Abstract
The structure of an intact glycosaminoglycan (GAG) chain of the bikunin proteoglycan (PG) was analyzed using a combined top-down and bottom-up sequencing strategy. PGs are proteins with one or more linear, high-molecular weight, sulfated GAG polysaccharides O-linked to serine or threonine residues. GAGs are often responsible for the biological functions of PGs, and subtle variations in the GAG structure have pronounced physiological effects. Bikunin is a serine protease inhibitor found in human amniotic fluid, plasma, and urine. Bikunin is posttranslationally modified with a chondroitin sulfate (CS) chain, O-linked to a serine residue of the core protein. Recent studies have shown that the CS chain of bikunin plays an important role in the physiological and pathological functions of this PG. While no PG or GAG has yet been sequenced, bikunin, the least complex PG, offers a compelling target. Electrospray ionization Fourier transform-ion cyclotron resonance mass spectrometry (ESI FTICR-MS) permitted the identification of several major components in the GAG mixture having molecular masses in a range of 5505-7102 Da. This is the first report of a mass spectrum of an intact GAG component of a PG. FTICR-MS analysis of a size-uniform fraction of bikunin GAG mixture obtained by preparative polyacrylamide gel electrophoresis, allowed the determination of chain length and number of sulfo groups in the intact GAGs.
Introduction
Many physiological and pathological processes such as fertilization, embryonic development, immune response, inflammation, angiogenesis, tumorogenesis, and metastasis are mediated by proteoglycans (PGs).1–6 PGs comprise a protein core and one or more glycosaminoglycan (GAG) chains. GAG components of PGs are linear, sulfated polysaccharides containing hexosamine and uronic acid (or hexose) disaccharide repeating sequence and include keratan sulfate (KS), heparin (HP), heparan sulfate (HS), chondroitin sulfate (CS), and dermatan sulfate (DS).7 PGs are ubiquitously present in the extracellular matrix (ECM) and on cell surfaces in animal tissues where they serve as structural elements and participate in a number of regulatory and signaling events.7 High specificity of interactions between PGs and other components of ECM and/or cell surface receptors is defined by the backbone structure and modification pattern of the GAG chain.1,4,7 Biosynthesis of GAGs is a nontemplate-driven process and takes place in the Golgi compartment of eukaryotic cells in response to an external stimulus or at a particular stage of development.8
While the importance of PGs and their potential as new therapeutic agents, targets, and disease markers are well recognized,3,9–12 sequence determination of GAGs remains a challenging task. Methods for GAG sequencing reported to date rely on a bottom-up approach which involves characterization of depolymerized fragments of GAGs that can be assembled like pieces of a puzzle into motifs or domains.13,14 The aim of the present work was to combine a new top-down approach that takes advantage of state-of-the-art mass spectrometric techniques with bottom-up approaches in one sequencing strategy and apply it in the characterization of a PG.
Bikunin, used in the present study, is a small PG, a member of the Kunitz-type, or kunin family of serine protease inhibitors.15-17 Bikunin is encoded by an α-microglobulin-bikunin precursor gene, AMBP (Swiss-Prot entry AMBP_HUMAN, P02760). The precursor protein is proteolyzed releasing α-1-microglobulin and bikunin, two proteins with apparently unrelated functions.15–18 Before secretion, bikunin is posttranslationally modified in the Golgi with both an N-glycan and a CS GAG chain.19-21 The CS GAG chain of bikunin is O-linked to the Ser10 of the core polypeptide (Ser215 in AMBP) through a xylopyranose (Xyl) residue and belongs to the chondroitin 4-sulfate (CS-A) family.19,22 The linkage region hexasaccharide of the bikunin GAG chain has been characterized by Sugahara and co-workers through a combination of radiolabeling, enzymatic depolymerization, high performance liquid chromatography (HPLC), and fast-atom bombardment mass spectrometry.23 The linkage region hexasaccharide, derived from pooled human plasma bikunin, reportedly was the only linkage region-derived structure present in the sample and had a sequence ΔUA(1→3)-β-D-GalNAc4S(1→4)-β-D-GlcA(1→3)-β-D-Gal4S-(1→4)-β-D-Gal(1→4)-D-Xylol (where ΔUA = 4-deoxy-α-L-threo-hex-4-enopyranosyluronic acid; GlcA = glucopyranosyluronic acid; Gal = galactopyranose; and S = sulfate).23 These workers reported that the linkage region hexasaccharide in urinary bikunin is also homogeneous and has the same structure as that in plasma bikunin.23 These results conflict with an earlier report by Toyoda et al.24 in which an unsulfated linkage region tetrasaccharide was found in the CS-A chain of urinary bikunin. Enghild and co-workers examined human plasma bikunin obtained from multiple volunteers.22,25 Based on the results of gradient polyacrylamide gel electrophoresis (PAGE) analysis of fluorophor-labeled linkage region oligosaccharides, this study suggested that both monosulfated and unsulfated linkage region hexasaccharides were present in plasma bikunin.22 Reports also vary in their determination of the average chain length of bikunin GAG and the number and location of sulfo groups,15,16,22,24,26,27 which can be explained by the fact that many studies examined pooled samples from different biological fluids. Bikunin GAG has been reported to contain 12–18 GlcAGalNAc disaccharides22,26 and 5 sulfo groups all clustered at the reducing end.15,22,24,26
During the past decade, the interest in structural analysis of bikunin GAG has increasingly shifted from purely academic to diagnostic and therapeutic.15,16,28–33 The bikunin GAG structure has been demonstrated to vary, depending on the physiological or pathophysiological conditions. A disaccharide composition analysis of the urinary bikunin GAGs obtained from a septic shock patient showed a significantly lower ratio of sulfated to unsulfated disaccharides: 1:6.2 as opposed to 1:1.8 in normal plasma.26 The GAG chain length was increased in the bikunin sample from the septic shock patient, compared to that in the normal bikunin sample.26 Bikunin GAG is important in solubilizing urinary calcium oxalate.34 Urinary bikunin concentration has been found to decrease in patients with renal stones.34–38 Bikunin PG inhibits a number of proteases including matrix proteases involved in cancer metastasis.31 The addition of bikunin treatment to standard chemotherapy improves the survival of patients with surgically treated advanced epithelial ovarian cancer.31
While the protein component of bikunin is well characterized,30,39 its GAG structure has received less attention, due to the technical difficulty associated with GAG analysis. In a few publications describing structural analysis of bikunin GAG, both the chain length and sulfation degree were determined indirectly. The chain length was calculated from matrix-assisted laser desorption ionization (MALDI) mass spectra as a difference in mass between intact bikunin PG and enzymatically or chemically deglycosylated protein core.22,26 The number of sulfo groups in the GAG chain was calculated from disaccharide composition as a ratio of unsulfated disaccharides to sulfated disaccharides and varies from 1.8:126 to 3:1.22 Since in both cases the source of bikunin GAG was human plasma, the variability could be attributed to different quantification methods. Nonetheless, the absolute number of sulfo groups in bikunin GAG is impossible to determine from the disaccharide ratio unless the chain length is known.
The present work describes the first direct mass spectral analysis of an intact GAG chain both as a mixture and after a separation step. The high resolving power of Fourier transform ion cyclotron resonance (FTICR)-MS analysis is applied to the determination of the number of disaccharides and sulfo groups in individual bikunin GAG chains.
Experimental Section
Materials
Bikunin (lot C170) was from Mochida Pharmaceuticals (Tokyo, Japan). Actinase E was from Kaken Biochemicals (Tokyo, Japan). Chondroitinase ABC (EC 4.2.2.4 from Proteus vulgaris), chondroitin AC endolyase (EC 4.2.2.5 from Flavobacterium heparinum), chondro-4-sulfatase, chondro-6-sulfatase (EC 3.1.6.9 and 3.1.6.10, from P. vulgaris) were from Seikagaku (East Falmouth, MA). Chondroitinase B (from F. heparinum) and chondroitin AC exolyase (EC 4.2.2.5, from Arthrobacter aurescens) were from Sigma (St. Louis, MO). Materials, equipment, and reagents for PAGE were from Bio-Rad (Hercules, CA). All other chemicals were from Fisher; all solvents were HPLC grade.
Bikunin Proteoglycan
Bikunin PG was purified from excipients and buffer salts in drug formulation by dialysis against distilled water using 3500 molecular weight cutoff (MWCO) membranes. The purified PG was freeze-dried and stored at −20 °C.
Bikunin Peptidoglycan
Bikunin peptidoglycan (pG) was prepared by the treatment of purified bikunin PG with actinase, a nonspecific protease. A 1% solution of actinase in 50 mM Tris-acetate buffer, pH 8, was added to a 6 mg/mL aqueous solution of bikunin PG in a volumetric ratio of 1:5. After an overnight incubation at 45 °C, sufficient amounts of urea and 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) were added to the digestion mixture to obtain 8 M and 2% concentrations respectively.40 A strong anion-exchange (SAX) spin-column (Vivapure Q, VivaScience, Hannover, Germany) was equilibrated with 8 M urea, 2% CHAPS, pH 8.5, and the digestion mixture containing8M urea and 2% CHAPS was loaded on the SAX membrane by centrifugation at 500g for 15 min. The sample bound to the membrane was washed once with the urea/CHAPS solution and 5 times with 0.1 M NaCl, after which bikunin pG was eluted with three 1-mL portions of 16% aqueous NaCl. Peptidoglycan was desalted by precipitation from 80% methanol at 4 °C overnight, followed by centrifugation. The precipitate was dissolved in water, freeze-dried, and stored at −20 °C.
Bikunin GAG Mixture
GAG component of bikunin pG was released by β-elimination under reducing conditions. Bikunin pG was dissolved in a 0.4 M NaOH solution containing 0.3 M NaBH4 at ~10mg/mL concentration of pG. The reaction was allowed to proceed for 24 h at 4 °C, after which it was carefully neutralized with 1 M HCl. The resulting bikunin GAG mixture was separated from low-molecular-weight contaminants by centrifugal filtration using a 3000 MWCO membrane (Microcon YM-3, Millipore, Billerica, MA).
Isolation and Analysis of Size-Similar GAG Fractions
Bikunin GAG chains were first separated using isocratic polyacrylamide gel electrophoresis (PAGE). The resolving gel contained 15% total acrylamide, and the stacking gel contained 5% total acrylamide.41 Bikunin GAG mixture (3–5 mg/mL) was loaded on a 20 cm × 20 cm gel and run in a Protean II electrophoresis system (Bio-Rad, Hercules, CA) for 30 min at 200 V, followed by 6 h at 400 V. The separated GAG chains were immediately transferred from the polyacrylamide gel to a positively charged nylon membrane (Zeta-Probe, Bio-Rad) using a vertical blotting apparatus (Criterion Blotter, Bio-Rad).41 Electrotrans- fer was carried out at 25 V for 12 min in a 50 mM Tris, 60 mM glycine, 20% methanol blotting buffer, after which the membrane was soaked in water for 10 min followed by 8 M urea, 2% CHAPS for 30 min and water again. A vertical strip was cut from the center of the nylon membrane, stained with Alcian blue, and used as a reference for locating the GAG bands on the unstained parts of the membrane. The membrane was cut into strips containing separated GAGs, and the GAGs were released by soaking the strips in 2 M NaCl solution. Separated GAG chains were desalted using the 3000 MWCO centrifugal filtration membranes (Microcon YM-3, Millipore).
NMR Spectroscopy
31P NMR spectra were recorded ona 300 MHz Varian instrument using sodium dibasic phosphate as external reference. 1H NMR spectra were recorded on a 500 MHz JEOL GSX500R instrument. After lyophilization from 99.9% 2H2O, bikunin GAG sample (13 mM based on a molecular mass of 6 kDa) was dissolved in 99.9% 2H2O for analysis and transferred to a Shigemi NMR microtube. Presaturation of the residual HO2H peak was achieved during the relaxation delay. The spectra were recorded at room temperature and at 333 K, with a flip angle of 90°, spectral width of 2806 Hz, and a minimum of 64 accumulation pulses. Line broadening of 1 Hz was applied in the data processing.
Enzymatic Analysis
Two 60-μL aliquots of1 mg/mL bikunin GAG mixture were treated with 0.2 U chondroitinase B and 0.2 U chondroitinase AC overnight at 37 °C. Products were analyzed by PAGE using Alcian blue and silver staining.42
Two aliquots of the chondroitinase AC digestion products were treated with 0.1 U of chondro-4-sulfatase and 0.1 U of chondro-6-sulfatase overnight at 37 °C. Resulting mixtures were analyzed by reverse-phase high-performance liquid chromatography (RP HPLC) with postcolumn fluorescence detection.
Disaccharide Analysis
Bikunin GAG mixture (50–100 μg) was treated with a mixture of 0.1 U chondroitinase AC II and 0.2 U chondroitinase ABC overnight at 37 °C and analyzed by RP HPLC with postcolumn fluorescence detection. The digestion mixture was separated on a 5 μm, 4.6 mm × 250 mm Supelco Discovery C18 column (Sigma) using a linear gradient of 0-100% B over 35 min and a flow rate of 1.0 mL/min. Mobile phase A contained 8% acetonitrile and 3 mM tetrabutylamine, pH 5 adjusted with HCl. Mobile phase B contained 8% acetonitrile, 3 mM tetrabutylamine, and 200 mM NaCl, pH 5 adjusted with HCl. Column temperature was kept at 55 °C. Fluorescent derivatization was achieved by introducing mobile phases C containing 1% 2-cyanoacetamide and D containing 750 mM NaOH into the postcolumn flow at a rate of 0.2 mL/min. Derivatization was carried out at 120 °C, and the products were detected using λex 346 nm and λem 410 nm.
Mass Spectrometry
UV-MALDI-TOF mass spectra of bikunin PG and pG were acquired on a TofSpec E mass spectrometer (Micromass) in linear mode with positive ion detection. Spectra were internally calibrated with bovine insulin, hen egg lysozyme, and cytochrome C. Sinapinic acid was used for the analysis of bikunin PG, and bis-1,1,3,3-tetramethylguanidinium R-cyano-4-hydroxycinnamate (G2CHCA) was used for the analysis of bikunin pG.43
The negative-ion ESI-FTICR mass spectra of bikunin GAG mixture were acquired on a 7T Bruker Apex IV QeFTMS instrument fitted with an Apollo II electrospray ion source. Bikunin GAG sample was dissolved in 50% aqueous methanol to a concentration of 0.1 mg/mL and directly infused using nanospray through a pulled fused silica tip (model FS360-75-15-D-5, New Objective, Woburn, MA) at a flow rate of 10 μL/h.
For electron-detachment dissociation (EDD)44 MS analysis of the nonreducing end (NRE) monosulfated trisaccharide, precursor ions were isolated in the external quadrupole and accumulated for 1–2 s before the injection into the FTMS cell. The isolation/cell fill cycle was repeated up to six times. The selection of the precursor ion was further refined by using in-cell isolation with a coherent harmonic excitation frequency (CHEF)45 event. The precursor ions were then irradiated with electrons for 1 s. For electron irradiation, the cathode bias was set to −19 V, the ECD lens was set to −17.5 V ± 0.5 V, and the cathode heater was set to 1.6 A. Each mass spectrum was a result of averaging 24 acquisitions. For each mass spectrum, 512 k points were acquired, padded with one zero fill, and apodized using a sinebell window. Background spectra were acquired by leaving all parameters the same but setting the cathode bias to 0 V to ensure that no electrons reached the analyzer cell.
ESI-LC-MS and tandem MS analyses of oligosaccharides derived from bikunin GAG were performed on an Agilent 1100 LC-MSD instrument equipped with an ion trap mass analyzer and an Agilent 1100 series HPLC system. The oligosaccharides were separated on a Zorbax SB C-18 capillary column (5 μm, 0.5 mm × 250 mm, Agilent, Palo Alto, CA). A binary mobile phase was delivered at a flow rate of 10 μL/min, and the sample was introduced through a micro-spray nebulizer. Mobile phase A contained 15 mM tributylamine (in place of the nonvolatile tetrabutylammonium chloride used in RP HPLC), 50 mM ammonium acetate, and 15% acetonitrile. Mobile phase B contained 15 mM tributylamine, 50 mM ammonium acetate, and 65% acetonitrile. The separation was achieved using the following gradient: 0% B 20 min, 0–50% B 20–40 min, 50–70% B 40–60 min. All mass spectra were acquired in the negative ionization mode.
Results and Discussion
Bikunin Proteoglycan and Peptidoglycan
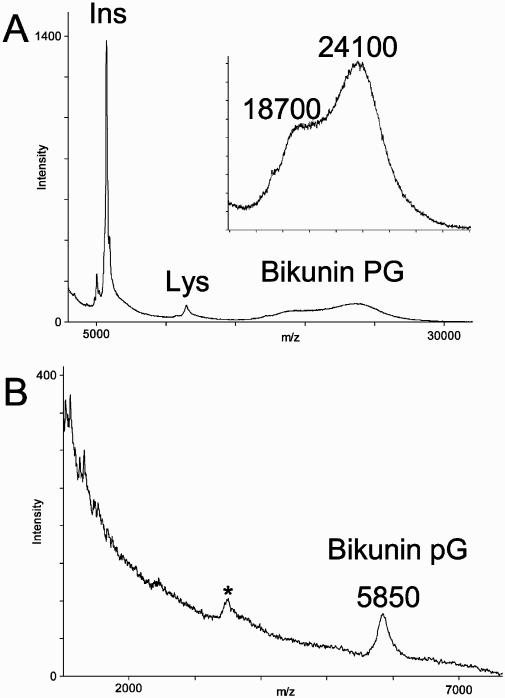
Bikunin drug formulation contained 46% PG and 54% excipients and buffer salts that were removed by dialysis. Recovered bikunin PG was analyzed by MALDI-TOF-MS in the linear positive-ion mode using sinapinic acid and G2CHCA as matrices. MALDI mass spectra of bikunin PG contained a peak at 24,100 m/z, attributed to the bikunin PG, and a peak at 18,700 m/z, corresponding to a bikunin PG with a truncated or missing GAG chain (Figure 1A). These results are consistent with previous MALDI-MS analyses of bikunin PG.22,26 Proteolysis of bikunin PG, followed by SAX purification,40 afforded bikunin peptidoglycan (pG) in a 21% yield. Purified pG was analyzed by MALDI-TOF-MS in the linear mode with negative-ion detection. A MALDI mass spectrum acquired using G2CHCA as a matrix showed a broad peak at ~6,000 m/z (Figure 1B). The shape of the pG peak reflected a high degree of polydispersity in the sample. The use of sinapinic acid and 2,5-dihydroxybenzoic acid matrices did not result in a pG signal.
Figure 1.
(A) Positive-ion, linear MALDI-TOF mass spectrum of bikunin PG. Calibrant peaks are labeled Lys (chicken egg lysozyme) and Ins (bovine insulin). (B) Negative-ion, linear MALDI-TOF mass spectrum of bikunin peptidoglycan. Unassigned peak is marked with an asterisk.
Bikunin GAG Mixture
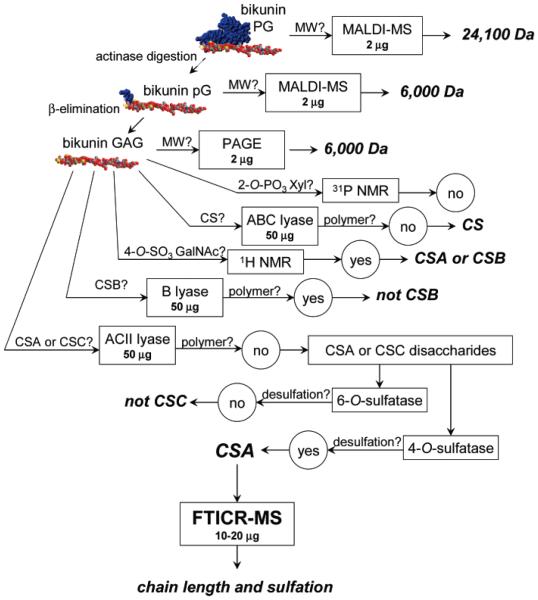
Bikunin GAG mixture, obtained by reductive β-elimination, was characterized by PAGE, 1H and 31P NMR, enzymatic studies, and ESI-FTICR-MS (Figure 2). Using heparin oligosaccharide standards, the average molecular mass of the bikunin GAG chains was estimated ~6 kDa by PAGE analysis.46
Figure 2.
Outline of the analytical strategy, showing amounts of sample in micrograms used in each experimental step.
1H NMR analysis of the GAG mixture was performed at 60 °C to avoid overlapping of the H-4 proton peak of 4-O-sulfo-GalNAc and the HO2H peak observed at room temperature. At 60 °C, the upfield shift of the HO2H peak permitted the H-4 signal of 4-O-sulfo-GalNAc residue to be observed at 4.8 ppm. The 1H NMR spectrum of bikunin GAG was consistent with a chondroitin 4-sulfate (CS-A) structure containing both →3)-β-D-GalNAc(1→4)-β-D-GlcA(1→ and →3)-β-D-GalNAc4S-(1→4)-β-D-GlcA(1→ disaccharide repeating sequences.47
A linkage hexasaccharide in shark cartilage chondroitin 6-sulfate (CS-C) has been reported to contain a 2-O-phospho-Xyl residue.48 To examine the possibility that bikunin GAG might also be phosphorylated, 31P NMR analysis was performed on the bikunin GAG mixture using sodium dibasic phosphate as an internal reference. No peak corresponding to a phospho group was observed, with detection sensitivity of 1.25 mM HPO42−, indicating that there was less than one phospho group substituent per ten bikunin GAG chains.
The C-5 epimerization of uronic acid residues in bikunin GAG was determined by exhaustive digestion of the GAG mixture with chondroitinase ABC, endolytic chondroitinase AC, and chondroitinase B. Chondroitinase ABC converts CS-A, CS-C, and dermatan sulfate (CS-B) completely to disaccharide products.49,50 Endolytic chondroitinase AC acts exclusively on the β-1,4-galactosaminidic bonds between GalNAc and GlcA.51 In contrast, chondroitinase B only cleaves β-1,4-galactosaminid- ic bonds between 4-O-sulfo-GalNAc and IdoA residues, acting exclusively on CS-B.50,51 The sequential use of these chondroitinases followed by PAGE analysis confirmed that the bikunin was a CS GAG, containing no IdoA residues. The disaccharide products of exhaustive depolymerization of bikunin GAG with chondroitinase ABC and endolytic chondroitinase AC were analyzed by RP-IP-HPLC using postcolumn fluorescence detection. The resulting chromatogram contained two peaks with retention times corresponding to those of the unsulfated disaccharide standard ΔUA(1→3)-D-GalNAc and monosulfated disaccharide standard ΔUA (1→3)-D-GalNAc4S. Based on the areas under the chromatographic peaks bikunin GAG mixture contained ΔUA (1→3)-D-GalNAc and ΔUA (1→3)-D-GalNAc4S in a ratio of 1.7:1. This ratio is consistent with the bikunin GAG disaccharide composition reported by Capon and co-workers.26
Since the retention times for ΔUA (1→3)-D-GalNAc4S and ΔUA (1→3)-D-GalNAc6S are close, it was important to eliminate the possibility that a small amount of ΔUA (1→3)-D-GalNAc6S was present. The disaccharide mixture was treated with 4-O-sulfatase, an enzyme that selectively desulfates 4-O-sulfo-GalNAc, and with 6-O-sulfatase, which selectively desulfates 6-O-sulfo-GalNAc.52 Analysis of the enzymatic reaction mixture by RP-IP-HPLC with postcolumn fluorescence detection indicated that the GAG component of bikunin does not contain 6-O-sulfo-GalNAc.
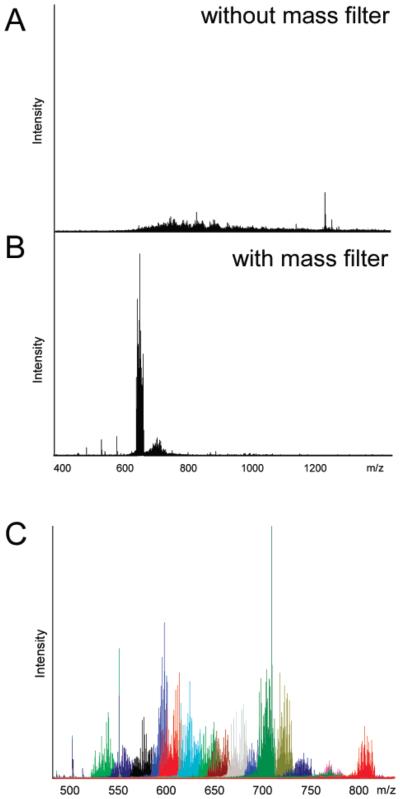
The molecular weight determination of intact bikunin GAGs was performed using ESI-FTICR-MS. Initial MS analysis afforded a very complex mass spectrum containing peaks corresponding to the multiple charge states and Na/H hetero-geneity products of each polysaccharide chain in the mixture (Figure 3A). The signal-to-noise ratio (S/N) observed in the initial FTICR-MS experiment was inadequate for a confident peak assignment due to the sample polydispersity. An improvement in the quality of mass spectral information was achieved by isolating ions within a narrow m/z range using a quadrupole mass filter (Figure 3B). Combining a series of mass spectra acquired over narrow, overlapping m/z regions, a full mass spectrum of the bikunin GAG mixture was afforded with greatly enhanced S/N (Figure 3C). Mass deconvolution of the several hundreds of m/z values observed in the full spectrum yielded 54 major neutral masses that were arranged into nine groups (Table 1). Each group of masses corresponds to a unique saccharide composition with a mass M in which n protons are replaced by n sodium cations, resulting in a mass difference of 22 mass units between successive members of a group. The Na/H exchange is commonly observed in mass spectra of acidic compounds.53,54 ESI-FTICR-MS data interpretation was based on information obtained from the PAGE, 1H and 31P NMR, and enzymatic experiments.
Figure 3.
Improvement in S/N in the FTICR mass spectrum of the bikunin CS mixture (a) achieved by acquiring MS data over narrow m/z regions using quadrupole mass filter (b). Combining mass spectra acquired over narrow overlapping m/z regions afforded a full spectrum shown in panel (c).
Table 1.
Neutral Masses Obtained in the FTICR-MS Analysis of Bikunin GAG
| I | II | III | IV | V | VI | VII | VIII | IX |
|---|---|---|---|---|---|---|---|---|
| 6453 | 7131 | 7190 | ||||||
| 5571 | 6074 | 6475 | 6752 | 6876 | 7153 | 7212 | ||
| 5594 | 5972 | 6096 | 6497 | 6774 | 6898 | 7175 | 7234 | |
| 5615 | 5994 | 6118 | 6351 | 6519 | 6796 | 6920 | 7198 | 7256 |
| 5637 | 6016 | 6142 | 6372 | 6541 | 6818 | 6942 | 7220 | 7278 |
| 5659 | 6038 | 6164 | 6417 | 6563 | 6840 | 6964 | 7242 | 7300 |
| 5681 | 6060 | 6184 | 6439 | 6585 | 6862 | 6986 | 7264 | 7322 |
First, both PAGE and FTICR-MS analyses place the molecular mass of bikunin GAGs in the 5–7 kDa range, which corresponds to the chain lengths of 12–17 disaccharides. Second, each CS-A GAG chain must contain a linkage region tetrasaccharide β-D-GlcA(1→3)-β-D-Gal(1→4)-β-D-Gal(1→4)-D-Xylol with a residue mass of 634 amu.23,55 The chain could be terminated with either a GlcA residue and consist of an even number of monosaccharides, or a GalNAc residue and consist of an odd number of monosaccharides. Finally, a GAG chain could be modified by sulfation on one or more GalNAc residues and/or a Gal residue of the linkage tetrasaccharide. Thus, the mass of a bikunin GAG chain can be calculated using expression 1 or 2, where Modd is the mass of GalNAc-terminated GAG chain; Meven is the mass of GlcA-terminated GAG chain. Each chain contains the linkage tetrasaccharide (634 amu), di number of disaccharides (Δ379 amu), s number of SO3 groups (Δ80 amu), and n number of Na+ replacing the H+ (Δ22 amu). A total of 18 amu is added to the chain termini to account for H and OH absent from the residue formulas, and 203 mass units are added to the chains containing odd numbers of sugars to account for the NRE GalNAc residue.
| (1) |
| (2) |
Expressions 1 and 2 make chemical sense only if condition 3 is satisfied: the number of sodium-cationized acidic groups does not exceed the total number of acidic groups in a chain, which includes GlcA and SO3.
| (3) |
From expressions 1 and 2, a master list containing all possible neutral masses in the 5−7 kDa range was generated and searched for matches to the experimental neutral masses satisfying condition 3. Table 2 illustrates the assignment of masses in groups VI, VII, VIII, and IX from Table 1. Masses in group VI were assigned to a 33-saccharide hexasulfated polysaccharide chain, but not to a 34-saccharide hexasulfated chain, because in the latter case, experimental masses 6797, 6775, and 6753 would imply negative values of n, which is impossible under the given experimental conditions. The assignment of experimental neutral masses resulted in a list of nine polysaccharide compositions summarized in Table 3. Simulated mass spectra generated for each of the nine compositions were in agreement with the experimental mass spectra (Figure 4). Interestingly, all assigned masses corresponded to GalNAc-terminated polysaccharides consisting of odd numbers of residues, and only two sulfation states were prominent, hexasulfated and heptasulfated.
Table 2.
Example of Master List of Possible Neutral Masses Generated on the Basis of the Results of the Oligosaccharide (Bottom-Up) Analysisa
| di = 14, GalNAc-terminated | di = 14, GlcA-terminated | di = 15, GalNAc-terminated | di = 15, GlcA-terminated | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | s = 4 | s = 5 | s = 6 | s = 7 | s = 8 | n | s = 4 | s = 5 | s = 6 | s = 7 | s = 8 | n | s = 4 | s = 5 | s = 6 | s = 7 | s = 8 | n | s = 4 | s = 5 | s = 6 | s = 7 | s = 8 |
| 0 | 6483 | 6563 | 6643 | 6723 | 6803 | 0 | 6280 | 6360 | 6440 | 6520 | 6600 | 0 | 6862 | 6942 | 7022 | 7102 | 7182 | 0 | 6659 | 6739 | 6819 | 6899 | 6979 |
| 1 | 6505 | 6585 | 6665 | 6745 | 6825 | 1 | 6302 | 6382 | 6462 | 6542 | 6622 | 1 | 6884 | 6964 | 7044 | 7124 | 7204 | 1 | 6681 | 6761 | 6841 | 6921 | 7001 |
| 2 | 6527 | 6607 | 6687 | 6767 | 6847 | 2 | 6324 | 6404 | 6484 | 6564 | 6644 | 2 | 6906 | 6986 | 7066 | 7146 | 7226 | 2 | 6703 | 6783 | 6863 | 6943 | 7023 |
| 3 | 6549 | 6629 | 6709 | 6789 | 6869 | 3 | 6346 | 6426 | 6506 | 6586 | 6666 | 3 | 6928 | 7008 | 7088 | 7168 | 7248 | 3 | 6725 | 6805 | 6885 | 6965 | 7045 |
| 4 | 6571 | 6651 | 6731 | 6811 | 6891 | 4 | 6368 | 6448 | 6528 | 6608 | 6688 | 4 | 6950 | 7030 | 7110 | 7190 | 7270 | 4 | 6747 | 6827 | 6907 | 6987 | 7067 |
| 5 | 6593 | 6673 | 6753 | 6833 | 6913 | 5 | 6390 | 6470 | 6550 | 6630 | 6710 | 5 | 6972 | 7052 | 7132 | 7212 | 7292 | 5 | 6769 | 6849 | 6929 | 7009 | 7089 |
| 6 | 6615 | 6695 | 6775 | 6855 | 6935 | 6 | 6412 | 6492 | 6572 | 6652 | 6732 | 6 | 6994 | 7074 | 7154 | 7234 | 7314 | 6 | 6791 | 6871 | 6951 | 7031 | 7111 |
| 7 | 6637 | 6717 | 6797 | 6877 | 6957 | 7 | 6434 | 6514 | 6594 | 6674 | 6754 | 7 | 7016 | 7096 | 7176 | 7256 | 7336 | 7 | 6813 | 6893 | 6973 | 7053 | 7133 |
| 8 | 6659 | 6739 | 6819 | 6899 | 6979 | 8 | 6456 | 6536 | 6616 | 6696 | 6776 | 8 | 7038 | 7118 | 7198 | 7278 | 7358 | 8 | 6835 | 6915 | 6995 | 7075 | 7155 |
| 9 | 6681 | 6761 | 6841 | 6921 | 7001 | 9 | 6478 | 6558 | 6638 | 6718 | 6798 | 9 | 7060 | 7140 | 7220 | 7300 | 7380 | 9 | 6857 | 6937 | 7017 | 7097 | 7177 |
| 10 | 6703 | 6783 | 6863 | 6943 | 7023 | 10 | 6500 | 6580 | 6660 | 6740 | 6820 | 10 | 7082 | 7162 | 7242 | 7322 | 7402 | 10 | 6879 | 6959 | 7039 | 7119 | 7199 |
| 11 | 6725 | 6805 | 6885 | 6965 | 7045 | 11 | … | 6602 | 6682 | 6762 | 6842 | 11 | 7104 | 7184 | 7264 | 7344 | 7424 | 11 | … | 6981 | 7061 | 7141 | 7221 |
| 12 | … | 6827 | 6907 | 6987 | 7067 | 12 | … | … | 6704 | 6784 | 6864 | 12 | … | 7206 | 7286 | 7366 | 7446 | 12 | … | … | 7083 | 7163 | 7243 |
| 13 | … | … | 6929 | 7009 | 7089 | 13 | … | … | … | 6806 | 6886 | 13 | … | … | 7308 | 7388 | 7468 | 13 | … | … | … | 7185 | 7265 |
| 14 | … | … | … | 7031 | 7111 | 14 | … | … | … | … | 6908 | 14 | … | … | … | 7410 | 7490 | 14 | … | … | … | … | 7287 |
| 15 | … | … | … | … | 7133 | … | … | … | … | … | … | 15 | … | … | … | … | … | 15 | … | … | … | … | … |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 23 | … | … | … | … | 23 | … | … | … | … | … | 23 | … | … | … | … | … | 23 | … | … | … | |||
Experimentally observed masses (bold type) that satisfy condition 0 ≤ n ≤ di + s + 1 are enclosed by rectangles.
Table 3.
Bikunin CS Chain Compositions Determined by FTICR-MS Analysis of the Mixture
| composition | number of possible sequencesa |
M odd b | |
|---|---|---|---|
| I | GalNAc(GlcAGalNAc)11 (SO3)6GlcAGalGalXylol | 924 | 5505 |
| II | GalNAc(GlcAGalNAc)12(SO3)6GlcAGalGalXylol | 1716 | 5884 |
| III | GalNAc(GlcAGalNAc)12(SO3)7GlcAGalGalXylol | 1716 | 5964 |
| IV | GalNAc(GlcAGalNAc)13(SO3)6GlcAGalGalXylol | 3003 | 6263 |
| V | GalNAc(GlcAGalNAc)13(SO3)7GlcAGalGalXylol | 3432 | 6343 |
| VI | GalNAc(GlcAGalNAc)14(SO3)6GlcAGalGalXylol | 5005 | 6643 |
| VII | GalNAc(GlcAGalNAc)14(SO3)7GlcAGalGalXylol | 6435 | 6723 |
| VIII | GalNAc(GlcAGalNAc)15(SO3)6GlcAGalGalXylol | 8008 | 7022 |
| IX | GalNAc(GlcAGalNAc)15(SO3)7GlcAGalGalXylol | 11440 | 7102 |
Number of possible sequences was calculated as nCr, where n is the number of sulfation sites, and r is the number of sulfo groups
Monoiso-topic mass of fully protonated GAG chain rounded off to the nearest unit
Figure 4.
Experimental (a) and simulated (b) negative-ion FTICR mass spectra of the bikunin CS-A mixture showing [M − 15H + 5Na]−10 ion which was assigned to GalNAc(GlcAGalNAc)11(SO3)6GlcAGalGalXylol.
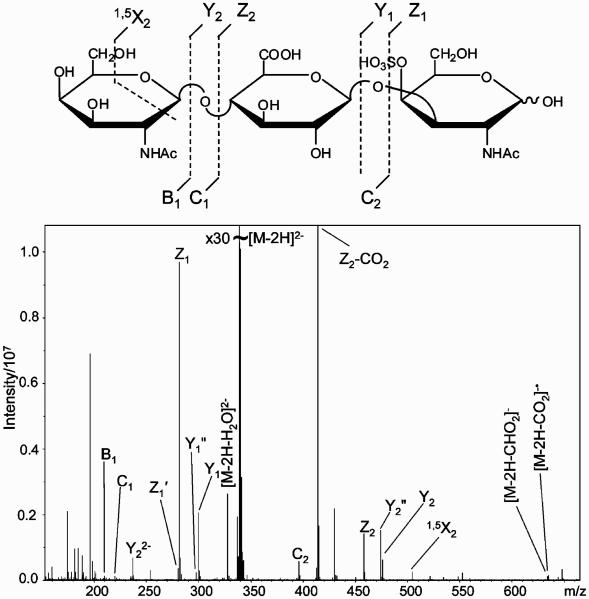
ESI-MS and CID tandem MS characterization of the non-reducing end (NRE) and linkage region oligosaccharides allowed us to determine the positions of two sulfo groups. Bikunin GAG mixture was exhaustively digested with a combination of two endolyases, chondroitinase ABC and endolytic chondroitinase AC. The LC-MS profile of the resulting mixture indicated the presence of three oligosaccharides: ΔUAGalNAc (m/z 379), ΔUAGalNAc4S (m/z 458), and ΔUAGalGalXylol(S) (m/z 713), and a terminal GalNAc residue (m/z 220). The ion corresponding to the monosulfated linkage region tetrasaccharide (m/z 713) was subjected to a tandem MS (MS2) experiment and yielded a major fragment resulting from a glycosidic bond cleavage between ΔUA and GalGalXylol(S) (m/z 555). The MS3 fragmentation of the ion at m/z 555 resulted in the cleavage of a glycosidic bond between two Gal residues and detection of a fragment GalXylol (m/z 241), which indicated that the sulfo group was located on the Gal residue linked to ΔUA (data not shown). Thus, the linkage region tetrasaccharide structure was determined as GlcAGal4SGalXylol, which is consistent with the structure of the bikunin linkage region hexasaccharide described by Sugahara and co-workers.23
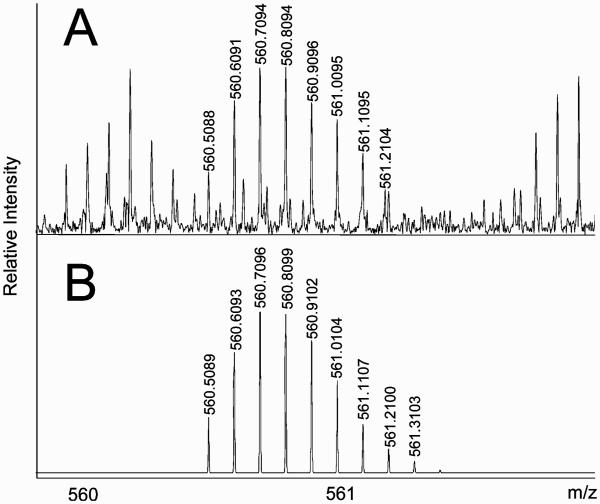
The GAG mixture was depolymerized with the exolytic chondroitinase AC50 to determine the structure of the NRE. LC–MS analysis of the digestion mixture detected two products: GalNAc and GalNAcGlcAGalNAc(S), m/z 220 and m/z 679, respectively. Fragmentation of the monosulfated trisaccharide (m/z 679) by EDD (Figure 5) unambiguously identified the sulfate position as C4 of midchain GalNAc residue. Considering the action pattern of exolytic chondroitinase AC and presence of GalNAc residue in the digestion mixture, it is reasonable to expect an unsulfated trisaccharide, β-D-GalNAc(1→4)-β-D-GlcA(1→3)-β-D-GalNAc(1→ at the NRE of some bikunin GAG chains. Thus, two structures of the NRE are possible: β-D-GalNAc(1→4)-β-D-GlcA(1→3)-β-D-GalNAc(1→ and β-D-Gal-NAc(1→4)-β-D-GlcA(1→3)-β-D-GalNAc4S(1→.
Figure 5.
EDD mass spectrum of the NRE monosulfated trisaccharide obtained through the endolytic chondroitinase AC digestion of the bikunin CS.
Isolation and Analysis of Size-Similar GAG Fractions
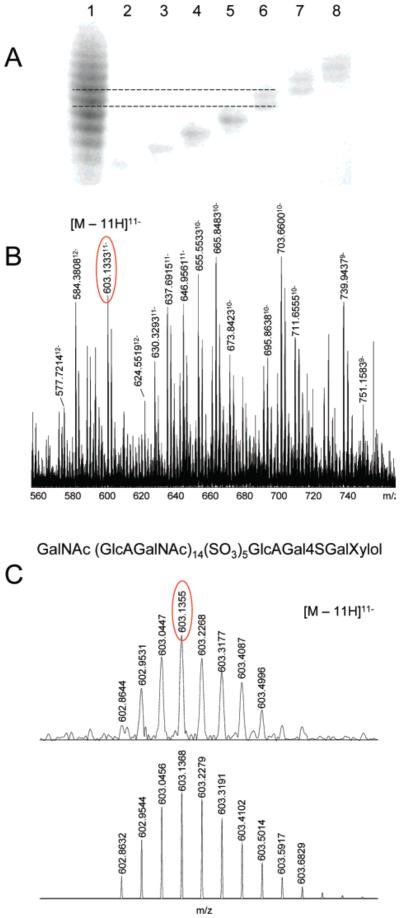
According to FTICR-MS analysis, bikunin GAG mixture consists of long-chain linear polysaccharides (average molecular mass 6000 Da) with polydispersity arising from differences in sulfation (80 Da, 1% of average molecular mass) and/or chain length (379 Da, 6% of average molecular mass). The small differences in charge and size between the chains compared to their overall charge and size make the isolation of a single chain difficult. The separation of bikunin GAG mixture was carried out using preparative PAGE,41 which affords better resolution than any other method available for separation of polyanionic biomolecules. The nine major components in the mixture were immediately transferred onto a positively charged membrane by electroelution to avoid diffusion of GAGs through the gel. The membrane was washed and cut into strips each of which contained a size-uniform fraction of the bikunin GAG mixture. The GAGs eluted from the membrane with 2 M NaCl solution were desalted and analyzed by PAGE (Figure 6A) and FTICR-MS (Figure 6B). PAGE analysis indicated that each fraction contained 1–3 species, and a more efficient separation was achieved for the low-molecular-weight (low-MW)components of the mixture (Figure 6A).
Figure 6.
(A) PAGE analysis of bikunin CS. Direction of migration is from top to bottom. Lane 1, bikunin CS mixture; lanes 2–8, fractions of the mixture isolated by preparative PAGE followed by electroblotting. (B) FTICR-MS analysis of the fraction in lane 6: portion of the spectrum showing peaks corresponding to several components with multiple charge states. (C) Experimental and simulated FTICR mass spectra of the 603.13 m/z peak, corresponding to [M – 11H]11− ion of a hexasulfated, 33-monosaccharide CS chain.
FTICR-MS analysis of bikunin GAG fraction 5 (Figure 6A, lane 6) showed that even after extensive separation, a polydisperse mixture of GAG chains is still present. In addition to the previously identified hexasulfated and heptasulfated GAG chains (Table 3), sulfation states as low as four were detected (Figure 6B; Table 4). Similarly, the results of FTICR-MS analysis of a higher-molecular weight fraction, 6 (Figure 6A, lane 7) indicated that it contains GAGs consisting of 33, 35, 37, and 39 monosaccharides, each having 4–7 sulfo groups. Simulated mass spectra generated for validation of the peak assignment were in good agreement with the experimental mass spectra (Figure 6C).
Table 4.
Bikunin CS Chain Compositions Determined by FTICR-MS Analysis of tine Isolated Fraction (Figure 6, Lane 6)
| composition | M odd a | |
|---|---|---|
| VI-A | GalNAc(GlcAGalNAc)14(SO3)4GlcAGalGalXylol | 6483 |
| VI-B | GalNAc(GlcAGalNAc)14(SO3)5GlcAGalGalXylol | 6563 |
| VI | GalNAc(GlcAGalNAc)14(SO3)6GlcAGalGalXylol | 6643 |
| VII | GalNAc(GlcAGalNAc)14(SO3)7GlcAGalGalXylol | 6723 |
| VIII-A | GalNAc(GlcAGalNAc)15(SO3)5GlcAGalGalXylol | 6942 |
| VIII | GalNAc(GlcAGalNAc)15(SO3)6GlcAGalGalXylol | 7022 |
| IX | GalNAc(GlcAGalNAc)15(SO3)7GlcAGalGalXylol | 7102 |
Monoisotopic mass of fully protonated GAG chain rounded off to the nearest unit
The comparison of FTICR mass spectra obtained from size-fractionated bikunin GAGs and those from the mixture suggests that the mixture analysis is biased toward lower-MW chains, presumably owing their higher ionization efficiency under the conditions used. In contrast, a separation step prior to the FTICR-MS affords mass spectra that more accurately represent the mixture composition.
Conclusion
The present work demonstrates for the first time a combined bottom-up and top-down approach in sequencing a GAG component of a PG. The top-down approach relies on the high resolving power and sensitivity of FTICR-MS and complements the bottom-up approach which employs the time-tested enzymatic analysis in conjunction with LC using spectroscopic or mass spectrometric detection.
The analytical strategy presented here facilitates the molecular weight determination of intact CS chains derived from pooled human urinary bikunin. The bikunin-derived CS GAGs characterized by FTICR-MS are terminated with GalNAc residues and comprised 27–39 monosaccharides with 4–7 sulfo groups distributed along the polysaccharide chain. Enzymology in conjunction with LC-MS and tandem MS analyses allowed us to determine positions of two sulfo groups: in the linkage region tetrasaccharide, β-D-GlcA(1→3)-β-D-Gal4S(1→4)-β-D-Gal(1→4)-D-Xylol; and in the NRE trisaccharide, β-D-GalNAc-(1→3)-β-D-GlcA(1→4)-β-D-GalNAc4S(1→. The results of FT-ICR-MS analysis of the bikunin GAGs separated by PAGE indicated that the median size of the bikunin CS used in this study is 33 monosaccharides (Figure 6, Table 4). The combined FTICR-MS and PAGE data suggest that the size of bikunin CS chains varies between 23 and 45 monosaccharides. Interesting structural features of the bikunin CS such as the odd number of monosaccharides, sulfation of NRE trisaccharide, and the presence of more than 5 sulfo groups were not described previously.
Beginning the study, we expected to find a tremendously complex GAG mixture. There could be 14 chain sizes and 34 compositions satisfying the ratios of unsulfated to sulfated disaccharides (1.8 and 3) and chain lengths of 12–18 disaccharides as reported in previous studies. On the basis of this information, more than 200,000 structures of bikunin CS GAGs could be written. We established that more than 90% of bikunin CS chains in a large pooled sample fall within a relatively narrow size distribution, 27–35 monosaccharides, which reduced the number of possible structures 2-fold. Determining that each CS chain contains an odd number of monosaccharides and characterizing the structures of the non-reducing end trisaccharide and the linkage region tetrasaccharide further reduced the number of possible structures by an order of magnitude.
The chain length and sulfation of the urinary bikunin GAG are altered in inflammation and renal stone disease,26,27,34 serving as the disease markers. These characteristics can be assessed using FTICR-MS, which should facilitate diagnostic studies involving bikunin. The amount of bikunin PG used in the present study for the FTICR-MS molecular weight determination of the bikunin GAG did not exceed 100 μg, a physiological amount, considering a 1–5 μg/mL range concentration of bikunin in human urine15,36 and >100 μg/mL in plasma.30 Thus, the described strategy has a potential application in characterization of bikunin CS obtained from individual patients. Future work will be directed toward further structural analysis of the bikunin CS, particularly, the characteristics and distribution of sulfated domains.
Supplementary Material
Acknowledgment
This work was supported by NIH Grant GM38060.
Footnotes
Supporting Information Available: 1H NMR of bikunin GAG and additional PAGE and FTICR-MS data. This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- (1).Bulow HE, Hobert O. Annu. Rev. Cell Dev. Biol. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- (2).Handel TM, Johnson Z, Crown SE, Lau EK, Sweeney M, Proudfoot AE. Annu. Rev. Biochem. 2005;74:385–410. doi: 10.1146/annurev.biochem.72.121801.161747. [DOI] [PubMed] [Google Scholar]
- (3).Iozzo RV. Nat. Rev. Mol. Cell Biol. 2005;6:646–656. doi: 10.1038/nrm1702. [DOI] [PubMed] [Google Scholar]
- (4).Lamanna WC, Kalus I, Padva M, Baldwin RJ, Merry CLR, Dierks T. J Biotechnol. 2007;129:290–307. doi: 10.1016/j.jbiotec.2007.01.022. [DOI] [PubMed] [Google Scholar]
- (5).Lin X. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- (6).Timar J, Lapis K, Dudas J, Sebestyen A, Kopper L, Kovalszky I. Semin. Cancer Biol. 2002;12:173–186. doi: 10.1016/S1044-579X(02)00021-4. [DOI] [PubMed] [Google Scholar]
- (7).Capila I, Linhardt RJ. Angew. Chem., Int. Ed. 2002;41:390–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- (8).Sasisekharan R, Venkataraman G. Curr. Opin. Chem. Biol. 2000;4:626–631. doi: 10.1016/s1367-5931(00)00145-9. [DOI] [PubMed] [Google Scholar]
- (9).Gesslbauer B, Kungl AJ. Curr. Opin. Mol. Ther. 2006;8:521–528. [PubMed] [Google Scholar]
- (10).Powell AK, Yates EA, Fernig DG, Turnbull JE. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- (11).Proudfoot AEI. Biochem. Soc. Trans. 2006;34:422–426. doi: 10.1042/BST0340422. [DOI] [PubMed] [Google Scholar]
- (12).Raman R, Sasisekharan R. Chem. Biol. 2005;12:267–277. doi: 10.1016/j.chembiol.2004.11.020. [DOI] [PubMed] [Google Scholar]
- (13).Merry CLR, Lyon M, Deakin JA, Hopwood JJ, Gallagher JT. J. Biol. Chem. 1999;274:18455–18462. doi: 10.1074/jbc.274.26.18455. [DOI] [PubMed] [Google Scholar]
- (14).Venkataraman G, Shriver Z, Raman R, Sasisekharan R. Science. 1999;286:537–542. doi: 10.1126/science.286.5439.537. [DOI] [PubMed] [Google Scholar]
- (15).Fries E, Blom AM. Int. J. Biochem. Cell Biol. 2000;32:125–137. doi: 10.1016/s1357-2725(99)00125-9. [DOI] [PubMed] [Google Scholar]
- (16).Fries E, Kaczmarczyk A. Acta Biochim. Pol. 2003;50:735–742. [PubMed] [Google Scholar]
- (17).Zhuo L, Salustri A, Kimata K. Glycoconjugate J. 2003;19:241–247. doi: 10.1023/A:1025331929373. [DOI] [PubMed] [Google Scholar]
- (18).Diarra-Mehrpour M, Bourguignon J, Sesboue R, Salier JP, Leveillard T, Martin JP. Eur. J. Biochem. 1990;191:131–139. doi: 10.1111/j.1432-1033.1990.tb19102.x. [DOI] [PubMed] [Google Scholar]
- (19).Enghild JJ, Salvesen G, Hefta SA, Thogersen IB, Rutherfurd S, Pizzo SV. J. Biol. Chem. 1991;266:747–751. [PubMed] [Google Scholar]
- (20).Morelle W, Capon C, Balduyck M, Sautiere P, Kouach M, Michalski C, Fournet B, Mizon J. Eur. J. Biochem. 1994;221:881–888. doi: 10.1111/j.1432-1033.1994.tb18803.x. [DOI] [PubMed] [Google Scholar]
- (21).Zhuo L, Hascall VC, Kimata K. J. Biol. Chem. 2004;279:38079–38082. doi: 10.1074/jbc.R300039200. [DOI] [PubMed] [Google Scholar]
- (22).Enghild JJ, Thogersen IB, Cheng F, Fransson LA, Roepstorff P, Rahbek-Nielsen H. Biochemistry. 1999;38:11804–11813. doi: 10.1021/bi9908540. [DOI] [PubMed] [Google Scholar]
- (23).Yamada S, Oyama M, Kinugasa H, Nakagawa T, Kawasaki T, Nagasawa S, Khoo K-H, Morris HR, Dell A, Sugahara K. 1995;5:335–341. doi: 10.1093/glycob/5.3.335. [DOI] [PubMed] [Google Scholar]
- (24).Toyoda H, Kobayashi S, Sakamoto S, Toida T, Imanari T. Biol. Pharm. Bull. 1993;16:945–947. doi: 10.1248/bpb.16.945. [DOI] [PubMed] [Google Scholar]
- (25).Enghild JJ, Thogersen IB, Pizzo SV, Salvesen G. J. Biol. Chem. 1989;264:15975–15981. [PubMed] [Google Scholar]
- (26).Capon C, Mizon C, Lemoine J, Rodie-Talbere P, Mizon J. Biochimie. 2003;85:101–107. doi: 10.1016/s0300-9084(03)00066-x. [DOI] [PubMed] [Google Scholar]
- (27).Mizon C, Mairie C, Balduyck M, Hachulla E, Mizon J. Eur. J.Biochem. 2001;268:2717. doi: 10.1046/j.1432-1327.2001.02168.x. [DOI] [PubMed] [Google Scholar]
- (28).Bergsland KJ, Kelly JK, Coe BJ, Coe FL. Am. J. Physiol. Renal Physiol. 2006;291:F530–536. doi: 10.1152/ajprenal.00370.2005. [DOI] [PubMed] [Google Scholar]
- (29).Jessen TE, Odum L. Reproduction. 2003;125:27–31. doi: 10.1530/rep.0.1250027. [DOI] [PubMed] [Google Scholar]
- (30).Josic D, Brown MK, Huang F, Lim YP, Rucevic M, Clifton JG, Hixson DC. Proteomics. 2006;6:2874–2885. doi: 10.1002/pmic.200500563. [DOI] [PubMed] [Google Scholar]
- (31).Kobayashi H, Suzuki M, Hirashima Y, Terao T. Biol. Chem. 2003;384:749–754. doi: 10.1515/BC.2003.083. [DOI] [PubMed] [Google Scholar]
- (32).Kobayashi H, Yagyu T, Inagaki K, Kondo T, Suzuki M, Kanayama N, Terao T. Int. J. Cancer. 2004;110:134–139. doi: 10.1002/ijc.20082. [DOI] [PubMed] [Google Scholar]
- (33).Kobayashi H, Yagyu T, Inagaki K, Kondo T, Suzuki M, Kanayama N, Terao T. Cancer. 2004;100:869–877. doi: 10.1002/cncr.20034. [DOI] [PubMed] [Google Scholar]
- (34).Suzuki M, Kobayashi H, Kageyama S, Shibata K, Fujie M, Terao T. J. Urol. 2001;166:268–274. [PubMed] [Google Scholar]
- (35).Iida S, Peck AB, Johnson-Tardieu J, Moriyama M, Glenton PA, Byer KJ, Khan SR. J. Am. Soc. Nephrol. 1999;10:986–996. doi: 10.1681/ASN.V105986. [DOI] [PubMed] [Google Scholar]
- (36).Medetognon-Benissan J, Tardivel S, Hennequin C, Daudon M, Drueke T, Lacour B. Urol. Res. 1999;27:69–75. doi: 10.1007/s002400050091. [DOI] [PubMed] [Google Scholar]
- (37).Tardivel S, Médétognon J, Randoux C, Kébédé M, Drüeke T, Daudon M, Hennequin C, Lacour B. Urol. Res. 1999;27:243–249. doi: 10.1007/s002400050117. [DOI] [PubMed] [Google Scholar]
- (38).Verkoelen CF. J. Am. Soc. Nephrol. 2006;17:1673–1687. doi: 10.1681/ASN.2006010088. [DOI] [PubMed] [Google Scholar]
- (39).Delaria KA, Muller DK, Marlor CW, Brown JE, Das RC, Roczniak SO, Tamburini PP. J. Biol. Chem. 1997;272:12209–12214. doi: 10.1074/jbc.272.18.12209. [DOI] [PubMed] [Google Scholar]
- (40).Zhang F, Sun P, Munoz E, Chi L, Sakai S, Toida T, Zhang H, Mousa S, Linhardt RJ. Anal. Biochem. 2006;353:284–286. doi: 10.1016/j.ab.2006.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Al-Hakim A, Linhardt RJ. Electrophoresis. 1990;11:23–28. doi: 10.1002/elps.1150110106. [DOI] [PubMed] [Google Scholar]
- (42).Al-Hakim A, Linhardt RJ. Appl. Theor. Electrophor. 1991;1:305. [PubMed] [Google Scholar]
- (43).Laremore TN, Zhang F, Linhardt RJ. Anal. Chem. 2007;79:1604–1610. doi: 10.1021/ac061688m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Wolff JJ, Amster IJ, Chi L, Linhardt RJ. J. Am. Soc. Mass Spectrom. 2007;18:234–244. doi: 10.1016/j.jasms.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Heck AJR, de Koning LJ, Pinkse FA, Nibbering NMM. Rapid Commun. Mass Spectrom. 1991;5:406–414. [Google Scholar]
- (46).Edens RE, Al-Hakim A, Weiler JM, Rethwisch DG, Fareed J, Linhardt RJ. J. Pharm. Sci. 1992;81:823–827. doi: 10.1002/jps.2600810821. [DOI] [PubMed] [Google Scholar]
- (47).Toida T, Toyoda H, Imanari T. Anal. Sci. 1993;9:53–58. [Google Scholar]
- (48).Sugahara K, Ohi Y, Harada T, de Waard P, Vliegenthart JF. J. Biol. Chem. 1992;267:6027–6035. [PubMed] [Google Scholar]
- (49).Hamai A, Hashimoto N, Mochizuki H, Kato F, Makiguchi Y, Horie K, Suzuki S. J. Biol. Chem. 1997;272:9123–9130. doi: 10.1074/jbc.272.14.9123. [DOI] [PubMed] [Google Scholar]
- (50).Jandik K, Gu K, Linhardt RJ. Glycobiology. 1994;4:289–296. doi: 10.1093/glycob/4.3.289. [DOI] [PubMed] [Google Scholar]
- (51).Gu K, Liu J, Pervin A, Linhardt RJ. Carbohydr. Res. 1993;244:369–377. doi: 10.1016/0008-6215(83)85014-9. [DOI] [PubMed] [Google Scholar]
- (52).Yamagata T, Saito H, Habuchi O, Suzuki S. J. Biol. Chem. 1968;243:1523–1535. [PubMed] [Google Scholar]
- (53).Gal J-F, Maria PC, Raczynska ED. J. Mass Spectrom. 2001;36:699–716. doi: 10.1002/jms.202. [DOI] [PubMed] [Google Scholar]
- (54).Zaia J. Mass Spectrom. Rev. 2004;23:161–227. doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]
- (55).de Waard P, Vliegenthart JF, Harada T, Sugahara K. J. Biol. Chem. 1992;267:6036–6043. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.