterations in the cell-wall integrity) (Hara et al., 1991; Kumru et al., 2011; Seoane et al., 1992). In addition, a number of Gram-negative bacteria, including E. coli, Brucella suis, Chlamydia trachomatis, Salmonella typhimurium, Pseudomonas aeruginosa and Burkholderia mallei, demonstrate reduced virulence upon inactivation of CTP (Bandara et al., 2005, 2008; Bäumler et al., 1994; Lad et al., 2007; Seo & Darwin, 2013; Wang et al., 2012). An E. coli prc mutant demonstrates decreased resistance to complement-mediated killing, while in Brucella suis, Burkholderia mallei and S. typhimurium, CtpA has been implicated in the ability of bacteria to survive in the intracellular environment. CT441, one of two CTPs in C. trachomatis, is involved in cleavage of host p65 protein, which interferes with the NF-κB pathway and may suppress the immune response to infection.
Although encoded within the genome of many Gram-positive bacteria, to date CTPs have only been studied in Bacillus subtilis. Bacillus subtilis encodes two CTPs, CtpA and CtpB, which display a high degree of homology, but are functionally distinct, and do not have overlapping roles (Pan et al., 2003; Shestakov et al., 1994). CtpB, the better studied of the two, plays a critical role in sporulation. It is produced in the mother-cell where it is thought to contribute to activation of the transcription factor σK (Pan et al., 2003). As a result, Bacillus subtilis ctpB− mutants demonstrate reduced sporulation efficiency. Much less is known about CtpA in Bacillus subtilis. No phenotypic differences have been reported for ctpA− mutants, and the abrogation of CtpA activity does not affect sporulation (Pan et al., 2003; Shestakov et al., 1994). The CTPs of Bacillus subtilis remain the only characterized members of this protease family in Gram-positive bacteria, and therefore it is unknown what role (if any) CTPs play in the virulence of Gram-positive organisms.
Staphylococcus aureus is a Gram-positive bacterium that is both a commensal and pathogen of humans. It causes a range of diseases with varying degrees of severity at multiple locations throughout the host. Its versatility is due, in part, to a large arsenal of virulence factors that includes adhesins, toxins and superantigens. Previously we have demonstrated that both secreted and intracellular proteases can influence disease causation by S. aureus (Carroll et al., 2012, 2013; Kolar et al., 2013). Our recent discovery that an N-terminal processing peptidase is required for virulence was the first such demonstration in a Gram-positive bacterium. Based upon these findings, and the recent emergence of CTPs as virulence-affecting entities in Gram-negative bacteria, we investigated the conservation, expression and role of CTPs in S. aureus. Our results identify a single S41 family member (CtpA) that is highly conserved in all sequenced strains. CtpA is localized to the bacterial cell wall where it may play a role in maintaining cell-wall stability. In addition, we demonstrate a role for CtpA in stress tolerance and show that expression of ctpA is maximal in conditions encountered during infection. Finally, we show that a ctpA mutant is attenuated for virulence in a murine model of sepsis, demonstrating, for the first time, to our knowledge, that an S41 family member is required for disease causation in a Gram-positive pathogen.
Methods
Bacterial strains, plasmids and growth conditions.
Bacterial strains, plasmids and oligonucleotides used in this study are listed in Table 1. Routinely, E. coli was grown in Luria–Bertani medium (LB) and S. aureus in tripicase soy broth (TSB) with shaking at 37 °C. Antibiotics were used at the following concentrations for S. aureus: chloramphenicol 5 µg ml−1, erythromycin 5 µg ml−1 and lincomycin 25 µg ml−1; and for E. coli: ampicillin 100 µg ml−1.
Table 1. Strains, plasmids and primers.
| Name | Characteristics | Source |
| S. aureus | ||
| RN4220 | Restriction-deficient transformation recipient | Kreiswirth et al. (1983) |
| USA300 HOU | Sequenced USA300-HOU-MRSA isolate cured of pUSA300-HOU-MRSA | Kolar et al. (2011) |
| LNS621 | RN4220 pLES604 ctpA− | This work |
| LNS1516 | USA300 HOU pLES604 ctpA− | This work |
| LNS1788 | USA300 HOU pLES605 ctpA–lacZ | This work |
| LNS1789 | RN4220 pLES1689 | This work |
| LNS1790 | LNS1516 pLES1689 ctpA+ | This work |
| JAI1570 | USA300 HOU pOS1sGFP : : PsarA | Weiss et al. (2014) |
| LNS1893 | LNS1516 (USA300 HOU ctpA−) pOS1sGFP : : PsarA | This work |
| LNS1887 | RN4220 pLNS1888 | This work |
| E. coli | ||
| Dh5α | Routine cloning strain | Invitrogen |
| LNS604 | Dh5α pLES604 | This work |
| LNS605 | Dh5α pLES605 | This work |
| LNS1689 | Dh5α pLES1689 | This work |
| Plasmids | ||
| pAZ106 | Promoterless lacZ suicide vector ermR | Kemp et al. (1991) |
| pMK4 | Shuttle vector cmR | Sullivan et al. (1984) |
| pLES604 | pAZ106 containing 471 bp ctpA fragment | This work |
| pLES605 | pAZ106 containing 1656 bp ctpA promoter fragment | This work |
| pLES1689 | pMK4 containing 2365 bp ctpA-his6 fragment | This work |
| pOS1sGFP : : PsarA | sarA promoter controlling expression of gfp | Benson et al. (2012) |
| Primers | ||
| OL1958 | 5′-CTGGAGAATTCGTGAAAATTGAAG-3′ | |
| OL1959 | 5′-GCGCGGATCCTTAATGATGATGATGATGATGTTTTAAAATATTAATCAACTTATCG-3′ | |
| OL255 | 5′-ACTGGATCCGCTGTCATCACAGTTGTTGC-3′ | |
| OL256 | 5′-ATGGAATTCCACTACCTCGTTGAACAG-3′ | |
| OL257 | 5′-ACTGGATCCGAAAGTGGCGGCTTAAGA-3′ |
Bioinformatics.
Searches via the blast program were performed using the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Percentage identity matrix of S41 family member proteases was performed using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Secretion signal prediction was performed using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/). Transmembrane domain prediction was performed using memsat (Jones et al., 1994). An alignment and heat map showing conserved residues was generated using CLC Sequence viewer 6 (CLC).
Construction of ctpA− mutant.
An internal fragment of the ctpA gene, amplified using primer pair OL255/OL256, was cloned into the suicide vector pAZ106 using restriction enzymes BamHI and EcoRI. The resulting plasmid, pLES604, was transformed into S. aureus RN4220, generating strain LNS621, with clones confirmed by PCR. The disrupted ctpA gene, marked with an erythromycin resistance cassette, was transduced into USA300 HOU, using bacteriophage Φ11, generating strain LNS1516, with clones again confirmed by PCR analysis.
Heat killing.
To determine the effects of increased heat on bacterial survival, heat killing assays were performed as follows. Bacterial strains were grown in TSB at 37 °C to mid-exponential phase and subcultured into fresh TSB at an OD600 of 0.05. Following 1 h of incubation at 37 °C cultures were immediately incubated at 55 °C for 15 min. Samples from each culture were taken before and after incubation at 55 °C and the number of bacteria was enumerated by serially diluting and plating on tripticase soy agar (TSA). Percentage survival was calculated by dividing the number of bacteria recovered by the number of bacteria in each flask immediately prior to incubation at 55 °C. Assays were performed in triplicate and means of the data are presented.
Survival in human serum.
To determine survival in human serum, bacterial cultures were grown in TSB to mid-exponential phase, washed twice with PBS, and used to inoculate human serum at an OD600 of 0.05. To determine bacterial counts (c.f.u. ml−1), samples were serially diluted and plated onto TSA. Percentage survival was calculated by dividing the number of bacteria recovered by the number of bacteria in the inoculated serum cultures. Assays were performed in triplicate.
Co-infection of whole human blood.
The USA300 HOU wild-type and ctpA− mutant strains were grown to mid-exponential phase, washed twice with PBS, and equal quantities of each strain were used to inoculate 1 ml aliquots of pooled, whole human blood (Bioreclamation) in a 1 : 1 ratio. Blood samples were inoculated with bacteria corresponding to an OD600 of 0.025 of each strain. At various time points post-inoculation the total number of bacterial c.f.u. per millilitre of blood was determined by serially diluting and plating onto TSA. Simultaneously, the number of ctpA− mutant bacteria in each sample was determined by serially diluting and plating onto TSA containing erythromycin. The percentage of ctpA− mutant cells was determined at each time point by dividing the number of ctpA− mutant cells by the total number of bacterial c.f.u. present in the sample. The assay was performed using six independent replicates.
THP-1 macrophage infection assay.
THP-1 cells were grown at 37 °C in a 5 % CO2 atmosphere in RPMI 1640 with l-glutamine (Cellgro) supplemented with 10 % heat-inactivated FBS (Gibco). To induce the cells to differentiate into adherent macrophages they were treated with 80 nM phorbol 12-myristate 13-acetate (PMA). PMA treatment was carried out 48 h prior to infection with S. aureus. PMA-treated THP-1 cells were seeded into six-well plates at a density of 106 cells per well. Wells were infected with bacteria at an m.o.i. of 1 (corresponding to 106 bacteria per well). Following the addition of bacteria, plates were centrifuged at 450 g for 10 min and incubated for 1 h at 37 °C in a 5 % CO2 atmosphere to allow phagocytosis to occur. At this point the cells were washed with PBS and culture medium containing 30 µg gentamicin ml−1 was added to kill any remaining extracellular bacteria. Following 1 h of incubation at 37 °C in 5 % CO2 the medium was replaced with medium containing 5 µg gentamicin ml−1, and the cells were returned to the incubator for the remainder of the experiment. At the time points indicated, cells were washed three times with PBS and THP-1 macrophages were lysed by adding 500 µl of 0.5 % Triton X-100 in PBS. The lysates were diluted and plated on TSA to determine the number of surviving intracellular bacteria. Data shown is the mean of three independent replicates.
Phagocytosis assay.
FACS-based phagocytosis assays were performed using whole human blood and bacterial strains constitutively expressing GFP, as described by us previously (Kolar et al., 2013). The GFP-expressing plasmid pOS1sGFP : : PsarA (Benson et al., 2012) was transduced into the ctpA− mutant using bacteriophage Φ11 to create strain LNS1893. The number of GFP-positive granulocytes and macrophages was determined following 30 min of incubation of bacteria in human blood.
Construction of ctpA–lacZ reporter fusion.
A 1656 bp region of DNA containing the ctpA promoter was amplified using primers OL257/OL256 and cloned into suicide vector pAZ106 upstream of a promoterless lacZ gene. The resulting plasmid, pLES605, was transformed into RN4220, and the ctpA–lacZ fusion transduced into USA300 HOU using bacteriophage Φ11, creating strain LNS1788.
β-Galactosidase assays.
β-Galactosidase assays were performed on cultures grown in TSB using 4-MUG as described by us previously (Carroll et al., 2012). Experiments were carried out in triplicate. β-Galactosidase assays to determine the effect of sublethal concentrations of antibiotics were carried out using overnight cultures.
Expression and detection of plasmid-encoded/histidine-tagged CtpA.
To construct a plasmid-encoded, histidine-tagged copy of CtpA, the ctpA gene and its promoter were amplified using primers OL1958/OL1959. OL1959 introduced 18 additional nucleotides at the 3′ end of the ctpA gene that, when translated, result in an additional six C-terminal histidine residues with S. aureus codon preference. The resulting fragment was cloned into pMK4, generating pLES1689. This was then transformed into RN4220 (generating strain LNS1789) and subsequently transduced into the USA300 HOU ctpA− mutant strain, to generate LNS1790. Strains were confirmed by PCR analysis. Western immunoblots to detect CtpA-his were performed as described previously using a monoclonal anti-histidine antibody (Covance) (Carroll et al., 2012, 2013). S. aureus cells expressing the plasmid-encoded CtpA-his were grown in TSB overnight and bacteria were collected by centrifugation. Culture supernatants were passed through a 0.22 µm filter, TCA precipitated and used in Western blot analysis. Bacterial lysis was achieved by resuspending the pellet and incubating at 37 °C for 30 min with 20 µg lysostaphin ml−1. Lysates were centrifuged and the supernatant containing intracellular proteins was collected. The insoluble pellet, representing cell membrane and cell-wall proteins, was resuspended in 6 M urea.
Murine sepsis model of infection.
A murine sepsis model of infection was used as described previously (Carroll et al., 2012). Mice were infected via tail vein injection with 1×108 c.f.u. Survival was monitored over a 7-day period and the data were analysed using Kaplan–Meyer plots. For mice that survived the 7-day infection, the bacterial burden in the brain, heart, lungs, liver, kidneys and spleens was determined by homogenizing organs in PBS followed by serial dilution and plating on TSA. Statistical analyses were performed using SAS software (version 9.2, SAS Institute). The distribution of data was determined in SAS through tests for normality (SAS proc univariate) and equality of variance (SAS proc ttest). The statistical significance of bacterial recovery from the murine model of sepsis was evaluated using a Mann–Whitney test. For all statistical analyses the significance level was set at α = 0.05. This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the University of South Florida (Permit Number: A-4100-01). Infection was carried out twice with comparable results obtained on each occasion. The data presented are representative of both outcomes.
Results
Identification of ctpA in S. aureus
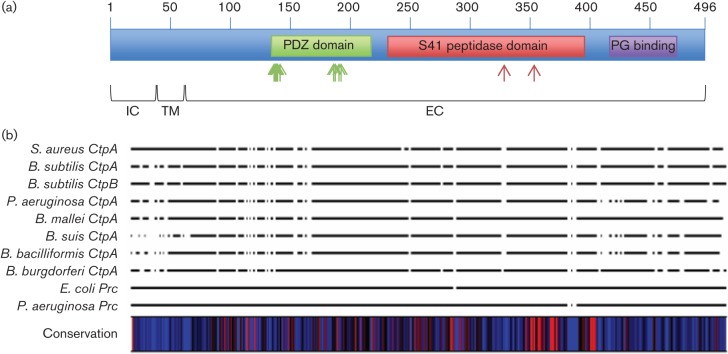
Recent work by ourselves and others has shown that amino terminal peptidases and CTPs can play an important role in bacterial virulence (Bandara et al., 2008; Carroll et al., 2012, 2013; Lad et al., 2007; Wang et al., 2012). These findings, along with the observation that C-terminal processing peptidases are poorly understood in Gram-positive pathogens, led us to investigate the role of C-terminal processing peptidases in S. aureus. While common, CTPs are not ubiquitous, and therefore we performed a blast search using the Bacillus subtilis CtpA protein sequence and the community-associated meticillin-resistant S. aureus (MRSA) strain USA300 FPR as a reference. From this analysis we identified a single CTP in S. aureus, designated SAUSA300_1313, and annotated as ctpA. Interestingly, interrogating the S. aureus genome with the sequence of the Bacillus subtilis CtpB protein returned the same results, demonstrating that S. aureus seemingly encodes a lone CTP. Next, we examined the genomes of all sequenced S. aureus strains in the NCBI database and found that ctpA is highly conserved. All 41 complete sequenced genomes contain the ctpA gene, suggesting that it may play an important role in the cell. A blast search performed using the S. aureus CtpA protein sequence demonstrates that it has a similar domain organization to CtpA and CtpB from Bacillus subtilis (Figs 1 and S1A, available in the online Supplementary Material). A protein binding PDZ domain is located between amino acid residues 135 and 218 while an S41 CTP peptidase domain is located between residues 231 and 395. Interestingly, a peptidoglycan binding domain was identified (residues 417–473), which is only present in CTPs from Gram-positive bacteria, and not found in those from Gram-negative bacteria (Fig. S1). The presence of this domain suggests it plays a unique structural and/or functional role in CTPs of Gram-positive bacteria.
Fig. 1.
Domain structure and multiple sequence alignment of CtpA and S41 family members. (a) Domain structure of S. aureus CtpA. Three putative conserved domains were identified by blast search of the S. aureus CtpA protein sequence. A PDZ domain is located from amino acids 135 to 218 (green box), an S41 peptidase domain from 231 to 395 (red box) and a peptidoglycan (PG) binding domain from 417 to 473 (purple box). The position of amino acid residues in the PDZ domain that are predicted to be involved in protein binding (G137, I138, G139, A140, M142, V187, V188, V191 and R192) are indicated by green arrows. The S41 protease catalytic dyad (consisting of residues S329 and K354) in the S41 domain is indicated by red arrows. Regions of CtpA that are predicted to be intracellular (IC), transmembrane (TM) and extracellular (EC) are indicated. (b) Multiple sequence alignment of S41 family members from S. aureus, Bacillus subtilis, P. aeruginosa, Burkholderia mallei, Brucella suis, Bartonella bacilliformis, E. coli and Borrelia burgdorferi. A heat map was generated to display similar/identical residues (red, 100 %) revealing an area with a high degree of conservation located from amino acids 321 to 395 of the S. aureus CtpA sequence.
Multiple sequence analysis of S. aureus CtpA and S41 family members
To investigate the degree of conservation between S. aureus CtpA and other S41 protease family members, a multiple sequence alignment was performed. The S. aureus CtpA sequence was aligned with studied S41 family members from Bacillus subtilis, P. aeruginosa, Burkholderia mallei, Brucella suis, Bartonella bacilliformis, E. coli and Borrelia burgdorferi. To identify regions in the alignment with a high degree of conservation, a heat map of conserved residues was generated (Fig. 1b). This analysis identified a region with a high degree of conservation located between residues 321 and 395 (amino acid positions relative to the S. aureus CtpA sequence). This area of high conservation lies in the S41 CTP peptidase domain and contains the Ser and Lys residues that form the catalytic dyad in S41 family members. The high degree of conservation around the catalytic site suggests that S. aureus CtpA has enzymic conservation with other S41 family members. The lack of significant homology elsewhere in the alignment (including in the PDZ domain) suggests that the substrates/targets of each protease may vary, leading to the broad range of biological functions observed for S41 family members.
Using the multiple sequence alignment data, a percentage identity matrix was generated for the ten S41 family members used (Table S1). Levels of identity for S. aureus CtpA compared with the Gram-negative S41 family members ranged from 23.61 % (for E. coli Prc) to 31.12 % (for P. aeruginosa CtpA). Identities compared with the two Gram-positive S41 family members were 42.45 % for Bacillus subtilis CtpA and 31.76 % for Bacillus subtilis CtpB. Interestingly, the level of identity with Bacillus subtilis CtpB is only slightly higher that that observed with Gram-negative S41 family members, whilst a much higher degree of similarity is observed with Bacillus subtilis CtpA. This suggests the in vivo role of S. aureus CtpA may be more closely related to that of Bacillus subtilis CtpA than CtpB. This hypothesis is supported by previous work demonstrating that the primary role for Bacillus subtilis CtpB is in sporulation, a process that does not occur in S. aureus (Campo & Rudner, 2007; Pan et al., 2003). Currently a role for CtpA in Bacillus subtilis has yet to be identified (Marasco et al., 1996).
Cellular localization of CtpA
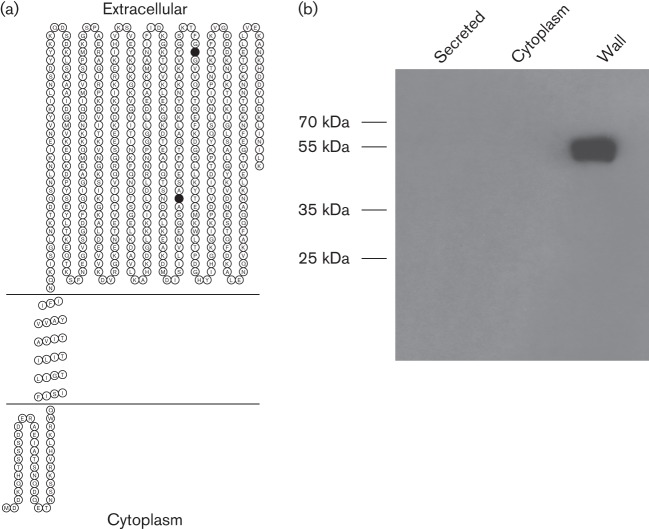
To identify the cellular location of CtpA we first performed an in silico analysis of the CtpA protein sequence. No secretion signal sequence was identified using signalP. A search for transmembrane helices identified one stretch of hydrophobic amino acids that, with high probability, transverses the membrane. The model predicts that the N-terminal portion of CtpA (amino acids 1–38) is located in the bacterial cytosol, residues 39–61 constitute a transmembrane helix and the C-terminal portion (amino acids 62–496) are located extracellularly, in the cell wall (Figs 1a and 2a). The PDZ domain, S41 protease domain (including the catalytic dyad) and peptidoglycan binding domain are all predicted to be located in the extracellular region of the protein, suggesting that the enzymic activity of CtpA is localized to the bacterial cell wall.
Fig. 2.
CtpA is localized to the bacterial cell wall. (a) A topology model for CtpA localization was generated using memsat (Jones et al., 1994). N-terminal residues 1–38 are located in the bacterial cytosol while a hydrophobic stretch of amino acids from 38 to 61 forms a transmembrane helix. Residues 61–455 are extracellular, located in the cell wall. The catalytic Ser and Lys residues are indicated as filled circles. (b) Western immunoblot analysis of CtpA localization. Histidine-tagged CtpA was expressed from a plasmid in S. aureus. Intracellular, extracellular and bacterial cell-wall protein samples were probed with an anti-histidine antibody.
To confirm the predicted localization of CtpA, a histidine-tagged copy of CtpA was expressed in trans from a plasmid in a USA300 ctpA− mutant background. Using an anti-histidine monoclonal antibody, Western immunoblots were performed using culture supernatants, cell lysates and cell-wall fractions to identify the location of CtpA. Western blot analysis revealed that no CtpA-his was detected in culture supernatants or cytoplasmic fractions (Fig. 2b). A band corresponding in size to CtpA-his was detected in the cell-wall fraction. These data confirm the predicted location of CtpA in the bacterial cell wall.
Analysis of ctpA gene expression
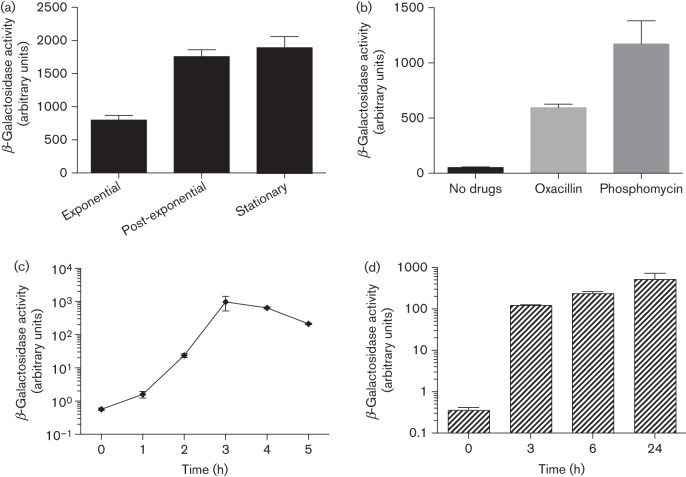
To examine ctpA gene expression, a ctpA–lacZ reporter fusion was constructed in USA300 HOU. This strain was grown in TSB, and β-galactosidase assays were performed on samples taken from exponential (3 h), post-exponential (6 h) and stationary phase (10 h). Results demonstrated clear growth-phase-dependent expression of ctpA, with approximately 2.4-fold higher expression detected in the post-exponential and stationary phases of growth than in the exponential phase (Fig. 3a). This pattern (i.e. higher expression in post-exponential phase) mirrors that of a number of S. aureus secreted virulence factors (Rivera et al., 2012).
Fig. 3.
Analysis of ctpA expression. (a) The USA300 ctpA–lacZ reporter fusion strain was grown in TSB and samples were taken for β-galactosidase assays at 3 h (exponential phase), 6 h (post-exponential phase) and 10 h (stationary phase). (b) Overnight cultures of USA300 ctpA–lacZ containing subinhibitory concentrations of antibiotics were used for β-galactosidase assays. (c) The USA300 ctpA–lacZ reporter fusion strain was grown in TSB for 3 h and used to inoculate human serum. Samples were taken for β-galactosidase assays at various time points post-inoculation. The 0 h time point corresponds to 3 h growth in TSB. (d) RAW264.7 cells were infected with the USA300 ctpA–lacZ reporter fusion strain and samples were taken for β-galactosidase assays at various time points post-infection. All data shown are the mean of three independent replicates, with error bars representing ±sd.
Previously we have demonstrated that activation of gene expression in response to stress can provide insight into the role of protein products within the cell (Kolar et al., 2011; Miller et al., 2012). Therefore, we utilized the ctpA–lacZ promoter fusion in a plate-based screen for compounds that activate expression (Shaw et al., 2008). Two compounds, oxacillin and phosphomycin, resulted in increased ctpA–lacZ expression, as indicated by a green ring in disc diffusion assays. To confirm and quantify these findings, β-galactosidase assays were performed using the ctpA–lacZ reporter fusion strain grown in liquid media containing subinhibitory concentrations of the two compounds. We observed an 11-fold increase in ctpA expression in the presence of oxacillin, and a 22-fold increase in expression in the presence of phosphomycin (Fig. 3b). Both of these agents target the bacterial cell wall, raising the possibility that activation of ctpA expression may occur in response to cell-wall damage.
To examine ctpA gene expression under physiologically relevant conditions we performed profiling during growth in human serum. The ctpA–lacZ reporter strain was grown to mid-exponential phase and used to inoculate human serum. Samples were taken and assays performed on aliquots from the TSB inoculum (0 h) and serum at 1–5 h post-inoculation. We determined a 2.8-fold activation of ctpA expression after 1 h in human serum (Fig. 3c). This increase is similar in magnitude to that observed between 3 and 10 h growth in TSB (2.4-fold), demonstrating that the level of ctpA expression in TSB in stationary phase is similar to that following 1 h in serum (Fig. 3a). Expression of ctpA in serum further increased 41-fold at 2 h, and a remarkable 1704- (3 h) and 1124-fold (4 h) at later time points (approximately 710 and 468 times higher than the level of ctpA expression in TSB in stationary phase). These data, demonstrating that expression of ctpA is highly induced in human serum, suggest that CtpA may be required in this environmental niche, which could have important consequences during infection.
To test if induction of ctpA expression is observed in additional in vivo-like niches, we performed β-galactosidase assays using the ctpA–lacZ reporter fusion in the intracellular environment. RAW246.7 cells were infected with the ctpA–lacZ fusion strain, and samples were collected pre-infection and at 3, 6 and 24 h post-infection. Similar to results from human serum, a large induction of ctpA expression was observed upon phagocytosis (Fig. 3d). A 345-fold increase in expression was observed at 3 h, a 661-fold increase at 6 h and a 1462-fold increase at 24 h. These data support the contention that ctpA expression is activated under conditions that are encountered during infection, raising the possibility that CtpA may be important for disease causation.
An S. aureus ctpA− mutant demonstrates increased sensitivity to heat
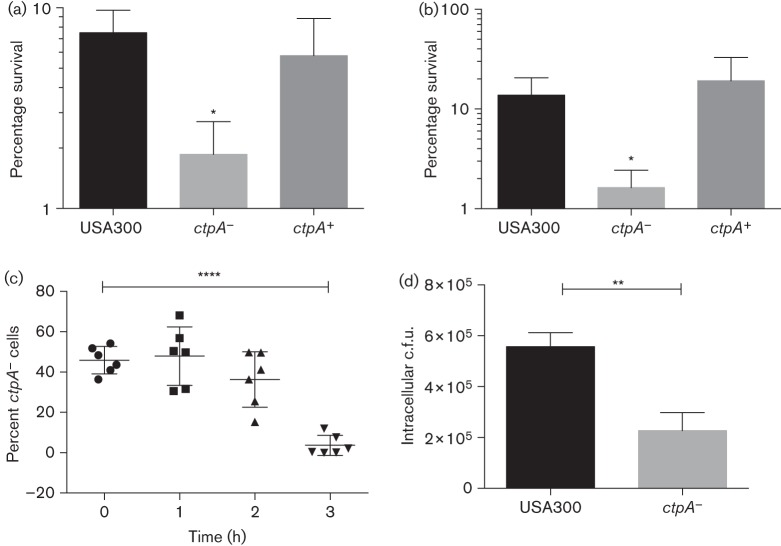
Strains deficient in ctpA have been reported to exhibit defects in survival when exposed to osmotic and environmental stress/heat shock. To examine the role of CtpA in stress tolerance we constructed a ctpA− mutant strain. Analysis of RNA-seq data from the USA300 Houston strain demonstrate that the ctpA− gene is monocistronic and therefore can be disrupted without causing polar effects on downstream genes (Fig. S2) (Weiss et al., 2014). The mutant generated did not exhibit a growth defect under any laboratory growth conditions tested (data not shown). We performed osmotic stress tests to assess the ability of the mutant strain to grow in the presence of high salt concentrations, and again no difference was observed between the wild-type and ctpA− mutant. These results suggest CtpA does not play a role in resistance to osmotic stress in S. aureus. Next, a heat killing analysis was performed using the wild-type, ctpA− and ctpA+ (complement) strains. Strains were grown in TSB at 37 °C for 1 h and then transferred to 55 °C. At 15 min intervals the number of surviving bacteria was determined by serial dilution and plating on TSA. Results show the number of bacterial c.f.u. surviving immediately following 15 min incubation at 55 °C was threefold higher in the WT than in the ctpA− mutant (Fig. 4a). The decrease in bacterial survival was mitigated in the ctpA+ complemented strain. At later time points (i.e. 30, 45 and 60 min of incubation at 55 °C) similar results were obtained (data not shown).
Fig. 4.
CtpA is required for stress tolerance and during interaction with the human immune system. (a) The wild-type, ctpA− and ctpA+ strains were grown in TSB at 37 °C and subsequently transferred to 55 °C for 15 min. Bacterial counts (c.f.u. ml−1) were calculated before and after incubation and the number of surviving bacteria is expressed as a percentage of the inocula. Data shown are the mean of three independent replicates, with error bars representing ±sd. (b) WT, ctpA− and ctpA+ strains were incubated in human serum for 1 h and the number of bacteria (c.f.u. ml−1) was calculated before and after incubation. The percentage survival of each strain is shown. Data are the mean of three independent replicates, with error bars representing ±sd. (c) Aliquots of whole human blood were inoculated with equal quantities of the WT and ctpA− mutant strains. Samples were incubated at 37 °C and the relative proportion of ctpA− mutant cells in each was determined over time by serial dilution and replicate plating on TSA and TSA containing erythromycin. Individual data as well as mean and sd are indicated for each time point. (d) THP-1 human derived macrophages were infected with the WT and ctpA− mutant strains. Twenty-four hours post-infection the number of surviving intracellular bacteria was enumerated. Data shown are the mean of three independent replicates. Statistical significance was determined for all experiments using Student’s t-test: *P≤0.05, **P≤0.01, ****P≤0.0001.
The ctpA− mutant demonstrates decreased survival when exposed to components of the human immune system
Recent reports have linked S41 family members with virulence in Gram-negative bacteria. Several of these studies have shown that mutation of ctpA leads to impaired interactions with components of the host immune system (Bandara et al., 2005, 2008; Lad et al., 2007; Wang et al., 2012). To investigate if mutation of ctpA affects the interaction of S. aureus with components of the human immune system, we examined the survival of the ctpA− mutant in the presence of a variety of immune components.
First we performed survival analysis of the WT, ctpA− and ctpA+ strains in human serum. Results show that following 1 h of incubation the proportion of surviving WT bacteria was 13.8 % (Fig. 4b). In contrast, the number of ctpA− mutant cells recovered at the same time point was 8.5-fold lower (1.6 % recovery). Complementation restored survival to WT levels (19 %). This stark difference in bacterial survival indicates strongly that the ctpA− mutant is more susceptible to components of the humoral immune system than the WT strain.
To further explore the interaction of CtpA with components of the human immune system, a competition assay was performed to compare the survivability of the USA300 HOU WT and ctpA− mutant in whole human blood. Aliquots of human blood were co-infected with both strains at a 1 : 1 ratio. At various time points post-infection the surviving bacteria were enumerated and the ratio of WT to ctpA− mutant was calculated. No significant differences were observed at 1 and 2 h, but a 22.2-fold increase in the ratio (WT/ctpA−) was observed at 3 h post-infection, with the number of ctpA− cells comprising only 3.7 % of the samples (Fig. 4c). These data collectively demonstrate that the mutant strain is at a competitive disadvantage in human blood and is probably more susceptible to killing by components of the host immune system.
To investigate if the ctpA− mutant strain was more sensitive to engulfment by professional phagocytes we performed a phagocytosis assay. WT and ctpA− mutant strains, constitutively expressing GFP from a plasmid, were used to infect aliquots of whole human blood and the number of GFP-positive leukocytes was determined by FACS analysis. Results demonstrate equal amounts of WT and ctpA− mutant bacteria inside granulocytes and macrophages after 30 min, indicating that there is no difference in the rate of phagocytosis (Fig. S3).
Finally to investigate whether the ctpA− mutant demonstrates decreased ability to survive inside professional phagocytes we infected THP-1 human macrophages with WT and ctpA− mutant bacteria, and assayed the ability of each strain to survive in the intracellular environment. One hour post-phagocytosis no difference in the number of intracellular bacteria (WT versus ctpA− mutant) was observed (data not shown). However, 24 h post-infection results show a 2.5-fold decrease in the number of surviving ctpA− mutant bacteria compared with the WT (Fig. 4d). Together these data suggest that while the rate of phagocytosis is similar for both strains, the ctpA− mutant is less tolerant of the macrophage intracellular environment than the WT strain.
CtpA contributes to virulence in S. aureus
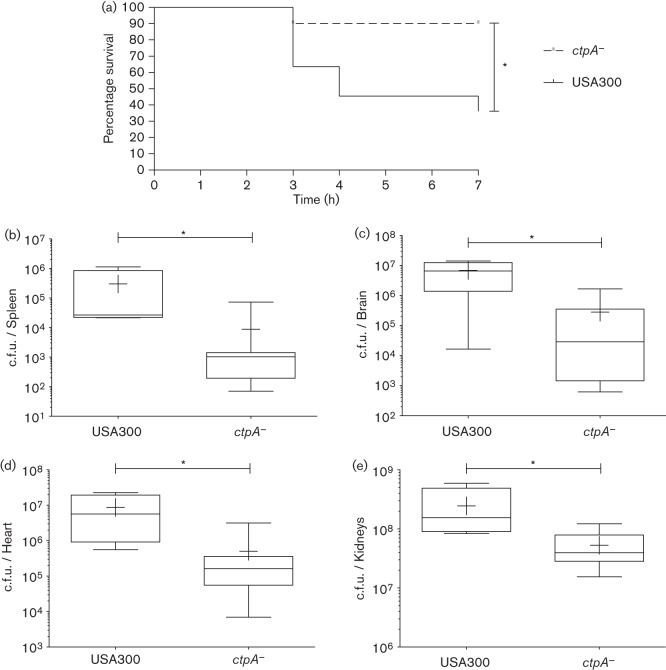
Our data demonstrating that a ctpA− mutant has increased sensitivity to killing by human immune system components, and that ctpA− expression is induced upon ex vivo interaction with the immune system, suggests strongly a role for this enzyme during infection. To test this hypothesis we utilized a murine sepsis model of infection and our USA300 wild-type and ctpA− mutant strains. CD-1 Swiss outbred mice were infected with either the WT or its ctpA− mutant (n = 10 each). Survival was monitored over 7 days and the data were used to generate a Kaplan–Meier curve (Fig. 5a). Results show a significant increase in survival for mice infected with the ctpA− mutant, with a lone mutant infected mouse not surviving the infection period, whilst six animals infected with the WT died. All surviving mice were killed after 7 days and the bacterial burden of internal organs (brain, heart, lungs, liver, kidneys and spleen) was determined. A significant decrease in the median bacterial c.f.u. per organ was observed in the spleen (28-fold), brain (228-fold), heart (65-fold) and kidneys (sixfold) of ctpA− mutant infected mice (Fig. 5b–e). These data show that the severity of disease in ctpA− mutant infected mice is markedly less than that of WT infected mice, confirming that CtpA is required for optimal virulence.
Fig. 5.
CtpA is required for virulence in S. aureus. Two groups, each consisting of ten mice, were infected with the WT or ctpA− mutant strain via tail vein injection. Mouse survival was monitored over a 7-day period (a), and analysed using a Kaplan–Meier survival curve. Statistical significance was determined using the log rank test (*P≤0.05). Following this, surviving mice were killed and the bacterial burden of internal organs was determined (b–e). Statistical significance was determined using a Mann–Whitney test (WT n = 4, ctpA− mutant n = 9; *P≤0.05).
Discussion
Our previous work, identifying a role in virulence for an S. aureus amino-terminal protease (Carroll et al., 2012, 2013), led us to investigate the role of the solitary C-terminal processing enzyme in this important human pathogen. Previous studies on this unusual and somewhat cryptic family of enzymes in prokaryotes have been carried out either in Gram-negative bacteria (where many have been shown to affect virulence) or in non-pathogenic Gram-positive species (where one example is known to affect sporulation) (Bandara et al., 2008; Lad et al., 2007; Marasco et al., 1996; Ostberg et al., 2004; Pan et al., 2003). In this work we have, for the first time, to our knowledge, identified and characterized a C-terminal protease from a Gram-positive, pathogenic bacterial species.
Experimental evidence from assays performed in human serum, whole human blood and cultured human macrophages demonstrates that the ctpA− mutant is more susceptible to killing by elements of the human immune system. The exact mechanism behind this increased susceptibility remains unclear, although certain inferences can be made. Human serum lacks components of cell-mediated immunity, and therefore components of humoral immunity must be responsible for the decreased survival observed for the ctpA− mutant. Recently, it was shown in E. coli that a prc mutant is more susceptible to killing by the complement membrane attack complex (Wang et al., 2012), although this clearly cannot be the explanation for the decreased survival observed in this study. Therefore, we speculate that the decrease in viability in serum results from the action of additional elements, such as antimicrobial peptides. To test this hypothesis we examined the sensitivity of the WT and ctpA− mutant strains to the human cathelicidin LL-37, but no difference in sensitivity was observed under the conditions tested (data not shown). While these data demonstrate that the decreased survival of the ctpA− mutant is not due to an increased sensitivity to LL-37, additional antimicrobial peptides may be responsible. It is also possible that a number of factors, such as nutrient and iron availability, may contribute to the decrease in ctpA− mutant survival in serum. However, because the defect is observed following a relatively short period in human serum (1 h), it appears likely that the reduction is a direct result of action by the immune system on the bacteria. The decrease in survival of the ctpA− mutant in whole blood is likely to have many underlying causes. As discussed above, the ctpA− mutant is sensitive to components of humoral immunity but it also demonstrates decreased survival inside professional phagocytes. Importantly, the decrease in ctpA− mutant bacteria recovered from the intracellular environment is not a result of decreased uptake as similar levels of phagocytosis were observed for the WT and ctpA− mutant strains.
One important goal of this study was to determine whether, as is the case for Gram-negative bacteria, CTPs can influence disease causation in Gram-positive species. The results from the murine sepsis model clearly demonstrate disease caused by the ctpA− mutant was less severe than that caused by the WT strain. The reasons behind this attenuation are unclear; however, because dissemination of S. aureus occurs via the bloodstream, the decrease in bacterial burden observed in organs may be a consequence of increased sensitivity of the ctpA− mutant to host immune system components encountered during dissemination. Alternatively, the absence of CtpA in the cell wall may result in aberrant processing/activation of cell-wall-associated virulence factors, which in turn could impact the ability of the mutant to cause disease. The study presented herein focused exclusively on the epidemic community-associated MRSA strain USA300 Houston, but the high degree of conservation of CtpA across all sequenced isolates of S. aureus (meticillin-sensitive and hospital-acquired MRSA strains) suggests that its role in virulence may also be conserved in these strains.
Frequently, insight into the function of a protein can be gained by identifying the conditions under which the corresponding gene is expressed. Expression of ctpA is highest under conditions likely to be encountered by the bacteria during infection (i.e. in human serum and in the intracellular environment). These data (along with the results of the virulence assay) further confirm that CtpA function is required in vivo. In addition, we demonstrate induction of ctpA expression in the presence of two antimicrobial agents, phosphomycin and oxacillin, both of which target peptidoglycan biosysthesis. Although ctpA expression was induced in the presence of these two compounds, no increase in sensitivity to either was observed in the ctpA mutant (data not shown), which indicates that CtpA is not required for meticillin resistance in S. aureus. Phosphomycin targets MurA at the first committed step in peptidoglycan biosynthesis, while oxacillin prevents transpeptidation by binding to and inhibiting PBPs. Interestingly, PBP-3 is a known substrate of the E. coli CTP, raising the intriguing possibility that PBPs may be a target for S. aureus CtpA. The S. aureus homologue of PBP-3 (named PBP-1) displays 67 % similarity to E. coli PBP-3 and contains a C-terminal 154 aa region not found in PBP-3. Currently, it is unknown if S. aureus PBP-1 is processed prior to activation, although this represents one interesting potential target for CtpA activity. The presence of a C-terminal peptidoglycan binding domain in CtpA homologues from Gram-positive bacteria increases the likelihood that their function may be related to this crucial cell-wall constituent.
In the absence of a periplasmic space (where Gram-negative CTPs are located) we sought to determine the location of the S. aureus CtpA. Results demonstrate that it is membrane anchored, with the majority of the protein, including the catalytic site, located in the bacterial cell wall. Known targets of Gram-negative CTPs, including PBP-3 and P13, are located either in the bacterial periplasm or in the outer membrane (Hara et al., 1991; Noppa et al., 2001). This suggests that in Gram-negative species CTPs co-localize with their target substrates (Hoge et al., 2011). If this hypothesis holds true for Gram-positive bacteria then it seems likely that the proteolytic targets of S. aureus CtpA-mediated hydrolysis are located in the bacterial cell wall. In Gram-negative bacteria, many of the phenotypes associated with ctpA− mutants, including altered cell morphology and increased sensitivity to heat and osmotic shock, are proposed to be a consequence of decreased cell-wall integrity (Hara et al., 1991; Kumru et al., 2011; Ostberg et al., 2004; Seoane et al., 1992). Studies in E. coli have demonstrated periplasmic protein leakage in a prc mutant, suggesting increased permeability of the outer membrane in this strain (Hara et al., 1991). Together, these studies suggest a role for CTPs in maintaining cell-wall stability and integrity.
Due to the considerable differences in cell-wall architecture between Gram-negative and Gram-positive bacteria we investigated the role of CtpA in S. aureus to determine whether alterations in cell-wall stability were evident. Similar to results obtained for Gram-negative bacteria, differences in heat tolerance were observed with S. aureus (Hara et al., 1991; Seoane et al., 1992). This, together with the CtpA localization data, and the induction of expression observed in the presence of peptidoglycan-targeting antibiotics, suggests strongly that CtpA is located in the bacterial cell wall where it functions as a protease to maintain cell-wall integrity.
In summary, we have demonstrated, for the first time, to our knowledge, a CTP that is required for virulence in a Gram-positive bacterial pathogen. Expression of ctpA in S. aureus is induced in stationary phase, with optimal expression observed under conditions likely to be encountered by bacteria during infection. The specific targets of CtpA remain unknown, although it is likely that the action of this protease helps maintain cell-wall stability and aids in defence against components of the host immune system.
Acknowledgements
This study was supported in part by grants from the National Institute of Allergies and Infectious Diseases (AI080626 and AI109389, both L. N. S.).
Abbreviations:
- CTP
carboxy terminal peptidase
- MRSA
meticillin-resistant Staphylococcus aureus
- PBP
penicillin-binding protein
Footnotes
One supplementary table and three supplementary figures are available with the online version of this paper.
References
- Bandara A. B., Sriranganathan N., Schurig G. G., Boyle S. M. (2005). Carboxyl-terminal protease regulates Brucella suis morphology in culture and persistence in macrophages and mice. J Bacteriol 187, 5767–5775. 10.1128/JB.187.16.5767-5775.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara A. B., DeShazer D., Inzana T. J., Sriranganathan N., Schurig G. G., Boyle S. M. (2008). A disruption of ctpA encoding carboxy-terminal protease attenuates Burkholderia mallei and induces partial protection in CD1 mice. Microb Pathog 45, 207–216. 10.1016/j.micpath.2008.05.005 [DOI] [PubMed] [Google Scholar]
- Bäumler A. J., Kusters J. G., Stojiljkovic I., Heffron F. (1994). Salmonella typhimurium loci involved in survival within macrophages. Infect Immun 62, 1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson M. A., Lilo S., Nygaard T., Voyich J. M., Torres V. J. (2012). Rot and SaeRS cooperate to activate expression of the staphylococcal superantigen-like exoproteins. J Bacteriol 194, 4355–4365. 10.1128/JB.00706-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo N., Rudner D. Z. (2007). SpoIVB and CtpB are both forespore signals in the activation of the sporulation transcription factor σK in Bacillus subtilis. J Bacteriol 189, 6021–6027. 10.1128/JB.00399-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll R. K., Robison T. M., Rivera F. E., Davenport J. E., Jonsson I. M., Florczyk D., Tarkowski A., Potempa J., Koziel J., Shaw L. N. (2012). Identification of an intracellular M17 family leucine aminopeptidase that is required for virulence in Staphylococcus aureus. Microbes Infect 14, 989–999. 10.1016/j.micinf.2012.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll R. K., Veillard F., Gagne D. T., Lindenmuth J. M., Poreba M., Drag M., Potempa J., Shaw L. N. (2013). The Staphylococcus aureus leucine aminopeptidase is localized to the bacterial cytosol and demonstrates a broad substrate range that extends beyond leucine. Biol Chem 394, 791–803. 10.1515/hsz-2012-0308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H., Yamamoto Y., Higashitani A., Suzuki H., Nishimura Y. (1991). Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J Bacteriol 173, 4799–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoge R., Laschinski M., Jaeger K. E., Wilhelm S., Rosenau F. (2011). The subcellular localization of a C-terminal processing protease in Pseudomonas aeruginosa. FEMS Microbiol Lett 316, 23–30. 10.1111/j.1574-6968.2010.02181.x [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. (1994). A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33, 3038–3049. 10.1021/bi00176a037 [DOI] [PubMed] [Google Scholar]
- Kemp E. H., Sammons R. L., Moir A., Sun D., Setlow P. (1991). Analysis of transcriptional control of the gerD spore germination gene of Bacillus subtilis 168. J Bacteriol 173, 4646–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar S. L., Nagarajan V., Oszmiana A., Rivera F. E., Miller H. K., Davenport J. E., Riordan J. T., Potempa J., Barber D. S. & other authors (2011). NsaRS is a cell-envelope-stress-sensing two-component system of Staphylococcus aureus. Microbiology 157, 2206–2219. 10.1099/mic.0.049692-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar S. L., Ibarra J. A., Rivera F. E., Mootz J. M., Davenport J. E., Stevens S. M., Horswill A. R., Shaw L. N. (2013). Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. MicrobiologyOpen 2, 18–34. 10.1002/mbo3.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth B. N., Löfdahl S., Betley M. J., O’Reilly M., Schlievert P. M., Bergdoll M. S., Novick R. P. (1983). The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712. 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- Kumru O. S., Bunikis I., Sorokina I., Bergström S., Zückert W. R. (2011). Specificity and role of the Borrelia burgdorferi CtpA protease in outer membrane protein processing. J Bacteriol 193, 5759–5765. 10.1128/JB.05622-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad S. P., Yang G., Scott D. A., Wang G., Nair P., Mathison J., Reddy V. S., Li E. (2007). Chlamydial CT441 is a PDZ domain-containing tail-specific protease that interferes with the NF-κB pathway of immune response. J Bacteriol 189, 6619–6625. 10.1128/JB.00429-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco R., Varcamonti M., Ricca E., Sacco M. (1996). A new Bacillus subtilis gene with homology to Escherichia coli prc. Gene 183, 149–152. 10.1016/S0378-1119(96)00543-4 [DOI] [PubMed] [Google Scholar]
- Miller H. K., Carroll R. K., Burda W. N., Krute C. N., Davenport J. E., Shaw L. N. (2012). The extracytoplasmic function sigma factor σS protects against both intracellular and extracytoplasmic stresses in Staphylococcus aureus. J Bacteriol 194, 4342–4354. 10.1128/JB.00484-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noppa L., Ostberg Y., Lavrinovicha M., Bergström S. (2001). P13, an integral membrane protein of Borrelia burgdorferi, is C-terminally processed and contains surface-exposed domains. Infect Immun 69, 3323–3334. 10.1128/IAI.69.5.3323-3334.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostberg Y., Carroll J. A., Pinne M., Krum J. G., Rosa P., Bergström S. (2004). Pleiotropic effects of inactivating a carboxyl-terminal protease, CtpA, in Borrelia burgdorferi. J Bacteriol 186, 2074–2084. 10.1128/JB.186.7.2074-2084.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Losick R., Rudner D. Z. (2003). A second PDZ-containing serine protease contributes to activation of the sporulation transcription factor σK in Bacillus subtilis. J Bacteriol 185, 6051–6056. 10.1128/JB.185.20.6051-6056.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N. D., Barrett A. J., Bateman A. (2010). MEROPS: the peptidase database. Nucleic Acids Res 38 (Database issue), D227–D233. 10.1093/nar/gkp971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera F. E., Miller H. K., Kolar S. L., Stevens S. M., Jr, Shaw L. N. (2012). The impact of CodY on virulence determinant production in community-associated methicillin-resistant Staphylococcus aureus. Proteomics 12, 263–268. 10.1002/pmic.201100298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J., Darwin A. J. (2013). The Pseudomonas aeruginosa periplasmic protease CtpA can affect systems that impact its ability to mount both acute and chronic infections. Infect Immun 81, 4561–4570. 10.1128/IAI.01035-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane A., Sabbaj A., McMurry L. M., Levy S. B. (1992). Multiple antibiotic susceptibility associated with inactivation of the prc gene. J Bacteriol 174, 7844–7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L. N., Lindholm C., Prajsnar T. K., Miller H. K., Brown M. C., Golonka E., Stewart G. C., Tarkowski A., Potempa J. (2008). Identification and characterization of σS, a novel component of the Staphylococcus aureus stress and virulence responses. PLoS ONE 3, e3844. 10.1371/journal.pone.0003844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakov S. V., Anbudurai P. R., Stanbekova G. E., Gadzhiev A., Lind L. K., Pakrasi H. B. (1994). Molecular cloning and characterization of the ctpA gene encoding a carboxyl-terminal processing protease. Analysis of a spontaneous photosystem II-deficient mutant strain of the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem 269, 19354–19359. [PubMed] [Google Scholar]
- Silber K. R., Keiler K. C., Sauer R. T. (1992). Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc Natl Acad Sci U S A 89, 295–299. 10.1073/pnas.89.1.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. A., Yasbin R. E., Young F. E. (1984). New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene 29, 21–26. 10.1016/0378-1119(84)90161-6 [DOI] [PubMed] [Google Scholar]
- Wang C. Y., Wang S. W., Huang W. C., Kim K. S., Chang N. S., Wang Y. H., Wu M. H., Teng C. H. (2012). Prc contributes to Escherichia coli evasion of classical complement-mediated serum killing. Infect Immun 80, 3399–3409. 10.1128/IAI.00321-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A., Ibarra J. A., Paoletti J., Carroll R. K., Shaw L. N. (2014). The δ subunit of RNA polymerase guides promoter selectivity and virulence in Staphylococcus aureus. Infect Immun 82, 1424–1435. 10.1128/IAI.01508-14 [DOI] [PMC free article] [PubMed] [Google Scholar]