Abstract
Major histocompatibility complex (MHC) class I polymorphisms are known to influence outcomes in a number of infectious diseases, cancers and inflammatory diseases. Human MHC class I heavy chains are encoded by the HLA-A, HLA-B and HLA-C genes. These genes are highly polymorphic, with the HLA-B locus being the most variable. Each HLA class I protein binds to distinct set of peptide antigens, which are presented to CD8+ T cells. HLA-disease associations have been shown in some cases to link to the peptide binding characteristics of individual HLA class I molecules. Here we show that polymorphisms at the HLA-B locus profoundly influence the assembly characteristics of HLA-B molecules and the stabilities of their peptide-deficient forms. In particular, dependence on the assembly factor tapasin is highly variable, with frequent occurrence of strongly tapasin-dependent or independent allotypes. Several polymorphic HLA-B residues located near the C-terminal end of the peptide are key determinants of tapasin-independent assembly. In vitro refolded forms of tapasin-independent allotypes assemble more readily with peptides compared to tapasin-dependent allotypes that belong to the same supertype, and during refolding, reduced aggregation of tapasin-independent allotypes is observed. Paradoxically, in HIV-infected individuals, greater tapasin-independent HLA-B assembly confers more rapid progression to death, consistent with previous findings that some HLA-B allotypes shown to be tapasin-independent are associated with rapid progression to multiple AIDS outcomes. Together, these findings demonstrate significant variations in the assembly of HLA-B molecules, and indicate influences of HLA-B folding patterns upon infectious disease outcomes.
Introduction
MHC class I molecules bind and present antigenic peptides to CD8+ T cells and thus mediate immune responses against intracellular pathogens and cancers (reviewed in (1)). MHC class I molecules are also important regulators of the activities of natural killer (NK) cells (reviewed in (2). MHC class I molecules comprise a heavy chain, a light chain called β2-microglobulin (β2m) and a peptide, which are assembled in the endoplasmic reticulum (ER) of cells. The heavy chain is highly polymorphic. There are hundreds of variants of the human leukocyte antigens (HLA) HLA-A, HLA-B and HLA-C genes, which encode human MHC class I heavy chains. The polymorphisms influence the specificities of peptide binding in the assembled MHC class I proteins in order to allow for the presentation of a distinct and diverse pool of antigenic peptides by each HLA class I molecule.
HLA class I molecules are known to exert profound influences on disease progression in a number of infectious diseases and cancers (reviewed in (3–6)). Among all genetic factors known to influence outcome to HIV infection, the strongest associations link to HLA class I genes. The peptide-binding characteristics of individual MHC class I proteins are shown to be a major factor that determines immune control of HIV (7, 8), but other characteristics of the HLA molecules, such as those relating to variation in the assembly and stability of individual HLA class I molecules may also have an influence on disease outcomes. By virtue of their highly polymorphic nature, the MHC class I molecules present unique challenges to the cellular protein folding machinery. Thousands of variants (across the population) must be correctly assembled for immunity to be effective at the individual level. Folding and assembly of MHC class I molecules is critically dependent on rare and transient peptides within the ER lumen. MHC class I molecules that are sub-optimally assembled are either retained in the ER, or rendered unstable at the cell surface (9). Thus, the assembly and stability characteristics of individual HLA class I allotypes in addition to their peptide binding specificities may also exert influences on disease outcomes.
The assembly of MHC class I molecules occurs in the ER lumen, with the help of a multiprotein peptide loading complex (PLC) (reviewed in (10)). The transporter associated with antigen processing (TAP) is responsible for translocation of peptides from the cytosol into the lumen of ER and also serves as a scaffold for the PLC assembly (reviewed in (11)). Tapasin, a key component of the PLC (12, 13), bridges a physical interaction between MHC class I and TAP to localize MHC class I in the vicinity of an incoming pool of TAP-translocated peptides. Tapasin also recruits the oxidoreductase ERp57 (14, 15) and the associated ER chaperone calreticulin (16–21) to facilitate MHC class I-peptide assembly in the ER. Although there may be multiple levels of quality control exerted on sub-optimally assembled MHC class I proteins, tapasin has a propensity to interact with peptide-deficient forms of MHC class I molecules (19, 22). Tapasin stabilizes the peptide-free conformation of MHC class I molecules, quantitatively increasing peptide associations with MHC class I molecules (19, 23, 24). Tapasin also optimizes the MHC class I peptide repertoire in favor of high affinity sequences (19, 24–26). Some HLA-B molecules are known to vary in their dependencies on tapasin for assembly and antigen presentation. For example, HLA-B*4402 and HLA-B*4405, which differ by a single residue at position 116 of the peptide binding groove, are highly tapasin-dependent or independent, respectively, for their assembly (25, 27, 28), which in turn affects intracellular trafficking (28–30). Tapasin-dependent or tapasin-independent HLA class I assembly is expected to influence the stability of antigenic peptide associations with MHC class I molecules. Additionally, tapasin-dependent or tapasin-independent HLA class I assembly could reflect intrinsic structural features of empty forms of MHC class I proteins. Since tapasin is a key determinant of the assembly and cell surface stability of several MHC class I proteins, in this study, we examined tapasin dependencies of several of the most frequent HLA-B allotypes observed in North American populations. To further examine potential relationships between intrinsic assembly and stability of empty HLA class I proteins and tapasin dependencies of their intracellular assembly, we also compared refolding and assembly efficiencies of soluble forms of the most highly tapasin-dependent or independent HLA-B allotypes. These studies revealed important differences between HLA-B molecules at the level of assembly and stability. Finally, we examined the influences of tapasin-dependent or independent assembly on progression to various AIDS outcomes.
Materials and Methods
Construction of LIC-pMSCVneo
The plasmid pMSCVneo (Clontech) was used as the parent vector for construction of a ligation-independent-cloning (LIC) variant. This was accomplished by inserting a unique sequence that could be digested with the restriction enzyme Pme I. LIC overhangs could then be generated by treatment of the linearized vector with T4 DNA polymerase exonuclease activity. The inserted sequence was constructed by annealing two oligos to generate a fragment with overhangs complimentary to an EcoR I restriction site. The oligo sequences used were 5’-AATTAGAGAGTTTAAACTTCCAC-3’ and 5’-AATTGTGGAAGTTTAAACTCTCT-3’. The lyophilized oligos were resuspended in distilled water at a concentration of 20 µM. Prior to annealing, the oligos were phosphorylated using T4 polynucleotide kinase (New England Biolabs). The 20 µl reactions contained 2 µl of 10 mM ATP, 12.5 µl of 20 µM oligo, 2 µl of 10X reaction buffer, 1 µl of enzyme and 2.5 µl of distilled water. Reactions were incubated for 1 hour at 37 °C then shifted to 95 °C for an additional 10 minutes. The two oligo reactions were then mixed together at 95 °C and slow cooled to 37 °C. A 2.5 µl aliquot was then added to a 20 µl ligation reaction composed of 1 µl of 45 ng/ml EcoR I-digested pMSCVneo, previously treated with calf intestinal phosphatase, 2 µl 10X reaction buffer, and 13.5 µl of distilled water. The reaction was slow cooled from 37 °C to 16 °C and 1 µl of 1unit/ml T4 DNA ligase (New England Biolabs) was added. Incubation was continued at 16 °C for approximately 16 hours. A 2 µl aliquot of the ligation reaction was used to transform 20 µl of competent XL1-Blue cells. These were plated on Luria Broth (LB) agar with ampicillin (Amp) selection and incubated at 37 °C overnight. Colonies were picked and grown in 5 ml of LB-Amp and plasmid DNA was isolated using Qiagen minispin columns. Positive clones were identified by restriction digestion with Pme I and Cla I and confirmed by DNA sequencing.
Cloning of HLA-B cDNAs into LIC pMSCVneo and LIC pMCSG7
Total RNA was extracted from HLA-typed B lymphoblastoid cell lines (Laboratory of Experimental Immunology, NCI) using TRIzol® Reagent (Invitrogen, USA) according to manufacturer’s instruction. The total RNA was purified by the PureLink® RNA Mini Kit (Invitrogen, USA) according to the manufacturer’s instruction to remove any DNA or protein contaminant. cDNA was synthesized from the purified total RNA using SuperScript™ II Reverse Transcriptase kit (Invitrogen, USA) and random hexamer primers. The cDNA was then used as template for further amplification of HLA-B sequences using HLA-B specific primers. The following oligo sequences were used for insertion of HLA-B genes into LIC pMSCVneo; 5’-GGAATTAGAGAGTTTCACCATGCTGGTCATGGCA-3’ or 5’-GGAATTAGAGAGTTTCACCATGCGGGTCACGGC-3’ for the start sequences and 5’GAATTGTGGAAGTTTCCTAAGCTGTGAGAGACACATC3’ for the stop sequence. Some HLA-B sequences were previously described (30). HLA-B*3701 was from Dr. Catia Traversari (MolMed S. p. A), HLA-B*1501 and HLA-B*1518 were from Dr. Hildebrand (Department of Microbiology and Immunology, University of Oklahoma Health Sciences Center,) and HLA-B*3801 was from Dr. Claire Gardiner (NK Cell Research Group, School of Biochemistry and Immunology, Trinity College, Dublin 2, Ireland). Irrespective of their source, all HLA-B sequences were cloned into retroviral vector MSCVneo using LIC. The oligos used for screening MSCVneo constructs were forward, 5’-CACCCTAAGCCTCCGCCTCC-3’and reverse, 5’-AATGTGTGCGAGGCCAGAGGCC-3’.
LIC was also used to transfer all HLA-B constructs into the pMCSG7 vector (31) for bacterial expression. DNA constructs encoding soluble HLA-B molecules containing N-terminal hexahistidine tags and tobacco etch virus (TEV) protease cleavage sites (and lacking signal sequences and transmembrane domains) were amplified by PCR using 5’-TACTTCCAATCCAATGCTGGCTCCCACTCCATGA-3’ for the start sequence and TTATCCACTTCCAATGTTACGGCTCCCATCTCAGG-3’ for the stop sequence. HLA-B-MSCVneo vectors were used as templates and PCR products were cloned into pMCSG7 vector (31).
For LIC, overhangs of 15 base pairs were generated in the insert DNA by processing 0.2 pmol of the PCR products with 0.5 U LIC-qualified T4 DNA polymerase (Novagen) in the presence of 4 mM dCTP (for ligation with pMCSG7) or 4 mM dGTP (for ligation with pMSCV-neo) and 5 mM DTT, 20 µl final reaction volume. Reactions were incubated for 30 minutes at 22 °C followed by 20 minutes at 75 °C. The pMCSG7 or pMSCV-neo vectors were linearized by restriction digestion with SspI (for pMCSG7) or PmeI (for pMSCV-neo) (New England Biolabs) at a concentration of 5 U/µg DNA at 37 °C for 1.5 hours. The proteins were removed using a Qiagen PCR minispin kit. Linearized vector DNA was then processed by T4 DNA polymerase to yield single-stranded ends for annealing. Reactions, 60 µl total, were set up with 1.6-2. 0 µg of linear DNA and 3.75 U LIC-qualified T4 polymerase (Novagen) in the presence of 4 mM dGTP (for pMCSG7) or 4 mM dCTP (for pMSCV-neo) and 5 mM DTT. Reactions were mixed on ice and incubated as described above for inserts. Insert and vector DNA were annealed in 96-well plates by combining 1 µl of processed vector and 2 µl of processed insert per well, followed by incubation in a PCR machine at 22 °C for 15 minutes with addition of 1 µl of 25 mM EDTA after 10 minutes of incubation. Annealed DNA was used to transform competent XL1-Blue cells. All HLA-B constructs in the pMSCV or pMCSG7 vectors were sequenced by the University of Michigan DNA Sequencing Core.
Cell lines
A human melanoma cell line M553 (obtained from Dr. N. Bangia, Rosewell Park Cancer Institute (32) and CEM cells were grown in RPMI 1640 (Life Technologies) supplemented with 2 mM glutamine. BOSC cells (obtained from Dr. K. Collins, University of Michigan) were grown in DMEM (Life Technologies). All growth media were supplemented with 10% (v/v) FBS (Life Technologies), 100 µg/ml streptomycin and 100 U/ml penicillin (Life Technologies).
Viruses and cell infections
Retroviruses were generated as previously described using BOSC cells, and used to infect M553 cells (30). Cells were infected with retroviruses encoding the HLA-B molecules, and selected by treatment with 1 mg/ml G418 (Life Technologies), and maintained in 0.5 mg/ml G418. Exogenous MHC class I expression was verified in M553 cells by flow cytometric analyses using the W6/32 antibody (33) and by immunoblotting analyses of cell lysates using the HC10 (34) or 171.4 antibodies (35). M553 cells expressing HLA-B molecules were infected with the tapasin retrovirus and selected by treatment with 1 µg/ml puromycin (Sigma) and cells were maintained in 0.5 µg/ml puromycin. Tapasin expression in M553 cells was verified by immunoblotting analysis of cell lysates using rabbit anti-tapasin antisera (generated against an N-terminal peptide of tapasin; obtained from Dr. Ted Hansen, Washington University). For immunoblotting analysis, the cells were lysed in Trition-X100 lysis buffer (1% Triton-X100 in PBS containing EDTA-free protease inhibitors [Roche] [pH 7.4]) for 1 hour on ice. The lysates were centrifuged at 4°C for 30 minutes to remove cell debris, and protein concentration in lysates was determined by a bicinchoninic acid protein assay (Pierce, Thermo Scientific). Equal microgram amounts of cell lysates were separated on 10% SDS-PAGE and transferred to Immobilon membranes (Millipore) for immunoblotting. Membranes were blocked in 5% milk in TBS for 1 hour at room temperature, followed by an overnight incubation with primary antibody in TBS–Tween 20 at 4°C. Western blotting analysis was done using HC10 or 171.4 antibody to detect MHC class I heavy chain and using rabbit anti-tapasin antisera to detect tapasin. Membranes were washed for 2 hours in TBS– Tween-20, incubated for 60 minutes with secondary antibody, and washed again for 2 hours. The secondary antibodies were GAM-HRP, GAR-HRP (Jackson ImmunoResearch laboratories) or GAM-IRDye 800CW (LI-COR Biosciences. The western blots were developed for chemiluminescence using the GE Healthcare ECL Plus kit or scanned for IRDye fluorescence using Odyssey System (LI-COR Biosciences).
Flow Cytometric analysis to assess MHC class I cell surface expression
A total of 1×105–1×106cells were washed with fluorescence-activated cell sorting (FACS) buffer (PBS [pH 7.4] containing 1% FBS) and then incubated with W6/32 antibody at 1:250 dilutions for 30–60 minutes on ice. Following this incubation, the cells were washed three times with FACS buffer and incubated with PE-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) at 1:250 dilutions for 30–60 minutes on ice. Following incubations, the cells were washed three times with FACS buffer and analyzed using a FACSCanto cytometer. The FACS data were analyzed with WinMDI software (J. Trotter, The Scripps Institute, Flow Cytometry Core Facility) or FlowJo software (Tree Star).
MHC class I refolding and gel filtration analysis
The bacterially expressed HLA-B constructs lacked signal sequences but contained an N-terminal MHHHHHHSSGVDLGTENLYFQSNA fusion sequence, including a histidine tag for nickel affinity chromatography and a tobacco etch virus (TEV) protease site for removal of the majority of the N-terminal tag (leaving a tripeptide sequence (SNA) prior to the start of the mature HLA-B sequences). All soluble HLA-B constructs were truncated prior to the transmembrane domain after amino acid 276. The HLA-B-pMCSG7 vectors were transformed into BL21 cells for protein preparation. Inclusion body preparations of selected HLA-B heavy chains and β2m were undertaken as previously described (36). The inclusion bodies were first solubilized in 6 M guanidine hydrochloride and diluted in 6 M guanidine hydrochloride to 200 or 25 µM stocks for β2m and heavy chains respectively. Refolding reactions were initiated with 0.45 ml refolding buffer (100 mM Tris pH 8.0, 400 mM L-arginine, 2mM EDTA, 0.5 mM oxidized glutathione, 5 mM reduced glutathione) combined in some experiments with 5–10 µg TEV protease. Heterodimers were typically refolded by sequential dilution of β2m (10 µl of the 200 µM stock) followed by heavy chains (40 µl of the 25 µM stock) into the refolding buffer with adequate stirring (using micro-stir bars), to final concentrations of 4 µM β2m and 2 µM heavy chains. The refolding reactions were conducted at room temperature for 1 hour or at 4°C overnight, in the absence or presence of TEV respectively as indicated. Following refolding, insoluble proteins were removed by centrifugation in a microfuge at 13,000 rpm and 4°C for 30 minutes. Refolded soluble proteins were analyzed by gel filtration chromatography on a FPLC system using Superdex 75 10/300 GL column (GE Healthcare). TEV-digested samples were incubated with nickel beads to remove uncleaved proteins, and the unbound fraction used for gel filtration analyses. Indicated gel filtration fractions (peaks 1 and 2) were pooled and tested for peptide binding. For the HLA-B7 supertype, pooled peak 1 and peak 2 fractions (10–20 nM) were directly used in binding assays. For the HLA-B44 supertype, peak 1 and peak 2 fractions were typically concentrated to 200–400 nM prior to use in binding assays, but similar results were obtained if protein was used at 10–20 nM in binding assays, without a prior concentration step. Protein fractions were incubated with 10 fold molar excess of purified β2m and peptide at 37°C for 2h or at room temperature for 24h. Following incubations, the samples were separated by 15% Native-PAGE, and gels were scanned using a Typhoon scanner (GE healthcare) for fluorimaging analyses.
Statistical analysis
For Figures 1, 2, 4 and 5, statistical analyses were performed using the GraphPad Prism 6 (GraphPad Software). The p values were calculated using the paired or unpaired Student’s t test or one-way analysis of variance with Tukey’s multiple comparison test. A p value of 0.05 was considered significant. Pearson correlation and linear regression analyses were also done using GraphPad Prism 6.
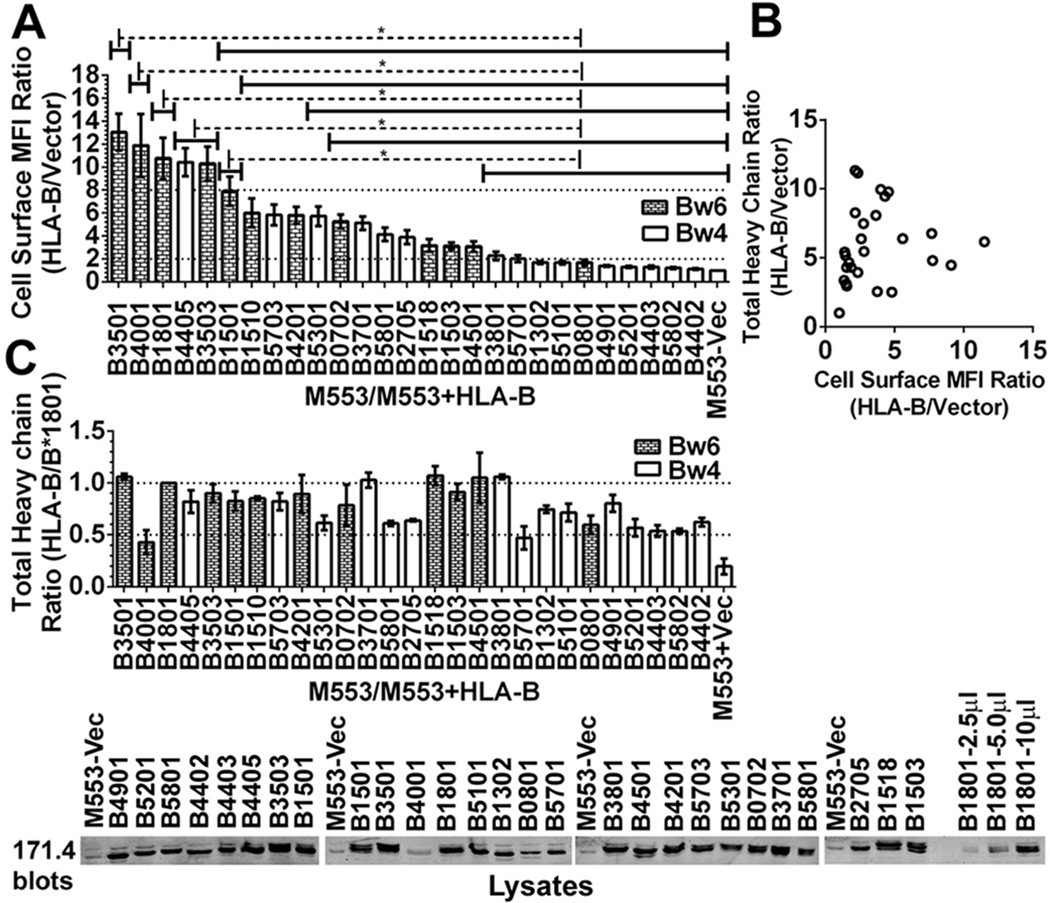
Figure 1. Variable expression of HLA-B molecules in tapasin-deficient cells.
A) The y-axes show cell surface expression ratios (mean fluorescence intensity (MFI ratios)) of MHC class I staining in M553 cells (a tapasin-deficient melanoma cell line) infected with retroviral constructs encoding the indicated HLA-B allotypes relative to M553 cells that were infected with a virus lacking MHC class I (vector). MHC class I surface expression was analyzed by flow cytometry using the W6/32 antibody. Statistical analyses were done using a one-way ANOVA test, followed by a Tukey’s multiple comparisons procedure for all pairwise differences of means. Significant differences are indicated (with an asterisk) on the graph (P< 0. 05). Data represents averaged MFI ratios derived from 10–15 independent flow cytometric analyses from five independent infections (infections 1–5) of M553 cells. B) In infection 5, a Pearson analysis was used to examine correlation between cell surface MFI ratios assessed by flow cytometry and total cellular expression ratios assessed by immunoblotting of cell lysates. A significant correlation was not observed. C) Top panel: Cell lysates from infection 5 of M553 cells were tested by quantitative fluorescence-based immunoblotting with the heavy chain-specific 171. 4 antibody. The bar graphs show total cellular expression ratios (relative to cells expressing HLA-B*1801) of the indicated HLA-B molecules. Averaged values from three independent sets of immunoblotting analyses of infection 5 are shown. Lower panel: Representative quantitative immunoblots with the 171. 4 antibody of indicated cell lysates from infection 5. 10 µl of total cell lysate was loaded in each lane, unless otherwise indicated. The last three lanes show HLA-B*1801 heavy chain expression in M553-HLA-B*1801 lysates (incremental loads of 2. 5–10 µl).
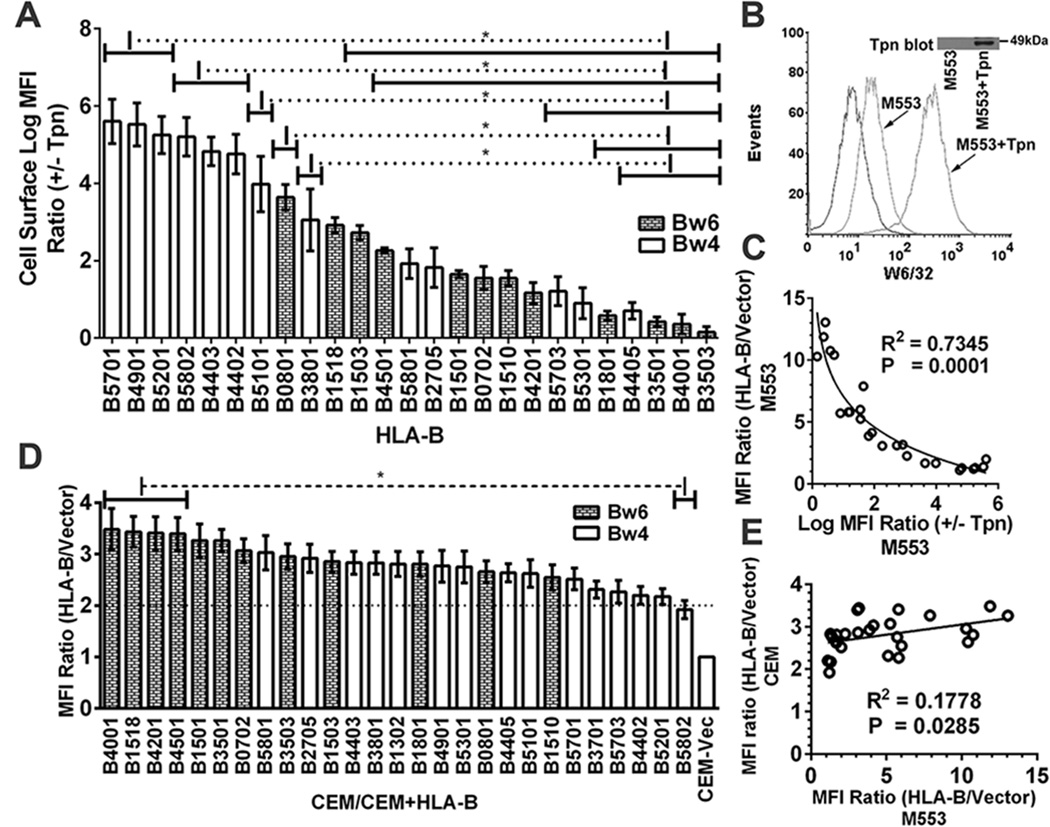
Figure 2. Reduced variability of HLA-B expression in tapasin-sufficient cells.
A) Cells from one of the infections described in Fig. 1A were subsequently infected with a tapasin-encoding virus. Following subtractions of the vector alone backgrounds the MFI ratios in the presence and absence of tapasin (+tapasin/−tapasin MFI ratios) were calculated for each HLA-B-expressing cell line. B) Representative histograms of M553 cell infections with a tapasin-encoding or control retrovirus. Flow cytometric analyses with the W6/32 antibody of cell surface expression of the endogenous MHC class I of M553 cells that were infected with retroviruses encoding or lacking tapasin. C) A Pearson analysis was used to examine correlation between averaged cell surface MFI ratios (HLA-B/vector) in M553 (Fig. 1A) vs. Log MFI (+Tapasin/−Tapasin) ratios (Fig. 2A). D) CEM cells (a CD4 T cell line) were infected with retroviruses encoding indicated HLA-B allotypes or a control virus lacking HLA-B (Vector). MHC class I cell surface expression was analyzed by flow cytometry using the W6/32 antibody, and HLA-B/vector MFI ratios were determined. E) A Pearson analysis was used to examine correlation between averaged MFI ratios (HLA-B/vector) in M553 and CEM cells. Data represent averaged (A and D) MFI values derived from 2–4 (A) or 10–12 (D) independent flow cytometric analyses from one (A) or three (D) independent infections. Statistical analyses (A and D) were done using a one-way ANOVA test, followed by a Tukey’s multiple comparisons procedure for all pairwise differences of means. Pearson R square (R2) and significantly different values (P) are indicated on the graph. P< 0. 05 is considered significant.
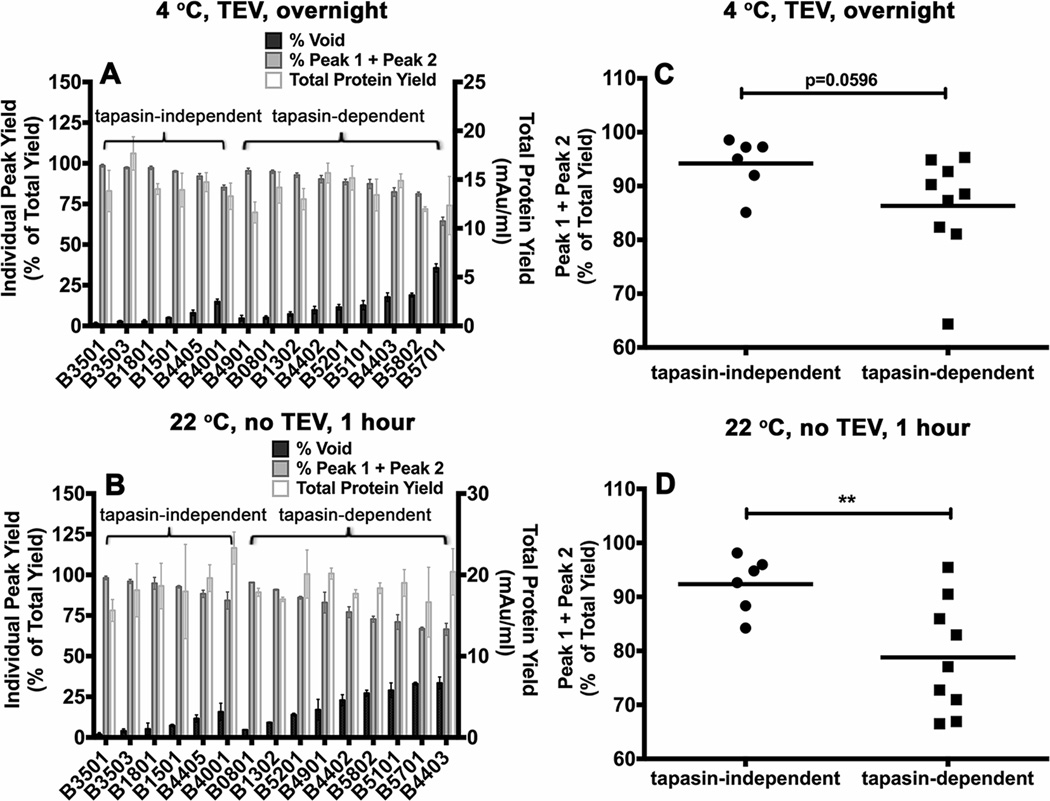
Figure 4. Variable levels of aggregation of soluble empty HLA-B molecules following refolding.
Soluble forms of the indicated MHC class I heavy chains were refolded with β2m A) overnight at 4 °C with TEV or B) at room temperature (22 ° C) for 1 hour without TEV. Following refolding, soluble proteins were analyzed by gel filtration chromatography. Percentages of fractions corresponding to the void volume and Peak1 + Peak 2 (peptide binding competent fractions) of different allotypes are shown (left Y axis) and total protein yields (right Y axis) are also shown. Data represent averaged values from three independent refolding reactions with two different inclusion body preparations for all allotypes except HLA-B*0801 and HLA-B*1302, which represent averaged values of two independent refolding reactions with a single inclusion body preparation. C and D) Percentages of Peak 1+ Peak 2 fractions, derived from data in A and B, were compared for the tapasin-independent and tapasin-dependent allotypes. Differences between two groups are significant when refolding is conducted under the more stringent conditions (1 hour at 22 °C). Statistical analyses were performed using an unpaired t-test. ** Indicates that the P value was < 0. 01.
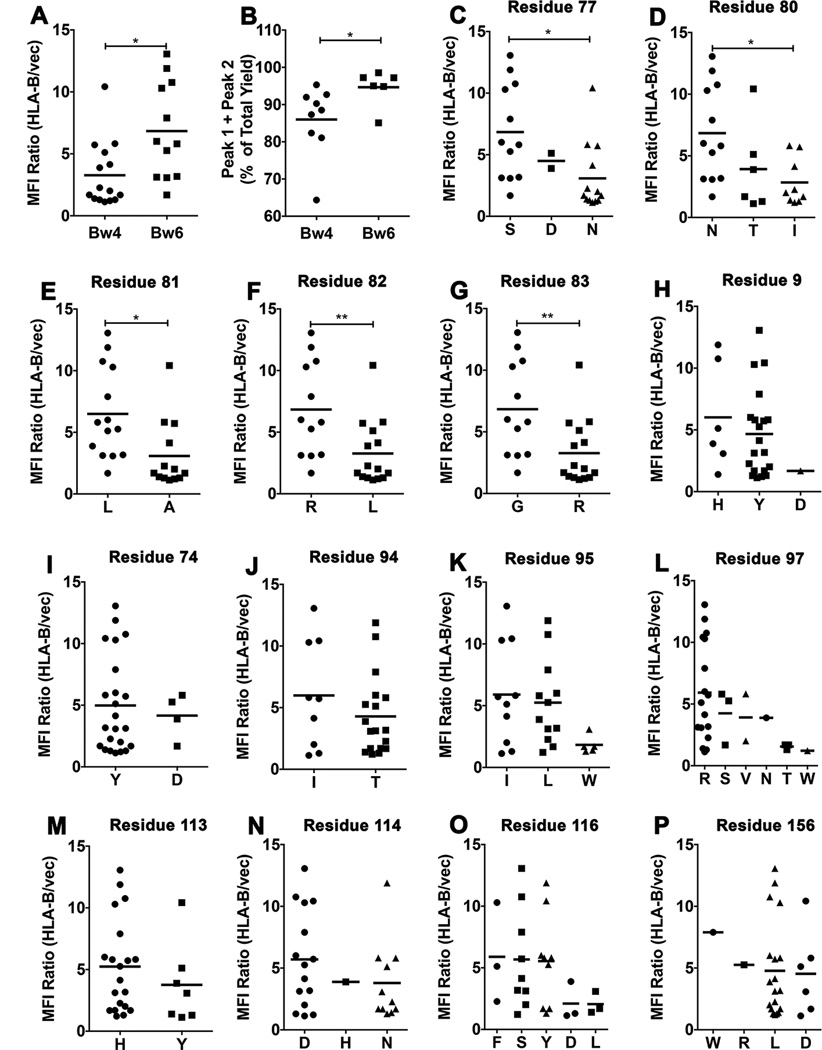
Figure 5. Residue contributions to tapasin-independent assembly.
A) Averaged MFI ratios derived in Fig. 1A were compared for HLA-Bw4 and HLA-Bw6 allotypes. Statistical analyses are based on an unpaired t-test. B) Peak 1+ Peak 2 yields, derived from the 4 ° C refolding data in Fig. 4A were compared for HLA-Bw4 and HLA-Bw6 allotypes. Statistical analyses are based on an unpaired t-test. More significant differences were observed for the 1 hour/room temperature refolding data. C–P) HLA-B allotypes were grouped based on the presence of dimorphic or polymorphic residues at the indicated heavy chain sequence positions. Averaged MFI ratios derived in Fig. 1A are compared for allotypes with a given amino acid residue at each of the indicated dimorphic or polymorphic sites. Statistical differences between groups were assessed based on unpaired t-tests for dimorphic residues. For polymorphic residues, a one-way ANOVA test was used, with a Tukey’s multiple comparisons procedure for all pairwise differences of means. Statistically significant differences are indicated by asterisks on the graphs (* indicates P< 0. 05; ** indicates P<0. 01).
AIDS Progression studies
496 HIV-1–infected subjects for whom the dates of seroconversion were known were derived from three cohorts: the Multicenter AIDS Cohort Study (MACS) (37), the Multicenter Hemophilia Cohort Study (MHCS) (38), and the DC gay cohort study (DCG) (39). SAS (vision 9.2, SAS Institute) procedure PROC PHREG was used for Cox proportional hazards regression analyses. Four AIDS-related outcomes were considered in the regression models: a CD4+ T-cell count of <200 cells/mm3, progression to AIDS according to the 1993 definition of the US Centers for Disease Control (CDC), progression to AIDS according to the more stringent 1987 CDC definition, and death from an AIDS-related cause. Statistical significance refers to two-sided P values of <0.05. PROC PHREG with STEPWISE selection was performed with including mean fluorescence intensity values as a continuous variable and presence versus absence of all individual HLA-B alleles with ≥ 3% frequency in the Cox model. The significance level for selecting a variable to stay in the model was P<0.05.
Results
Tapasin dependencies of HLA-B assembly
Using immortalized B cell lines from HLA typed donors, cDNAs were generated for several of the most frequent HLA-B allotypes observed in North American populations. The sequences were cloned into retroviral vectors, verified, and the different HLA-B alleles were expressed in a tapasin deficient human melanoma cell line (M553) (32). Cell surface expression of HLA-B allotypes was analyzed by flow cytometry (Fig. 1A). HLA-B allotypes showed large variations in cell surface expression in M553 cells (Fig. 1A). Cell surface expression of HLA-B*3501, B*4001, B*1801, B*4405, B*3503, and B*1501 was high, and about 7–15 fold higher than the cell surface expression of the endogenous MHC class I of M553 cells (Fig. 1A), which are HLA-A*28, HLA-B*5001 and HLA-B*5701 (32). Cell surface expression of B*3801, B*5701, B*1302, B*5101, B*0801, B*4901, B*5201, B*4403, B*5802 and B*4402 was very low, and on average either undetectable or less than two-fold above endogenous MHC class I cell surface expression (Fig. 1A). Other HLA-B allotypes, such as HLA-B*1510, B*5703, B*4201, B*5301, B*0702, B*3701, B*5801, B*2705, B*1518, B*1503 and B*4501 showed intermediate phenotypes, which were 2.5–8 fold higher than endogenous MHC class I cell surface expression (Fig. 1A). The levels of HLA-B allotype expression shown in Figure 1 are averaged from five independently infected sets of cell lines. Thus, the observed effects are not due to differences in individual retroviral vector preparations.
In general, there was poor correlation between extracellular MHC class I expression assessed by flow cytometry (Fig. 1A) and total cellular expression assessed by quantitative immunoblotting analyses for HLA class I heavy chains (Fig. 1B). The latter analyses indicated that the intracellular expression of the majority of HLA-B alleles were within two fold of each other and of HLA-B*1801 (one of the higher expressing HLA-B allotypes) (Fig. 1C). Serial dilutions of HLA-B*1801-expressing cell lysates (Fig. 1C, last three lanes on the right) verified that the protein loads used were within the linear ranges of the quantitative immunoblotting assays. Some tapasin-dependent HLA-B allotypes consistently displayed lower total cellular expression compared to tapasin-independent allotypes (for example, HLA-B*4403 and HLA-B*4402 compared to HLA-B*4405 (representative data are shown in Fig. 1C)), suggesting the possibility of enhanced ER-associated degradation (ERAD) of some HLA-B allotypes.
We next examined cell surface expression of MHC class I molecules in each of the HLA-B-expressing M553 cells following further infections with a tapasin-encoding virus (using a separate retroviral vector with a different drug selection vehicle) (Fig. 2A). Similar levels of tapasin were expressed in most cell lines as assessed by immunblotting analyses (data not shown). Expression of tapasin strongly induced cell surface expression of endogenous MHC class I of M553 cells (>20 fold; Fig. 2B). The MFI ratios (+tapasin/−tapasin) were compared for the different HLA-B-expressing cell lines, after subtracting background signals from the corresponding M553 cells lacking exogenous HLA-B expression (Fig. 2A). There was a strong inverse correlation between low cell surface expression under tapasin-deficient conditions and the extent of tapasin-mediated induction (Fig. 2C).
Major differences were observed upon expression of HLA-B allotypes in tapasin-deficient M553 cells (Fig. 1A); however, such differences were not observed following expression of the HLA-B allotypes in other cells such as a tapasin-expressing CD4 T cell line, CEM (Fig. 2D). Although a positive correlation between cell surface expression in M553 cells and CEM cells was noted (Fig. 2E), the extent of variation in HLA-B cell surface expression was much greater in M553 cells compared to that observed in CEM cells (Fig. 2D compared to 1A). Together, the findings of Figs. 1 and 2 indicated that tapasin-deficiency limits expression of some HLA-B molecules in M553 cells.
The overall tapasin-independence/dependence profiles established with untagged HLA-B molecules (Fig. 1A and 2A) are similar to those previously obtained with influenza hemagglutinin-tagged versions of HLA-B*3501, HLA-B*3503, HLA-B*4402, HLA-B*4405 and HLA-B*5701 (30). Additionally, the stronger tapasin dependence of HLA-B*4402 compared to HLA-B*4405 and HLA-B*2705 is consistent with previous findings (25, 27). The strong tapasin dependence of HLA-B*0801, HLA-B*4402, HLA-B*4403, HLA-B*5101 and HLA-B*5701, and the strong tapasin-independence of HLA-B*3501 is also consistent with previous findings (32, 40, 41). HLA-B molecules are grouped into one of two serotypes, HLA-Bw4 and HLA-Bw6, which differ at residues 77–83 of the heavy chain α1 domain (42). Only HLA-Bw4 molecules engage the three domain receptors (KIR3D) of NK cells (reviewed in (2)). It is noteworthy that some of the most tapasin-dependent allotypes are of the Bw4 serotype, whereas several of the most highly tapasin-independent allotypes are of the Bw6 serotype (Fig. 1A).
Intrinsic assembly and refolding efficiencies of HLA-B molecules
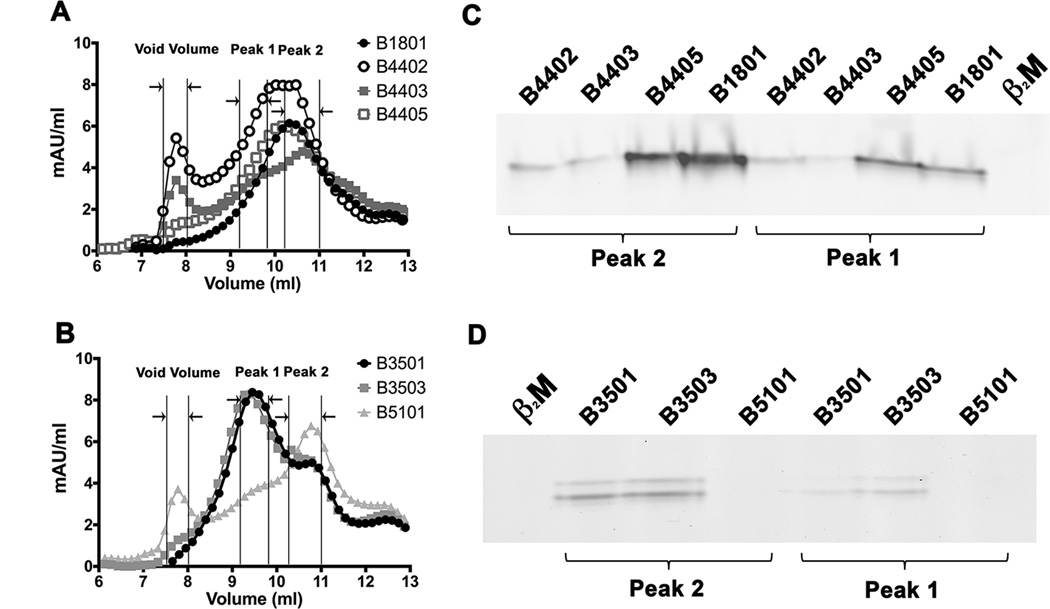
We further examined whether tapasin-independent assembly reflects the intrinsic abilities of HLA-B molecules to assemble with peptides or the conformational stabilities of the refolded proteins. We analyzed assembly profiles of HLA-B allotypes at the extreme ends of tapasin-independence (Figure 1A), following expression of soluble versions of the heavy chains and β2m in. E. coli. Purified inclusion bodies of heavy chain and β2m were refolded overnight at 4 °C in the absence of peptides and in the presence of the TEV protease to remove the N-terminal hexahistidine epitope tag. Recoveries of soluble heterodimeric or aggregated proteins were compared by gel filtration chromatography on a Superdex 75 10/300 GL column, and the assembly competencies of the refolded proteins were also compared using fluorescent peptides (Fig. 3). Three heavy chain-containing peaks were generally observed (Figs. 3A and 3B). The first peak corresponds to the column void volume (>100 KDa). The second peak (peak 1) corresponds to the elution volume for bovine serum albumin (67 kDa). The third peak (peak 2), is close to the elution volume for a folded HLA-A2 heterodimer purified from insect cell supernatants. HLA class I molecules belonging to a “supertype” share similarities in peptide binding (43). We compared peptide binding by HLA-B molecules with low or high tapasin-independence that belong to the B44 and B7 supertypes. Within the B44 supertype, HLA-B*1801 and HLA-B*4402 share similar peptide binding specificities at the P2 and P9 anchor residues (44). Glutamic acid at P2 is a primary anchor residue for both B*1801 and HLA-B*4402, and phenylalanine at P9 is a dominant secondary anchor residue for both. HLA-B*4402 is also closely related to HLA-B*4405 and HLA-B*4403, differing only at positions 156 or 116 respectively. EEFGRAFSF is a dominant self-peptide that was previously eluted from HLA-B*4402, HLA-B*4403 and HLA-B*4405 (45). We used a fluorescently-modified version of EEFGRAFSF (EEFGKFITCAFSF), to compare peptide binding by the refolded allotypes, following normalizations of the relative protein concentrations and the addition of excess β2m. These binding studies revealed that the refolded forms of allotypes with high tapasin-independence (HLA-B*1801 and HLA-B*4405) bound more efficiently to the peptide than those with low tapasin-independence (HLA-B*4402 and HLA-B*4403) (Fig. 3C). Within the B7 supertype, the HLA-B*3501 and HLA-B*3503 allotypes share similarities with each other and with HLA-B*5101 (which is strongly tapasin-dependent for its assembly) in peptide binding, with common P2 and some common P9 anchor residues (46–49). The HIV-1 Pol-derived peptide IPLTEEAEL elicits cytotoxic T cell responses in the context of both HLA-B*3501 and HLA-B*5101(50). We used a modified version of IPLTEEAEL (IPLKFITCEEAEL) to compare peptide binding by the refolded HLA-B35 and HLA-B*5101 allotypes. Again the highly tapasin-independent HLA-B35 allotypes bound to the peptide more efficiently, compared to HLA-B*5101 (Figure 3D).
Figure 3. Higher assembly competence of soluble refolded tapasin-independent HLA-B allotypes.
A soluble form of the indicated HLA class I heavy chains (2 µM) was refolded with β2m (4 µM) overnight at 4 °C in the presence of TEV. Following refolding, the soluble fractions were analyzed by gel filtration chromatography using a Superdex 75 10/300 GL column. Representative gel filtration analyses of A) B1801, B4402, B4403 and B4405 (B44 supertype) and B) B3501, B3503 and B5101 (B7 supertype) are shown. The vertical lines show peak position ranges of the gel filtration column void volume (left), Peak 1 (middle) and Peak 2 (right). C and D) Peak 1 and Peak 2 fractions from A and B were assessed for their peptide binding competence. Peak 1 and Peak 2 fractions were normalized for protein concentration (to 200 nM (Peak 1) or 400 nM (peak 2) for B44 supertypes, and 20 nM (peaks 1 and 2) for B7 supertypes) and incubated with a 10-fold excess of refolded β2m and FITC labeled peptide (peptide EEFGKFITCAFSF for B44 supertypes and peptide IPLKFITCEEAEL for B7 supertypes). Peptide bound heterodimers were separated by native gel electrophoresis and visualized by fluorescence scanning. Both Peak 1 and Peak 2 fractions are competent for peptide binding. Representative data of four independent measurements are shown.
For the tested HLA-B allotypes, both the peak 1 and 2 fractions were competent for binding fluorescent peptides in the presence of excess β2m, with slightly reduced assembly competence for peak 1 compared to peak 2, as assessed by native-PAGE gels and fluorimaging analyses (Figs. 3C and 3D). In comparison, the protein fraction recovered in the void volume was poorly assembly competent (data not shown). Comparisons of the refolding patterns of the most tapasin-dependent and tapasin-independent allotypes revealed a hierarchy of refolding efficiencies (Figs. 4A and 4B). Although not absolute, there was a trend towards increased recovery of assembly-competent (peak 1 + peak 2) fractions for the peptide-deficient forms of tapasin-independent allotypes compared to their tapasin-dependent counterparts (Figs. 4C and 4D). Compared to overnight refolding at 4 °C, refolding differences between tapasin-dependent and tapasin-independent allotypes were more pronounced following room temperature refolding for 1 hour, which generally increased the level of aggregation (Figs. 4C and 4D; TEV was not included in the 1 hour refolding reactions, as an overnight reaction is recommended for completion of TEV digestions). Together, the findings of Figs. 3 and 4 are consistent with the model of decreased conformational heterogeneity of peptide-deficient forms of several tapasin-independent allotypes compared to their tapasin-dependent counterparts, which results in greater assembly competence and reduced aggregation of the peptide-deficient forms of tapasin-independent allotypes.
Determinants of tapasin-independent assembly
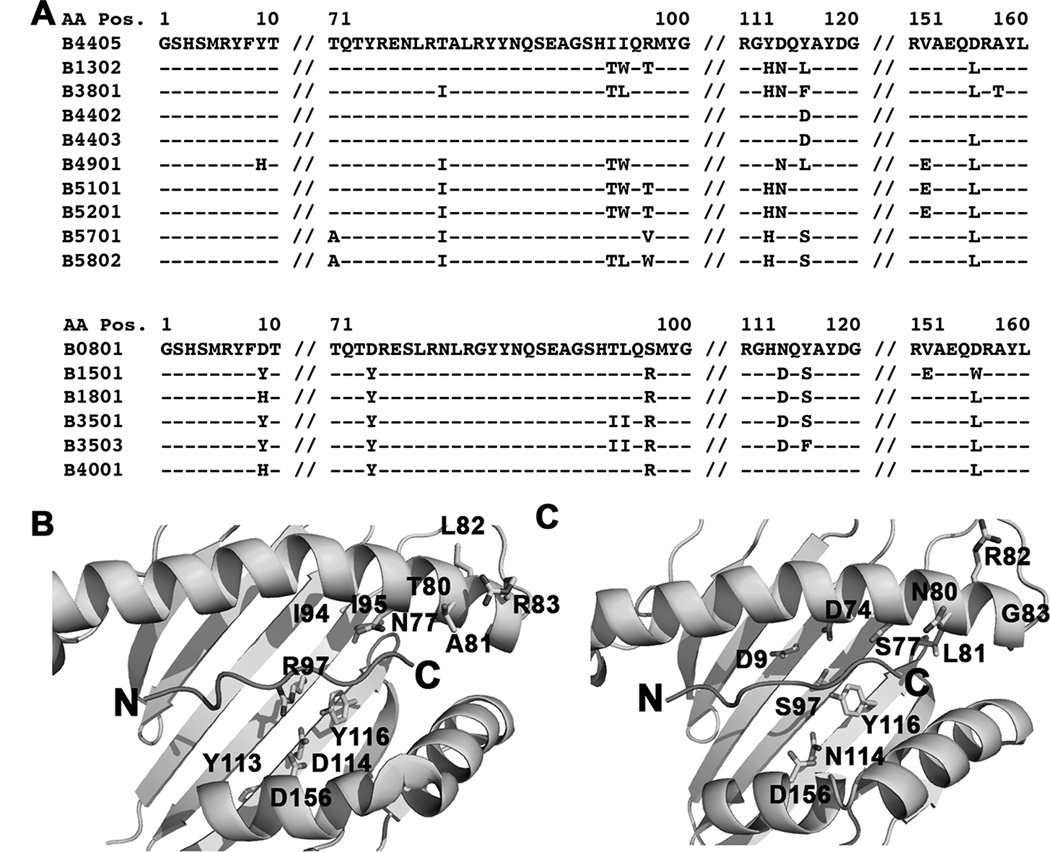
The Bw4/6 antibody epitope is contained within residues 77–83 of the HLA-B heavy chains, and is a key determinant of NK cell inhibitory receptor engagement by HLA-B molecules (51). Expression studies in M553 cells indicated that the most strongly tapasin-independent allotypes were of the HLA-Bw6 serotype (with the exception of HLA-B*4405), whereas allotypes that were poorly expressed in M553 cells were of the HLA-Bw4 serotype (with the exception of HLA-B*0801) (Fig. 1A). HLA-Bw4. vs. HLA-Bw6 groups are significantly different for tapasin-independent assembly (Fig. 5A). Additionally, in the refolding analyses, the yields of the folding competent fractions are significantly different for Bw6 epitopes compared to Bw4 epitopes (Fig. 5B), although a hierarchy of assembly efficiencies is also observed within each group (Fig. 4). Together, these findings suggested that the region corresponding to the Bw4/6 epitopes, a known determinant of NK recognition (51), is also a key determinant of the assembly and stability characteristics of HLA-B molecules. To further examine individual residue contributions to tapasin-independent assembly, HLA-B allotypes were grouped based on dimorphic or polymorphic residue distributions at each position of their extracellular domain sequences, and averaged MFI ratios derived in Figure 1A were compared between groups. A total of 47 dimorphic or polymorphic HLA-B residues were compared (corresponding to positions 9, 11, 12, 24, 30, 32, 41, 45, 46, 62, 63, 65, 66, 67, 69, 70, 71, 74, 77, 80, 81, 82, 83, 94, 95, 97, 99, 103, 113, 114, 116, 131, 143, 145, 147, 152, 156, 158, 163, 167, 171, 177, 178, 180, 194, 199 and 282 of the heavy chain sequences). Statistically significant averaged MFI differences were observed for polymorphisms corresponding to positions 77 (S vs. N), 80 (N vs. I), 81 (L vs. A), 82 (R vs. L) and 83 (G vs. R) (Fig. 5C–5H), residues define the Bw4/Bw6 epitope differences.
Despite significant influences of residues within the HLA-Bw4/Bw6 region upon tapasin-independent assembly (Figures 5C–5H), these residues alone are insufficient to fully determine tapasin-independence. Within the HLA-Bw4 group, N77A81L82R83 is present within the most tapasin-dependent HLA-Bw4 allotypes, but also in the strongly tapasin-independent HLA-B*4405 (Fig. 6A). Residue 116 is the only site of difference between HLA-B*4405 and HLA-B*4402 (D in HLA-B*4402 and Y in HLA-B*4405). In HLA-B*4402 and HLA-B*4405, residue 116 is involved in a network of interactions within the F-pocket of the peptide-binding groove, with residues 114, 156 and 97 and with the side chain of the carboxy-terminal peptide (P9) residue (28, 52) (Fig. 6B). Residues 114, 116, 156 and 97, as well as residues 80 (which interacts with the peptide carboxy-terminus), 95 (which bounds the wall of the F-pocket), 94 and 113 (adjacent to 94 and 114 respectively), are sites of variation between HLA-B*4405 and several strongly tapasin-dependent HLA-Bw4 molecules (Figs. 6A and 6B). Within the Bw6 group, S77N80L81R82G83 is present in the most tapasin-independent HLA-Bw6 allotypes as well as in HLA-B*0801, which is strongly tapasin-dependent (Fig. 6A). Similar to the HLA-Bw4 group, amino acids within/near the F-pocket region may constitute the determinants of assembly differences between HLA-B*0801 and tapasin-independent HLA-Bw6 molecules, including residues 9, 74, 97, 114, 116 and 156 (Figs. 6A and 6C). Although residues variations at positions 9, 74, 94, 95, 97, 113, 114, 116 and 156 do not result in significant differences in tapasin independencies between the tested HLA-B allotypes, (Figs. 5H–5P), these residues are expected to influence the structure and conformation of the peptide binding groove in the vicinity of the F-pocket, and thus also determine the nature of interactions with the peptide C-terminus.
Figure 6. Determinants of tapasin-independent assembly.
A) Sequence alignments of HLA-B*4405 with tapasindependent HLA-Bw4 molecules and of HLA-B*0801 with tapasin-independent HLA-Bw6 molecules. B and C) Based on the sequence alignments, residues expected to contribute to tapasin-dependent and tapasin-independent HLA-B assembly are highlighted on the structures of HLA-B*4405 (pdb 1SYV) and HLA-B*0801 (pdb 1AGD).
Influences of tapasin-independent assembly on progression to various AIDS outcomes
We examined whether tapasin-dependent or independent assembly influences progression to AIDS and survival in HIV infections. Individuals were assigned tapasin-independence scores based on their HLA-B genotypes. The MFI ratio in M553 cells (Fig. 1A) was used to compute mean tapasin-independent expression of the two relevant HLA-B allotypes in each individual. Cox models were used to examine the influences of mean tapasin-independent expression as a continuous variable upon progression to three AIDS outcomes in Caucasian subjects infected with HIV. Significant effects of MFI values on progression to different AIDS outcomes were observed (Death, N=496, p= 0.001, hazard ratio (HR) = 1.11; AIDS87, N=496, p=0.02, HR=1.06; CD4<200 and AIDS 93, N=458, p=0.02, HR=1.06), where the HR represents an increase in 1 MFI unit. Thus, the difference between homozygotes for the most extreme tapasin independent and dependent alleles, B*3501 and B*4402, is 11.94 units with HR = 2.0. When all HLA class I alleles with frequencies ≥ 3% were included in the model, the effect of MFI values upon progression to death remained significant (N=496; p= 0.001; HR=1.12). These data suggest that greater tapasin-independent assembly confers more rapid progression and that the effect of tapasin-independent assembly is primarily a late effect in AIDS progression.
Discussion
Low and high tapasin-independence were prevalent among 37% (ten allotypes) and 22% (six allotypes), respectively (Fig. 1A), of the 27 HLA-B allotypes considered in this study. Based on the extent of assembly (Fig. 3) or the extent of aggregation of soluble refolded proteins (Fig. 4), tapasin-independent allotypes were found to be more assembly competent and more stable in their empty forms compared to tapasin-dependent allotypes. Higher intrinsic assembly competence of tapasin-independent allotypes is consistent with findings from other recent in vitro refolding/assembly studies (41), and with previous findings of more rapid intracellular trafficking of tapasin-independent allotypes compared to their tapasin-dependent counterparts belonging to the same supertype (for example HLA-B*4405 compared to HLA-B*4402 and HLA-B*4403, and HLA-B*3501 compared to HLA-B*5101 (28–30, 53, 54)). These findings support the view that peptide-binding grooves of tapasin-independent HLA-B allotypes are in a more stable peptide-receptive conformation compared to tapasin-dependent HLA-B molecules. Molecular dynamics simulation studies have suggested significant differences in the dynamics of the F-pocket regions of HLA-B*4402 and HLA-B*4405, with greater differences between the peptide-bound and peptide-free forms of HLA-B*4402 compared to those of HLA-B*4405 (55). Thus, differences in the structure and flexibility of MHC class I molecules in the absence of bound peptide may play a key role in determining the requirement for tapasin during peptide loading.
The studies described here point to several residues in the vicinity of the C-terminal end of the peptide, including those comprising Bw4/Bw6 epitope differences, as key determinants of tapasin-independent assembly (Fig. 5 and Fig. 6). Consistent with these findings, tapasin-mediated peptide selection is suggested to disrupt hydrogen bonds near the peptide C-terminus (24), and peptides bound to MHC class I molecules of tapasin-deficient cells are elongated and contain a broader set of C-terminal residues compared to those derived from wild type cells (56). Near the peptide C-terminus, polymorphic Bw4/Bw6 determinants 77 and 80 and invariant residue 84 interact with the peptide main chain (for example, (28, 52, 57)). Residue 80 form hydrogen bonds with the peptide C-terminal carboxyl group. Additionally, based on previous structural comparisons (57), S77 could allow for bulky/hydrophobic P9 side chains to be more readily accommodated within the F-pocket compared to N77, and polymorphic residue 81 (L vs. A) could alter the nature of interactions with the peptide P9 residue. Furthermore, residues 82 and 83 could determine the overall conformational stability of interactions mediated by residues 77, 80, 81 and 84 with the peptide main chain and side chain. The combination of interactions with the peptide C-terminus could intrinsically be more favorable for tapasin-independent allotypes. Additionally, the significant Bw4/Bw6 differences in tapasin-independence suggest that tapasin is particularly critical for guiding the expression or peptide repertoires of HLA-B molecules that are recognized by NK receptors.
Our findings show that HLA-B molecules vary in their requirements for tapasin for their assembly (Fig. 1A), and additionally that refolded empty HLA-B molecules have distinct assembly competencies and patterns of stability of their empty forms (Fig. 3 and Fig. 4). Such differences can influence the stabilities of antigenic peptide associations with HLA-B molecules, as well as interactions with ER quality control factors, and thus the abilities of different HLA-B molecules to mediate immune responses during infections. Our findings indicate that the presence of tapasin-independent allotypes links to a greater hazard ratio for death following HIV infections. Consistent with these findings, allotypes such as HLA-B*5701 and HLA-B*2705 that are associated with slow progression to AIDS (reviewed in (3, 4)) display low or intermediate tapasin-independence (Figure 1A). Some studies also suggest protective effects of HLA-Bw4 homozygosity in HIV infections (58), and low tapasin-independence is strongly prevalent within the HLA-Bw4 serotype (Figs. 1 and 5). Conversely, some allotypes such as HLA-B*3503 and HLA-B*3501 that are associated with more rapid AIDS progression (4) are highly tapasin-independent for their assembly. Tapasin is suggested to function as a peptide editor, facilitating MHC class I occupancy with high affinity peptide (19, 24–26). MHC class I molecules loaded with a slow-dissociating peptide are expected to be more stable at the cell surface and thus may be able to present antigens to the CD8+ T cells over longer durations. In contrast, the tapasin-independent allotypes may be loaded with fast-dissociating sub-optimal peptides. Sub-optimal epitope selection could result in more transient and sub-optimal antigen presentation to the CD8+ T cells. Although more detailed mechanistic studies are needed to understand the cellular and molecular basis for the differences in disease progression, these findings suggest that assembly characteristics of HLA-B molecules can and do influence survival outcomes following infections. While the presence of tapasin-independent HLA-B molecules enhances progression to death in HIV infections, it is possible that tapasin-independent assembly confers advantages in other infectious contexts, particular those that interfere with MHC class I assembly in the ER. Further studies are needed to better understand the influences of HLA-B assembly/stability characteristics upon disease outcomes in other disease contexts and upon global CD8+ T cell responses during infections.
Acknowledgments
We thank Dr. Naveen Bangia for the M553 cell line, Dr. Kathleen Collins for BOSC cells and retroviral protocols, Dr. Ted Hansen for an antibody directed against the tapasin N-terminus, and indicated laboratories for HLA-B cDNA. We are grateful to Dr. Clay Brown (University of Michigan High Throughput Protein Laboratory) for assistance with the generation of HLA-B-encoding retroviral constructs, to Dr. James Goedert for the MHCS and DCG data, to Dr. Sanjeeva Wijeyesakere for assistance with and Figure 4 and Sukhmani Bedi for her contributions to the project. We thank the University of Michigan DNA sequencing core for sequencing analyses, and the University of Michigan hybridoma core for antibody production. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U. S. Government.
This work was funded by a NIH grant AI044115 (to MR). This work was supported in part by the University of Michigan Medical School Protein Folding Disease Initiative. The work utilized the DNA sequencing and the Hybridoma core of the University of Michigan. This research was also supported in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E and by the Intramural Research Program of the NIH, Frederick National Laboratory, Center for Cancer Research. Some data in this manuscript were collected by the Multicenter AIDS Cohort Study (MACS) with centers (Principal Investigators) at: Johns Hopkins University Bloomberg School of Public Health (Joseph Margolick), U01-AI35042; Northwestern University (Steven Wolinsky), U01-AI35039; University of California, Los Angeles (Roger Detels), U01-AI35040; University of Pittsburgh (Charles Rinaldo), U01-AI35041; the Center for Analysis and Management of MACS, Johns Hopkins University Bloomberg School of Public Health (Lisa Jacobson), UM1-AI35043. The MACS is funded primarily by the National Institute of Allergy and Infectious Diseases (NIAID), with additional co-funding from the National Cancer Institute (NCI).
Abbreviations
- β2m
β2-microglobulin
- ER
Endoplasmic reticulum
- PLC
Peptide loading complex
- TAP
Transporter associated with antigen processing
- ERAD
ER-associated degradation
- HR
Hazard ratio
- MFI
Mean fluorescence intensity
References
- 1.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 2.Campbell KS, Purdy AK. Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology. 2011;132:315–325. doi: 10.1111/j.1365-2567.2010.03398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrington M, Walker BD. Immunogenetics of spontaneous control of HIV. Annu Rev Med. 2012;63:131–145. doi: 10.1146/annurev-med-062909-130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goulder PJ, Walker BD. HIV and HLA class I: an evolving relationship. Immunity. 2012;37:426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jamil KM, Khakoo SI. KIR/HLA interactions and pathogen immunity. J Biomed Biotechnol. 2011;2011:298348. doi: 10.1155/2011/298348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reveille JD. Genetics of spondyloarthritis--beyond the MHC. Nat Rev Rheumatol. 2012;8:296–304. doi: 10.1038/nrrheum.2012.41. [DOI] [PubMed] [Google Scholar]
- 7.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O'Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, Allen TM, Altfeld M, Henn MR, Le Gall S, Streeck H, Haas DW, Kuritzkes DR, Robbins GK, Shafer RW, Gulick RM, Shikuma CM, Haubrich R, Riddler S, Sax PE, Daar ES, Ribaudo HJ, Agan B, Agarwal S, Ahern RL, Allen BL, Altidor S, Altschuler EL, Ambardar S, Anastos K, Anderson B, Anderson V, Andrady U, Antoniskis D, Bangsberg D, Barbaro D, Barrie W, Bartczak J, Barton S, Basden P, Basgoz N, Bazner S, Bellos NC, Benson AM, Berger J, Bernard NF, Bernard AM, Birch C, Bodner SJ, Bolan RK, Boudreaux ET, Bradley M, Braun JF, Brndjar JE, Brown SJ, Brown K, Brown ST, Burack J, Bush LM, Cafaro V, Campbell O, Campbell J, Carlson RH, Carmichael JK, Casey KK, Cavacuiti C, Celestin G, Chambers ST, Chez N, Chirch LM, Cimoch PJ, Cohen D, Cohn LE, Conway B, Cooper DA, Cornelson B, Cox DT, Cristofano MV, Cuchural G, Jr, Czartoski JL, Dahman JM, Daly JS, Davis BT, Davis K, Davod SM, DeJesus E, Dietz CA, Dunham E, Dunn ME, Ellerin TB, Eron JJ, Fangman JJ, Farel CE, Ferlazzo H, Fidler S, Fleenor-Ford A, Frankel R, Freedberg KA, French NK, Fuchs JD, Fuller JD, Gaberman J, Gallant JE, Gandhi RT, Garcia E, Garmon D, Gathe JC, Jr, Gaultier CR, Gebre W, Gilman FD, Gilson I, Goepfert PA, Gottlieb MS, Goulston C, Groger RK, Gurley TD, Haber S, Hardwicke R, Hardy WD, Harrigan PR, Hawkins TN, Heath S, Hecht FM, Henry WK, Hladek M, Hoffman RP, Horton JM, Hsu RK, Huhn GD, Hunt P, Hupert MJ, Illeman ML, Jaeger H, Jellinger RM, John M, Johnson JA, Johnson KL, Johnson H, Johnson K, Joly J, Jordan WC, Kauffman CA, Khanlou H, Killian RK, Kim AY, Kim DD, Kinder CA, Kirchner JT, Kogelman L, Kojic EM, Korthuis PT, Kurisu W, Kwon DS, LaMar M, Lampiris H, Lanzafame M, Lederman MM, Lee DM, Lee JM, Lee MJ, Lee ET, Lemoine J, Levy JA, Llibre JM, Liguori MA, Little SJ, Liu AY, Lopez AJ, Loutfy MR, Loy D, Mohammed DY, Man A, Mansour MK, Marconi VC, Markowitz M, Marques R, Martin JN, Martin HL, Jr, Mayer KH, McElrath MJ, McGhee TA, McGovern BH, McGowan K, McIntyre D, McLeod GX, Menezes P, Mesa G, Metroka CE, Meyer-Olson D, Miller AO, Montgomery K, Mounzer KC, Nagami EH, Nagin I, Nahass RG, Nelson MO, Nielsen C, Norene DL, O'Connor DH, Ojikutu BO, Okulicz J, Oladehin OO, Oldfield EC, 3rd, Olender SA, Ostrowski M, Owen WF, Jr, Pae E, Parsonnet J, Pavlatos AM, Perlmutter AM, Pierce MN, Pincus JM, Pisani L, Price LJ, Proia L, Prokesch RC, Pujet HC, Ramgopal M, Rathod A, Rausch M, Ravishankar J, Rhame FS, Richards CS, Richman DD, Rodes B, Rodriguez M, Rose RC, 3rd, Rosenberg ES, Rosenthal D, Ross PE, Rubin DS, Rumbaugh E, Saenz L, Salvaggio MR, Sanchez WC, Sanjana VM, Santiago S, Schmidt W, Schuitemaker H, Sestak PM, Shalit P, Shay W, Shirvani VN, Silebi VI, Sizemore JM, Jr, Skolnik PR, Sokol-Anderson M, Sosman JM, Stabile P, Stapleton JT, Starrett S, Stein F, Stellbrink HJ, Sterman FL, Stone VE, Stone DR, Tambussi G, Taplitz RA, Tedaldi EM, Theisen W, Torres R, Tosiello L, Tremblay C, Tribble MA, Trinh PD, Tsao A, Ueda P, Vaccaro A, Valadas E, Vanig TJ, Vecino I, Vega VM, Veikley W, Wade BH, Walworth C, Wanidworanun C, Ward DJ, Warner DA, Weber RD, Webster D, Weis S, Wheeler DA, White DJ, Wilkins E, Winston A, Wlodaver CG, van't Wout A, Wright DP, Yang OO, Yurdin DL, Zabukovic BW, Zachary KC, Zeeman B, Zhao M. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, Luca ADe, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ljunggren HG, Stam NJ, Ohlen C, Neefjes JJ, Hoglund P, Heemels MT, Bastin J, Schumacher TN, Townsend A, Karre K, et al. Empty MHC class I molecules come out in the cold. Nature. 1990;346:476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- 10.Blum JS, Wearsch PA, Cresswell P. Pathways of Antigen Processing. Annu Rev Immunol. 2013 doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abele R, Tampe R. The TAP translocation machinery in adaptive immunity and viral escape mechanisms. Essays Biochem. 2011;50:249–264. doi: 10.1042/bse0500249. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Sjogren HO, Hellman U, Pettersson RF, Wang P. Cloning and functional characterization of a subunit of the transporter associated with antigen processing. Proc Natl Acad Sci U S A. 1997;94:8708–8713. doi: 10.1073/pnas.94.16.8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ortmann B, Copeman J, Lehner PJ, Sadasivan B, Herberg JA, Grandea AG, Riddell SR, Tampe R, Spies T, Trowsdale J, Cresswell P. A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science. 1997;277:1306–1309. doi: 10.1126/science.277.5330.1306. [DOI] [PubMed] [Google Scholar]
- 14.Dong G, Wearsch PA, Peaper DR, Cresswell P, Reinisch KM. Insights into MHC class I peptide loading from the structure of the tapasin-ERp57 thiol oxidoreductase heterodimer. Immunity. 2009;30:21–32. doi: 10.1016/j.immuni.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peaper DR, Wearsch PA, Cresswell P. Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex. Embo J. 2005;24:3613–3623. doi: 10.1038/sj.emboj.7600814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Cid N, Jeffery E, Rizvi SM, Stamper E, Peters LR, Brown WC, Provoda C, Raghavan M. Modes of calreticulin recruitment to the major histocompatibility complex class I assembly pathway. J Biol Chem. 2010;285:4520–4535. doi: 10.1074/jbc.M109.085407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rizvi SM, Cid NDel, Lybarger L, Raghavan M. Distinct functions for the glycans of tapasin and heavy chains in the assembly of MHC class I molecules. J Immunol. 2011;186:2309–2320. doi: 10.4049/jimmunol.1002959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizvi SM, Raghavan M. Mechanisms of function of tapasin, a critical major histocompatibility complex class I assembly factor. Traffic. 2010;11:332–347. doi: 10.1111/j.1600-0854.2009.01025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wearsch PA, Cresswell P. Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nat Immunol. 2007;8:873–881. doi: 10.1038/ni1485. [DOI] [PubMed] [Google Scholar]
- 20.Wearsch PA, Peaper DR, Cresswell P. Essential glycan-dependent interactions optimize MHC class I peptide loading. Proc Natl Acad Sci U S A. 2011;108:4950–4955. doi: 10.1073/pnas.1102524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solheim JC, Harris MR, Kindle CS, Hansen TH. Prominence of beta 2-microglobulin, class I heavy chain conformation, and tapasin in the interactions of class I heavy chain with calreticulin and the transporter associated with antigen processing. J Immunol. 1997;158:2236–2241. [PubMed] [Google Scholar]
- 22.Rizvi SM, Raghavan M. Direct peptide regulatable interactions between tapasin and MHC class I molecules. Proc. Natl. Acad. Sci. 2006;103:18220–18225. doi: 10.1073/pnas.0605131103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zarling AL, Luckey CJ, Marto JA, White FM, Brame CJ, Evans AM, Lehner PJ, Cresswell P, Shabanowitz J, Hunt DF, Engelhard VH. Tapasin is a facilitator, not an editor, of class I MHC peptide binding. J Immunol. 2003;171:5287–5295. doi: 10.4049/jimmunol.171.10.5287. [DOI] [PubMed] [Google Scholar]
- 24.Chen M, Bouvier M. Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. Embo J. 2007;26:1681–1690. doi: 10.1038/sj.emboj.7601624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams AP, Peh CA, Purcell AW, McCluskey J, Elliott T. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. 2002;16:509–520. doi: 10.1016/s1074-7613(02)00304-7. [DOI] [PubMed] [Google Scholar]
- 26.Howarth M, Williams A, Tolstrup AB, Elliott T. Tapasin enhances MHC class I peptide presentation according to peptide half-life. Proc Natl Acad Sci U S A. 2004;101:11737–11742. doi: 10.1073/pnas.0306294101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peh CA, Burrows SR, Barnden M, Khanna R, Cresswell P, Moss DJ, McCluskey J. HLA-B27-restricted antigen presentation in the absence of tapasin reveals polymorphism in mechanisms of HLA class I peptide loading. Immunity. 1998;8:531–542. doi: 10.1016/s1074-7613(00)80558-0. [DOI] [PubMed] [Google Scholar]
- 28.Zernich D, Purcell AW, Macdonald WA, Kjer-Nielsen L, Ely LK, Laham N, Crockford T, Mifsud NA, Bharadwaj M, Chang L, Tait BD, Holdsworth R, Brooks AG, Bottomley SP, Beddoe T, Peh CA, Rossjohn J, McCluskey J. Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion. J Exp Med. 2004;200:13–24. doi: 10.1084/jem.20031680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thammavongsa V, Raghuraman G, Filzen TM, Collins KL, Raghavan M. HLA-B44 polymorphisms at position 116 of the heavy chain influence TAP complex binding via an effect on peptide occupancy. J Immunol. 2006;177:3150–3161. doi: 10.4049/jimmunol.177.5.3150. [DOI] [PubMed] [Google Scholar]
- 30.Thammavongsa V, Schaefer M, Filzen T, Collins KL, Carrington M, Bangia N, Raghavan M. Assembly and intracellular trafficking of HLA-B*3501 and HLA-B*3503. Immunogenetics. 2009;61:703–716. doi: 10.1007/s00251-009-0399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 32.Belicha-Villanueva A, McEvoy S, Cycon K, Ferrone S, Gollnick SO, Bangia N. Differential contribution of TAP and tapasin to HLA class I antigen expression. Immunology. 2008;124:112–120. doi: 10.1111/j.1365-2567.2007.02746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnstable CJ, Bodmer WF, Brown G, Galfre G, Milstein C, Williams AF, Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 34.Stam NJ, Spits H, Ploegh HL. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J Immunol. 1986;137:2299–2306. [PubMed] [Google Scholar]
- 35.Kao KJ, Riley WJ. Genetic predetermination of quantitative expression of HLA antigens in platelets and mononuclear leukocytes. Hum Immunol. 1993;38:243–250. doi: 10.1016/0198-8859(93)90551-b. [DOI] [PubMed] [Google Scholar]
- 36.Garboczi DN, Hung DT, Wiley DC. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc Natl Acad Sci U S A. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phair J, Jacobson L, Detels R, Rinaldo C, Saah A, Schrager L, Munoz A. Acquired-Immune-Deficiency-Syndrome Occurring within 5 Years of Infection with Human-Immunodeficiency-Virus Type-1 - the Multicenter Aids Cohort Study. J Acq Immun Def Synd. 1992;5:490–496. [PubMed] [Google Scholar]
- 38.Goedert JJ, Kessler CM, Aledort LM, Biggar RJ, Andes WA, White GC, Drummond JE, Vaidya K, Mann DL, Eyster ME, Ragni MV, Lederman MM, Cohen AR, Bray GL, Rosenberg PS, Friedman RM, Hilgartner MW, Blattner WA, Kroner B, Gail MH. A Prospective-Study of Human Immunodeficiency Virus Type-1 Infection and the Development of Aids in Subjects with Hemophilia. New Engl J Med. 1989;321:1141–1148. doi: 10.1056/NEJM198910263211701. [DOI] [PubMed] [Google Scholar]
- 39.Goedert JJ, Biggar RJ, Winn DM, Mann DL, Byar DP, Strong DM, Digioia RA, Grossman RJ, Sanchez WC, Kase RG, Greene MH, Hoover RN, Blattner WA. Decreased Helper Lymphocytes-T in Homosexual Men .1. Sexual Contact in High-Incidence Areas for the Acquired Immunodeficiency Syndrome. Am J Epidemiol. 1985;121:629–636. doi: 10.1093/aje/121.5.629. [DOI] [PubMed] [Google Scholar]
- 40.Badrinath S, Saunders P, Huyton T, Aufderbeck S, Hiller O, Blasczyk R, Bade-Doeding C. Position 156 influences the peptide repertoire and tapasin dependency of human leukocyte antigen B*44 allotypes. Haematologica. 2012;97:98–106. doi: 10.3324/haematol.2011.046037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geironson L, Thuring C, Harndahl M, Rasmussen M, Buus S, Roder G, Paulsson KM. Tapasin facilitation of natural HLA-A and -B allomorphs is strongly influenced by peptide length, depends on stability, and separates closely related allomorphs. J Immunol. 2013;191:3939–3947. doi: 10.4049/jimmunol.1201741. [DOI] [PubMed] [Google Scholar]
- 42.Muller CA, Engler-Blum G, Gekeler V, Steiert I, Weiss E, Schmidt H. Genetic and serological heterogeneity of the supertypic HLA-B locus specificities Bw4 and Bw6. Immunogenetics. 1989;30:200–207. doi: 10.1007/BF02421207. [DOI] [PubMed] [Google Scholar]
- 43.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. jBMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hillen N, Mester G, Lemmel C, Weinzierl AO, Muller M, Wernet D, Hennenlotter J, Stenzl A, Rammensee HG, Stevanovic S. Essential differences in ligand presentation and T cell epitope recognition among HLA molecules of the HLA-B44 supertype. Eur J Immunol. 2008;38:2993–3003. doi: 10.1002/eji.200838632. [DOI] [PubMed] [Google Scholar]
- 45.Macdonald W, Williams DS, Clements CS, Gorman JJ, Kjer-Nielsen L, Brooks AG, McCluskey J, Rossjohn J, Purcell AW. Identification of a dominant self-ligand bound to three HLA B44 alleles and the preliminary crystallographic analysis of recombinant forms of each complex. FEBS Lett. 2002;527:27–32. doi: 10.1016/s0014-5793(02)03149-6. [DOI] [PubMed] [Google Scholar]
- 46.Falk K, Rotzschke O, Grahovac B, Schendel D, Stevanovic S, Jung G, Rammensee HG. Peptide motifs of HLA-B35 and -B37 molecules. Immunogenetics. 1993;38:161–162. doi: 10.1007/BF00190906. [DOI] [PubMed] [Google Scholar]
- 47.Steinle A, Falk K, Rotzschke O, Gnau V, Stevanovic S, Jung G, Schendel DJ, Rammensee HG. Motif of HLA-B*3503 peptide ligands. Immunogenetics. 1996;43:105–107. doi: 10.1007/BF00186615. [DOI] [PubMed] [Google Scholar]
- 48.Sakaguchi T, Ibe M, Miwa K, Kaneko Y, Yokota S, Tanaka K, Takiguchi M. Binding of 8-mer to 11-mer peptides carrying the anchor residues to slow assembling HLA class I molecules (HLA-B*5101) Immunogenetics. 1997;45:259–265. doi: 10.1007/s002510050201. [DOI] [PubMed] [Google Scholar]
- 49.Kubo H, Ikeda-Moore Y, Kikuchi A, Miwa K, Nokihara K, Schonbach C, Takiguchi M. Residue 116 determines the C-terminal anchor residue of HLA-B*3501 and -B*5101 binding peptides but does not explain the general affinity difference. Immunogenetics. 1998;47:256–263. doi: 10.1007/s002510050355. [DOI] [PubMed] [Google Scholar]
- 50.Tomiyama H, Yamada N, Komatsu H, Hirayama K, Takiguchi M. A single CTL clone can recognize a naturally processed HIV-1 epitope presented by two different HLA class I molecules. Eur J Immunol. 2000;30:2521–2530. doi: 10.1002/1521-4141(200009)30:9<2521::AID-IMMU2521>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 51.Vivian JP, Duncan RC, Berry R, O'Connor GM, Reid HH, Beddoe T, Gras S, Saunders PM, Olshina MA, Widjaja JM, Harpur CM, Lin J, Maloveste SM, Price DA, Lafont BA, McVicar DW, Clements CS, Brooks AG, Rossjohn J. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature. 2011;479:401–405. doi: 10.1038/nature10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macdonald WA, Purcell AW, Mifsud NA, Ely LK, Williams DS, Chang L, Gorman JJ, Clements CS, Kjer-Nielsen L, Koelle DM, Burrows SR, Tait BD, Holdsworth R, Brooks AG, Lovrecz GO, Lu L, Rossjohn J, McCluskey J. A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition. J Exp Med. 2003;198:679–691. doi: 10.1084/jem.20030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hill A, Takiguchi M, McMichael A. Different rates of HLA class I molecule assembly which are determined by amino acid sequence in the alpha 2 domain. Immunogenetics. 1993;37:95–101. doi: 10.1007/BF00216831. [DOI] [PubMed] [Google Scholar]
- 54.Khanna R, Burrows SR, Neisig A, Neefjes J, Moss DJ, Silins SL. Hierarchy of Epstein-Barr virus-specific cytotoxic T-cell responses in individuals carrying different subtypes of an HLA allele: implications for epitope-based antiviral vaccines. J Virol. 1997;71:7429–7435. doi: 10.1128/jvi.71.10.7429-7435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sieker F, Springer S, Zacharias M. Comparative molecular dynamics analysis of tapasin-dependent and -independent MHC class I alleles. Protein Sci. 2007;16:299–308. doi: 10.1110/ps.062568407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanaseki T, Lind KC, Escobar H, Nagarajan N, Reyes-Vargas E, Rudd B, Rockwood AL, Kaer LVan, Sato N, Delgado JC, Shastri N. ERAAP and tapasin independently edit the amino and carboxyl termini of MHC class I peptides. J Immunol. 2013;191:1547–1555. doi: 10.4049/jimmunol.1301043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith KJ, Reid SW, Stuart DI, McMichael AJ, Jones EY, Bell JI. An altered position of the alpha 2 helix of MHC class I is revealed by the crystal structure of HLA-B*3501. Immunity. 1996;4:203–213. doi: 10.1016/s1074-7613(00)80429-x. [DOI] [PubMed] [Google Scholar]
- 58.Flores-Villanueva PO, Yunis EJ, Delgado JC, Vittinghoff E, Buchbinder S, Leung JY, Uglialoro AM, Clavijo OP, Rosenberg ES, Kalams SA, Braun JD, Boswell SL, Walker BD, Goldfeld AE. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc Natl Acad Sci U S A. 2001;98:5140–5145. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]