Abstract
Assessment of health in large population studies has increasingly incorporated measures of blood-based biomarkers based on the use of dried blood spots (DBS). The validity of DBS assessments made by labs used by large studies is addressed by comparing assay values from DBS collected using conditions similar to those used in the field with values from whole blood samples. The DBS approach generates values that are strongly related to whole blood levels of HbA1c, cystatin C, and C-reactive protein. Assessing lipid levels reliably with DBS appears to be a greater challenge. However, even when DBS values and values from venous blood are highly correlated, they are often on a different scale, and using conventional cutoffs may be misleading.
Introduction
Large population-based studies are increasingly collecting data that allow the estimation of biopsychosocial models of health. One of the major innovations in large population studies over the past decade has been the introduction of measured biomarkers. This technology has been used by a number of studies, including the Health and Retirement Study (HRS); the National Social Life Health and Aging Project (NSHAP); the National Longitudinal Study of Adolescent Health (ADD Health); the Chinese Health and Retirement Longitudinal Study (CHARLS); the Mexican and Indonesian Family Life Studies (MxFLS and IFLS, respectively); the Indian Longitudinal Study of Aging (LASI); and the Survey of Health, Ageing and Retirement in Europe (SHARE). The rationale for adding biomarkers to these studies was that they validate and add nuance to self-reports of health, allow richer modeling of pathways of influence between socioeconomic and physical variables, and may capture aspects of health unknown to survey participants (Weir 2008). This article describes in some detail a comparison of the assay results for five important analytes obtained from dried blood spots (DBS) and from venous blood drawn from a volunteer sample that was not part of a major study. Because values resulting from assays of DBS samples can differ from values resulting from assays derived from the more conventionally used venous blood, it is important to establish their scaling relative to conventional assays. Assay results may differ across laboratories because of the use of different procedures, analytes, or equipment/technology. We compared results from all the HRS laboratories used from 2006 to 2012 that are still operating (one went out of business). These labs are used by many large health studies. This study was partially supported and planned in conjunction with HRS, but it was carried out by the University of Southern California (USC)/University of California, Los Angeles (UCLA), Center on Biodemography and Population Health (CBPH).
Background
Because research has increasingly clarified that many of the important health outcomes associated with aging (e.g., mortality, loss of both physical and cognitive functioning, and cardiovascular disease) have similar risk factors, the value of collecting a set of biomarkers related to these health outcomes along with socioeconomic and psychological information in population surveys has become widely recognized (Crimmins and Seeman 2001; Crimmins and Seeman 2004; Crimmins et al. 2008). Aging is accompanied by an increasing likelihood of dysregulation within physiological systems that may precede the diagnosis of disease and disability. Biomarkers have been chosen in HRS to reflect these important age-related changes in cardiovascular and metabolic functioning, levels of inflammation, and organ reserve or frailty. Biomarkers such as cholesterol and glycosylated hemoglobin may indicate preclinical or premorbid dysregulation that is unknown to survey participants, especially those without regular preventive health care, and they are an important addition to self-reports in determining health risks within groups in the United States (White et al. 2012), as well as international differences in health risks (Crimmins, Garcia, and Kim 2010; Goldman et al. 2011). These markers can also be used with information on therapeutic medication usage to determine differential levels of control within population subgroups (Chiu and Wray 2011) and changes in population risk over time (Crimmins, Vasunilashorn, and Kim 2010). Markers of inflammation such as C-reactive protein (CRP) and markers related to organ deterioration such as cystatin C are predictors of a number of major health outcomes, including mortality.
Collection of DBS has become increasingly common in surveys as a low-cost alternative to venipuncture (McDade, Williams, and Snodgrass 2007; Williams and McDade 2009). DBS are drops of blood deposited on specially manufactured paper after a finger prick that can be collected in the home as part of a regular interview without the assistance of medically trained personnel. Their development has markedly changed the feasibility of collecting blood samples in large populations (Lindau and McDade 2008; McDade, Williams, and Snodgrass 2007). The HRS DBS samples have been assayed for five biomarkers: total and HDL cholesterol, which provide indicators of lipid levels; glycosylated hemoglobin (HbA1c), which is an indicator of glycemic control over the past two to three months; C-reactive protein, which is a general marker of systemic inflammation; and cystatin C, which is an indicator of kidney function. The choice of which assays to perform was based on the importance of the indicators as documentation of health status or as predictors of subsequent health events; however, the choice was both limited and influenced by the availability of DBS assays that had at least been validated in the lab and adapted for the high throughput needed by large studies.
Studies that relate the DBS values to standard metrics, comparable to what one would observe with venous blood, are essential for examining biomarkers over time in situations in which instruments, assays, and materials change and for comparing results across studies. In this study, blood samples from volunteers, collected in a UCLA clinical setting, were used to compare the values from DBS to those from venous blood, and to compare results across laboratories conducting DBS assays. The USC/UCLA Center on Biodemography and Population Health collected the samples, arranged for and shipped the material to labs for the assays, and analyzed and reported the results.
Methods
Specimen Collection
DBS and venous blood samples were collected from 92 volunteer subjects aged 50 and older during the period from June through December 2010. Participants provided informed consent and were given a $25 gift card to a popular retailer as an incentive to participate.
DBS specimens from University of Vermont and University of Washington laboratories were collected on Whatman 903 cards, and specimens from Heritage Laboratories and Geonostics were collected on two specially treated cards.
Laboratories
DBS were sent to Heritage Laboratory for assay of HbA1c, total cholesterol, and HDL cholesterol; the University of Vermont Laboratory for Clinical Biochemistry Research for assay of CRP and cystatin C; the University of Washington Department of Laboratory Medicine for assay of total cholesterol, HDL cholesterol, HbA1c, CRP, and cystatin C; and Geonostics Laboratory for assay of HbA1c, performed at the FlexSite Laboratory. Details on the laboratories, the assays used, and the laboratory assessment of the reliability and validity of their assays are provided in Crimmins et al. (2013).
DBS for the University of Vermont, the University of Washington, and Geonostics were sent frozen. For Heritage, procedures were designed to mimic the HRS procedures that were in use at the time of the validation study. DBS were sent by regular postal service to the HRS office in Michigan, frozen upon arrival, and then sent from HRS to Heritage in regular shipments with HRS survey subject cards.
HbA1c based on venous samples was assayed from fresh whole blood at a local lab. For other assays from venous samples, serum was frozen and sent to the University of Vermont laboratory headed by Russell Tracy for assay of total and HDL cholesterol, CRP, and cystatin C.
Comparisons were made between whole blood values of HbA1c and DBS assays from the University of Washington, Heritage, and Geonostics (using the FlexSite assay). Serum blood comparisons were made with DBS assays for total and HDL cholesterol from the University of Washington and Heritage labs. DBS assays for the University of Washington and Heritage were compared for HbA1c and total and HDL cholesterol. Comparisons of CRP assays from DBS and serum values from the University of Vermont and the University of Washington were done. These DBS assays were also compared to each other. Cystatin C from serum at the University of Vermont was compared to the assays from DBS at the University of Vermont and the University of Washington.
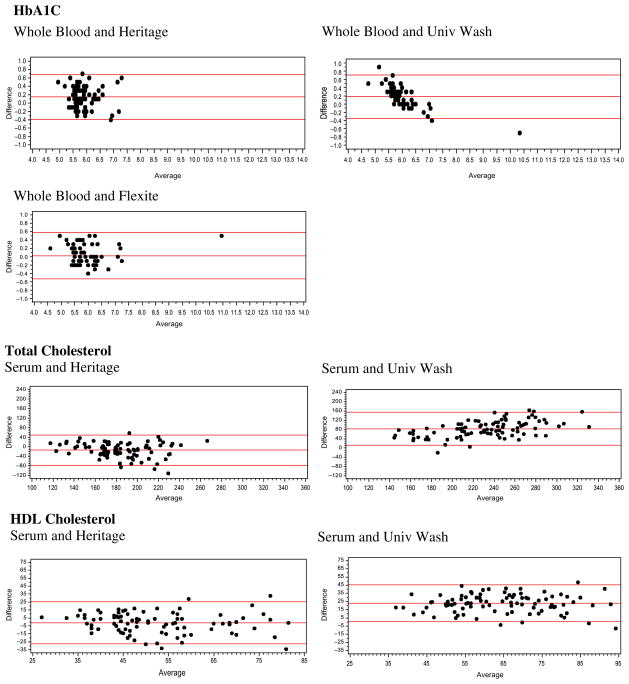
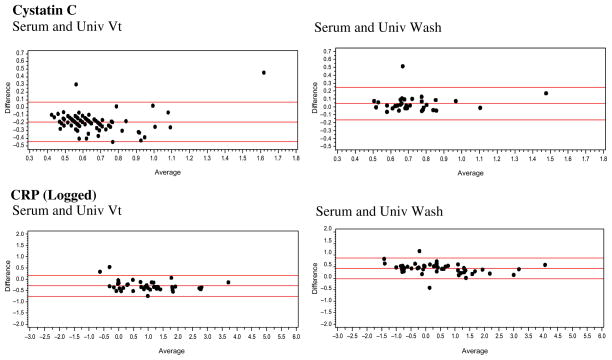
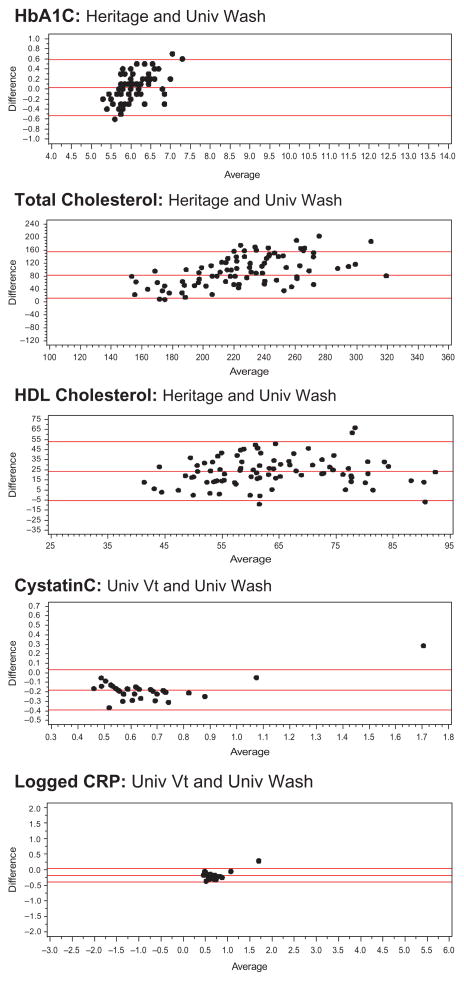
For each pairing of assays, we show descriptive statistics (e.g., mean, standard deviation, range), the correlation between the values, and the equation linking the two. We also provide Bland Altman plots for the whole blood or serum and DBS comparisons, as well as the DBS assays, to provide a method of comparing the differences between two assays across the range of values and to see how frequently outliers occur (Bland and Altman 1986; Altman and Bland 1983). Bland Altman plots map the difference between two assays on the Y-axis against the average on the X-axis. For our assays, we used the formulas Average = (DBS + Whole Blood)/2 and Difference = DBS − Whole blood; we also calculated the 95 percent confidence intervals for the difference. We examined the Bland Altman plots for DBS to DBS comparisons with the same type of formula.
Results
For HbA1c, the average values from the DBS and venous blood appear quite close, although the DBS values are significantly higher than the whole blood values (Table 1). All the DBS values are reasonably highly correlated with the venous blood values (for Heritage, r = .86; for the University of Washington, r = .97; and for FlexSite, r = .96). When comparing the DBS assays for HbA1c, the FlexSite DBS assay values are significantly lower than the other two DBS values. The correlations between the DBS values are also high (r = .84–.97).
Table 1.
HbA1c percentages: Descriptive information on matched DBS and venous blood samples and matched DBS samples
| Average | SD | Range | a | b1 | Correlation coefficient | |
|---|---|---|---|---|---|---|
| Whole blood/DBS | ||||||
| Whole blood % | 5.84 | 0.51 | 4.7–7.3 | 0.524 | +0.887 | 0.86 |
| Heritage % | 5.99 | 0.49 | 5.2–7.6 | <.0001 | ||
| N = 64 | <.0001 | |||||
| Whole blood % | 5.92 | 0.84 | 4.5–10.7 | −1.904 | +1.283 | 0.97 |
| Univ Wash % | 6.09 | 0.64 | 5.0–10.0 | <.0001 | ||
| N = 58 | <.0001 | |||||
| Whole blood % | 5.89 | 0.80 | 4.5–10.7 | 0.288 | +0.940 | 0.96 |
| FlexSite % | 5.96 | 0.82 | 4.7–11.2 | <.0001 | ||
| N = 66 | <.0001 | |||||
| Matched DBS samples | ||||||
| Univ Wash % | 6.04 | 0.35 | 5.4–7.0 | 0.597 | +0.044 | 0.84 |
| Heritage % | 6.07 | 0.50 | 5.2–7.6 | <.0001 | ||
| N = 75 | .4570 | |||||
| Heritage % | 6.13 | 0.96 | 5.2–13.8 | −0.198 | +1.062 | 0.93 |
| FlexSite % | 5.96 | 0.85 | 5.0–12.4 | <.0001 | ||
| N = 87 | <.0001 | |||||
| Univ Wash % | 6.08 | 0.59 | 5.0–10.0 | 1.805 | +0.718 | 0.97 |
| FlexSite % | 5.95 | 0.80 | 4.7–11.2 | <.0001 | ||
| N = 76 | <.0001 | |||||
Y value in equation is top row, x value is bottom row; regression results from regression of whole blood on DBS value and regression of DBS values.
The Bland Altman results indicate that the differences between the University of Washington DBS assay for HbA1c and the venous blood assay vary with the level of the assay (Figure 1). At low values, the difference is positive; at high values, it is negative. Differences between the venous assay and the Heritage assay and the FlexSite assay do not seem to systematically vary across the range (Figure 1). Only a few points are outside the 95 percent confidence intervals for any of the assays. Comparing Heritage and University of Washington DBS assays using the Bland Altman plots reveals an upward pattern, with higher values showing a greater difference (Figure 2).
Figure 1.
Bland Altman plots comparing DBS assays and whole blood of serum assays.
Figure 2.
Bland Altman plots comparing DBS assays.
Even when the mean values of the samples are quite similar, as they are for these HbA1c assays, using conventional cutoff values for high-risk levels (≥6.5%) leads to widely varying results (11% for whole blood, 21% for Heritage, 13% for the University of Washington, and 16% for FlexSite).
Total cholesterol values differ markedly across the assays (Table 2). The University of Washington DBS value is very high relative to the serum values and the Heritage DBS value; the Heritage value is closer to but lower than the serum value. The University of Washington distribution of DBS assays shows that there are a significant number of very high values. While the mean level of the University of Washington assays differs more from the venous level than from the Heritage mean difference, the University of Washington DBS is more closely related to venous blood values than to the Heritage DBS (r = .74 vs. r = .55). The relationship between the University of Washington and Heritage values is low (r = .48). Using conventional cutoff levels (≥240 mg/dL), three-quarters of the sample is high (76%) according to the University of Washington DBS assay, and almost no one is using the Heritage assay (4.5%) or the serum assay (8%).
Table 2.
Total Cholesterol mg/dL: Descriptive information on matched DBS and venous blood samples and matched DBS samples
| Average | SD | Range | a | b1 | Correlation coefficient | |
|---|---|---|---|---|---|---|
| Serum/DBS | ||||||
| Serum mg/dL | 191.38 | 35.52 | 110–285 | 78.883 | +0.635 | 0.55 |
| Heritage mg/dl | 177.14 | 30.79 | 114–279 | <.0001 | ||
| N = 86 | <.0001 | |||||
| Serum mg/dL | 190.22 | 35.34 | 110–285 | 54.486 | +0.4972 | 0.74 |
| Univ Wash mg/dL | 273.02 | 52.68 | 167–403 | <.0001 | ||
| N = 90 | <.0001 | |||||
| Matched DBS samples | ||||||
| Univ Wash mg/dL | 274.60 | 52.37 | 167–403 | 128.66 | +0.8205 | 0.48 |
| Heritage mg/dL | 177.87 | 30.42 | 114–279 | <.0001 | ||
| N = 86 | <.0001 | |||||
Y value in equation is top row, x value is bottom row; regression results from regression of whole blood on DBS value and regression of DBS values.
Differences between both University of Washington DBS assays and the serum assays get larger when the values are higher (Figure 1). The spread of the differences gets wider at higher values for the Heritage assay, indicating less reliability at higher values. Comparison of the DBS assays also indicates larger differences at higher levels along with more cases outside the confidence intervals (Figure 2).
The HDL results are similar to the total cholesterol results in that the University of Washington DBS assay produces average values much higher than those produced by the others (Table 3). The means for the serum and Heritage DBS assays are quite similar. On the other hand, the association between the University of Washington DBS values and the venous blood values is stronger (r = .70) than that between the Heritage DBS values and venous values (r = .56). The association between the two DBS assay values is relatively low (r = .45). Using cutoffs with these values is problematic. High levels of HDL are good, and low levels are a risk for poor health (defining <40 mg/dL as high risk). None of the University of Washington values are below the normal cutoff. For the serum, 18 percent are below cutoff, and for the Heritage assays, 20 percent are below cutoff. Differences between both DBS assays and the serum assay (Figure 1) and between two DBS assays (Figure 2) do not appear to differ much by level of the assay.
Table 3.
HDL cholesterol mg/dL: Descriptive information on matched DBS and venous blood samples and matched DBS samples
| Average | SD | Range | a | b1 | Correlation coefficient | |
|---|---|---|---|---|---|---|
| Serum/DBS | ||||||
| Serum mg/dL | 52.85 | 14.65 | 24–98 | 20.635 | +0.628 | 0.56 |
| Heritage mg/dL | 51.81 | 13.23 | 30–94 | <.0001 | ||
| N = 89 | 0.4542 | |||||
| Serum mg/dL | 53.04 | 14.47 | 24–98 | 0.042 | +0.699 | 0.70 |
| Univ Wash mg/dL | 75.78 | 14.38 | 46 – 112 | <.0001 | ||
| N = 90 | <.0001 | |||||
| Matched DBS samples | ||||||
| Univ Wash mg/dL | 75.63 | 14.53 | 46 – 112 | 49.827 | +0.4955 | 0.45 |
| Heritage mg/dL | 52.07 | 13.21 | 30–94 | <.0001 | ||
| N = 89 | <.0001 | |||||
Y value in equation is top row, x value is bottom row; regression results from regression of whole blood on DBS value and regression of DBS values.
For cystatin C, the mean of the serum values is considerably higher than that of the Vermont DBS assays (Table 4). The mean value from the University of Washington DBS is close to that of the serum value. The N is lower for the University of Washington DBS assay because the cystatin C and CRP assays were not available when the study began, and the laboratory finished validating the assays in the lab while the study was ongoing. The assays were done from extra spots available only for a subset of cases. The correlation of the serum and DBS values is fairly high (r = .78 and .80, respectively). The values of the two DBS assays are strongly correlated (r = .91). The serum value from the University of Vermont assay does not have the same range as cystatin C assays in the literature; no one would have high cystatin C using the traditional cutoff (>1.55 mg/l). Only 1 percent of the University of Vermont DBS values and 3 percent of the University of Washington values are in this range. The differences between the DBS assays and the serum blood assays are generally consistent throughout the range, and only a few points are outside the 95 percent confidence interval (Figure 1). A similar pattern is observed in comparing the two DBS assays, with only one point outside the 95 percent confidence interval (Figure 2).
Table 4.
Cystatin C mg/l: Descriptive information on matched DBS and venous blood samples and matched DBS samples
| Average | SD | Range | a | b1 | Correlation coefficient | |
|---|---|---|---|---|---|---|
| Serum/DBS | ||||||
| Serum mg/l | 0.75 | 0.18 | 0.41–1.39 | 0.355 | +0.700 | 0.78 |
| Univ Vt mg/l | 0.56 | 0.20 | 0.34–1.85 | <.0001 | ||
| N = 82 | ||||||
| Serum mg/l | 0.71 | 0.18 | 0.41–1.39 | 0.148 | +0.748 | 0.80 |
| Univ Wash mg/l | 0.75 | 0.20 | 0.52–1.56 | <.0001 | ||
| N = 33 | <.0001 | |||||
| Matched DBS samples | ||||||
| DBS Wash mg/l | 0.75 | 0.19 | 0.52–1.56 | 0.355 | 0.693 | 0.91 |
| DBS Univ VT mg/l | 0.57 | 0.26 | 0.34–1.85 | <.0001 | ||
| N = 34 | <.0001 | |||||
Y value in equation is top row, x value is bottom row; regression results from regression of whole blood on DBS value and regression of DBS values.
The CRP assays done at the University of Vermont had a lower detectable limit of .63 mg/L, resulting in about half of the assays being in the nondetectable range (Table 5). For this anlysis, we impute values to the nondetectable DBS cases (.428) using the average value of a set of nondetectable cases that were assayed again using ELISA from surplus HRS DBS. The lower detection limit for the serum assay was .16 mg/L. Three cases fall below this level and are assigned a level of .10. The means and standard deviations of the serum sample are higher than those from the University of Vermont DBS; however, the values for the University of Washington DBS assays are much higher than those of either of the other assays. The association between the Vermont DBS assays and the serum assays is very high (r = .99); the value for the University of Washington assay is not quite as high (r = .87). The association between the two DBS assays is very strong. The values of the Vermont serum assays are lower than those of many other serum assays of CRP in the literature. Defining high-risk CRP using the conventional cutoff of >3.0 mg/L results in 24 percent of the serum sample being high, compared to 17 percent of the University of Vermont DBS. Comparing the serum samples to the University of Washington DBS, 19 percent of the serum samples and 26 percent of the DBS assays are at high-risk levels. The differences between the DBS and serum values in absolute values are larger at higher levels (not shown). When both values are log transformed, as is often the case in CRP analysis, the differences are similar across the assay range (Figure 1). The differences are also quite similar across the range for the two logged DBS assays, with only one point outside the 95 percent confidence interval (Figure 2)
Table 5.
C-reactive protein mg/L: Descriptive information on matched DBS and venous blood samples and matched DBS samples
| Average | SD | Range | a | b1 | Correlation coefficient | |
|---|---|---|---|---|---|---|
| Serum/DBS | ||||||
| Serum mg/L | 2.91 | 6.00 | 0.10 – 43.80 | 0.99 | ||
| DBS Univ VT mg/L | 2.18 | 4.79 | 0.428 – 38.14 | 0.215 | 1.237 | <.0001 |
| N = 82 | <.0001 | |||||
| Serum mg/L | 2.95 | 6.78 | 0.10 – 43.80 | 0.87 | ||
| DBS Wash mg/L | 4.41 | 11.37 | 0.096 – 78.32 | 0.355 | 0.589 | <.0001 |
| N = 54 | 0.0024 | |||||
| Matched DBS samples | ||||||
| DBS Univ Wash mg/L | 4.42 | 11.37 | 0.096 – 78.32 | 0.997 | ||
| DBS Univ VT mg/L | 2.32 | 5.60 | 0.428 – 38.14 | −0.281 | 2.022 | <.0001 |
| N = 54 | 0.0036 | |||||
Y value in equation is top row, x value is bottom row. DBS assigned .428 mg/L for <.63 (−333), serum assigned .10 for (−333 for three cases); regression results from regression of whole blood on DBS value and regression of DBS values.
Discussion and Conclusion
Comparison of results from assays on DBS collected using conditions similar to those that would be used in the field for large studies and venous blood samples shows that the values for CRP, HbA1c, and cystatin C are highly correlated with venous blood levels. Cholesterol measures, both for total cholesterol and HDL, appear to be less reliably measured relative to venous blood levels. Our results are novel in the breadth of comparisons provided; we can conclude that these generalizations are true of results from multiple laboratories. Similar findings on the strength of the relationships between values from DBS assays and those from venous samples have been reported in the literature for other CRP assays (McDade, Burhop, and Dohnal 2004; Brindle et al. 2010), cholesterol (Lakshmy et al. 2010), and HbA1c (Fokkema et al. 2009).
Our results make clear that indicators of biomarkers from DBS-based assays in large population studies will be quite appropriate for use in studies in which the relative level of an assay value is important; the use of absolute levels is more problematic. However, there are many ways to transform the values of the assays to make their levels more comparable to those of whole blood–based assays. The equations provided in this article are one of those ways. We should note, however, that this transformation will generally result in a marked change in the variability of the values and is not necessarily recommended. HRS has chosen to benchmark its 2006 and 2008 assays to the NHANES distribution to maintain the variability while transforming the distribution (Crimmins et al. 2013).
One limitation of our assessment of the similarity of the assay values is that with a relatively small sample, it is not clear that we always get the full distribution of the analyte. In addition, our participants were not drawn to constitute a representative sample. However, our results should prove useful to many surveys that have collected DBS for these analytes and especially to those whose results have been harmonized to results from one of the labs used in this project.
Acknowledgments
Funding
This work was supported by the National Institute on Aging through grants to the USC/UCLA Center on Biodemography and Population Health (P30 AG17265), the Network on Measurement of Biological Risk (R24 AG037898), and the Health and Retirement Study (U01 AG009740).
References
- Altman DG, Bland JM. Measurement in medicine: the analysis of method comparison studies. Statistician. 1983;32:307–317. [Google Scholar]
- Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;327:307–310. [PubMed] [Google Scholar]
- Brindle E, Fujita M, Shofer J, O’Connor KA. Serum, plasma, and dried blood spot high-sensitivity C-reactive protein enzyme immunoassay for population research. J Immunol Methods. 2010;362:112–120. doi: 10.1016/j.jim.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CJ, Wray LA. Gender differences in functional limitations in adults living with type 2 diabetes: biobehavioral and psychosocial mediators. Ann Behav Med. 2011;41:71–82. doi: 10.1007/s12160-010-9226-0. [DOI] [PubMed] [Google Scholar]
- Crimmins E, Faul J, Kim JK, Guyer H, Langa K, Ofstedal MB, Sonnega A, Wallace R, Weir D. [Accessed March 19, 2014];Documentation of biomarkers in the Health and Retirement Study. 2013 at http://hrsonline.isr.umich.edu/index.php?p=userg.
- Crimmins E, Garcia K, Kim JK. Are international differences in health similar to international differences in life-expectancy? In: Crimmins E, Preston S, Cohen B, editors. International differences in mortality at older ages: dimensions and sources. Washington, DC: National Academies Press; 2010. pp. 68–101. [PubMed] [Google Scholar]
- Crimmins E, Seeman T. Integrating biology into demographic research on health and aging (with a focus on the MacArthur Study of Successful Aging) In: Finch C, Vaupel J, Kinsella K, editors. Cells and surveys: should biological measures be included in social science research? Washington, DC: National Academies Press; 2001. pp. 9–41. [PubMed] [Google Scholar]
- Crimmins E, Seeman T. Integrating biology into the study of health disparities. Popul Dev Rev. 2004;30:89–107. [Google Scholar]
- Crimmins E, Vasunilashorn S, Kim JK. Biodemography: new approaches to understanding trends and differences in population health and mortality. Demography. 2010;47(suppl):S41–S64. doi: 10.1353/dem.2010.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimmins E, Vasunilashorn S, Kim JK, Alley D. Biomarkers of aging. Annu Rev Clin Chem. 2008;46:161–215. doi: 10.1016/s0065-2423(08)00405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkema MR, Bakker AJ, de Boer F, Kooistra J, de Vries S, Wolthuis A. HbA1c measurements from dried blood spots: validation and patient satisfaction. Clin Chem Lab Med. 2009;47:1259–1264. doi: 10.1515/CCLM.2009.274. [DOI] [PubMed] [Google Scholar]
- Goldman N, Turra CM, Rosero-Bixby L, Weir D, Crimmins EM. Do biological measures mediate the relationship between education and health: a comparative study. Soc Sci Med. 2011;72:307–315. doi: 10.1016/j.socscimed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmy R, Gupta R, Prabhakaran D, Snehi U, Reddy S. Utility of dried blood spots for measurement of cholesterol and triglycerides in a surveillance study. J Diabetes Sci Tech. 2010;4:258–262. doi: 10.1177/193229681000400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindau ST, McDade TW. Minimally invasive and innovative methods for biomeasure collection in population-based research. In: Weinstein M, Vaupel JW, Wachter KW, editors. Biosocial surveys. Washington, DC: National Academies Press; 2008. pp. 252–277. [PubMed] [Google Scholar]
- McDade TW, Burhop J, Dohnal J. High-sensitivity enzyme immunoassay for C-reactive protein in dried blood spots. Clin Chem. 2004;50:652–654. doi: 10.1373/clinchem.2003.029488. [DOI] [PubMed] [Google Scholar]
- McDade TW, Williams S, Snodgrass JJ. What a drop can do: dried blood spots as a minimally-invasive method for integrating biomarkers into population-based research. Demography. 2007;44:899–925. doi: 10.1353/dem.2007.0038. [DOI] [PubMed] [Google Scholar]
- Weir D. Elastic powers: the integration of biomarkers into the Health and Retirement Study. In: Weinstein M, Vaupel JW, Wachter KW, editors. Biosocial surveys. Washington, DC: National Academies Press; 2008. pp. 78–95. [PubMed] [Google Scholar]
- White K, Avendano M, Capistrant BD, Moon JR, Liu SY, Glymour MM. Self-reported and measured hypertension among older US- and foreign-born adults. J Immigr Minor Health. 2012;14:721–726. doi: 10.1007/s10903-011-9549-3. [DOI] [PubMed] [Google Scholar]
- Williams SR, McDade TW. The use of dried blood spot sampling in the National Social Life, Health, and Aging Project. J Geront B Psychol Sci Soc Sci. 2009;64(suppl 1):i131–i136. doi: 10.1093/geronb/gbn022. [DOI] [PMC free article] [PubMed] [Google Scholar]