SUMMARY
dsRNA can function as genetic information and may have served as genomic material before the existence of DNA-based life. By developing a method to purify dsRNA, we have investigated the diversity of dsRNA in microbial populations. We detect large dsRNAs in multiple microbial populations. Analysis of an aquatic microbial population reveals some dsRNA sequences match metagenomic DNA suggesting that microbes contain pools of sense-antisense transcripts. In addition, ~30% of the dsRNA sequences are not present in the corresponding DNA pool, and are strongly biased toward encoding novel proteins. Of these “dsRNA unique” sequences, only a small percentage share similarity to known viruses, a large fraction assemble into RNA-virus-like contigs, and the remaining fraction has an unexplained origin. These results have uncovered dsRNA virus-like elements and underscore that dsRNA potentially represents an additional reservoir of genetic information in microbial populations.
Keywords: microbial metagenomics, dsRNA, virus, aquatic
INTRODUCTION
Microbial communities are important to ecosystems, including human associated microbiomes impacting human health (Falkowski et al.,, 1998, DeLong 2009, Madsen 2011; Clemente et al., 2012, Sommer and Backhed, 2013). Moreover, DNA sequencing has revealed that microbial communities have much richer diversity than anticipated (e. g. Pace et al., 1986, Hugenholtz and Pace 1996, Venter et al., 2004, Eckburg et al., 2005, Human Microbiome Project Consortium, 2012). Metagenomic analyses allow for the full spectrum of metabolic pathways present in a community to be identified thus increasing our understanding of ecosystems. To date, metagenomic analyses of microbial communities have primarily focused on DNA as the source of genomic information; however, RNA can also serve as genetic material. Sequencing of microbial metatranscriptomes has identified RNAs not observed in the corresponding DNA metagenomes (Frias-Lopez et al., 2008, Shi et al., 2011, Baker et al., 2013), although the significance of this observation is not clear.
One possible explanation is that microbial RNA, independent of DNA, is serving as genetic information in microbial populations
A potential source of genetic material in microbial populations is dsRNA. dsRNA is used as genomic material by some viruses that infect bacterial (Mindich 2006) and eukaryotic microbial hosts (e.g. Gallimore et al., 1995; Strauss et al., 2000, Naitow et al., 2001, Jiang and Ghabrial 2004; Hacker et al., 2005; Fukuhara 2006). Although novel dsRNA viruses have been identified by sequencing of RNA from viral populations (Culley et al., 2006, Djikeng et al., 2009, Cantalupo et al., 2011, Steward et al., 2013), our understanding of dsRNA viruses in the environment is far from complete. Indeed, dsRNA viruses may be under represented in RNA sequencing experiments since many cDNA libraries are made under conditions that bias against the reverse transcription of double-stranded RNAs. Furthermore, examination of viral particles does not allow for the detection of dsRNA viruses that lack an extracellular phase. The prevalence of such endogenous viruses in microbial communities is not known though they are found in fungi where they can be associated with satellite dsRNA elements and alter the phenotype of their hosts (Schmitt and Breinig 2006,Pearson et al., 2009). Whether there are additional dsRNA genetic elements in microbial ecosystems, and their diversity, has not been examined in a systematic manner.
To examine dsRNA in microbial ecosystems, we developed methods to determine if microbial populations contain dsRNA and to specifically purify dsRNA from total nucleic acids. We observe dsRNA molecules in multiple microbial communities. Sequencing of the purified dsRNA from one microbial sample demonstrates that approximately one third of it is unrelated to the DNA from the same microbial community. The ”dsRNA unique” sequences encode a higher percentage of unknown proteins than the DNA pool and has little overlap with known viral sequences. Moreover, a substantial fraction of the dsRNA unique sequences can be assembled into discrete viral-like elements that encode proteins with no significant similarity to known viruses or to RNA viral metagenomic sequences. These findings demonstrate that dsRNA isolated from the cellular fraction of microbial communities represents an unexplored pool of what could be genetic information.
RESULTS
Biochemical detection of dsRNA in microbial populations
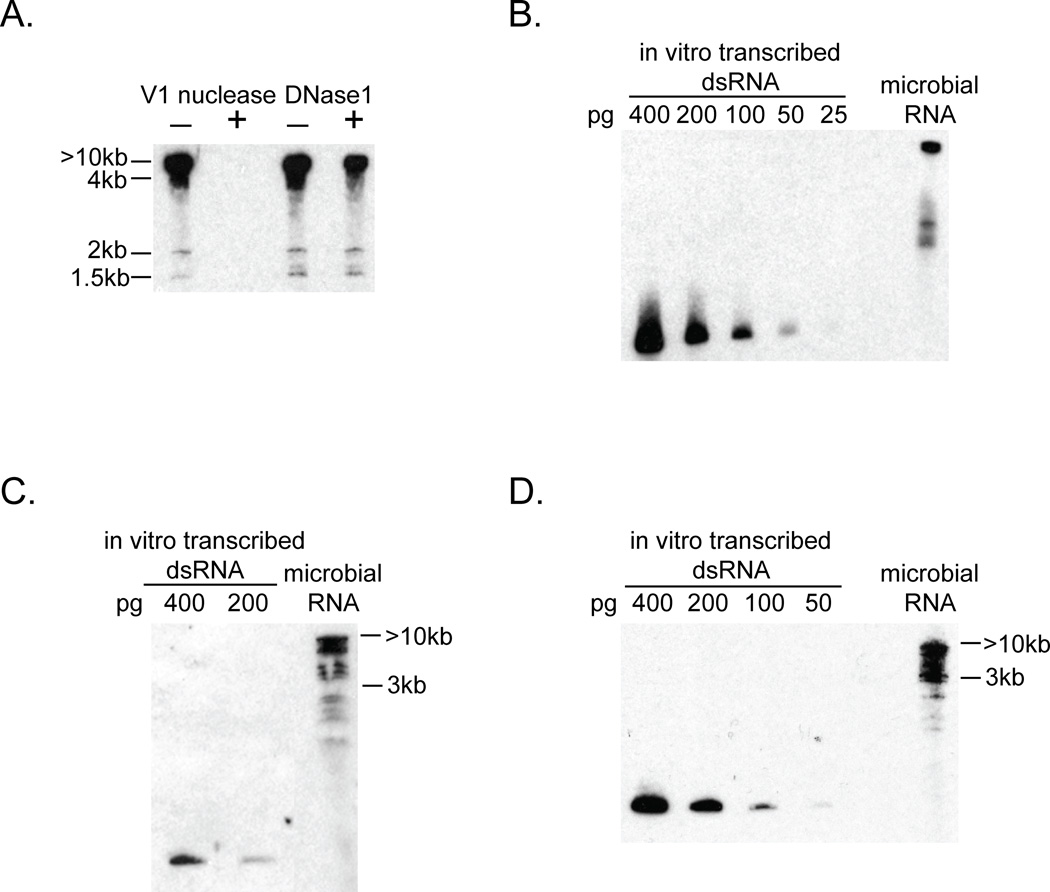
To determine if microbial populations contain detectable dsRNA we performed western analysis using an antibody that is specific for dsRNA (Schonborn et al., 1991) on total RNA isolated from microbes collected from a wetland. We prepared total RNA from the microbial fraction (2.7 to 0.2 micron) and then analyzed the RNA by gel electrophoresis followed by western analysis using the anti-dsRNA specific antibody (Figure 1A). The majority of the dsRNA ran at the exclusion limit of the gel ≥10kb, although some discrete bands were detected at approximately 1.5 and 2 kb. Additional evidence that this signal is due to dsRNA is that it is abolished by treatment with V1 nuclease, which is specific for dsRNA (Lockard and Kumar 1981), but not by treatment with DNase 1 (Figure 1A). By comparison with known amounts of an in vitro transcribed dsRNA (Figure 1B), we determined that ~0.025% of the total RNA from the wetland sample was dsRNA as detected by western analysis. dsRNA of similar size distributions was also detected in the two additional aquatic microbial samples we tested (Figure 1C and D) suggesting that dsRNA is commonly present in microbial populations.
Figure 1. Biochemical detection of dsRNA in microbial samples isolated from different aquatic environments.
Western analysis to detect dsRNA using anti-dsRNA antibody against total microbial RNA. A. Detection of dsRNA in a microbial sample isolated from Sweetwater Wetlands. Total RNA was treated with or without eitherV1 nuclease, a dsRNA-specific nuclease, or DNase1, a DNA-specific nuclease, separated by electrophoresis on a polyacrylamide gel, transferred then probed with anti-dsRNA antibody. Approximate size of dsRNA species was determined by comparison to migration of DNA ladder on same gel. B. dsRNA in the wetland microbial sample compared to dilution series of an in vitro transcribed dsRNA. C. dsRNA in a microbial sample isolated from a coastal seawater sample collected at Scripps Institute of Oceanography, La Jolla, CA. D. dsRNA in a microbial sample isolated from the artificial ocean at the Biosphere 2, Oracle, AZ.
Identification of dsRNA sequences that are not present in microbial DNA
To analyze the composition of dsRNA in microbial populations, we developed a method to purify dsRNA from complex biological samples. We developed this protocol by using it to purify a 0.9kb dsRNA expressed in E. coli from a plasmid with convergent T7 promotors. First, we digested total E. coli RNA with DNase 1 and RNase 1 in the presence of 100 mM NaCl, which limits the digestion of RNA by RNase 1 to ssRNA. To specifically enrich for dsRNA we used the sequence nonspecific dsRNA binding protein DRB4 fused to GST as an affinity reagent to select the dsRNA (Kobayashi et al., 2009). Applying this method to samples in which total RNA from E. coli that express the dsRNA was mixed with a 200 fold excess of total RNA from E. coli that did not express dsRNA demonstrated that the expected 0.9 kb dsRNA was recovered efficiently (~50% recovery) from a large excess of total RNA (Figure S1A). Moreover, RT-PCR using primers complementary to E. coli ribosomal RNA indicated that rRNA was depleted from the purified dsRNA (Figure S1A). We interpret these results to indicate that we can specifically purify dsRNA from a mixed population of nucleic acid, which can then be used for sequencing. Additional evidence that this method was effective at purifying dsRNA is the detection of dsRNA elements by RT-PCR in a purified microbial dsRNA sample (see below).
Using this method, we purified dsRNA from the wetland microbial community. If the sequences in the dsRNA pool represent antisense-sense hybrids produced from conventional transcripts from DNA, we would expect the dsRNA sequences to be present in DNA isolated from the same microbial sample. Conversely, if the dsRNA represents genetic elements that are distinct from DNA then the dsRNA sequence should not be present in the DNA sequence population. To address this possibility, we sequenced cDNA obtained from dsRNA purified from the total microbial RNA (Experimental Procedures). We then comprehensively compared each sequence from the dsRNA pool (23.3Mb) to a 900-fold excess of microbial DNA (22.6Gb) using a “shared k-mer” analysis (Experimental Procedures; Hurwitz et al., 2013). This analysis involved comparing all the 20-mers present in each of the dsRNA reads to determine if there was an exact match to any of the 20mers present in the DNA. A dsRNA read was scored as being unique to the dsRNA sequence pool if none of the 20mers present in the read matched a 20mer in the DNA sequence. This analysis yielded two important observations.
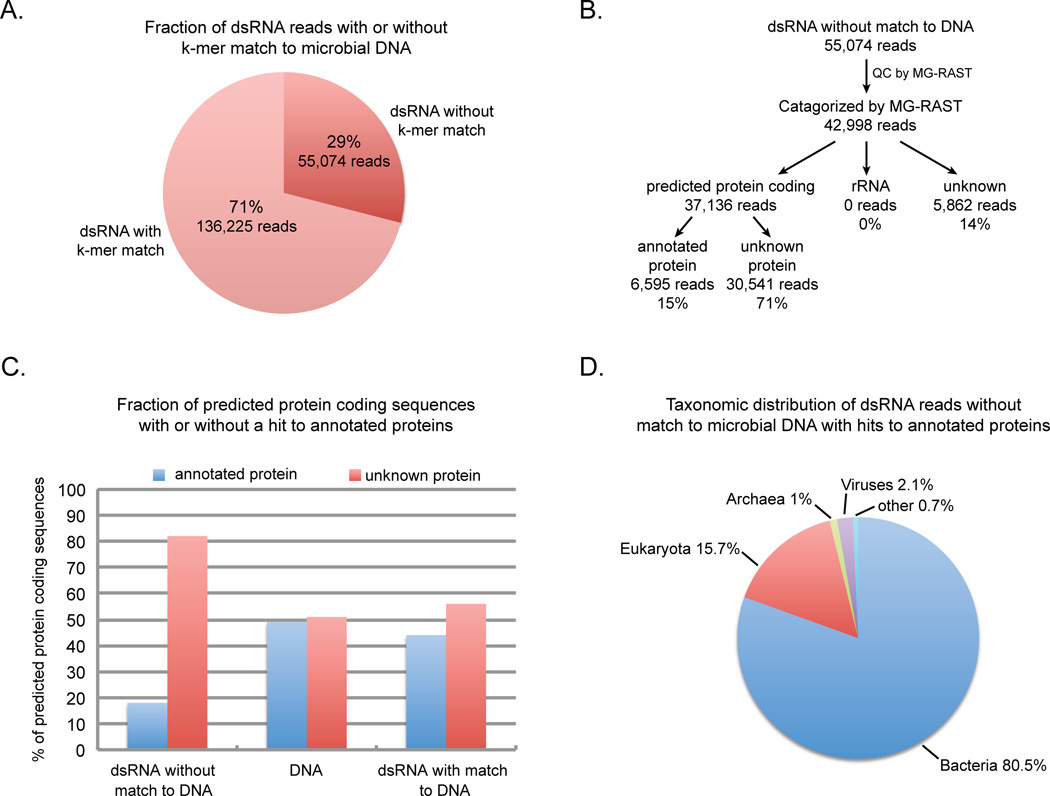
One interesting finding was that of the 191,299 dsRNA reads, 136,225 (~71%) reads shared at least one 20mer sequence in common with the DNA sequence (Figure 2A). Approximately one half of these reads are predicted to be rRNA sequences (Figure S1B) indicating that the dsRNA purification method did not completely remove ribosomal RNA. Contamination of the dsRNA with ribosomal RNA is likely due to its high abundance in total RNA as well as its highly structured nature leading to partial resistance to nuclease treatment. The overlap of the remaining dsRNA reads with the microbial DNA is consistent with this population of the dsRNA coming from antisense-sense hybrids of transcripts (e.g. Georg et el. 2009; Raghavan et al., 2012) or potentially from extensive stem-loop structures in transcripts that produce long dsRNA regions (e.g. Morse and Bass 1999) or highly structured RNAs (e.g. Weinberg et al., 2009). The presence of these sequences indicates that some microbes produce detectable pools of dsRNA that are encoded in their DNA genomes.
Figure 2. Analysis of microbial dsRNA sequences.
A. Results of shared k-mer analysis between dsRNA sequences and DNA sequences obtained from the same microbial sample. B. Flow diagram of the analysis of the dsRNA reads without a k-mer match to the microbial DNA. C. The percentage of reads that were predicted by MG-RAST to encode protein which were either similar to known annotated proteins or did not share similarity to a known protein sequence in the dsRNA unique reads that did not share any k-mer sequence with microbial DNA, microbial DNA reads (Ion Torrent) or dsRNA reads that shared at least one k-mer sequence with the microbial DNA. D. Taxonomic distribution of dsRNA unique reads with hits to annotated proteins. Pie chart of the percentage of reads with hits to annotated proteins that were assigned to major taxonomic groups including eukaryota, bacteria, archaea, viruses or other (eg. vector sequences) by MG-RAST. (See also Figure S1 and Table S1)
A second and more important observation was that 55,074 (~29%) of the microbial dsRNA reads were determined to be unique to the dsRNA based on the k-mer analysis (Figure 2A).We believe that it is unlikely that their absence is simply due to the DNA pool not having been sequenced deep enough given that, of the total number of 20mers present in the DNA sequence, 85% of the k-mers are represented over 15 fold (Figure S1C). This argues that the majority of sequences present in the microbial DNA population are covered in depth in the sequenced pool. Likewise, although it is possible that some of the dsRNA sequences appear to be unique to dsRNA because they are derived from DNA that is rare in the microbial population, the observation that 29% of the dsRNA reads do not share a single 20mer with the DNA whereas only 10.5% of the total 20mers in the DNA population are represented only once in the DNA (Figure S1C) argues that the dsRNA pool is enriched for sequences not present in the sequenced DNA pool. It is therefore likely that at least a substantial fraction of the dsRNA sequences identified as being unique to dsRNA are not encoded in microbial DNA.
dsRNA unique sequences are biased toward encoding unknown proteins
To characterize the sequences uniquely present in dsRNA, we submitted the dsRNA sequences to the Metagenomics RAST server, MG-RAST (Meyer et al., 2008). The MG-RAST server uses an ab-initio gene calling algorithm to identify coding regions within sequences and then uses the BLAT similarity search algorithm against the M5 non-redundant protein database to determine if the potential protein coding sequences resemble known annotated proteins. MG-RAST also searches for sequences with similarity to ribosomal RNA using BLAT .For comparison, we also analyzed a sample of microbial DNA sequences and the pool of dsRNA sequences that shared a k-mer match with the microbial DNA.
A striking observation was that the vast majority of the dsRNA unique sequences are not similar to known protein or rRNA sequences. Of the 42,998 dsRNA unique sequences that passed the MG-RAST quality control process, 37,136 were predicted to encode protein (Figure 2B). Of the potential protein coding reads, only 18% was predicted to encode proteins similar to annotated proteins, whereas 82% potentially encode unknown proteins (Figure 2C). In contrast, in the wetland microbial DNA sample, 49% of the reads predicted to encode protein were highly similar to known proteins (Figure 2C). The propensity to encode unknown proteins is not a feature of the microbial dsRNA per se given that only 56% of the potential protein coding reads in the dsRNA that shared k-mers in common with the DNA were predicted to encode unannotated proteins (Figure 2C). The difference between the dsRNA unique and the DNA samples in the number of potential coding sequences that did not share significant similarity to known proteins by BLAT is highly significant (Chi square 13,078 p value ≪0.0001). A striking difference between the dsRNA unique and the DNA sequences was also seen when Blastp was used to search for similarity with proteins in the NCBI non-redundant database. Only 8.9% of the dsRNA unique sequences shared significant similarity to known proteins (E value 10−3) in contrast to the 57.7% of the DNA sequences that had significant hits. Thus, in comparison to sequences from the same ecosystem, the dsRNA unique sequences are biased toward potentially encoding novel or divergent forms of proteins. It should be noted that while the clear prediction is that these nucleotide reads are translated into protein in cells, we do not yet have direct mass spectroscopy data evidence that these sequences are actually produced into proteins.
Another difference between the dsRNA unique sequences and the DNA population was that 14% of the dsRNA pool was of unknown sequences which were not predicted to encode protein or ribosomal RNA (Figure 2B) whereas only 2% of the microbial DNA was classified as unknown (Figure S1B). In addition, none of the dsRNA unique sequences shared similarity to known ribosomal RNAs (Figure 2B). Therefore, 85% of the dsRNA unique sequence pool was composed of previously unrecognized sequences underscoring that, relative to the microbial DNA from the same sample (Figure 2C and Figure S1B), the genetic information in the dsRNA may be highly diverged or novel compared to known DNA sequences.
The majority of dsRNA unique sequences with similarity to annotated proteins are not of known viral origin
One possible explanation for reads that are exclusively present in dsRNA is that they are derived from RNA viruses. To examine this possibility, the dsRNA unique reads with BLAT hits to annotated proteins were assigned by MG-RAST to taxonomic groups based on the annotations of the corresponding hits. This analysis reveals several important observations. First, most of the sequences were associated with cellular organisms (Figure 2D), with only 2.1% of the dsRNA unique reads with hits to annotated proteins predicted to encode viral proteins. Moreover, the Blastp analysis did not significantly increase the number of dsRNA unique reads with hits to viral proteins and the hits are primarily to dsRNA viruses (Table S1).This finding is in contrast to many RNA viral metagenomes where a high percentage of sequences with hits to known proteins are to viral proteins (Culley et al., 2006, Djikeng et al., 2009, Steward et al., 2013). These results indicate that the vast majority of the sequences that are exclusive to the dsRNA are not similar to known viral proteins, and therefore could come from previously undescribed dsRNA viruses or other dsRNA elements.
Analysis of dsRNA elements assembled from dsRNA unique sequences
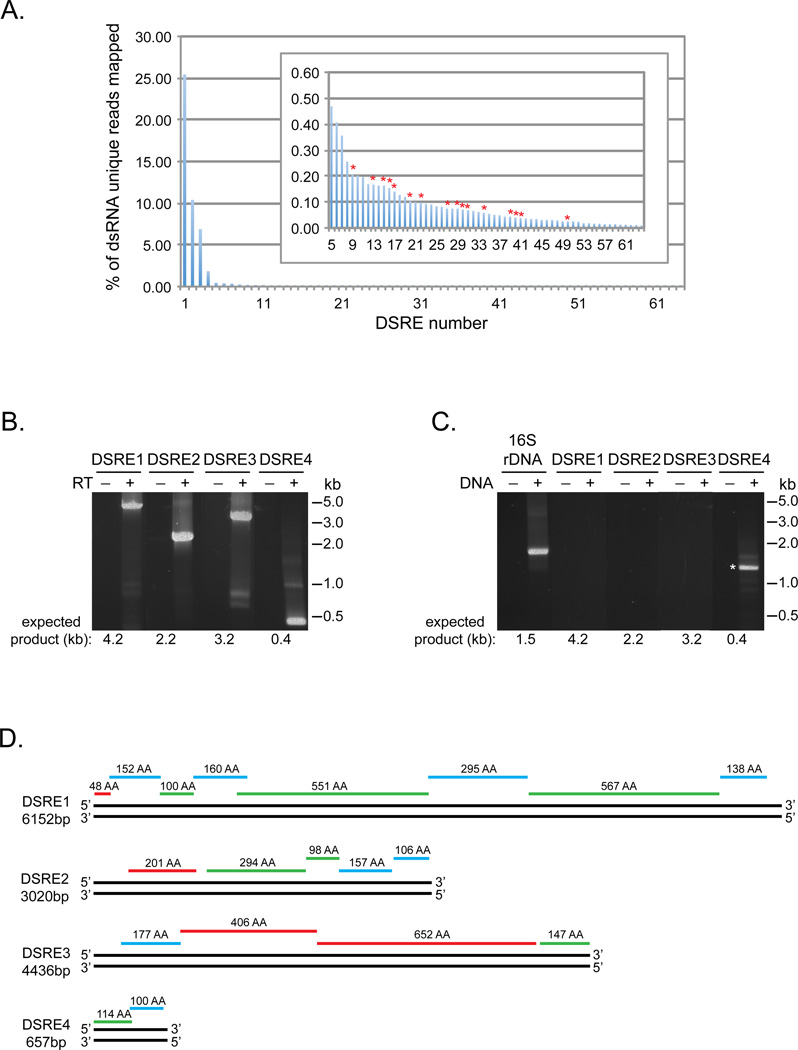
To understand what genetic elements might be encoded by dsRNA isolated from microbes we used Trinity, a program designed to assemble transcripts from short sequencing reads (Grabherr et al., 2011), to assemble the dsRNA unique sequences into 64 unique contigs from 500 to 4968 bp in length, which we refer to as DSREs (dsRNA elements). Using Bowtie2, a total of 48.1% of the dsRNA unique reads map to the 64 DSREs. 44.6% of the reads map to just four of the contigs (Figure 3A) indicating that these four elements, DSRE1 through 4, are abundant within the dsRNA unique sequence pool. In contrast, the remaining DSREs are not highly represented in the dsRNA unique sequences (Figure 3A insert).
Figure 3. Analysis of microbial dsRNA elements assembled from dsRNA unique sequences.
A. Percentage of the dsRNA reads without a k-mer match to the microbial DNA that mapped to each of the 64 dsRNA elements (DSREs) assembled from the dsRNA unique reads. Insert is blowup of the data for DSRE5 through DSRE64. DSREs with sequence similarity to known dsRNA viruses are highlighted with red asterisks. B. RT-PCR analysis of purified microbial dsRNA using primers corresponding to DSRE1, 2, 3 and 4. The presence or absence of reverse transcriptase (RT) in the reactions is indicated. C. PCR analysis of microbial DNA using the same primers to detect DSRE1, 2, 3 and 4 as used in (B) and universal primers to amplify 16S rDNA sequences. The presence or absence of microbial DNA in the reactions is indicated. A non-specific PCR product that has no sequence similarity to DSRE4 is marked with an asterisk. D. Diagram of DSRE1, 2, 3 and 4 illustrating the location and length of predicted protein coding sequences. Red lines indicate ORFs in frame 1, blue lines in frame 2 and green lines in frame 3 relative to the 5’ end of the top strand. (See also Figure S2 and Table S2.)
Examination of these four contigs validates that our analyses identify dsRNA elements not encoded in DNA. First, we can PCR amplify DSRE1 through 4 from the purified microbial dsRNA, but not from the microbial DNA, in a manner that is dependent on reverse transcriptase (Figure 3B and C). Thus, the contigs are present as RNA in the purified dsRNA and are not detectable in the corresponding DNA. Second, 5’ RACE products were observed for both ends of the three contigs we examined, DSRE1 through 3, verifying that they are present as dsRNA (data not shown). Third, we confirmed the sequences of the contigs by sequencing the RT-PCR products of DSRE1 though 4 and 5’ RACE products of DSRE1 through 3 and assembling the sequences together. This analysis revealed that the elements ranged in size from ~0.66 to 6.1kb.
Examination of the DSRE1, 2, 3 and 4 sequences indicate that they are not predicted to contain rRNA or tRNA genes, but are predicted to encode a total of 19 proteins ranging in size from 48 to 652 amino acids (Figure 3D). Each of the elements potentially encode multiple protein products with the predicted protein coding genes being tightly packed along the length of the elements. In some cases there is overlap between ORFs in different reading frames raising the possibility that longer protein products are produced from the elements by translational frame shifting. The tight packing of the ORFs and the observation that all of the predicted protein coding genes are encoded on a single strand, suggests that DSRE1, 2, 3 and 4 might represent unknown dsRNA viruses.
Comparison of the organization of the dsRNA elements and the genomic structure of known dsRNA virus families revealed that the elements are organized most similarly to Cystoviridae (Figure S2A), a family of dsRNA viruses that infect bacteria. The genomes of Cystoviridae are composed of multiple dsRNA segments (Figure S2A). The observations that DSRE1, DSRE2, DSRE3 and DSRE4 all share a similar GC content (58–60%), have similar synonomous codon usage bias (data not shown) and are all high in abundance in the dsRNA pool (Figure 3A) suggests that they may be components of the same viral genome.
Although DSRE1, 2, 3 and 4 resemble Cystoviridae in their genetic organization, the potential proteins encoded by these elements do not show any significant sequence similarity to previously described Cystoviridae proteins or to any other annotated proteins by Blastp analysis (Experimental Procedures, E value 10−3). In addition, we do not find any significant similarity (tblastx, E value 10−5) between these DSREs and viral metagenomic databases that contain RNA viruses (Experimental Procedures, Table S2). The simplest interpretation of these observations is that these elements represent members of one or more previously undescribed class(es) of dsRNA viruses.
Blast analysis of the potential proteins encoded in the remaining 60 dsRNA contigs revealed that 16 of the DSREs share significant similarity to proteins encoded in five different families of eukaryotic dsRNA viruses including picobirnaviridea, reoviridae, partitiviridae, totiviridae, and endornaviridae (Table 1). Thus, these DSREs are likely to be new members of known dsRNA viral families. These newly identified viruses are not very abundant in the dsRNA pool given that they contain only 1.5% of the dsRNA unique reads (Figure 3A insert). The remaining 44 dsRNA contigs are also not highly represented in the dsRNA unique sequences (Figure 3A insert) but these DSREs did not share significant similarity to any known RNA viral protein. These DSREs may be derived from dsRNA viruses but encode proteins that are too dissimilar to known viruses to be detected. Alternatively, they could represent novel non-viral dsRNA genetic elements.
Table 1.
Summary of dsRNA unique DSREs with similarity to dsRNA viral proteinsa
| # of DSREs | Viral Protein | Viral Family |
|---|---|---|
| 1 | RdRp | Picobirnaviridea |
| 3 | RdRp | Partitiviridae |
| 4 | polyprotein | Endornaviridae |
| 2 | RdRp | Totiviridae |
| 1 | RdRp | Reoviridae |
| 3 | S2 | Reoviridae |
| 1 | S3 | Reoviridae |
| 1 | P3/P4 | Reoviridae |
Blastx analysis against NCBI non-redundant protein database E value 10−4
The fact that reads corresponding to the DSRE contigs were over-represented in the dsRNA pool led us to consider the diversity of the dsRNA unique sequences and if there were any other over-represented sequences in this population that might represent novel types of dsRNA. Given this, we analyzed the dsRNA unique sequences, after removal of reads mapping to DSRE1, 2, 3 and 4, by examining the frequency of 20 nucleotide long k-mers in the population. This analysis led to two interesting observations. First, the majority (~76%) of k-mers are unique in this population, and the remaining k-mers are generally present at less than 4 copies (Figure S2B). This provides additional evidence that this dsRNA unique sequence population is a diverse pool of nucleotide information. Second, we observed that there was an over-represented 126 nucleotide sequence which made up ~0.8% of the total k-mers. This specific over-represented sequence (Figure S2C) is not related to any of the primers used to amplify the dsRNA, the dsRNA elements assembled from the dsRNA unique sequences or any known nucleic acid (data not shown) and its origin will be of interest in future work.
DISCUSSION
We present evidence that aquatic microbial communities contain dsRNAs. The key observation is that total RNA samples isolated from these communities contain dsRNA as judged by material being detected using a dsRNA specific antibody, and that material being sensitive to the dsRNA specific nuclease, V1 (Figure 1). In each of the microbial populations we detect relatively discrete-sized dsRNA species which likely represent dsRNA viruses infecting these communities (Figure 1) Interestingly, the majority of the dsRNA runs near the exclusion limit of the gel used and represents molecules of >8 kb, which suggests that some of the dsRNA present in microbes is found in large molecules that could represent complex genetic elements. dsRNA may be a common component of microbial populations given that we detected it in all of three of the environmental samples we tested.
To determine the potential types of dsRNA in microbial ecosystems, we developed a method to isolate dsRNA from total nucleic acids that involved enzymatic removal of ssRNA and DNA followed by affinity purification of dsRNA using a dsRNA binding protein. This approach differs from methods commonly used to identify viruses from plant and fungal samples that use differential binding of nucleic acid species to cellulose to enrich for dsRNA (Morris and Dodds 1979), which we found to be relatively inefficient at recovering dsRNA (data not shown). In contrast, our approach allows for the efficient recovery of small amounts of dsRNA from a large excess of nucleic acid (Figure S1A). Although ribosomal RNA was not entirely depleted from the microbial dsRNA sample we analyzed, we believe this method is effective at enriching for dsRNA for several reasons. First, sequences derived from the microbial dsRNA sample were assembled into contigs that encoded proteins with significant similarity to known dsRNA viruses and are therefore likely to be derived from dsRNA (Table 1). More importantly, we confirmed that both strands of three additional contigs, DSRE1, DSRE2 and DSRE3, were present as RNA in the dsRNA sample. This result indicates that these contigs, which represent 12% of the entire pool of dsRNA sequences, are indeed derived from dsRNA present in the purified microbial dsRNA. Therefore, given that only ~0.025% of the total RNA from the wetland water sample was estimated to be dsRNA, our purification method highly enriched for dsRNA.
An interesting finding is that ~30% of the microbial dsRNA sequences are unique to the dsRNA population and are not seen in the corresponding DNA metagenome. Strikingly, the potential protein coding sequences in the dsRNA unique reads are enriched in novel proteins suggesting that dsRNA encodes a largely previously undescribed pool of potential genetic information. Given this, a full understanding of the metagenomics and metabolic potential of a population will require an analysis of its unique dsRNA composition.
Through the analysis of the dsRNA unique sequences we have identified new members of known dsRNA virus families as well as elements that potentially represent a previously undescribed class of bacterial dsRNA viruses. A small subset of the dsRNA unique sequences (1.5%) assembled into contigs that encode proteins with significant similarity to proteins from families of known dsRNA viruses. These contigs represent new members of these viral families given that they do not share significant similarity at the nucleic acid level to known viruses (blastN, E value 10−4). In addition, 46% of the dsRNA unique sequences assemble into four predominant contigs we refer to as DSRE1, DSRE2, DSRE3 and DSRE4. Because these elements contain multiple ORFs that are closely packed and are all encoded on one strand, we suggest that DSRE1–4 represent a new family of dsRNA viruses with an organization most like Cystoviridae, although the predicted proteins encoded by these elements do not share any sequence similarity to known viral proteins including proteins from cystoviruses. Thus, the sequencing of dsRNA from microbes is likely to identify both new members of known viral families as well as new dsRNA viral groups.
Our knowledge of RNA viruses in nature is far from complete (Lang et al., 2008). However, recent analysis indicates that the abundance of RNA viruses may equal or exceed the number of DNA viruses in coastal seawater samples (Steward et al., 2013) suggesting that RNA viruses may have a significant impact on ecosystems. The analysis of microbial dsRNA could enhance our understanding of viral ecology and diversity in two ways. First, viral sequences in microbial dsRNA could reflect RNA viruses that are actively infecting their microbial hosts. In addition, several families of dsRNA viruses that infect fungi and other microbial eukaryotic hosts often do not have free viral forms, and thus would not be present in viral particle preparations. Interestingly, nine of the sixteen DSREs that encoded proteins with significant similiarity to known viruses were related to partitiviridae, totiviridae, and endornaviridae which frequently, or in the case of endornaviridae, totally lack extracellular viral forms. Thus, the analysis of microbial dsRNA provides a means to reveal classes of viruses that would be otherwise missed by the analysis of viral particles alone. Both approaches would complement each other in obtaining a more thorough understanding of the impact of RNA viruses on ecosystems.
The analysis of dsRNA isolated from microbial populations may also reveal novel dsRNA elements that are not of viral origin. In addition to the sequences that are likely derived from dsRNA viruses, approximately one half of the dsRNA unique sequences are of unexplained origin some of which potentially encode proteins with similarity to cellular proteins (Figure 2D). Several possibilities exist for the source of these dsRNA unique sequences. First, until we understand their source, it cannot be formally ruled out that they are encoded by DNA in the population and such DNA was either not represented in the sequenced DNA pool, or the dsRNA was extensively edited after transcription. Second, these dsRNA sequences could be derived from as yet uncharacterized viruses. Finally, a speculative possibility is that dsRNA genetic elements, rather than DNA, encode these predicted cellular components, in some microbial organisms. At a minimum, examining dsRNA isolated from the cellular fraction of natural microbial communities will lead to a better understanding of the impact of dsRNA viruses in ecosystems and in the future may reveal other types of dsRNA elements if they exist.
EXPERIMENTAL PROCEDURES
Sample Collection
Twenty liters of water collected from the surface of a reclaimed water wetland (Sweetwater Wetlands, Tucson, Arizona, USA latitude +32.278983, longitude −111.021591) was filtered using a Whatman GF/D (2.7micron) filter. Cells were collected from the filtrate by centrifugation in 500ml tubes in GSA rotor 6800g 20min. The majority of water was removed and cells resuspended in a total volume of ~700ml of remaining water then the sample was filtered a second time using a Whatman GF/D (2.7micron) filter. Microbial cells were collected by tangential flow on Memteq CT40 (0.2micron) filter units, recovered in 10mM TrisCl pH7.5, collected by centrifugation at 13,000g, and stored at −80C.
Total RNA Isolation and Detection of dsRNA by Immunoblotting with anti-dsRNA Antibody
Microbial cells were resuspended in sucrose lysis buffer (50mM TrisCl pH8, 40mM EDTA 0.75M sucrose) containing 1mg/ml lysozyme, incubated for 10 minutes at room temperature then lysed using Trizol LS (Invitrogen). RNA was isolated following the manufacturer ’s protocol except an additional chloroform extraction and ethanol precipitation was used to remove residual Trizol. RNA was resuspended in 10mM TrisCl pH7.5. Aliquots of 1µg total RNA were treated with either 0.05µ/ml DNase 1 (Ambion) in 10mM TrisCl pH7.5, 2.5mM MgCl2, 0.5mM CaCl2, 50mM NaCl or 0.0025u/µl RNase V1 (Ambion) in 10mM TrisCl pH7.5, 0.3Mm MgCl2, 100mM NaCl at 37°C. Treated and untreated samples of total microbial RNA and were separated on 5% nondenaturing acrylamide gels and transferred to Nytran in 0.5XTBE at 0.5 Amps for 3 hours. Gels included a dilution series of an in vitro transcribed ~0.9kb dsRNA produced from plasmid L4440-Y75B7AL.4 which contains converging T7 promotors. J2 anti-dsRNA monoclonal antibody (English&Scientific Consulting Kft) was used to detect dsRNA by immunoblot essentially as described in Schonborn et al., 1991 except 1xPBS with 5% nonfat dry milk and 50µg/ml salmon DNA in 1xPBS was used as a blocking solution and goat anti-mouse HRP antibody (Sigma A4416) was used as a secondary antibody.
Microbial dsRNA Purification and Sequencing
dsRNA was purified from 100 µg total microbial RNA by treatment with 0.008u/µl DNase 1 (Ambion) DNase 1 and 0.2u/µl RNase 1 (Ambion) in 0.1M TrisCl pH 7.0, 0.1M NaCl, 10mM MgCl2 at room temperature for 20 minutes followed by affinity purification using GST-DRB4* as described in Kobayashi et al., 2009. The eluted dsRNA was concentrated using RNA Clean-Up and Concentration Micro Kit (Norgen Biotek Corporation). The amount of dsRNA recovered was estimated to be 12ng based on immunoblotting using the anti-dsRNA antibody and comparison to a dilution series of an in vitro transcribed dsRNA.
The dsRNA was amplified using a TransPlex Complete Whole Transcriptome Amplification Kit (WTA2, Sigma) following the manufacturer’s protocol except the dsRNA was denatured at 95°C for 2 minutes, a lower amount of library synthesis primers was used by making a 1/8 dilution of the Library Synthesis Buffer in 5 mM dNTP mix and 23 cycles of amplification were performed. The WTA2 PCR product was purified using QIAquick PCR Purification kit (Qiagen). The dsRNA cDNA was sequenced at the University of Arizona Genetics Core using an Ion Torrent Personal Genome Machine (PGM) system and the Ion Sequencing 200 kit (Life Technologies) resulting in a library of 297,397 reads with a mean length of 163bp for a total of 48.6Mb.
The dsRNA cDNA reads were processed and filtered as follows: Primer sequences introduced during the Whole Transcriptome Amplification procedure were removed from the 5’ and 3’ ends of reads using TagCleaner (http://tagcleaner.sourceforge.net) (Schmieder et al., 2010). Reads were trimmed from the 3’ end if mean quality score was less than 15 using a sliding window size of 2bp and a step size of 1bp and reads with mean quality score below 15 or that were less than 15bp were removed using PRINSEQ (http://prinseq.sourceforge.net) (Schmieder and Edwards 2011). Contaminating sequences from the plasmid vector used to express GST-DRB4* were identified by blastn and removed. The resulting dsRNA cDNA library contained 201,865 reads with a mean length of 121bp for a total of 24.4Mb. After the k-mer comparison of the dsRNA cDNA sequences with the microbial DNA (see below) contaminating sequences derived from BL21 E. coli used to express GST-DRB4* were identified using Bowtie2 and removed resulting in 55,076 reads with average length of 122bp for a total of 6.75Mb in the dsRNA without a match to DNA pool and in 136,225 reads with average length of 122bp for a total of 16.7Mb in the dsRNA with a match to DNA pool.
Microbial DNA Isolation and Sequencing
DNA was isolated from microbial cells using a PowerWater DNA Isolation kit (MoBio) and the manufacturer’s protocol except an aliquot of cells were resuspended in PW1 and disrupted in a 2ml tube containing 0.1m beads (MoBio) by vortexing horizontally for 10 minutes. An aliquot of the microbial DNA was sequenced at the University of Arizona Genetics Core using an Ion Torrent Personal Genome Machine (PGM) system and the Ion Sequencing 200 kit (Life Technologies) producing a library of 375,426 reads, mean length 195bp for a total of 73.2Mb. To compare the Ion Torrent microbial DNA library to the dsRNA cDNA library, the reads were processed and filtered using PRINSEQ and sequences similar to BL21 E. coli were removed as described above for the dsRNA cDNA reads resulting in a library of 341,374 sequences with a mean length of 192bp for a total of 65.6Mb. For the k-mer comparison between the dsRNA cDNA library and microbial DNA, another aliquot of DNA was sequenced at the Tufts University Core Facility using a HiSeq 2500 system (Illumina) to obtain single-end 100bp reads resulting in a library of 226,542,350 reads with mean length of 100bp for a total of 22.6Gb (dataset will be available at NCBI SRA accession #). The Illumina HiSeq DNA library was processed and filtered using components of the SolexaQA package (solexaqa.sourceforge.net), DynamicTrim using the default settings was used to trim the reads and LengthSort was used to remove any reads less than 75bp in length.
K-mer Comparison between the Microbial dsRNA and DNA Sequences
The 20-mers present in the dsRNA cDNA reads were compared to those present in the Illumina HiSeq DNA read dataset by Bonnie Hurwitz at the University of Arizona using vmatch version 2.1.5 (http://www.vmatch.de/) similar to what is described in Hurwitz et al., 2013. Briefly, mkvtree was used to create a suffix array of all 20-mers present in each read in both of the samples. vmerstat was then used to search for the frequency of 20-mers between each of the datasets, a frequency of 1 for a 20-mer indicates that it was only found in one dataset and not the other. A PERL script was then used to parse the vmatch data to determine for each read in the dsRNA library the mode k-mer frequency of all its 20-mer subsequences. A mode frequency of 1 for a dsRNA read indicates that all of the 20-mers present in its sequence were only present in the dsRNA k-mer set and not in the DNA dataset. The dsRNA cDNA reads were then sorted into different pools based on whether they had a mode k-mer frequency of 1 (dsRNA without match to DNA) or greater than 1 (dsRNA with match to DNA).
Analysis of Metagenomic Sequences
The dsRNA sequences with no k-mer match to the microbial DNA, dsRNA sequences with a k-mer match to the microbial DNA and the processed Ion Torrent microbial DNA sequences were uploaded to the MG-RAST server (Meyer et al., 2008) and the default parameters were used for the taxonomic and functional assignments of reads.
In addition, Blastp was used to search for similarity between the predicted protein coding sequences in the dsRNA unique reads identified by MG-RAST and the NCBI nr database using soft masking and an E value 10−3. The MEtaGenome ANalyser (MEGAN version 4.64.1) was then used to assign dsRNA unique reads with Blastp hits to annotated proteins to taxonomic groups (Huson et al., 2011).
k-mer analysis of the dsRNA unique reads that did not map to DSRE1, 2, 3 or 4 was performed using tallymer (Kurtz et al., 2008) from genometools.org.
Contig Assembly and Analysis of dsRNA Elements (DSREs)
Trinity RNA-Seq assembler r2012-10-05 (Grabherr et al., 2011) was used to assemble contigs with a minimum length of 500bp from the dsRNA unique reads that had no match to the microbial DNA. Contigs that overlapped were manually assembled for a total of 64 unique contigs ranging in size from 500 to 4968bp. Blastx was used to search for significant similarity (E value 10−4) between the contig sequences and known proteins in the NCBI nr database. Bowtie2 v2.0.2 (Langmead and Salzberg, 2012) was used to map the dsRNA reads with no match to microbial DNA to the contigs. Four contigs with the highest number of reads mapped were selected for further study.
To test whether the four contigs were present in microbial dsRNA and/or DNA, purified dsRNA was reverse transcribed using random hexamer primers and Superscript III First Strand Synthesis Kit (Life Technologies) following the manufacturer’s protocol except the dsRNA was denatured at 95°C for 2 minutes. Nested PCR of the dsRNA cDNA and the microbial DNA was then performed using the primers listed in Table S3. The sequence of DSRE1, 2 and 3 was determined by sequencing their RT-PCR and 5’ RACE (5’RACE v2.0 kit, Life Technologies) products after cloning (Topo TA, Life Technologies). The full-length of DSRE4 was not determined by 5’RACE but its internal sequence was confirmed by sequencing its RT-PCR product.
Potential protein coding genes in the dsRNA elements were identified using a combination of three gene calling algorithms Glimmer v3.02 (Delcher et al., 2007) at http://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi, Metagene (Noguchi et al., 2006) at http://weizhong-lab.ucsd.edu/metagenomic-analysis/server/metagene/ and Prodigal (Hyatt et al., 2010) at http://prodigal.ornl.gov/server.html. Blastp was used to search for similarity between the predicted proteins and the NCBI nr database using an E value of 10−3. Workflows on Camera 2.0 (https://portal.camera.calit2.net) were used to screen the dsRNA elements for the presence of rRNA (Huang et al., 2009) and tRNAs (Lowe and Eddy, 1997) genes. tBlastx was used to search for similarity between the dsRNA elements and metagenomic datasets containing RNA viral sequences using a expect value of 10−5.
Supplementary Material
Highlights.
Developed method to purify dsRNA from complex environmental samples
Microbial populations contain dsRNA not encoded in corresponding DNA
dsRNA unique sequences potentially encode novel or divergent proteins
Fraction of dsRNA unique sequences assemble into new viral-like elements
ACKNOWLEDGEMENTS
We especially thank Bonnie Hurwitz for her computational analysis of the dsRNA and DNA sequences. We also thank Rob Knight for comments on the manuscript, Matthew Sullivan for many helpful discussions and other members of the Tucson Marine Phage Lab at the University of Arizona for their help in sample collection from the Scripps Institute of Oceanography and Biosphere 2 and Hanna Fares for plasmid L4440-Y75B7AL.4 for expressing dsRNA in E. coli and in vitro. This work was supported by funds from the Howard Hughes Medical Institute to C.J.D and R.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The MG-RAST ID numbers for the dsRNA sequences with no k-mer match to the microbial DNA, dsRNA sequences with at least one k-mer match to the microbial DNA and the microbial DNA sequences obtained by Ion Torrent sequencing are 451675.3, 4520281.3 and 4517766.3. The NCBI SRA accession number for the microbial DNA obtained by Illumina HiSeq sequencing is SRR1068156. The dsRNA elements, DSRE1 through DSRE64 were deposited as a Whole Genome Shotgun project. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession JFZN00000000. The version described in this paper is version JFZN01000000.
The authors declare that no competing interests exist.
REFERENCES
- Baker BJ, Sheik CS, Taylor CA, Jain S, Bhasi A, Cavalcoli JD, Dick GJ. Community transcriptomic assembly reveals microbes that contribute to deep-sea carbon and nitrogen cycling. ISME J. 2013;7:1962–1973. doi: 10.1038/ismej.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo PG, Calgua B, Zhao G, Hundesa A, Wier AD, Katz JP, Grabe M, Hendrix RW, Girones R, Wang D, Pipas JM. Raw sewage harbors diverse viral populations. MBio. 2011;2 doi: 10.1128/mBio.00180-11. e00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley AI, Lang AS, Suttle CA. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312:1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF. The microbial ocean from genomes to biomes. Nature. 2009;459:200–206. doi: 10.1038/nature08059. [DOI] [PubMed] [Google Scholar]
- Djikeng A, Kuzmickas R, Anderson NG, Spiro DJ. Metagenomic analysis of RNA viruses in a fresh water lake. PLoS ONE. 2009;4:e7264. doi: 10.1371/journal.pone.0007264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski PG, Barber RT, Smetacek V. Biogeochemical controls and feedbacks on ocean primary production. Science. 1998;281:200–206. doi: 10.1126/science.281.5374.200. [DOI] [PubMed] [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, DeLong EF. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara T, Koga R, Aoki N, Yuki C, Yamamoto N, Oyama N, Udagawa T, Horiuchi H, Miyazaki S, Higashi Y, Takeshita M, Ikeda K, Arakawa M, Matsumoto N, Moriyama H. The wide distribution of endornaviruses, large double-stranded RNA replicons with plasmid-like properties. Arch Virol. 2006;151:995–1002. doi: 10.1007/s00705-005-0688-5. [DOI] [PubMed] [Google Scholar]
- Gallimore CI, Green J, Casemore DP, Brown DW. Detection of a picobirnavirus associated with Cryptosporidium positive stools from humans. Arch Virol. 1995;140:1275–1278. doi: 10.1007/BF01322752. [DOI] [PubMed] [Google Scholar]
- Georg J, Voss B, Scholz I, Mitschke J, Wilde A, Hess WR. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BQ, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker CV, Brasier CM, Buck KW. A double-stranded RNA from a Phytophthora species is related to the plant endornaviruses and contains a putative UDP glycosyltransferase gene. J Gen Virol. 2005;86:1561–1570. doi: 10.1099/vir.0.80808-0. [DOI] [PubMed] [Google Scholar]
- Huang Y, Gilna P, Li W. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics. 2009;25:1338–1340. doi: 10.1093/bioinformatics/btp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Pace NR. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 1996;14:190–197. doi: 10.1016/0167-7799(96)10025-1. [DOI] [PubMed] [Google Scholar]
- Human MicrobiomeProject Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz BL, Deng L, Poulos BT, Sullivan MB. Evaluation of methods to concentrate and purify ocean virus communities through comparative replicated metagenomics. Environmental Microbiology. 2013;15:1428–1440. doi: 10.1111/j.1462-2920.2012.02836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Mitra A, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Ghabrial SA. Molecular characterization of Penicillium chrysogenumvirus: reconsideration of the taxonomy of the genus Chrysovirus. J Gen Virol. 2004;85:2111–2121. doi: 10.1099/vir.0.79842-0. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Tomita R, Sakamoto M. Recombinant plant dsRNA-binding protein as an effective tool for the isolation of viral replicative form dsRNA and universal detection of RNA viruses. J Gen Plant Pathol. 2009;75:87–91. [Google Scholar]
- Kurtz S, Narechania A, Stein JC, Ware D. A new method to compute K-mer frequencies and its application to annotate large repetitive plant genomes. BMC Genomics. 2008;9:517. doi: 10.1186/1471-2164-9-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AS, Rise ML, Culley AI, Steward GF. RNA viruses in the sea. FEMS Microbiol Rev. 2008;33:295–323. doi: 10.1111/j.1574-6976.2008.00132.x. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard RE, Kumar A. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 1981;9:5125–5140. doi: 10.1093/nar/9.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EL. Microorganisms and their roles in fundamental biogeochemical cycles. Curr Opin Biotechnol. 2011;22:456–464. doi: 10.1016/j.copbio.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L. Phages with segmented double-stranded RNA genomes. In: Calendar R, editor. The Bacteriophages. 2nd ed. New York: Oxford University Press; 2006. pp. 197–207. [Google Scholar]
- Morris TJ, Dodds JA. Isolation and analysis of double-stranded RNA from virus infected plant and fungal tissue. Phytopathology. 1979;69:854–858. [Google Scholar]
- Morse DP, Bass BL. Long RNA hairpins that contain inosine are present in Caenorhabdities elegans poly(A)+ RNA. Proc Natl Acad Sci USA. 1999;96:6048–6053. doi: 10.1073/pnas.96.11.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naitow H, Canady MA, Lin T, Wickner RB, Johnson JE. Purification, crystallization, and preliminary X-ray analysis of L-A: a dsRNA yeast virus. J Struct Biol. 2001;135:1–7. doi: 10.1006/jsbi.2001.4371. [DOI] [PubMed] [Google Scholar]
- Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequence. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NR, Stahl DA, Lane DJ, Olsen GJ. The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol. 1986;9:1–55. [Google Scholar]
- Pearson MN, Beever RE, Boine B, Arthur K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol. 2009;10:115–128. doi: 10.1111/j.1364-3703.2008.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan R, Sloan DB, Ochman H. Antisense transcription is pervasive but rarely conserved in enteric bacteria. MBio. 2012;3:e00156–e00212. doi: 10.1128/mBio.00156-12. ppii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Lim YW, Rohwer F, Edwards R. TagCleaner: Identification and removal of tag sequences from genomic and metagenomic datasets. BMC Bioinformatics. 2010;11:341. doi: 10.1186/1471-2105-11-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt MJ, Breinig F. Yeast viral killer toxins: lethality and self-protection. Nat Rev Microbiol. 2006;4:212–221. doi: 10.1038/nrmicro1347. [DOI] [PubMed] [Google Scholar]
- Schonborn J, Oberstrass J, Breyel E, Tittgen J, Schumacher J, Lukacs N. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991;19:2993–3000. doi: 10.1093/nar/19.11.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Tyson GW, Eppley JM, DeLong EF. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2011;5:999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Backhed F. The gut microbiota---masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Strauss EE, Lakshman DK, Tavantzis SM. Molecular characterization of the genome of a partitivirus from the basidiomycete Rhizoctonia solani. J Gen Virol. 2000;81:549–555. doi: 10.1099/0022-1317-81-2-549. [DOI] [PubMed] [Google Scholar]
- Steward GF, Culley AI, Mueller JA, Wood-Charlson EM, Belcaid M, Poisson G. Are we missing half of the viruses in the ocean? ISME J. 2013;7:672–679. doi: 10.1038/ismej.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Weinberg Z, Perreault J, Meyer MM, Breaker RR. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature. 2009;462:656–659. doi: 10.1038/nature08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.