Abstract
Structural specialisations enable von Willebrand factor (VWF) to assemble during biosynthesis into helical tubules in Weibel-Palade bodies (WPB). Specialisations include a pH-regulated dimeric bouquet formed by the C-terminal half of VWF and helical assembly guided by the N-terminal half that templates inter-dimer disulphide bridges. Orderly assembly and storage of ultra-long concatamers in helical tubules, without crosslinking of neighboring tubules, enables unfurling during secretion without entanglement. Length regulation occurs post-secretion, by hydrodynamic force-regulated unfolding of the VWF A2 domain, and its cleavage by the plasma protease ADAMTS13 (a disintegrin and metalloprotease with a thrombospondin type 1 motif, member 13). VWF is longest at its site of secretion, where its haemostatic function is most important. Moreover, elongational hydrodynamic forces on VWF are strongest just where needed, when bound to the vessel wall, or in elongational flow in the circulation at sites of vessel rupture or vasoconstriction in haemostasis. Elongational forces regulate haemostasis by activating binding of the A1 domain to platelet GPIbα, and over longer time periods, regulate VWF length by unfolding of the A2 domain for cleavage by ADAMTS13. Recent structures of A2 and single molecule measurements of A2 unfolding and cleavage by ADAMTS13 illuminate the mechanisms of VWF length regulation. Single molecule studies on the A1-GPIb receptor-ligand bond demonstrate a specialised flex-bond that enhances resistance to the strong hydrodynamic forces experienced at sites of haemorrhage.
Introduction
Von Willebrand factor (VWF) is central in haemostasis and thrombosis in the rapid flow of the arteriolar circulation [1–4]. Different domains within VWF bind clotting factor VIII, collagen, platelet glycoprotein Ib (GPIb), and integrins αIIbβ3 and αVβ3 (Fig. 1A). VWF multimers are organised as long linear concatamers, in which each monomer disulphide bonds tail-to-tail at its C-terminus, and head-to-head at its N-terminus, with adjacent monomers (Fig. 1). “Multimer” denotes both branched and unbranched polymers; because the biology and physics of VWF are intertwined with the linear nature of its polymer, I prefer the more specific term “concatamer” for a linear polymer. The enormous length of VWF concatamers brings them into a range where the hydrodynamic forces acting on them in the circulation are highly significant, like on blood cells in the circulation, whereas such forces on other plasma proteins and cell surface receptors are negligible. These forces activate both the haemostatic function and thrombotic pathology of VWF. I review recent advances in our understanding of the biosynthesis, activation by elongational flow, mechanoenzymatic length regulation, and biophysics of the A1 and A2 domains of VWF. Mutations in von Willebrand disease (VWD) both contribute to and are illuminated by understanding of VWF biology.
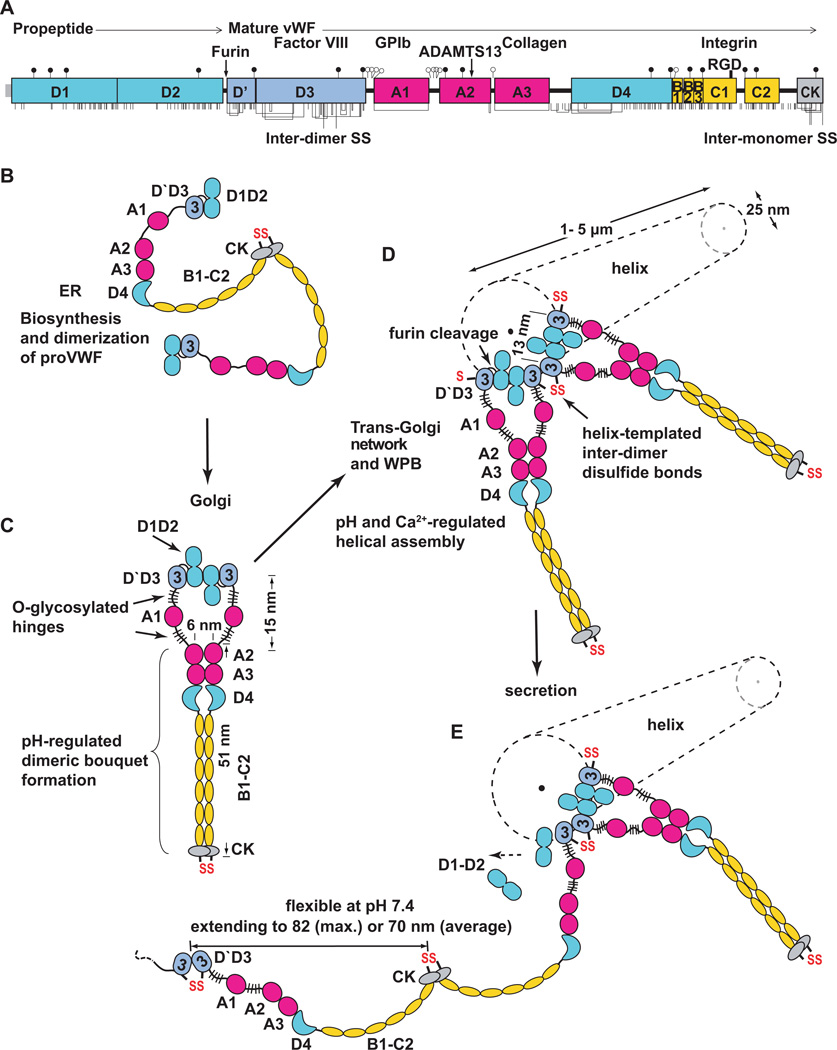
Figure 1.
Biosynthesis, helical assembly, and secretion of VWF concatamers. A. Primary structure and domain organisation of VWF [66]. Cysteines are vertical lines and are connected for chemically determined disulphide bonds [67, 68]. N and O-linked glycans are closed and open lollipops, respectively [9]. B–E. Scheme for biosynthesis, helical assembly, and secretion. EM evidence for C-terminal pH-regulated dimeric bouquet and domain shapes is from Y.-F. Zhou, E., Eng, N. Nishida, C. Lu, T. Walz, and T.A. Springer, unpublished. EM evidence for N-terminal D1–D2 homodimerisation and the helical assembly is from [8].
Biosynthesis
Multiple biological specialisations are required for the biosynthesis of the largest known soluble vertebrate protein [2, 5–7]. VWF glycoprotein is biosynthesised as a preproprotein; the signal sequence is removed during translocation into the endoplasmic reticulum (ER) (Fig. 1A). N-linked glycans are added in the ER, most disulphide bonds are formed, and proVWF is dimerised through formation of inter-monomer disulphide bonds in the C-terminal cystine-knot (CK) domain (Fig. 1B). ProVWF dimers are then transported from the ER (pH ~ 7.4) to the Golgi (pH ~ 6.2). N-linked glycans are processed, and O-linked glycans are added.
A previously undescribed dimeric bouquet forms at pH 6.2 in the C-terminal portion of VWF (Y.-F. Zhou, E. Eng, N. Nishida, C. Lu, T. Walz, and T.A. Springer, unpublished) (Fig. 1C). The term dimeric bouquet describes the appearance in EM of the A2, A3, and D4 domains as six flowers in a raceme, and C-terminal domains as a stem. pH-dependent homotypic association involves specific pairing between the A2, A3, D4 and six smaller domains between D4 and CK (Fig. 1C). To our knowledge, the pH-dependent ordering of such a large protein segment, encompassing two segments in each monomer from residue 1490 at the N-terminus of the A2 domain to residue 2722 at the beginning of the CK domain, is unprecedented.
D1–D2 monomers and D`D3 dimers assemble in vitro in Ca2+ and pH 6.2 into helices that have the same dimensions and three-dimensional shape as VWF tubules in Weibel-Palade bodies (WPB) in vivo [7, 8] (Fig. 1D and 2). Neither dimeric bouquets nor VWF helices form at pH ~7.4 characteristic of the ER or plasma, but do so at pH ~6.2 characteristic of the trans-Golgi. Each contributes to overcoming the unique challenges of biosynthesising an ultralong disulphide-linked polymer, coiling it into a compact form for storage, and enabling orderly unfurling during secretion.
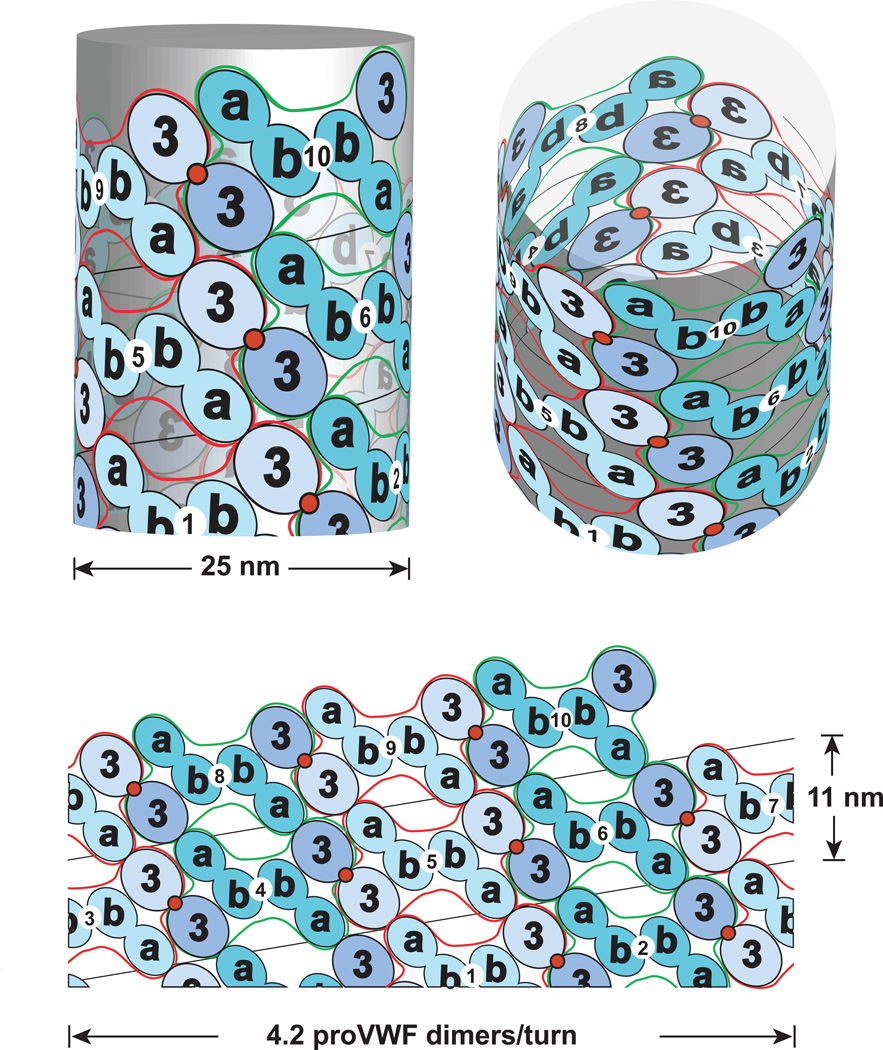
Figure 2.
Helical assembly of the N-terminal domains of VWF in the tubules of WPB. Modified from [8]. Helical assembly is shown as progressing from bottom to top. Each successive proVWF dimer is numbered and shown alternately as lighter domains outlined in red or darker outlined in green. The density for the D’D3 domains (3) is assigned based on larger size than the D1 and D2 domains (a and b). Inter-dimer disulphide crosslinks form at the 2-fold symmetry axis between D’D3 domains (red circles). The D1 and D2 domains are denoted a and b since it is not known which is in which position in the helical assembly. Noncovalent, homomeric interactions within each dimer are mediated by either the D1 or D2 domain, in the b position [8].
Knowledge of the structure of the VWF C-terminal dimeric bouquet and N-terminal helical assembly at pH 6.2, together with electron micrographs and tomograms of endothelial cells [5–7], allows the steps in VWF concatamer biogenesis to be reconstructed (Fig. 1B–E). D1D2 monomers dimerise at pH 6.2 in Ca2+ [8]. Thus, proVWF dimeric bouquets are hypothesised to form in the Golgi that are zipped at the N-terminus by D1D2 homodimerisation, and zipped C-terminally from A2 to the CK domain (Fig. 1C). Assembly of proVWF dimeric bouquets into helical tubules nucleates in the trans-Golgi network [5]. Further assembly occurs in nascent WPB (Fig. 1D), with growing ends of multiple tubules adjacent to one another at the end of the WPB that connects to the trans-Golgi network [5]. VWF tubules are aligned with one another in nascent WPB as in mature WPB, but the spacing between them is larger, ~80 nm centre-to-centre [5].
C-terminal bouquets are important for the assembly of proVWF dimers onto the ends of helical tubules for several reasons. Zipping from the C-terminus up to the A2 domains ensures that the D3 domains in a dimer are no more than 30 nm away from one another (Fig. 1C). Dimerisation has second order dependence on monomer concentration, and this increase in proximity of D1–D2 monomers within a proVWF dimer will thus significantly contribute to D1–D2 association within a dimer (Fig. 1C) rather than between different dimers. Furthermore, without this zipping up, a proVWF dimer could easily extend (Fig. 1E) more than the 80 nm distance between two nascent tubules, enabling subsequent disulphide cross-linking between tubules, termination of disulphide linkage within a tubule, and even N-terminal disulphide crosslinking within a proVWF dimer.
Moreover, the C-terminal dimeric bouquet helps position the D`D3 domains for helix assembly (Fig. 1C, D). Within the dimeric bouquet, the centre-to-centre distance between A2 domains is 6 nm (Fig. 1C), whereas that between the two D3 domains in the same dimer should be 13 nm in the assembling helical VWF tubule (Fig. 1D). The two A1 domains and their linkers easily enable the transition between the 13 nm D`D3 spacing in the helix and the 6 nm spacing between the A2 domains in the dimeric bouquet (Fig. 1C).
The disulphide bonds that link two VWF dimers toward the N-terminal end in the D3 domain are templated by assembly onto the ends of growing helical tubules in nascent WPB. Deposition of neighbouring proVWF dimers at growing tubule ends places one D`D3 domain of the most recently assembled dimer adjacent to one D`D3 domain of the previously assembled proVWF dimer (Fig. 1D and 2). Helical assembly templates inter-dimer disulphide formation and also enables cleavage by furin between the D2 and D`D3 domains (Fig. 1D). Covalent linkage between VWF dimers is thus co-linear with assembly into helical tubules. Whether disulphide isomerases and furin promote VWF maturation by binding to the ends of growing tubules, or the sides of assembled helices, remains to be determined.
The organisation of VWF dimers in WPB tubules has been deduced [8] (Fig. 2). The only ambiguity is whether the D1 or D2 domains mediate homodimerisation; these are labelled a and b in Fig. 2 where the b domains homodimerise. In contrast to DNA, with two molecules per helix, the VWF helices are one-start; i.e. there is one VWF concatamer per helix [8]. However, like DNA there is 2-fold symmetry, so that the helical structure remains identical when flipped end-for-end. In VWF this is a consequence of the two-fold dyad axis about the D1, D2, D`D3 dimer in the helix (Fig. 2).
The A1 domain and its N and C linkers, the most heavily O-glycosylated segments in VWF [9 16878] (Fig. 1), may act as hinges between the N-terminal helical assembly and the C-terminal dimeric bouquet. The A1 domain is not ordered in dimeric bouquets, and does not contribute strong density to in vitro assembled tubules [8]. Thus, the ~ 51 nm long dimeric bouquets need not obey helical 2-fold symmetry and extend radially from the 25 nm diameter tubules, but could instead extend more axially and avoid interdigitation during biogenesis when the centre-to-centre tubule spacing is 80 nm [5]. Compaction of the dimeric bouquet region of VWF must become extreme in mature WPB, where the 25 nm tubules pack with centre-to-centre spacings of 28.4 ± 3 nm [7]. No doubt this is assisted by the reduction in pH in mature WPB to 5.45 ± 0.26, which approaches the isoelectric point of VWF protein [10].
Secretion of VWF
Orderly coiling of VWF in tubules in WPB is also important for rapid unfurling of VWF during secretion. Experimental elevation of pH within WPB, which results in their rounding, inhibits subsequent release of VWF fibrils. Thus, entanglement is thought to inhibit VWF secretion [11]. The first step in secretion is formation of a narrow constriction between the plasma membrane and the WPB, which then experiences a rapid rise in pH. Alkalinisation rapidly propogates from one end of a WPB to the other [10]. Secretion is from one end of a WPB, as shown by EM [7].
Early after fusion with the plasma membrane, there is a great expansion in width of WPB and increase in average spacing in the tubules [7]. VWF is by far the predominant protein constituent of WPB [12]. This suggests that upon alkalinisation, dissociation of the dimeric bouquet, and charge repulsion between domains above their pI’s, causes the observed increase in spacing between WPB tubules [7]. Maintenance of the helical patterns in Fourier transforms of cryoelectron tomograms after expansion (and presumed pH increase to ~ 7.4) demonstrates that the inter-dimer disulphide crosslinks stabilise VWF helical structure, and suggests that the D1–D2 propiece remains associated [7].
These findings further suggest that secretion begins by unfurling of VWF from the ends of tubules, where packing interactions are the weakest. Thus, both dissociation of the D1–D2 propiece, and unfurling of VWF concatamers are likely to proceed in parallel from the ends of the VWF tubules closest to the secretion pore (Fig. 1E).
Secretion might proceed in a spinneret-like fashion, in which multiple VWF concatamers emerge from the secretion pore in parallel, each unfurling from a separate tubule. VWF molecules can self-associate [4], and spinneret-like secretion might be responsible for the long VWF fibrils observed to extend from stimulated endothelial cells in the flow direction [13].
The conformation of VWF at physiologic pH
VWF applied to EM grids at plasma pH can appear either compact or extended [14, 15], apparently depending on whether it adsorbed in stasis or in shear during droplet spreading. Atomic force microscopy (AFM) has demonstrated VWF extension in shear flow [16].
Recent imaging of VWF N- and C-terminal dimeric fragments at both acidic and neutral pH (Y.-F. Zhou, E. Eng, N. Nishida, C. Lu, T. Walz, and T.A. Springer, unpublished) confirms and extends classic EM studies [14, 15]. Each A domain is a ~ 4.5 nm globule, D domains are larger, more irregular ~ 6–7 nm globules, and the D4 domain has a bulbous crescent shape (Fig. 1B–E). The six smaller domains between the D4 and CK domains are only well resolved in dimeric bouquets at acidic pH (Fig. 1C), but are likely to be present at neutral pH as well (Fig. 1E). These small domains correspond to the B and C repeats (Fig. 1A) and the thin flexible rod-like segment previously described in EM [14].
The overall domain organisation is of variably sized beads, mainly spaced close together on a flexible string, but with some thin interdomain segments, including the mucin-like O-glycosylated segments on either side of the A1 domain (Fig. 1A and E). Using the largest or average spacing observed between domains results in length estimates of 82 or 70 nm per monomer, respectively (Fig. 1E); compared to the previous estimate of 60 nm [14]. The high degree of flexibility of VWF at plasma pH of ~ 7.4 permits highly dynamic, rapid change in concatamer shape in flow.
The length of VWF
Many individual tubules in 3D-tomograms extend the full length of 1 µm long WPB [7]. Since WPB range from 1 to 5 µm in length, a single helical concatameric molecule may extend up to 5 µm. One helical turn of 4.2 dimers/11 nm [8] (Fig. 2) or 4.3 dimers/12 nm [7] yields 2.7 nm/VWF dimer, and an estimate of 3,700 monomers/5 µm tubule.
Measurements on VWF in plasma yield shorter estimates. SDS-agarose suggests larger concatamers have 14–20 dimers [14]. Estimates based on the longest molecules seen in EM suggest > 30 dimers per concatamer [14]. Fluorescent VWF molecules in shear flow extend up to 15 µm [17]. Together with the extended length of 60 to 80 nm/monomer in EM these measurements yield estimates of 60 to 250 monomers per concatamer, much less than the estimate of 3,700 monomers per helical tubule in WPB.
With the 50-fold expansion in length between stored VWF and the extended form seen in EM [8], a 5 µm long WPB tubule would yield a 250 µm long extended VWF molecule. This is consistent with 100–1000 µm strings of VWF seen after secretion from endothelial cells in vitro in shear flow [13]; however, such strings may include many VWF concatamers associated in parallel. Strings are released into shear flow after cleavage by ADAMTS13. A bolus of ultralarge VWF that appears in plasma after systemic stimulation of VWF secretion is cleaved within 2 h to the shorter, pre-existing, homeostatic size distribution [18]. I speculate that VWF freshly secreted at a site of vascular challenge may be an “ultra-ultralarge” form, and remain bound to endothelium or the extracellular matrix until cleavage by ADAMTS13 and release into the circulation as ultralarge VWF, followed by further trimming in plasma over 2 h.
Shear and elongational flow and the shape of VWF
Once released into the circulation, the overall shape of VWF is dynamic and dependent on the type of flow encountered [15–17, 19–21]. In shear flow, the rate of fluid flow increases from the wall toward the centre (Fig. 3). Flow is laminar, and the velocity of each lamina increases from 0 at the wall to maximal at the centre (Fig. 3A). Conversely, the shear rate is the change in velocity with change of distance between lamina, and is highest at the wall.
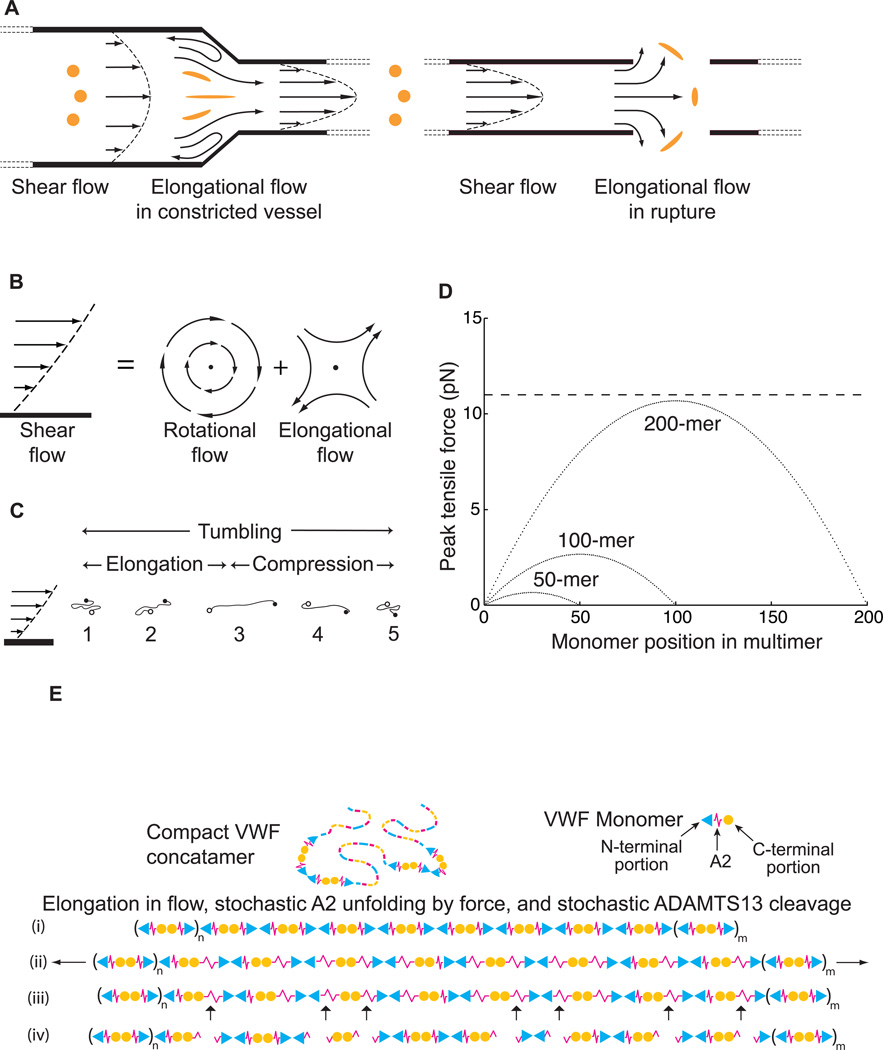
Figure 3.
Shear and elongational flow, VWF concatamer conformation, and mechanoenzymatic cleavage. Modified from [20]. A. Shear flow, and transition to elongational flow in vessels at sites of constricted or ruptured vessels. Round orange spheres show the effect of enlongational flow on the shape of a polymeric protein in the flow field (adapted from [21]). Elongation of VWF concatamers would occur in the indicated directions. B. Shear flow, which may be represented as elongational flow superimposed on rotational flow [22]. Arrows show stream lines and dots regions of no flow. C. Cartoon of VWF elongating, compressing, and tumbling in shear flow. D. Peak force as function of monomer position in a VWF multimer chain of 200, 100 or 50 monomers at 100 dyn/cm2. Dashed line shows the most likely unfolding force for the A2 domain at a loading rate of 25 pN/s. E. Schematic of VWF, with N-terminal end as triangle, A2 as spring, and C-terminal end as circle. Elongation results in stochastic unfolding of some A2 domains (ii), some of which are cleaved by ADAMTS13 (iii). The resulting fragments are shown (iv).
Shear flow can be conceptualised as the superposition of rotational flow and elongational flow [22] (Fig. 3B). Rotational flow causes particles to tumble (Fig. 3C). VWF multimers have an overall compact, bird’s nest shape in stasis [14, 15, 17, 19]. However, above a critical shear stress of 50 dyn/cm2, attractive forces between the monomers within each concatamer are overcome by hydrodynamic drag, and VWF free in flow elongates and tumbles [16, 17, 19, 23] (Fig. 3C). Simulations of shear flow and the weak attractive forces within a VWF concatamer suggest dynamic changes in each tumbling cycle [19] (Fig. 3C). A compact VWF molecule with its ends in lamina with differing velocities will elongate as one end moves faster than the other (Fig. 3C, 1–3). However, as VWF tumbles, the faster and slower moving ends will change lamina, so that the faster moving end will become the slower moving end and vice versa (Fig. 3C, 4). The hydrodynamic forces will then cause compaction of VWF, which will be aided by the shear-independent tendency of the domains within VWF to self-associate (Fig. 3C, 4–5). Because of this alternation between compaction and elongation, and the limited time available for VWF extension in each cycle of extension, extension of VWF is limited in shear flow.
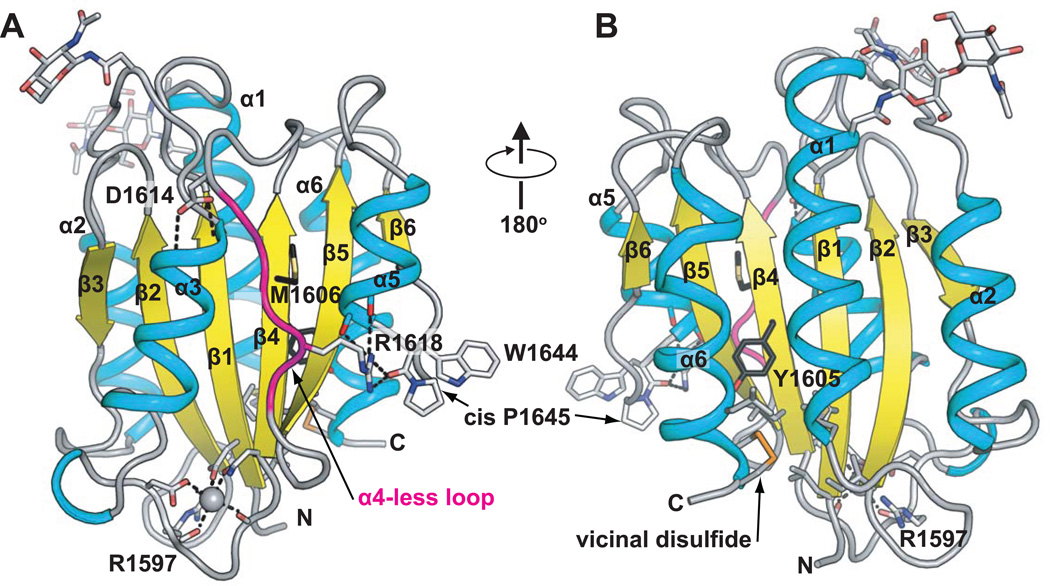
Figure 4.
The VWF A2 domain. Ribbon diagrams show α-helices (cyan), β-strands (yellow) and loops (grey). The α4-less loop is in magenta. Key sidechains are shown as sticks, and Ca2+ as a sphere. Carbons of the ADAMTS13 cleavage site residues Tyr1605 and Met1606 are black. The vicinal disulphide is in gold. Two Asn-linked carbohydrates are in stick near the top of the domain. N and C-termini are marked. Hydrogen and metal coordination bonds are dashed. The chimeric model was made by superimposing two A2 structures [30, 31], and grafting the Ca2+-bound α3-β loop [31] onto the mammalian wild type structure [30].
Figure 5.
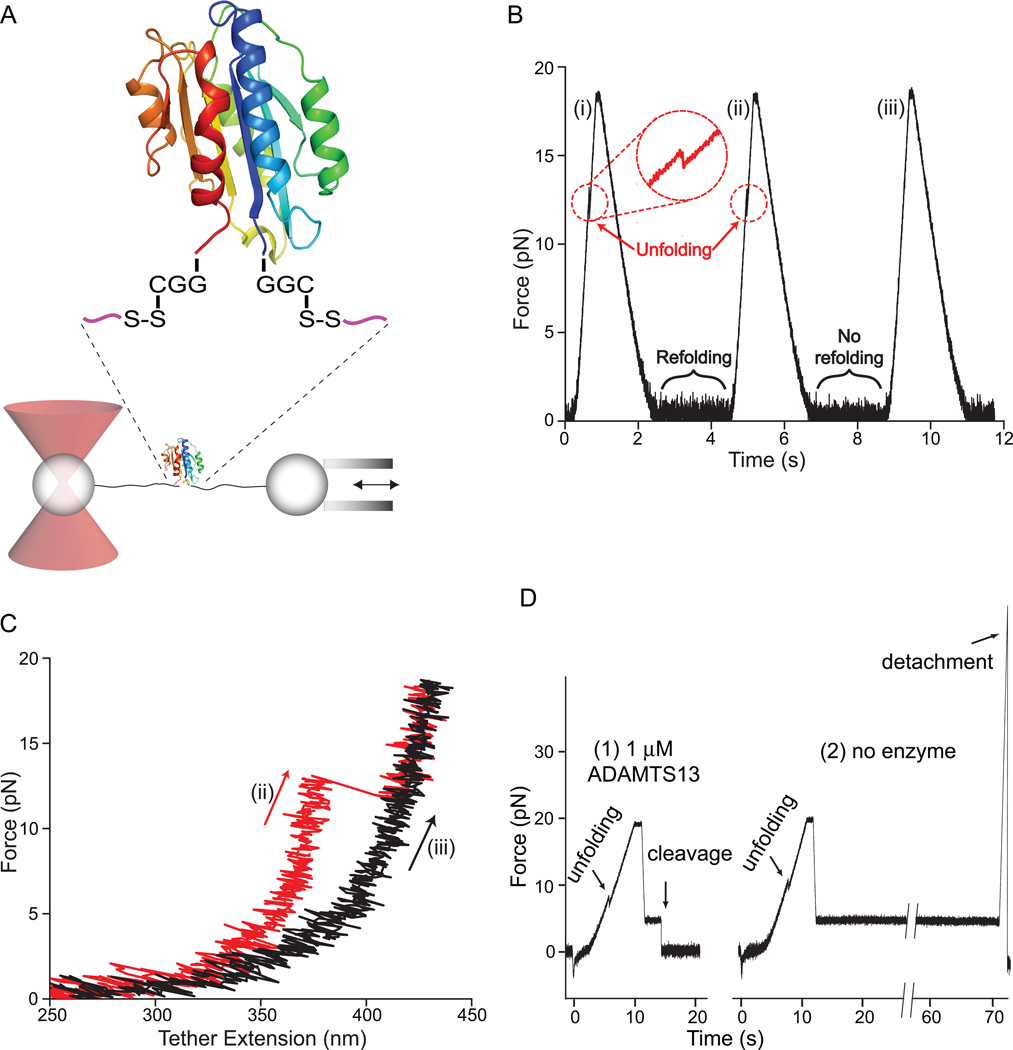
Single molecule demonstration of A2 domain unfolding, refolding, and mechanoenzymatic cleavage. Modified from [47]. A. Schematic diagram of the DNA handle-A2 domain construct (upper) and laser trap (lower). The A2 domain cartoon is in rainbow, blue to red from N-terminus to C-terminus, respectively. The A2 domain is coupled through Cys-Gly-Gly and Gly-Gly-Cys linkages and disulphides to double-stranded DNA handles, that have biotin and digoxygenin tags at the opposite ends. The tether is held by beads functionalised with streptavidin and digoxigenin Fab. Force is applied by micropipette movement (right), and measured by bead displacement in the laser trap (left). B. Three representative cycles of force increase, decrease, and clamping at a constant low level. C. Traces of force versus tether extension in the force increase phases of cycles ii and iii in B. An abrupt unfolding event is seen in ii and not iii. It is inferred that A2 was unfolded at the beginning of iii. D. Representative traces showing ADAMTS13 cleavage (1) and no cleavage (2). In each trace unfolding of A2 is seen, and A2 is returned to a clamped force of 5 pN to assess kinetics of cleavage. Cleavage of the tether returns force to 0. A new tether must be formed for each test of cleavage; cleavage is measured one A2 molecule at a time.
In elongational flow, an important concept in polymer physics recently introduced into the VWF field [20], concatamer extension will increase and be prolonged compared to shear flow (Fig.3A). Immediately after arterioles are cut, flow will increase, increasing both shear and elongational flow; moreover, the elongational flow component selectively increases at the site of rupture (Fig. 3A). Subsequent vasoconstriction will decrease flow and hence decrease the shear rate. However, elongational flow will increase precisely at the site of vasoconstriction, of utmost strategic importance in haemostasis (Fig. 3A).
The peak tensile force on a VWF concatamer at a site of vasoconstriction or stenosis can increase ~ 100-fold in elongational flow [21]. Moreover, the average extension of VWF increases with a much sharper threshold, and at about 100-fold lower levels of pure elongational flow, compared to shear flow [21]. In agreement, shear gradients, which correspond to sites where the elongational component of shear flow increases [21] trigger platelet-dependent thrombus formation [24]. These results are of profound importance for understanding haemostasis, thrombosis, and ADAMTS13-dependent shortening of VWF and bleeding diathesis associated with arterial stenosis [25].
Remarkably, VWF will be most extended in just those environments where its adhesive haemostatic functions are needed most. Shear stress is greatest at the vessel wall, and when stuck to endothelium at sites of secretion or to collagen where the extracellular matrix is exposed in injury, VWF cannot tumble. Thus the hydrodynamic forces on vessel-bound VWF are not only greater than when free in flow, but are purely elongational. Furthermore, platelets have a greater hydrodynamic surface area than VWF, so when they bind to vessel wall-bound VWF, elongational force on VWF increases further. This provides a positive feedback mechanism for amplifying haemostasis.
Thus, the physics of long VWF concatamers in flow, and the propensity of extended/elongated VWF to bind to ligands including collagen and platelet GPIb [17, 26] reveal an inherent mechanism by which the transition from shear to elongational flow at sites of haemorrhage can trigger haemostasis [20, 21]. Furthermore, VWF is longest precisely where it is most needed, where it is secreted locally by endothelial cells in vascular injury.
Relation between VWF extension and adhesiveness
There is a strong correlation between the effects of hydrodynamic flow on VWF elongation and VWF-dependent function including platelet aggregation; however, the discussion above must be tempered, because the relationship between VWF extension and haemostasis is a hypothesis rather than an established fact. Moreover, even how extension is mechanistically related to VWF adhesiveness is unclear. Early observations established a threshold hydrodynamic shear above which VWF aggregates platelets in stirred suspensions. Subsequently, when VWF was adsorbed to the wall of a flow chamber, platelets were found to adhere through GPIb, and to roll with a jerky motion on VWF, but only above a threshold shear. Furthermore, VWF itself in the fluid phase shows shear-dependent deposition on VWF substrates [4]. All of these observations suggest a correlation between VWF extension and adhesive function. This was taken a step further by demonstration that the shear thresholds for VWF extension and deposition on collagen substrates were superimposable [17]. However, VWF deposition on collagen can also be found in stasis [27].
The binding site for GPIb in the A1 domain appears to be cryptic in VWF in stasis, and exposed by flow. Moreover, haemostatic potency of VWF strongly correlates with its length. The D`D3 domain can shield recognition by the neighbouring A1 domain [28]. VWF length may potentiate binding to GPIb on platelets both by enhancing shear or elongational flow-dependent extension of VWF, and by increasing multivalent binding, i.e. avidity. Multivalent binding sites would be much better exposed in the extended than bird’s nest conformation of VWF. It is tempting to propose that the same mechanism that shields the A1 domain is also responsible for the weak attractive forces within a VWF multimer that favour the compact over the extended concatamer conformation.
The A domains
VWF A domains (VWA) are the defining members of the VWA protein fold and family [29]. They have a central hydrophobic β-sheet with 6 β-strands, surrounded typically by 6 amphipathic α-helices (Fig. 4). VWA domains are also found in integrins as ligand-binding αI and βI domains, in complement components, and in intracellular proteins with diverse functions. Some VWA domains contain a metal-ion dependent adhesion site, but those in VWF do not. VWA domains in integrins and complement undergo substantial conformational changes that regulate affinity for ligand. Conformational change in VWF A domains has not yet been seen, but circumstantial evidence suggests this might yet be found for the A1 domain. VWF A1 and integrins differ substantially in location of ligand binding sites on the surfaces of VWA domains. Most modules in extracellular proteins are αll-β, and have their N- and C-terminal ends at opposite ends of the domain. In contrast, VWA domains are α/β, and their N- and C-termini are adjacent to one another (Fig. 4), enabling greater flexibility when arranged as beads on a string as in VWF.
A2, the shearbolt domain of VWF
Unique among VWF domains, A2 lacks a long-range disulphide bond (Fig. 1A) and, therefore, is unprotected from unfolding by tensile force applied along the length of VWF concatamers. A shearbolt has a groove cut in it, so that when excessive force is applied, it breaks, and prevents damage to more valuable parts of a machine. The A2 domain has a similar, but much more elaborate function, by which the size distribution of VWF concatamers is regulated after secretion.
Two 1.9 Å crystal structures of A2 have revealed surprising evolutionary adaptations to its function as a shear bolt domain, and how mutations destabilise A2 in VWD [30, 31]. The structures are in excellent agreement overall, but each also reveals unique features. The wild-type mammalian A2 structure reveals two N-glycosylation sites and the vicinal disulphide bond [30]; the mutant E. coli A2 structure reveals a Ca2+-binding loop [31]. These physiologically relevant features are combined in the chimeric structure shown in Fig. 4. Structural specialisations of A2 are discussed in parallel with results from unfolding of single A2 domains in elongational force [20].
The structure of A2 predicts that unfolding will begin at its C-terminus. Elongational force in VWF multimers would be applied equally to the A2 N-terminus at the beginning of the β1-strand and the A2 C-terminus at the end of the α7-helix (Fig. 4). However, the β1-strand is in the middle of the fold and, thus, highly force resistant. In contrast, the α7-helix is on the periphery, and could easily be peeled away from the remainder of the domain to initiate folding.
The ADAMTS13 cleavage site at residues Tyr1605 and Met1606 could hardly be in a less accessible site in the folded A2 domain (Fig. 4). It is in the centre of the β4-strand, the central β-strand in the β-sheet, which lies in the centre of the fold, sandwiched between amphipathic α-helices and loops on either side (Fig. 4).
The A2 domain is distinguished from A1 and A3, and all other structurally characterised VWA domains [29], by its lack of an α4-helix (Fig. 4A). In place of the α4-helix is an idiosyncratic “α4-less loop”. The lesser number of residues in the α4-less loop than in α4-helices in other VWA domains, and the lack of α-helix backbone hydrogen bonds, suggest a weakening of the fold here similar in character to the break-off groove of shear bolts.
The regions around the cleavage site, and between it and the domain C-terminus, are poorly packed [30]. Multiple side chain conformations are clearly visible for cleavage site residue Met1606 and Leu1603. Disordered sidechains are not seen in similar buried positions in A1 and A3 domain crystal structures. Poor packing of A2’s hydrophobic core may contribute to unfolding.
Cys1669 and Cys1670 are linked in a vicinal disulphide bond, at the very C-terminus of the A2 domain (Fig. 4B). The hydrophobic disulphide is buried in a large interface with hydrophobic residues (Fig. 4B) suggesting an important role in stabilising the hydrophobic core. Deletion of Cys1669-Cys1670 markedly enhances cleavage of VWF multimers by ADAMTS13 [32].
Disulphide bonds between vicinal cysteines are rare, and when present, often are functionally important [33]. The unusual 8-membered ring formed by the mainchain and sidechains of vicinal disulphide-bonded cysteines imparts rigidity. The rigid vicinal disulphide may have a champagne cork-like function that enables resistance to force up to a point, beyond which popping of the cork enables substantial unfolding.
A cis-peptide bond between Trp1644 and Pro1645 is present in A2 (Fig. 4), whereas A1 and A3 have no cis-prolines. Intracellular cis-trans proline isomerisation acts as an on-off switch in some signalling proteins [34]. Proline isomerisation occurs with a rate on the order of 0.01 s−1 and is rate-limiting for protein folding [35]. Therefore, after unfolding of the A2 domain in the vasculature, conversion to trans-Pro could slowly occur. Conversion to trans-Pro would serve as a marker for A2 domains in VWF multimers that had been unfolded for unusually long periods of time, or been exposed to unusually high elongational forces, which can accelerate cis to trans peptide conversion [36].
The rate of subsequent conversion to cis-Pro would then limit the rate of A2 refolding. Consistent with this idea, in single-molecule experiments, A2 would sometimes cease to refold, then resume refolding [30].
A Ca2+-binding loop unique to A2 is present in the protruding α3-β4 loop near the N- and C-termini (Fig. 4), and contributes to stabilising A2 against ADAMTS13 cleavage [31].
Single-molecule studies showed that a subset of about 20% of unfolding events included a discernable pause at a partially unfolded intermediate state, 40% of the distance between the fully folded and unfolded states [30]. This would correspond to unfolding from the C-terminus up to the α3-β4 loop.
Strikingly, all A2 structural specialisations occur in this C-terminal portion, and this region is bounded by the β4-strand and the α6-helix, which contain the key recognition sites for ADAMTS13 defined with peptide substrates [37]. The crystal structure of an ADAMTS13 multi-module fragment demonstrates linearly distributed exosites that recognise distinct substrate regions [38]. It will be interesting to determine whether elongation of the substrate by force enhances recognition of such sites, which are present not only in the unfolded A2 domain but also in the more C-terminal D4 domain [39, 40].
The kinetics of A2 refolding, as well as force-induced unfolding, will be important for regulating cleavage by ADAMTS13. The time constant for refolding of the A2 domain, 1.9 s [20], is much slower than seen for many other domains. Since the single molecule KM of ADAMTS13 of ~160 nM [20] is higher than its plasma concentration of 6 nM [41], the length of time that A2 takes to refold, after elongational force is removed, is an important determinant of the amount of ADAMTS13 trimming. The cis-Pro could regulate the kinetics of refolding. The Ca2+-binding α3-β4 loop could be an important site for nucleating refolding from either completely or partially unfolded A2. Compared to an α-helix, which can refold independently of other secondary structure elements, folding of the α4-less loop will be dependent on folding of nearby structural elements and may limit refolding rate. However, once in place, the α4-less loop may accelerate refolding, as its Asp-1614 N-caps the α3-helix, and its Arg-1618 both C-caps the α5-helix and stabilises the cis-peptide backbone (Fig. 4).
VWD Type 2A
Type 2A VWD is associated with low haemostatic activity of VWF and marked decrease in the length of VWF concatamers as shown in SDS-agarose gels. Type 2A VWD almost always results from mutations in the A2 domain [1]. Type 2A can be subdivided. Group I mutations impair secretion and may also affect multimer assembly. Group II mutations do not affect secretion or multimer length prior to secretion, but result in cleavage by ADAMTS13 to aberrantly short lengths [42–44]. The groups may be distinguished by in vitro pulse-chase biosynthesis experiments in transfected cells. Alternatively, patient and healthy control VWF may be compared by SDS-agarose electrophoresis from plasma (concatamer size distribution after secretion), and from platelets (concatamer size distribution in α-granules prior to secretion).
A2 crystal structures show that group I type 2A mutations tend to be more buried and structurally disruptive than group II mutations [30]. Group I mutations include buried sidechains contributing important hydrogen bonds, and buried hydrophobic residues substituted with polar residues or Pro. Group II mutations include those that introduce mild packing defects in the protein interior and loss of hydrogen bonds that stabilise loops, α-helices, or buried water molecules [30]. Interestingly, the most commonly reported VWD mutation, Arg1597 Trp, a group II mutation, locates to the α3-β4 loop (Fig. 4). Its carbonyl backbone oxygen coordinates the Ca2+ ion, and its sidechain stabilises the Ca2+-coordinating loop [31].
Single molecule studies
Both A2 domain unfolding and the A1/GPIbα interaction have been characterised in single molecule AFM and laser tweezer experiments [20, 45–48]. Here, we focus on laser tweezer results that enabled fitting to biophysical theories that predict how force affects the kinetics of domain unfolding, refolding, and receptor-ligand unbinding, demonstrating the robustness of the data. The laser tweezer experiments were on proteins that were coupled to stiff DNA handles (Fig. 5 and 6). One key advantage of the DNA handles is that they provide an unmistakable signature that single molecule events are being studied: a plateau in the force-extension curve at 67 pN where B DNA transitions to stretched S DNA. A second key advantage is repeated measurements on a single “tether”, i.e. single molecule. Multiple cycles of unfolding and refolding, or unbinding and rebinding, are measured with each single molecule, enabling repeated measurements on what is actually the same molecule.
Figure 6.
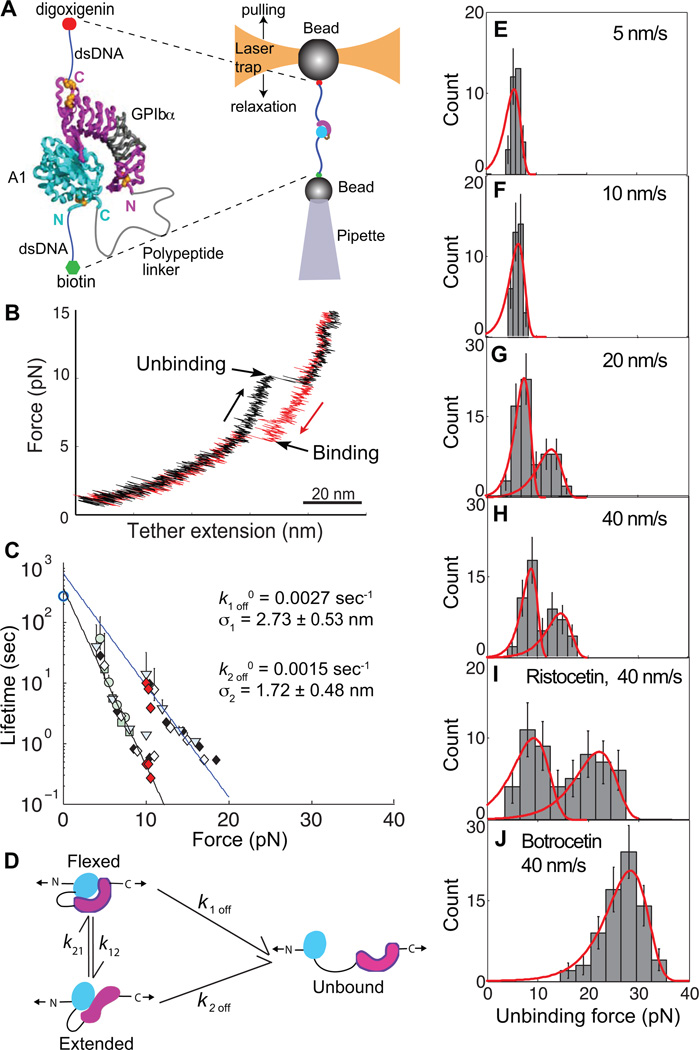
Repeated measurement of GPIbα and VWF A1 domain binding and unbinding in a single molecule. Modified from [47]. A. Schematic. The A1 domain and GPIbα complex crystal structure are shown as ribbon diagrams, with native disulphides in gold. The ReaLiSM fusion construct contains from N to C the A1 domain, a 43 or 26-residue polypeptide linker, and GPIbα, and is expressed as a secreted protein in mammalian cells. Cysteines are included at the N and C-termini for disulphide linkage to DNA handles, which are coupled to beads as in Fig. 5. The mini-laser tweezers differs from that in Fig. 5. Force is applied by movement of the laser trap, and measured as a change in light momentum. B. One representative cycle. Unbinding and rebinding are measured as abrupt changes in tether extension during pulling (black) and relaxation (red). C. Kinetic measurements. Data from each bin in E–H is a different symbol, and each symbol shape/fill is at a different pulling rate. Red diamonds are from hopping kinetics in force-clamp mode [47]. Open circle at 0 pN is bulk phase data from 125I-A1 domain dissociation from platelets [55]. D. Schematic model of a flex-bond. A1 (cyan) and GPIbα (magenta) are subjected to tensile force at the N and C termini of the ReaLiSM construct (arrows). k1off and k2off are dissociation rates from two different states. k21 and k12 are rates of equilibration between the two bound states. Measured dissociation kinetics reflect all four kinetic rates. E–H. Unbinding force distributions at four different indicated pulling rates. Dissociation at a higher force occurs at faster pulling rates. As predicted by theory and global fit of all data (red curves), the average force at which each pathway dissociates increases slightly with loading rate. I and J. Unbinding force distributions at the faster pulling rate in presence of ristocetin and botrocetin.
Unfolding, refolding, and mechanoenzymatic cleavage of single A2 domains
The A2 domain was covalently coupled through N- and C-terminal linkers to DNA handles (Fig. 5A). Tensile force was therefore transmitted to the A2 domain through a pathway that would be almost identical to that experienced by A2 in intact VWF, where hydrodynamic force acting on either end of the concatamer is transmitted as elongational force to A2 through linkers to the A1 and A3 domains (Fig. 1 and 4). A2 domains were subjected to cycles of force increase, force decrease, and clamping at a low force to enable refolding [20] (Fig. 5B). A2 domain unfolding was marked by abrupt tether extension events (Fig. 5B and C, cycles i and ii). The increase in length at different forces was fit to the worm-like chain model for an unfolded polymer chain, and yielded an A2 contour length of 57 nm ± 5 nm, in good agreement with length expected for unfolding of the 177-residue A2 domain.
The force at which A2 unfolded varied from one unfolding event to another. However, the most likely unfolding force increased with increase in the rate at which force was applied (loading rate). Plots of this relationship showed an excellent fit to theory, and were extrapolated to yield the unfolding rate in absence of force of 0.0007 s−1 [20]. Unfolding increases exponentially with force; with each 2.5 pN increment, the rate of A2 unfolding increases by 10-fold.
During the pause at a clamped force between each cycle of force decrease and increase, the A2 domain had the opportunity to refold (Fig. 5B). Subsequent unfolding revealed folding during the pause (Fig. 5B and C, cycle ii), while a lack of unfolding suggested an absence of refolding (Fig. 5B and C, cycle iii). Pausing for various durations at different clamped forces demonstrated fit to the expected exponential decrease of refolding rate with increase of force, and yielded an extrapolated rate of folding in absence of force of 0.54 s−1. Using ∆G = −RT ln (kfolding /kunfolding) the folding free energy of A2 was estimated as −3.9±0.9 kcal/mol, in good agreement with the ∆G of −3.5±0.5 kcal/mol estimated from urea-induced unfolding of an E. coli A2 fragment [49].
To test the hypothesis that A2 unfolding is required for cleavage by ADAMTS13, A2 was mechanically unfolded in the absence or presence of ADAMTS13, and relaxed to a clamped force of 5 pN (Fig. 5D). Cleavage by ADAMTS13 was detected as a drop in force on the tether to 0 pN. Spontaneous rupture at 5 pN, i.e. the background with no enzyme was rare. No cleavage of folded A2 at 5 pN with 100 nM or 1 µM enzyme was observed. Fits at three different enzyme concentrations to the single-molecule Michaelis-Menten equation yielded Kcat = 0.14±0.01 s−1, and KM = 0.16±0.04 µm. These are comparable to values from bulk phase experiments on cleavage of peptide substrates by ADAMTS13 [50, 51].
VWF length regulation in vivo and the second power dependence of elongational force on length
Unfolding of single A2 molecules showed at the rate of force loading on VWF concatamers in plasma a most likely unfolding force of 11 pN [20] (dashed line, Fig. 3D). Hydrodynamic force scales roughly with dimension. Longer VWF concatamers not only have longer dimension, but also cross proportionally more shear lamina when extended. Thus, tensile force on the centre of an extended VWF concatamer in shear or elongational flow scales with N2, where N is the number of monomers (Fig. 3D). This concept from polymer dynamics [52] has important implications for VWF [20]. Estimation of the force on VWF concatamers in vivo in shear flow (curves, Fig. 3D) showed that the force required for unfolding of single A2 domains in single molecule experiments could successfully predict the maximal length of VWF in plasma [20]. Furthermore, the second power dependence of force at the centre of a concatamer on number of monomers sharpens the distribution of maximum lengths. It also focuses cleavage toward the middle rather than ends of VWF multimers.
A flex-bond for adhesion in the vasculature
The interaction between GPIb on platelets and the A1 domain in VWF must resist substantial hydrodynamic force to fulfill its function in haemostasis in arterioles. Tests of the hypothesis that the A1-GPIbα bond is mechanochemically specialised for force resistance were enabled by a novel method for measuring binding and unbinding of a receptor and ligand in a single molecule (ReaLiSM). Crystal structures show that the VWF A1 domain interacts with the concave face of the leucine-rich repeat (LRR) domain of GPIbα [53, 54] (Fig. 6A and 7). The C-terminus of VWF A1 is close to the N-terminus of GPIbα, and neither terminus contributes to the complex interface. Therefore, these termini were connected with 43 or 26-residue polypeptide linkers (Fig. 6A).
Figure 7.
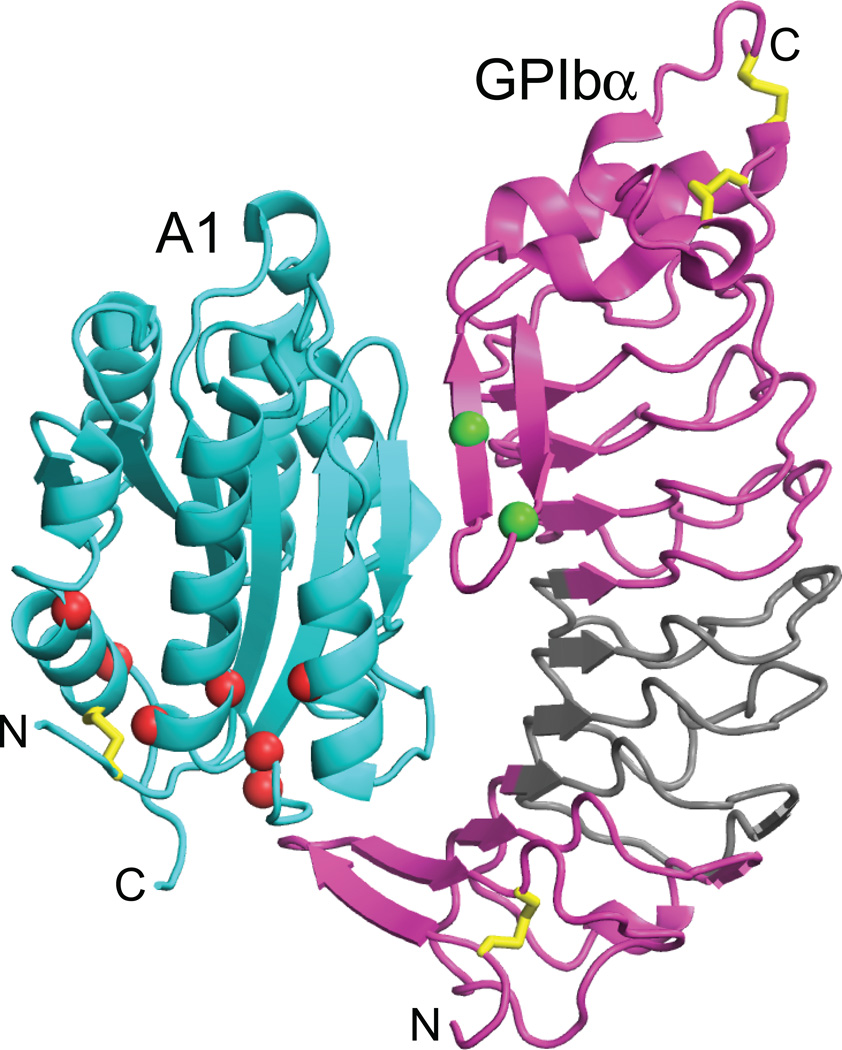
Crystal structure of the complex of GPIbα LRR domain and VWF A1, with mutations marked [53, 54]. Disulphides are shown with gold spheres. VWD type 2B mutations in A1 and platelet-type VWD mutations in GPIbα are shown with Cα atom spheres and ribbon in red and green, respectively. All mutations are gain-of-function, i.e. they increase adhesiveness. The A1 mutations map to regions that are buried and in the vicinity of the long-range disulphide bond, and are mostly not in contact with GPIbα. The GPIbα mutations are in its β-thumb, which changes conformation when bound to A1. LRR 2–4 of GPIbα are in grey. They are not in contact with A1 in the crystal structure, and are not important in binding to A1 in stasis, but become increasingly required for binding to A1 as shear is increased, as shown by exchange for canine sequence [64].
The A1-GPIbα ReaLiSM construct suspended between beads with DNA handles held in a laser trap (Fig. 6A) was subjected to cycles of increasing and decreasing force. In each cycle, an abrupt increase in the length of the tether between the two beads was observed during pulling, and an abrupt contraction was observed during relaxation (Fig. 6B). The abrupt extension events during unbinding, and contraction events during rebinding, were fit to the wormlike chain model. The experimental contour length closely corresponded to that expected for the linker between A1 and GPIbα domains, and provided strong support that single A1-GPIbα binding and unbinding events were measured [47].
The force at which A1 and GPIbα dissociated was determined at different pulling rates (Fig. 6E–H). At low pulling rates, bond rupture events had a narrow, unimodal distribution (Fig. 6E–F). At higher pulling rates, the distribution of dissociation forces was clearly bimodal (Fig. 6G–H). Thus, there are two pathways for dissociation of the GPIb-A1 bond, each of which excellently fit the predicted exponential decrease in bond lifetime with increase of force (Fig. 6C). Kinetics determined in force-clamp experiments, in which single receptor-ligand bonds hopped between the bound and unbound state hundreds of times, also showed two pathways for bond dissociation, and the same fit to theory (red diamonds in Fig. 6C). Importantly, the pathway at low force has an extrapolated k1off 0 in absence of force of 0.0027 s−1, in excellent agreement with previous bulk-phase measurements of 125I-A1 domain dissociation from platelets [55] (open circle at 0 pN, Fig. 6C).
In conclusion, the bond between A1-GPIbα has two states, and is therefore termed a “flex-bond” (Fig. 6D). The analogy is to muscles and to adhesion molecules such as selectins and integrins, that have distinct flexed, low-affinity and extended, high-affinity states [56–59]. Only one pathway was seen at lower force-loading rates, while at and above loading rates of 0.9 pN/s, or at constant force of ~10 pN, a second pathway was found, and the force-rupture distribution was bimodal. The rates of switching between the states, k12 and k21 (Fig. 6D), were also measured, and were increased by force. Thus, tensile force influences the lifetime of an A1-GPIbα bond not only by shifting the equilibrium between the two states, but also by lowering the barrier for conversion between the two states [47].
The effect of the flex-bond is to broaden the distribution of forces that A1-GPIbα can withstand. As force is applied, lifetime drops from 370 s in the absence of force, to 0.44 s at 10 pN in the first state (Fig. 6C). However, at 10 pN the second state kicks in, which has a lifetime of 9.2 s at 10 pN, thus increasing lifetime by 21-fold. The second state has the same lifetime at 17 pN as the first state at 10 pN, thus extending the working range of the receptor-ligand bond by an additional 7 pN (Fig. 6C). This broad force range is important, because the force experienced by the A1-GPIbα bond will vary widely in vivo, depending on the length of the VWF concatamer, whether the concatamer is bound to the vessel wall, the number and distribution of platelets bound to the VWF concatamer, and the shear and elongational forces at the site of bleeding.
In clinical diagnosis of VWD, ristocetin is used to mimic shear-enhanced activation of VWF [60]. Ristocetin is an antibiotic that adventitiously activates VWF; botrocetin is a snake venom protein that activates VWF by a different mechanism and does not mimic shear ([60, 61] and references therein). Ristocetin is thought to bind near a stretch of C-terminal A1 domain residues to which gain-of-function VWD mutations map ([60, 61] and references therein). Concordance between functional effects of antibodies and mutations suggest that ristocetin, and not botrocetin, induces a conformational state of the A1-GPIbα bond similar to its shear-induced configuration [60]. The force-rupture histogram with ristocetin remained bimodal, with a marked shift of the second peak from 14.8 to 22.4 pN at 40 nm/s (Fig. 6I). The second, extended state of the flex-bond induced by force corresponds to the shear-induced state, and the selective strengthening of this state by ristocetin agrees with these previous conclusions that ristocetin mimics the effect of shear on the GPIbα-A1 bond.
In the presence of botrocetin a qualitatively different, unimodal distribution was seen, and the force-rupture peak was shifted to an even higher level at 40 nm/s of 28.4 pN (Fig. 6J). A single state in botrocetin is consistent with lack of mimicry of shear. This state may correspond to the flexed state, strengthened by binding of botrocetin to both GPIbα and A1 [62].
The observation of two states of the GPIbα-A1 receptor-ligand bond is consistent with mutational data that suggest a second, ristocetin and force-stabilised state. The N-terminal and C-terminal LRR of GPIbα (magenta, Fig. 7), contact A1 in the crystal structure [53, 54]. Although the middle repeats 2–4 (grey, Fig. 7) make no contacts in the crystal structure and are unimportant in botrocetin-stimulated binding of VWF to GPIbα, they are required for ristocetin-stimulated binding [63]. Furthermore, the higher the shear, the greater the importance of repeats 2–4 [64]. Thus, ristocetin and tensile force stabilise a second conformational state of the A1-GPIbα complex in which LRR 2–4 of GPIbα appear to contact the A1 domain. This evidence for two conformational states corresponds closely with the observation that the A1-GPIbα bond is a flex-bond with two states.
VWD Type 2B
Type 2B VWD results from gain-of-function mutations in the A1 domain. Enhanced basal binding of A1 to platelet GPIbα depletes longer VWF concatamers and platelets, paradoxically leading to bleeding diatheses. Type 2B mutations map to the “bottom” face of the A1 domain, are often buried, and generally do not contact GPIbα (Fig. 7). In pseudo- or platelet-type VWD, gain-of-function mutations in the platelet GPIbα subunit cause similar clinical symptoms [1]. Platelet-type mutations map to a β-thumb region of GPIbα that contacts A1 (Fig. 7).
Mutations in the A1 domain in gain-of-function type 2B von Willebrand disease are suggestive of conformational change. These mutations enhance binding of A1 to GPIbα, yet map to sites that are buried in the A1 domain (Fig. 7) [65]. Interestingly, the Arg1306 Gln VWD 2B mutation lowers the force at which the GPIbα-A1 bond switches to the more force-resistant, extended state (J. Kim and T. A. Springer, unpublished). Together with the burial of most gain-of-function 2B mutations in the A1 domain, this finding suggests that a second conformational state of the GPIbα-A1 complex may exist that has not yet been seen in crystal structures. Although current structural studies on A1 and A1-GPIbα complexes, including those that have incorporated gain-of-function type 2B and platelet-type VWD mutations have thus far only revealed a flexed state [53, 54, 65], it will be interesting to determine whether further studies can reveal the hypothesised extended state of the A1-GPIbα complex.
Coda
VWF concatamers are beautiful molecules to biophysicists and fascinating to clinicians. pH and Ca2+-regulated dimeric bouquet and helix assembly enable disulphide linkage between monomers that is aligned with assembly in tubules, and orderly uncoiling in secretion with a 50-fold expansion in length. The enormous length and multivalency of VWF concatamers are evolutionary adaptations that enable their unique ability within the vascular system to mediate haemostasis in the rapid arteriolar flow environment. Concatamer extension and length strongly correlate with exposure of the A1 domain for binding to GPIbα on platelets, but more work is required for mechanistic understanding of these relationships. Remarkably, VWF is longest and most extended when secreted and bound to endothelium or the matrix at sites of vascular injury. More remarkably, extension and hence activation of haemostatic potential, is triggered by change from shear to elongational flow at sites of vasoconstriction and vessel rupture. Trimming by ADAMTS13 occurs locally when bound to the vessel wall, and subsequently in the circulation. Cleavage of the A2 domain by ADAMTS13 occurs after unfolding by elongational force. Parallel regulation of haemostatic potential and length distribution by elongational force provide safeguards against thrombosis within certain limits, which may be exceeded in stenosis. Biophysical studies of structure and response to force demonstrate a shearbolt-like function of the A2 domain. Two states of the A1- GPIbα flex-bond broaden its ability to resist a wide range of forces in vivo, while avoiding excessive bond strength at low force. The largely dominant effect of VWD mutations reflect incorporation of both healthy and mutant alleles into VWF concatamers. Loss-of-function VWD type 2A mutations in A2, and gain-of-function VWD type 2B mutations in A1, greatly contribute to, and are illuminated by, understanding of VWF biophysics.
Acknowledgements
I am grateful to collaborators in cited publications for discussion that contributed to the concepts presented here. Yan-Feng Zhou, Jongseong Kim, and Cheng-Zhong Zhang are thanked for preparation of special figures for this review. Author’s work was supported by NIH grants HL-48675 and HL-108248.
References
- 1.Sadler JE. New concepts in von Willebrand disease. Annu Rev Med. 2005;56:173–191. doi: 10.1146/annurev.med.56.082103.104713. [DOI] [PubMed] [Google Scholar]
- 2.Wagner DD. Cell biology of von Willebrand factor. Annu Rev Cell Biol. 1990;6:217–246. doi: 10.1146/annurev.cb.06.110190.001245. [DOI] [PubMed] [Google Scholar]
- 3.Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 4.Ruggeri ZM, Mendolicchio GL. Adhesion mechanisms in platelet function. Circ Res. 2007;100:1673–1685. doi: 10.1161/01.RES.0000267878.97021.ab. [DOI] [PubMed] [Google Scholar]
- 5.Zenner HL, Collinson LM, Michaux G, Cutler DF. High-pressure freezing provides insights into Weibel-Palade body biogenesis. J Cell Sci. 2007;120:2117–2125. doi: 10.1242/jcs.007781. [DOI] [PubMed] [Google Scholar]
- 6.Valentijn KM, Valentijn JA, Jansen KA, Koster AJ. A new look at Weibel-Palade body structure in endothelial cells using electron tomography. J Struct Biol. 2008;161:447–458. doi: 10.1016/j.jsb.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Berriman JA, Li S, Hewlett LJ, Wasilewski S, Kiskin FN, Carter T, Hannah MJ, Rosenthal PB. Structural organization of Weibel-Palade bodies revealed by cryo-EM of vitrified endothelial cells. Proc Natl Acad Sci U S A. 2009;106:17407–17412. doi: 10.1073/pnas.0902977106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang RH, Wang Y, Roth R, Yu X, Purvis AR, Heuser JE, Egelman EH, Sadler JE. Assembly of Weibel-Palade body-like tubules from N-terminal domains of von Willebrand factor. Proc Natl Acad Sci U S A. 2008;105:482–487. doi: 10.1073/pnas.0710079105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Titani K, Kumar S, Takio K, Ericsson LH, Wade RD, Ashida K, Walsh KA, Chopek MW, Sadler JE, Fujikawa K. Amino acid sequence of human von Willebrand factor. Biochemistry. 1986;25:3171–3184. doi: 10.1021/bi00359a015. [DOI] [PubMed] [Google Scholar]
- 10.Erent M, Meli A, Moisoi N, Babich V, Hannah MJ, Skehel P, Knipe L, Zupancic G, Ogden D, Carter T. Rate, extent and concentration dependence of histamine-evoked Weibel-Palade body exocytosis determined from individual fusion events in human endothelial cells. J Physiol. 2007;583:195–212. doi: 10.1113/jphysiol.2007.132993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michaux G, Abbitt KB, Collinson LM, Haberichter SL, Norman KE, Cutler DF. The physiological function of von Willebrand's factor depends on its tubular storage in endothelial Weibel-Palade bodies. Dev Cell. 2006;10:223–232. doi: 10.1016/j.devcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Ewenstein BM, Warhol MJ, Handin RI, Pober JS. Composition of the von Willebrand factor storage organelle (Weibel-Palade body) isolated from cultured human umbilical vein endothelial cells. J Cell Biol. 1987;104:1423–1433. doi: 10.1083/jcb.104.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong JF, Moake JL, Nolasco L, Bernardo A, Arceneaux W, Shrimpton CN, Schade AJ, McIntire LV, Fujikawa K, Lopez JA. ADAMTS-13 rapidly cleaves newly secreted ultralarge von Willebrand factor multimers on the endothelial surface under flowing conditions. Blood. 2002;100:4033–4039. doi: 10.1182/blood-2002-05-1401. [DOI] [PubMed] [Google Scholar]
- 14.Fowler WE, Fretto LJ, Hamilton KK, Erickson HP, McKee PA. Substructure of human von Willebrand factor. J Clin Invest. 1985;76:1491–1500. doi: 10.1172/JCI112129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slayter H, Loscalzo J, Bockenstedt P, Handin RI. Native conformation of human von Willebrand protein. J Biol Chem. 1985;260:8559–8563. [PubMed] [Google Scholar]
- 16.Siedlecki CA, Lestini BJ, Kottke-Marchant KK, Eppell SJ, Wilson DL, Marchant RE. Shear-dependent changes in the three-dimensional structure of human von Willebrand factor. Blood. 1996;88:2939–2950. [PubMed] [Google Scholar]
- 17.Schneider SW, Nuschele S, Wixforth A, Gorzelanny C, Alexander-Katz A, Netz RR, Schneider MF. Shear-induced unfolding triggers adhesion of von Willebrand factor fibers. Proc Natl Acad Sci U S A. 2007;104:7899–7903. doi: 10.1073/pnas.0608422104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batlle J, Lopez-Fernandez MF, Lopez-Borrasca A, Lopez-Berges C, Dent JA, Berkowitz SD, Ruggeri ZM, Zimmerman TS. Proteolytic degradation of von Willebrand factor after DDAVP administration in normal individuals. Blood. 1987;70:173–176. [PubMed] [Google Scholar]
- 19.Alexander-Katz A, Schneider MF, Schneider SW, Wixforth A, Netz RR. Shear-flow-induced unfolding of polymeric globules. Phys Rev Lett. 2006;97:138101. doi: 10.1103/PhysRevLett.97.138101. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Halvorsen K, Zhang CZ, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein, von Willebrand Factor. Science. 2009;324:1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sing CE, Alexander-Katz A. Elongational flow induces the unfolding of von Willebrand factor at physiological flow rates. Biophys J. 2010;98:L35–L37. doi: 10.1016/j.bpj.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith DE, Babcock HP, Chu S. Single-polymer dynamics in steady shear flow. Science. 1999;283:1724–1727. doi: 10.1126/science.283.5408.1724. [DOI] [PubMed] [Google Scholar]
- 23.Steppich DM, Angerer JI, Sritharan K, Schneider SW, Thalhammer S, Wixforth A, Alexander-Katz A, Schneider MF. Relaxation of ultralarge VWF bundles in a microfluidic-AFM hybrid reactor. Biochem Biophys Res Commun. 2008;369:507–512. doi: 10.1016/j.bbrc.2008.02.062. [DOI] [PubMed] [Google Scholar]
- 24.Nesbitt WS, Westein E, Tovar-Lopez FJ, Tolouei E, Mitchell A, Fu J, Carberry J, Fouras A, Jackson SP. A shear gradient-dependent platelet aggregation mechanism drives thrombus formation. Nat Med. 2009;15:665–673. doi: 10.1038/nm.1955. [DOI] [PubMed] [Google Scholar]
- 25.Vincentelli A, Susen S, Le Tourneau T, Six I, Fabre O, Juthier F, Bauters A, Decoene C, Goudemand J, Prat A, Jude B. Acquired von Willebrand syndrome in aortic stenosis. N Engl J Med. 2003;349:343–349. doi: 10.1056/NEJMoa022831. [DOI] [PubMed] [Google Scholar]
- 26.Ruggeri ZM, Orje JN, Habermann R, Federici AB, Reininger AJ. Activation-independent platelet adhesion and aggregation under elevated shear stress. Blood. 2006;108:1903–1910. doi: 10.1182/blood-2006-04-011551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanhoorelbeke K, Deckmyn H. The role of VWF-collagen interaction in acute platelet thrombus formation. Drugs of the Future. 2003;28:61–67. [Google Scholar]
- 28.Ulrichts H, Udvardy M, Lenting PJ, Pareyn I, Vandeputte N, Vanhoorelbeke K, Deckmyn H. Shielding of the A1 domain by the D'D3 domains of von Willebrand factor modulates its interaction with platelet glycoprotein Ib-IX-V. J Biol Chem. 2006;281:4699–4707. doi: 10.1074/jbc.M513314200. [DOI] [PubMed] [Google Scholar]
- 29.Springer TA. Complement and the multifaceted functions of VWA and integrin I domains. Structure. 2006;14:1611–1616. doi: 10.1016/j.str.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q, Zhou Y-F, Zhang C-z, Springer TA. Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor. Proc Natl Acad Sci U S A. 2009;106:9226–9231. doi: 10.1073/pnas.0903679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou M, Dong X, Baldauf C, Chen H, Zhou Y, Springer T, Lu X, Zhong C, Grater F, Ding J. A novel calcium-binding site of von Willebrand factor A2 domain-regulates its cleavage by ADAMTS13. Blood. 2011 doi: 10.1182/blood-2010-11-321596. in press. [DOI] [PubMed] [Google Scholar]
- 32.Luken BM, Winn LY, Emsley J, Lane DA, Crawley JT. The importance of vicinal cysteines, C1669 and C1670, for von Willebrand factor A2 domain function. Blood. 2010;115:4910–4913. doi: 10.1182/blood-2009-12-257949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruggles EL, Deker PB, Hondal RJ. Synthesis, redox properties, and conformational analysis of vicinal disulfide ring mimics. Tetrahedron. 2009;65:1257–1267. doi: 10.1016/j.tet.2008.11.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andreotti AH. Native state proline isomerization: an intrinsic molecular switch. Biochemistry. 2003;42:9515–9524. doi: 10.1021/bi0350710. [DOI] [PubMed] [Google Scholar]
- 35.Schmid FX. Prolyl isomerase: enzymatic catalysis of slow protein-folding reactions. Annu Rev Biophys Biomol Struct. 1993;22:123–142. doi: 10.1146/annurev.bb.22.060193.001011. [DOI] [PubMed] [Google Scholar]
- 36.Valiaev A, Lim DW, Oas TG, Chilkoti A, Zauscher S. Force-induced prolyl cis-trans isomerization in elastin-like polypeptides. J Am Chem Soc. 2007;129:6491–6497. doi: 10.1021/ja070147r. [DOI] [PubMed] [Google Scholar]
- 37.Gao W, Anderson PJ, Sadler JE. Extensive contacts between ADAMTS13 exosites and von Willebrand factor domain A2 contribute to substrate specificity. Blood. 2008;112:1713–1719. doi: 10.1182/blood-2008-04-148759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akiyama M, Takeda S, Kokame K, Takagi J, Miyata T. Crystal structures of the noncatalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor. Proc Natl Acad Sci U S A. 2009;106:19274–19279. doi: 10.1073/pnas.0909755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zanardelli S, Chion AC, Groot E, Lenting PJ, McKinnon TA, Laffan MA, Tseng M, Lane DA. A novel binding site for ADAMTS13 constitutively exposed on the surface of globular VWF. Blood. 2009;114:2819–2828. doi: 10.1182/blood-2009-05-224915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feys HB, Anderson PJ, Vanhoorelbeke K, Majerus EM, Sadler JE. Multi-step binding of ADAMTS-13 to von Willebrand factor. J Thromb Haemost. 2009;7:2088–2095. doi: 10.1111/j.1538-7836.2009.03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feys HB, Liu F, Dong N, Pareyn I, Vauterin S, Vandeputte N, Noppe W, Ruan C, Deckmyn H, Vanhoorelbeke K. ADAMTS-13 plasma level determination uncovers antigen absence in acquired thrombotic thrombocytopenic purpura and ethnic differences. J Thromb Haemost. 2006;4:955–962. doi: 10.1111/j.1538-7836.2006.01833.x. [DOI] [PubMed] [Google Scholar]
- 42.Lyons SE, Bruck ME, Bowie EJ, Ginsburg D. Impaired intracellular transport produced by a subset of type IIA von Willebrand disease mutations. J Biol Chem. 1992;267:4424–4430. [PubMed] [Google Scholar]
- 43.Lyons SE, Cooney KA, Bockenstedt P, Ginsburg D. Characterization of Leu777Pro and Ile865Thr type IIA von Willebrand disease mutations. Blood. 1994;83:1551–1557. [PubMed] [Google Scholar]
- 44.Ribba AS, Voorberg J, Meyer D, Pannekoek H, Pietu G. Characterization of recombinant von Willebrand factor corresponding to mutations in type IIA and type IIB von Willebrand disease. J Biol Chem. 1992;267:23209–23215. [PubMed] [Google Scholar]
- 45.Yago T, Lou J, Wu T, Yang J, Miner JJ, Coburn L, Lopez JA, Cruz MA, Dong JF, McIntire LV, McEver RP, Zhu C. Platelet glycoprotein Ibalpha forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. J Clin Invest. 2008;118:3195–3207. doi: 10.1172/JCI35754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu T, Lin J, Cruz MA, Dong JF, Zhu C. Force-induced cleavage of single VWF A1A2A3-tridomains by ADAMTS-13. Blood. 2010;115:370–378. doi: 10.1182/blood-2009-03-210369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J, Zhang C, Zhang X, Springer TA. A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature. 2010;466:992–995. doi: 10.1038/nature09295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ying J, Ling Y, Westfield LA, Sadler JE, Shao JY. Unfolding the A2 domain of von Willebrand factor with the optical trap. Biophys J. 2010;98:1685–1693. doi: 10.1016/j.bpj.2009.12.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Auton M, Cruz MA, Moake J. Conformational stability and domain unfolding of the Von Willebrand factor A domains. J Mol Biol. 2007;366:986–1000. doi: 10.1016/j.jmb.2006.10.067. [DOI] [PubMed] [Google Scholar]
- 50.Gao W, Anderson PJ, Majerus EM, Tuley EA, Sadler JE. Exosite interactions contribute to tension-induced cleavage of von Willebrand factor by the antithrombotic ADAMTS13 metalloprotease. Proc Natl Acad Sci U S A. 2006;103:19099–19104. doi: 10.1073/pnas.0607264104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zanardelli S, Crawley JT, Chion CK, Lam JK, Preston RJ, Lane DA. ADAMTS13 substrate recognition of von Willebrand factor A2 domain. J Biol Chem. 2006;281:1555–1563. doi: 10.1074/jbc.M508316200. [DOI] [PubMed] [Google Scholar]
- 52.Odell JA, Keller A. Flow-induced scission of isolated macromolecules. J Chem Phys. 1988;88:4022–4028. [Google Scholar]
- 53.Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, de Groot PG, Sixma JJ, Gros P. Structures of glycoprotein Iba and its complex with von Willebrand factor A1 domain. Science. 2002;297:1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- 54.Dumas JJ, Kumar R, McDonagh T, Sullivan F, Stahl ML, Somers WS, Mosyak L. Crystal structure of the wild-type von Willebrand factor A1-glycoprotein Ibalpha complex reveals conformation differences with a complex bearing von Willebrand disease mutations. J Biol Chem. 2004;279:23327–23334. doi: 10.1074/jbc.M401659200. [DOI] [PubMed] [Google Scholar]
- 55.Miura S, Li CQ, Cao Z, Wang H, Wardell MR, Sadler JE. Interaction of von Willebrand factor domain A1 with platelet glycoprotein Iba-(1–289). Slow intrinsic binding kinetics mediate rapid platelet adhesion. J Biol Chem. 2000;275:7539–7546. doi: 10.1074/jbc.275.11.7539. [DOI] [PubMed] [Google Scholar]
- 56.Phan UT, Waldron TT, Springer TA. Remodeling of the lectin/EGF-like interface in P- and L-selectin increases adhesiveness and shear resistance under hydrodynamic force. Nat Imm. 2006;7:883–889. doi: 10.1038/ni1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Astrof NS, Salas A, Shimaoka M, Chen JF, Springer TA. Importance of force linkage in mechanochemistry of adhesion receptors. Biochemistry. 2006;45:15020–15028. doi: 10.1021/bi061566o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Springer TA. Structural basis for selectin mechanochemistry. Proc Natl Acad Sci USA. 2009;106:91–96. doi: 10.1073/pnas.0810784105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas WE, Vogel V, Sokurenko E. Biophysics of catch bonds. Annu Rev Biophys. 2008;37:399–416. doi: 10.1146/annurev.biophys.37.032807.125804. [DOI] [PubMed] [Google Scholar]
- 60.Dong JF, Berndt MC, Schade A, McIntire LV, Andrews RK, Lopez JA. Ristocetin-dependent, but not botrocetin-dependent, binding of von Willebrand factor to the platelet glycoprotein Ib-IX-V complex correlates with shear-dependent interactions. Blood. 2001;97:162–168. doi: 10.1182/blood.v97.1.162. [DOI] [PubMed] [Google Scholar]
- 61.De Luca M, Facey DA, Favaloro EJ, Hertzberg MS, Whisstock JC, McNally T, Andrews RK, Berndt MC. Structure and function of the von Willebrand factor A1 domain: analysis with monoclonal antibodies reveals distinct binding sites involved in recognition of the platelet membrane glycoprotein Ib-IX-V complex and ristocetin-dependent activation. Blood. 2000;95:164–172. [PubMed] [Google Scholar]
- 62.Fukuda K, Doggett T, Laurenzi IJ, Liddington RC, Diacovo TG. The snake venom protein botrocetin acts as a biological brace to promote dysfunctional platelet aggregation. Nat Struct Mol Biol. 2005;12:152–159. doi: 10.1038/nsmb892. [DOI] [PubMed] [Google Scholar]
- 63.Shen Y, Romo GM, Dong JF, Schade A, McIntire LV, Kenny D, Whisstock JC, Berndt MC, Lopez JA, Andrews RK. Requirement of leucine-rich repeats of glycoprotein (GP) Ibalpha for shear-dependent and static binding of von Willebrand factor to the platelet membrane GP Ib-IX-V complex. Blood. 2000;95:903–910. [PubMed] [Google Scholar]
- 64.Shen Y, Cranmer SL, Aprico A, Whisstock JC, Jackson SP, Berndt MC, Andrews RK. Leucine-rich repeats 2–4 (Leu60-Glu128) of platelet glycoprotein Ibalpha regulate shear-dependent cell adhesion to von Willebrand factor. J Biol Chem. 2006;281:26419–26423. doi: 10.1074/jbc.M604296200. [DOI] [PubMed] [Google Scholar]
- 65.Celikel R, Ruggeri ZM, Varughese KI. von Willebrand factor conformation and adhesive function is modulated by an internalized water molecule. Nat Struct Biol. 2000;7:881–884. doi: 10.1038/79639. [DOI] [PubMed] [Google Scholar]
- 66.Sadler JE, Shelton-Inloes BB, Sorace JM, Harlan JM, Titani K, Davie EW. Cloning and characterization of two cDNAs coding for human von Willebrand factor. Proc Natl Acad Sci U S A. 1985;82:6394–6398. doi: 10.1073/pnas.82.19.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marti T, Rosselet SJ, Titani K, Walsh KA. Identification of disulfide-bridged substructures within human von Willebrand factor. Biochemistry. 1987;26:8099–8109. doi: 10.1021/bi00399a013. [DOI] [PubMed] [Google Scholar]
- 68.Katsumi A, Tuley EA, Bodo I, Sadler JE. Localization of disulfide bonds in the cystine knot domain of human von Willebrand factor. J Biol Chem. 2000;275:25585–25594. doi: 10.1074/jbc.M002654200. [DOI] [PubMed] [Google Scholar]