Abstract
A major goal of synthetic biology is to develop a deeper understanding of biological design principles from the bottom up, by building circuits and studying their behavior in cells. Investigators initially sought to design circuits “from scratch” that functioned as independently as possible from the underlying cellular system. More recently, researchers have begun to develop a new generation of synthetic circuits that integrate more closely with endogenous cellular processes. These approaches are providing fundamental insights into the regulatory architecture, dynamics, and evolution of genetic circuits and enabling new levels of control across diverse biological systems.
Cells use genetic circuits of interacting genes and proteins to implement diverse functions including growth and division, signaling, and differentiation. Most of our knowledge of these circuits comes from top-down approaches based on genetic or pharmacological perturbations of model systems. Despite the increasingly comprehensive interaction maps these approaches are producing, it remains challenging to answer fundamental questions about gene circuit design, such as why one circuit architecture may have been selected over another or how a given circuit will respond to changes in its inputs (1–3). In addition, it remains difficult to engineer circuits for use in biotechnological or biomedical applications (4).
Synthetic biology offers an alternative bottom-up approach to understanding biological circuits, based on designing and constructing simple synthetic gene circuits from well-characterized genes and proteins and then analyzing their behavior in living cells (1–5). It thus reflects a shift in genetic engineering from the level of an individual gene to the level of a gene circuit. Over the last decade, synthetic approaches have provided key insights into gene circuit design principles (1–3, 6, 7).
This field began with the goal of creating autonomous genetic circuits that could function as independently as possible from endogenous cellular circuitry or even functionally replace endogenous circuits. For example, early work demonstrated a working bistable switch, as well as self-sustaining oscillations (8, 9). The view was that underlying cellular processes could be used to support the synthetic circuits, for example, by providing gene expression machinery, but that the two layers could function independently.
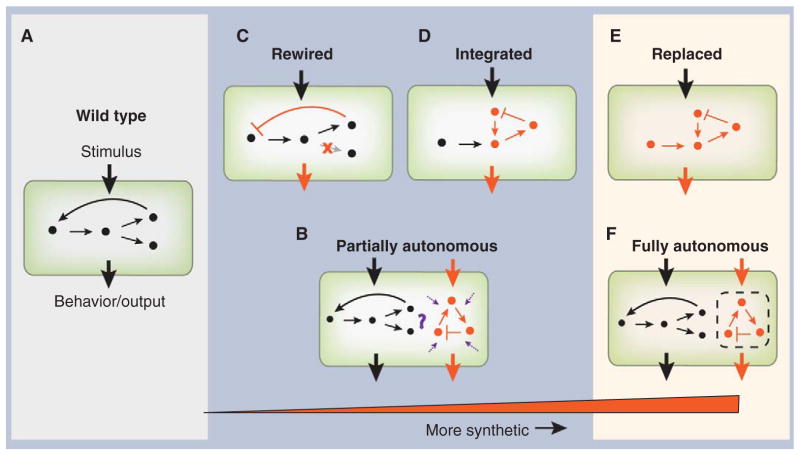
Recently, a new generation of synthetic biology experiments has moved toward tighter integration between endogenous and synthetic circuitry (Fig. 1). This has been driven both by difficulties in building autonomous synthetic gene circuits—”from scratch”—that behave predictably and by the need to engineer synthetic systems that control central biological processes in the host organism. Here, we discuss results that show how fundamental biological understanding can be obtained at the interface between the natural and the synthetic.
Fig. 1.
A continuum of synthetic biology. Wild-type cells (A) can be subject to two basic types of synthetic manipulation. (B) Autonomous synthetic circuits, consisting of ectopic components, may be introduced into the cell. Such circuits process inputs and implement functions (orange arrows) separate from the endogenous circuitry (black). However, unknown interactions with the host cell may affect their function (purple arrows). (C) An alternative is to rewire (orange lines) the endogenous circuits themselves to have new connectivity. (D) Extending this line of synthetic manipulation, synthetic circuits could be integrated into appropriately rewired endogenous circuitry to act as sensors and to implement additional functionality. Ultimate goals of this program are to be able to design and construct (E) synthetic circuits that can functionally replace endogenous circuits or (F) fully autonomous circuits that operate independently of the cellular mileu.
Effects of Cellular Milieu on Synthetic Gene Circuits
Does a synthetic circuit need to operate independently of its host to function reliably? Hasty and co-workers recently constructed a simple transcriptional oscillator that exhibited regular self-sustained oscillations in Escherichia coli. Their design, based on previous theoretical work (10), consisted of just two genes: an activator and a repressor. Expression of either gene could be enhanced by the activator protein but blocked by the repressor protein, as both were transcription factors. Small molecule inducers could be used to modulate the strength of these two transcription factors, enabling “tuning” of circuit parameters. In individual cell lineages, the oscillations were precise, with sister cells remaining in phase for multiple periods. They were also robust, as they occurred across a broad range of inducer concentrations (11).
In fact, the circuit performed almost too well. The model predicted oscillations in a much more limited range of parameters than observed experimentally. Careful analysis showed that this apparent discrepancy arose from two unexpected sources: First, time delays inherent in the process of gene expression, although much shorter than the overall period of the oscillator and hence initially ignored, were nevertheless critical for its robust operation. The authors confirmed the importance of these delays by demonstrating that even a one-gene synthetic oscillator based on auto-repression could generate oscillations—albeit not as strong, precise, or tunable as those in the two-gene circuit (11) [see also (12, 13)].
Second, there was an unintended, but critical, interaction with host cell components: Both the activator and repressor contained identical destabilization sequences that targeted them for proteolysis. High levels of either protein saturated the proteolytic machinery and effectively stabilized both, causing an indirect posttranslational coupling between the activator and repressor. This coupling, a consequence of unintended interactions with the host, helped to reduce phase drifts between the proteins and improved the precision of the oscillator (14).
Although such unanticipated interactions are often assumed to be disruptive, it is clear that they may also play more supportive roles in the functioning of synthetic circuits. More generally, this result provokes the questions of how such interactions can be identified and exploited to improve synthetic circuit performance (15).
Rewiring Endogenous Gene Circuits
Many important genetic circuits are either incompletely understood or tightly integrated into larger genetic systems that control complex processes. Although replacing or reconstructing such systems synthetically may be impractical, one can in many cases modify (“rewire”) parts of these circuits, providing insights into the design principles of the natural circuit architecture.
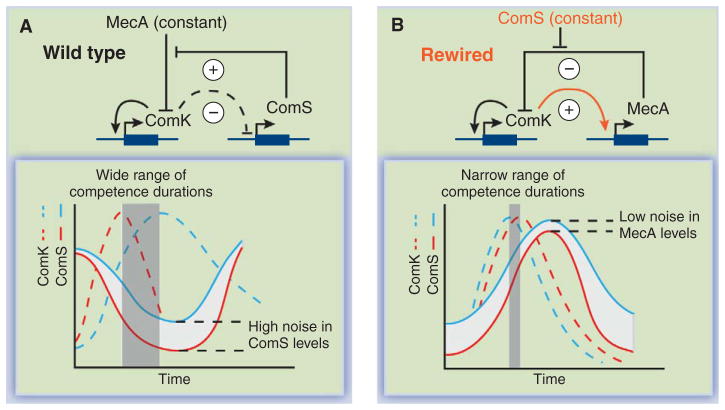
For example, Çağatay et al. recently rewired the Bacillus subtilis gene circuit that allows individual cells to sporadically and transiently differentiate into a genetically competent state, where they can take up DNA from the environment (16). A core feedback module enables the system to work in an excitable fashion, where fluctuations (noise) stochastically trigger episodes of competence (17). The system revolves around the master transcription factor ComK, which is sufficient to initiate and maintain competence but which eventually brings about its own destruction and exit from competence through a negative-feedback loop. In this loop, ComK indirectly represses expression of its stabilizing partner, ComS (Fig. 2A). Exit from competence occurs when ComS decreases to low levels, where it is more susceptible to stochastic fluctuations, which explains the broad distribution of competence durations observed in a wild-type population.
Fig. 2.
Rewiring an endogenous gene circuit. (A) (Top) Part of the natural competence circuit from B. subtilis. The MecA protease adaptor (assumed to be constant) degrades ComK; ComS inhibits this degradation and thus is an indirect activator. ComK indirectly represses ComS. (Bottom) Exit from competence depends on returning to low ComS levels. Noise in ComS (white region between red and blue curves, representing the extremes of the distribution of ComS profiles in a population) is significant at such low levels. The resulting distribution in ComK curves (red dashed and blue dashed)—and thus competence durations (vertical gray bar)—is wide. (B) (Top) The rewired competence circuit: Here the activation and repression loops have been switched. Competence exit occurs when MecA levels reach a high threshold. (Bottom) The resulting distribution of competence curves is narrow because variability in MecA is relatively low at high MecA concentrations.
But what if the negative-feedback loop were structured the opposite way—if ComK activated its own inhibitor, MecA (Fig. 2B), rather than inhibiting expression of its activator, ComS (Fig. 2A)? In that case, competence exit would occur at high MecA concentrations and therefore would be less sensitive to noise (Fig. 2B). To test this prediction, the negative-feedback loop was rewired to the alternative feedback architecture. As predicted, the rewired cells exhibited much greater precision in competence durations, while functioning normally in other ways (16). Why have cells evolved the inherently more variable design? In this case, variability is functional: At low external DNA concentrations, it allows some cells to stay competent long enough to take up DNA, while ensuring that other cells do not stay in the slow-growing competent state longer than necessary when DNA concentrations are high (16, 18).
Such rewiring can also provide insight into higher organisms, where circuit diagrams are complex and incomplete. For example, Lahav and co-workers recently rewired regulatory circuitry surrounding the mammalian tumor suppressor p53, which plays a central role in cancer and cell cycle regulation. In response to damaging radiation, the endogenous circuit displays sustained oscillations. With the rewired circuit, the authors showed how different features of the circuit, especially its feedbacks, tune the amplitude, frequency, and damping of p53 responses (19).
Rewiring Signal Transduction
A set of core signaling pathways allows cells to send, receive, and process information from the environment and other cells. Signaling pathways have undergone considerable diversification during evolution. Several synthetic experiments have exploited the evolutionary plasticity of signaling pathways to gain basic insights into their structure and mode of diversification and to elucidate their signal-processing capabilities.
Modifying signaling specificity
Mitogen-activated protein kinase (MAPK) pathways integrate information from a wide range of growth factors and other pathways and activate specific classes of targets. These input-output connections have changed over evolution. How is specificity encoded in these proteins, and can we learn to reprogram it?
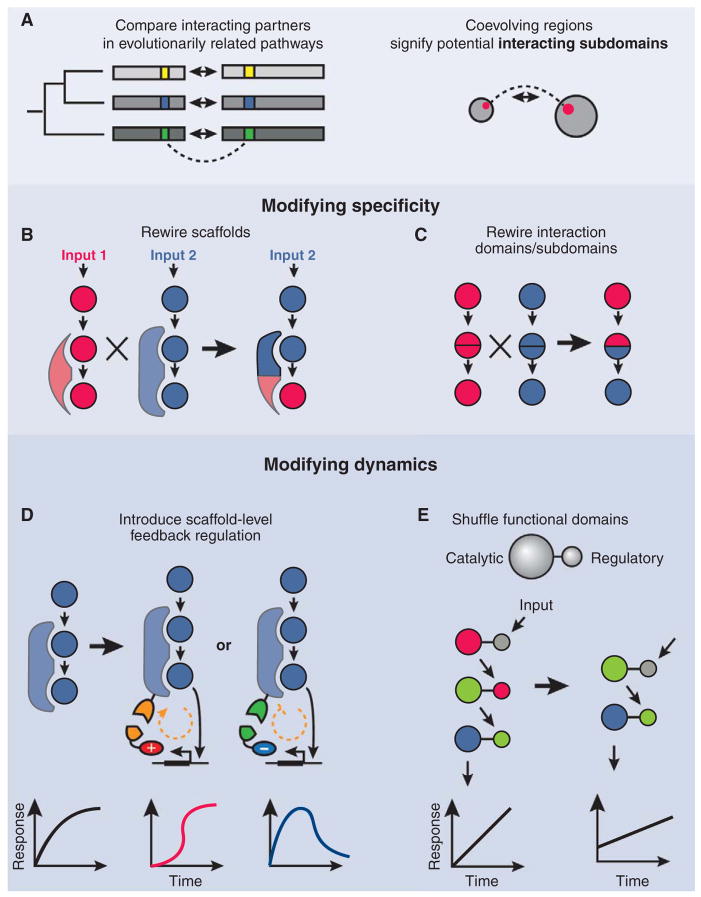
Pioneering work from Lim and co-workers demonstrated that specificity could be rewired by modifying the scaffold proteins (illustrated in Fig. 3B) that bring together multiple MAPK components (20). Even with scaffolds, however, MAP kinases still require correct molecular recognition for specific phosphorylation of substrates. To understand and reprogram these specificities, Mody et al. analyzed MAPKs from four different families, including orthologs of the yeast MAPKs Hog1 (high osmolarity) and Fus3 (mating), across diverse eukaryotic species (21). They identified distinct patches on the protein surface that had residues that were variable across families, but conserved within the same family, and that may determine interaction specificity. They reasoned, for example, that a residue in Hog1 that is conserved in its orthologs is likely to be important for the functioning of the pathway but can only be responsible for the interaction specificity of Hog1 if it differs in other families, such as Fus3.
Fig. 3.
Diversifying signaling pathways through rewiring. (A) Sequence correlations among proteins from a large family (left, SCA, also see text) can be used to identify interacting subdomains (right). Interaction specificity can be altered by rewiring scaffolds (B) or by shuffling specificity-determining domains or subdomains (C). The dynamic behavior of a pathway can be modified by (D) introducing new autoregulatory connections or by (E) altering regulation directly at the protein level by generating new combinations of catalytic and regulatory protein domains.
They synthesized a library of 64 Fus3-Hog1 hybrid proteins by dividing the primary sequence of each protein into six putative specificity-determining segments, with each synthetic protein incorporating one of the two variants for each segment. The library was highly enriched for functional proteins capable of rerouting signaling specificity (21) (Fig. 3C). For example, some variants activated the same pathway in response to either input; others activated both pathways in response to one input.
Similar questions occur in prokaryotes, which rely heavily on two-component systems for signaling. In the canonical two-component system, a sensor histidine kinase (HK) phosphorylates a corresponding response regulator (RR). Tens or even hundreds of such systems can occur in a single genome, and interactions are highly specific, with one HK almost always signaling to one RR (22). It had long remained unclear whether and how it would be possible to rationally and systematically reengineer their specificity.
To address these issues, Skerker et al. used statistical coupling analysis (SCA). SCA quantitatively examines evolutionary correlations between amino acid positions, by assuming that pairs of amino acids that functionally interact with each other are more likely to covary during evolution (23). Applying SCA to an alignment of many HK-RR sequence pairs and taking advantage of existing structural data, they identified potential specificity-determining residues in covarying patches on the interacting protein surfaces (Fig. 3A). By systematically mutating these residues in one HK to the corresponding amino acids in another HK, they created a new highly specific HK-RR pair (23). Evidently, these proteins have evolved an economical structure, where specificity determinants are concentrated into relatively compact regions of the proteins, facilitating functional diversification through minimal sequence evolution. More generally, evolutionary information is proving to be a powerful tool both for addressing fundamental questions in structural biology, such as the mechanisms of allostery (24), and for engineering new protein components for synthetic biology.
Programming signaling dynamics
Signaling pathways are characterized not just by their molecular interactions but also by their response dynamics. Recently, Peisajovich et al. showed that new signaling responses could be efficiently generated by systematically shuffling regulatory and catalytic domains within the yeast mating pathway (25). For example, in a 66-protein library containing domains from 11 yeast mating-pathway proteins, they observed several variants that exhibited qualitative changes in the mating pathway response dynamics (Fig. 3E). Remarkably, 6 out of the top 10 variant strains created by domain recombination mated more efficiently than wild type (25).
Feedback loops strongly modulate response dynamics in several pathways. In order to understand the role of feedback, Bashor et al. (26) rewired the yeast MAPK pathway by genetically modifying the scaffold protein to recruit positive or negative modulators. By expressing these modulators under the control of the pathway itself, they were able to create a variety of feedback structures, whose strength could be controlled with competing “decoy” proteins. The scaffold thus became a versatile synthetic “signal hub” that integrated regulatory information from multiple sources. A slew of nonnative dynamic responses, from adaptation to ultrasensitivity, could be generated by modulating the strength, timing, and sign of these synthetic feedbacks (Fig. 3D) (26).
Deciphering signal encoding
Feedback, crosstalk, and the induction of dramatic cellular changes like differentiation make signaling difficult to study in natural contexts. To circumvent this problem, researchers have begun to transplant signaling pathways from one organism to another and to divert the outputs of signaling pathways away from their native targets to reporter genes that permit quantitative readouts.
For instance, MAPK pathways display diverse behaviors, ranging from graded responses (25) to ultrasensitivity (27), in different contexts. To better understand this physiological plasticity, O’Shaughnessy et al. ported into yeast the core mammalian cascade, comprising the three kinases Raf, MAPK kinase, and ERK (extracellular signal–regulated kinase, a type of MAPK) (28). They also modified the upstream kinase Raf to make it directly controllable by β-estradiol. This system showed that ultrasensitivity was an inherent feature of the cascade and that the sharpness and amplitude of the ultrasensitive response could be independently controlled simply by varying the relative concentrations of kinase components. Thus, the core MAPK cascade acts as a tunable amplifier, whose behavior can be modulated by the cell to generate diverse responses (28).
In the Notch pathway, which is normally under elaborate regulation, diverting signaling has provided qualitative insights that would have been difficult or impossible to obtain in native systems. The membrane-bound Notch receptor and its ligand Delta together enable direct communication between neighboring cells in developmental patterning processes (29). Previous work showed that Delta inhibits Notch in its own cell, but activates Notch on neighboring cells (30). Activation involves the release of the cleaved Notch intracellular domain, which translocates to the nucleus to activate target genes (31).
To understand how Notch output depends on ligand levels in its own cell and neighboring cells, Sprinzak et al. sought to reconstruct the signaling pathway from the bottom up (32). They incorporated Notch-Gal4 hybrid receptors, which activate engineered nonnative target genes in response to signals (31), and analyzed their activation in individual cells. These studies revealed that, because of inhibitory interactions between Notch and Delta in the same cell, the pathway acts like a “walkie-talkie,” allowing cells to send or receive signals but not both at the same time. This property could facilitate many Notch-dependent developmental patterning processes, by helping to enforce sharp distinctions between neighboring cells (33).
Toward Functional Replacement
Could complicated endogenous circuits eventually be replaced by controllable synthetic counterparts that have altered functionality (Fig. 1E)? Coudreuse and Nurse took a step in this direction by replacing much of the eukaryotic cell cycle control network with a single gene (34).
Although much is known about regulation of cell cycle progression, the specific roles of many components, as well as aspects of the overall logic of the system, remain unclear. Working in fission yeast, Coudreuse and Nurse began by systematically deleting many of the regulatory components associated with the cell cycle, including the single cyclin-dependent kinase (CDK) and all the cyclins known to interact with it. In this background, they expressed a cyclin-CDK fusion protein, under the control of the endogenous cyclin promoter. This minimal replacement was sufficient to drive the cell cycle, from G1 through S, G2 and M, with no observable differences from the wild type under laboratory conditions.
To understand how the cell executes different cell cycle phases with a single cyclin-CDK fusion, the authors used an engineered inhibitor of the CDK. They found that different inhibitor concentrations were required for impairment, depending on the stage of the cell cycle. For instance, more inhibitor was required to delay the G2/M phase progression than the G1/M. This suggested that different checkpoints in the cell cycle were navigated simply by varying the concentration of active CDK. Moreover, the cycle could be reset and controlled arbitrarily by inhibiting the cyclin-CDK fusion at different points and to different extents. The cell cycle is one of the foundations of life; successfully replacing it with a synthetic module represents a fundamental advance in synthetic biology.
Integrated Synthetic Circuits
Sensing cell state
Several new “plug-and-play” synthetic devices can interface with cells as sensors (35, 36) to monitor dynamic changes in cell state. Burrill and Silver recently created a synthetic memory circuit that can remain on for several generations after an activating event, and they interfaced it with the natural DNA damage response in yeast (37). Using this system, they learned that DNA damage response was heterogeneous in the population and led to heritable pleiotropic effects in progeny. Cleverly designed sensors like these may prove to be useful in studies of cell differentiation and decision-making, where cells are thought to progress through a continuum of poorly understood cellular states.
Controlling multicellular development and genetic inheritance
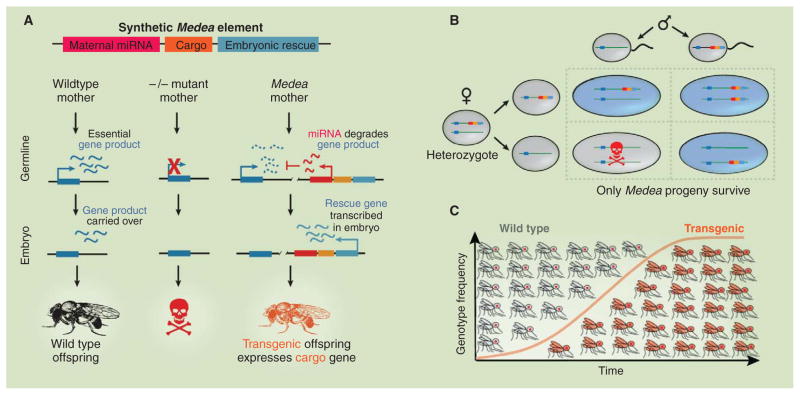
Recent work in Drosophila has shown that synthetic circuits can fundamentally alter the development and life cycle of a multicellular organism in a controlled way. Chen et al. created a synthetic selfish genetic element, named Medea, capable of spreading through a population (38). The synthetic Medea element (Fig. 4A) maternally expresses a microRNA (miRNA) that blocks expression of an essential protein normally produced by the mother and deposited in the egg. The element also expresses an “antidote” to this toxic miRNA, which consists of a second copy of the gene (with different codons) expressed by the embryo rather than the mother. Replacing the maternally expressed gene with its zygotically expressed Medea-based counterpart maintained normal development in offspring. Medea-positive mothers always express the toxic maternal miRNA. Thus, progeny of such mothers only survive if they inherit Medea from either or both parents—a dramatically non-Mendelian inheritance pattern.
Fig. 4.
An integrated synthetic circuit controls development and population dynamics. (A) In Drosophila, the synthetic Medea element (top) maternally expresses an miRNA (red) that silences a maternally expressed gene whose product is essential for embryogenesis (left column). Eggs from female flies mutant for this gene do not hatch (middle column). The Medea element also contains a rescue gene that is expressed only in the early embryo. The Medea element may also accommodate a cargo gene that is expressed in Medea progeny (right column). (B) Progeny of female Medea-positive flies will only survive if they receive the Medea element from either parent. (C) This super-Mendelian inheritance pattern can efficiently drive Medea into populations.
A key consequence is that Medea is capable of invading populations. When Medea-positive flies are introduced into a wild-type laboratory population, the Medea element rapidly takes over the whole population (38). A similar synthetic system in mosquitos could in principle be engineered to carry a “cargo” gene that would diminish the ability of malarial parasites to survive in the mosquito or to be transmitted to human hosts (Fig. 4C).
A striking aspect of the Medea system is that it works across multiple levels: At the circuit level, it rewires expression of a critical gene to alter the timing and genetic source of expression (Fig. 4A). At the developmental level, this leads to a selective killing of embryos that lack the Medea element (Fig. 4B). Finally, at the population level, this gives Medea transgenic organisms the ability to efficiently spread through a population (Fig. 4C). Although many challenges remain, this system and others [see (39, 40)] demonstrate the power of integrating synthetic biology approaches into the circuitry of a complex organism.
Conclusion: Exploring the Biology That Could Be
Synthetic biology opens up the possibility of creating circuits that would not survive in the natural world and studying their behaviors in living cells, expanding our notion of biology (41). The last decade has shown how even our first steps toward building and analyzing synthetic circuits can identify fundamental biological design principles and can produce useful new understanding. Future progress will require work across a range of synthetic levels (Fig. 1), from rewiring to building autonomous and integrated circuits de novo. Going forward, we anticipate that synthetic biology will become one of the primary tools we use to understand, control, imagine, and create biological systems.
Acknowledgments
The authors thank D. Sprinzak, J. Locke, J. Levine, P. Neveu, J. Young, and other members of the Elowitz lab for helpful discussions and critical reading of the manuscript. This work was supported by NIH grants 5R01GM086793, 5R01GM079771, and P50GM068763; NSF Career Award 0644463; and a Packard Fellowship.
References and Notes
- 1.Lim WA. Nat Rev Mol Cell Biol. 2010;11:393. doi: 10.1038/nrm2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mukherji S, van Oudenaarden A. Nat Rev Genet. 2009;10:859. doi: 10.1038/nrg2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sprinzak D, Elowitz MB. Nature. 2005;438:443. doi: 10.1038/nature04335. [DOI] [PubMed] [Google Scholar]
- 4.Lu TK, Khalil AS, Collins JJ. Nat Biotechnol. 2009;27:1139. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endy D. Nature. 2005;438:449. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 6.Xavier JB. Mol Syst Biol. 2011;7:483. doi: 10.1038/msb.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayer TS. Curr Biol. 2010;20:R772. doi: 10.1016/j.cub.2010.06.049. [DOI] [PubMed] [Google Scholar]
- 8.Elowitz MB, Leibler S. Nature. 2000;403:335. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 9.Gardner TS, Cantor CR, Collins JJ. Nature. 2000;403:339. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 10.Hasty J, Dolnik M, Rottschäfer V, Collins JJ. Phys Rev Lett. 2002;88:148101. doi: 10.1103/PhysRevLett.88.148101. [DOI] [PubMed] [Google Scholar]
- 11.Stricker J, et al. Nature. 2008;456:516. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swinburne IA, Miguez DG, Landgraf D, Silver PA. Genes Dev. 2008;22:2342. doi: 10.1101/gad.1696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. Nature. 2009;457:309. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]
- 14.Cookson NA, Tsimring LS, Hasty J. FEBS Lett. 2009;583:3931. doi: 10.1016/j.febslet.2009.10.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purnick PEM, Weiss R. Nat Rev Mol Cell Biol. 2009;10:410. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 16.Çağatay T, Turcotte M, Elowitz MB, Garcia-Ojalvo J, Süel GM. Cell. 2009;139:512. doi: 10.1016/j.cell.2009.07.046. [DOI] [PubMed] [Google Scholar]
- 17.Süel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. Nature. 2006;440:545. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 18.Eldar A, Elowitz MB. Nature. 2010;467:167. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toettcher JE, Mock C, Batchelor E, Loewer A, Lahav G. Proc Natl Acad Sci USA. 2010;107:17047. doi: 10.1073/pnas.1005615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SH, Zarrinpar A, Lim WA. Science. 2003;299:1061. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 21.Mody A, Weiner J, Ramanathan S. Nat Cell Biol. 2009;11:484. doi: 10.1038/ncb1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laub MT, Goulian M. Annu Rev Genet. 2007;41:121. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 23.Skerker JM, et al. Cell. 2008;133:1043. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, et al. Science. 2008;322:438. doi: 10.1126/science.1159052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peisajovich SG, Garbarino JE, Wei P, Lim WA. Science. 2010;328:368. doi: 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bashor CJ, Helman NC, Yan S, Lim WA. Science. 2008;319:1539. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 27.Huang CY, Ferrell JE., Jr Proc Natl Acad Sci USA. 1996;93:10078. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Shaughnessy EC, Palani S, Collins JJ, Sarkar CA. Cell. 2011;144:119. doi: 10.1016/j.cell.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinmaster G, Kopan R. Development. 2006;133:3277. doi: 10.1242/dev.02515. [DOI] [PubMed] [Google Scholar]
- 30.del Álamo D, Rouault H, Schweisguth F. Curr Biol. 2011;21:R40. doi: 10.1016/j.cub.2010.10.034. [DOI] [PubMed] [Google Scholar]
- 31.Struhl G, Adachi A. Cell. 1998;93:649. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 32.Sprinzak D, et al. Nature. 2010;465:86. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sprinzak D, Lakhanpal A, Lebon L, Garcia-Ojalvo J, Elowitz MB. PLOS Comput Biol. 2011;7:e1002069. doi: 10.1371/journal.pcbi.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coudreuse D, Nurse P. Nature. 2010;468:1074. doi: 10.1038/nature09543. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi H, et al. Proc Natl Acad Sci USA. 2004;101:8414. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fung E, et al. Nature. 2005;435:118. doi: 10.1038/nature03508. [DOI] [PubMed] [Google Scholar]
- 37.Burrill DR, Silver PA. Genes Dev. 2011;25:434. doi: 10.1101/gad.1994911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen CH, et al. Science. 2007;316:597. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- 39.Ruder WC, Lu T, Collins JJ. Science. 2011;333:1248. doi: 10.1126/science.1206843. [DOI] [PubMed] [Google Scholar]
- 40.Windbichler N, et al. Nature. 2011;473:212. doi: 10.1038/nature09937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elowitz M, Lim WA. Nature. 2010;468:889. doi: 10.1038/468889a. [DOI] [PMC free article] [PubMed] [Google Scholar]