SUMMARY
Chromosome banding analysis is the gold standard method for the identification of recurrent cytogenetic abnormalities in acute myeloid leukaemia (AML). It allows stratification of AML patients into subgroups with distinct responses to therapy and survival. Unfortunately a variety of issues hamper cytogenetic evaluation in ~10% of cases (unsuccessful cytogenetics [UC]) and the outcome of these patients is poorly understood. To better define the significance of unsuccessful cytogenetic in patients with AML, we compared the baseline characteristics and the prognostic impact of the 94 (6%) patients, whose standard metaphase analysis yielded unacceptable results, to the remaining 1403 AML patients with successful cytogenetic analysis treated on successive SWOG protocols. The incidence of UC increased with age, with peak incidence in patients older than 60 years. These patients had a lower response rate to induction chemotherapy (complete remission rate of 43%) and dismal 5-year survival rates (16%), which was especially poor in patients older than 60 years (<5%). The complete remission and survival rates were similar to those seen in patients with unfavorable karyotype. The early death rate was not increased. These results suggest that UC increases with age and predict for poor outcomes, similar to the outcomes of patients with unfavorable karyotype.
Keywords: Unsuccessful metaphase analysis, acute myeloid leukaemia, karyotype, unfavorable and overall survival
INTRODUCTION
Standard metaphase analysis is one of the most important tools for predicting initial response to therapy, duration of remission and survival in patients with acute myeloid leukaemia (AML). (Byrd JC et al, 2002 and Grimwade D et al, 2010) However, chromosome banding studies may be hindered by several factors, including low proliferative rate of the leukemic clone in tissue culture, insufficient number of metaphase cells, reduced cell viability or hypocellularity upon arrival to the reference laboratory, poor chromosome morphology or complexity of the karyotype. (Fischer K et al, 1996) Unsuccessful karyotyping may also reflect intrinsic biological properties of the leukemic clone that may be otherwise difficult to ascertain and that could reveal valuable prognostic features in these patients. Previous studies have demonstrated that the rate of unsuccessful karyotyping in patients with AML is approximately 10%. (Grimwade D et al, 2010 and Fischer K et al, 1996) We determine the characteristics of AML patients with unsuccessful cytogenetics (UC) and investigate its prognostic significance in AML patients treated on SWOG clinical trials.
METHODS
Patients and treatments
We used data from 1,623 patients with previously untreated AML registered between 1986 and 2009 in one of ten successive SWOG clinical trials (S8600, S9031, S9034, S9126, S9333, S9500, S9617, S9918, S0106, and S0112). (Medeiros BC et al, 2010) Centrally reviewed cytogenetic data from diagnosis were used. Patients were classified into one of 3 categories: normal, abnormal or UC. A result was regarded as normal karyotype only after successful analysis of 20 or more normal metaphases. Cytogenetic results were considered abnormal even if fewer than 20 metaphases were analyzed if at least two metaphases had the same aberration in the case of a structural abnormality or an extra chromosome, or if at least 3 shared the same abnormality in case of a monosomy. If no clonal abnormality was present and the total metaphases available for analysis were fewer than 20, the cytogenetic study was regarded as unacceptable. The central review process of the SWOG Cytogenetics Committee ensured proper culturing conditions for all patient samples, and results were reviewed for accuracy in description of the structural and numerical clonal chromosomal abnormalities, which were reported in accordance with the International System for Human Cytogenetic Nomenclature (International System for Human Cytogenetic Nomenclature, 2005) and were grouped according to published criteria adopted by SWOG. (Medeiros BC et al, 2010) Given the retrospective nature of this study, comprehensive assessment of known prognostic recurrent cytogenetic abnormalities by FISH or genomic mutations by PCR was not possible. Informed consent was obtained from all subjects involved in this study and research was conducted in accordance with the Declaration of Helsinki. Induction therapies were grouped into those: (1) with “standard” doses of cytarabine (100 mg/m2 daily for 7 days), (2) with higher than “standard” doses of cytarabine (typically at least 1 g/m2, per dose), (3) without cytarabine. For patients achieving a complete remission, consolidation therapy varied based on protocol design. Data on off-protocol allogeneic hematopoietic cell transplant (alloHSCT) was not routinely collected.
Definitions of end points
Complete remission (CR) was defined as bone marrow blasts < 5%; absence of blasts with Auer rods; absence of extramedullary disease; absolute neutrophil count > 1,000/μL; platelet count > 100,000/μL and independence of red cell transfusions. Complete remission with incomplete platelet recovery was not consistently captured in all studies and therefore not included in the analysis. Early death (ED) was defined as death within 28 days of initiating therapy. Overall survival (OS) was measured from the time of registration to death from any cause, with patients last known to be alive censored at date of last contact. Event-free survival (EFS) measures the time from registration to removal from protocol treatment without achieving a CR, relapse from a CR, or death, whichever occurred first, with patients last known to be alive without relapse censored at date of last contact. Relapse-free survival measures the time from CR to relapse from CR or death, with patients last known to be alive without relapse censored at date of last contact.
Definition of Unsuccessful Central Cytogenetic Review
The central review process of the SWOG Cytogenetics Committee rejects submitted results from local institutions for up to 5 separate reasons. These are classified as: 1) Inadequate banding; 2) Too few metaphases were available for analysis, which includes cases where no analyzable metaphases were obtained; 3) The local cytogenetics laboratory or the central reviewer deemed the specimen to be inadequate, which includes cases of pre-analytical errors where wrong anticoagulant used, insufficient mix with even the correct anticoagulant leading to clotting, too much time spent in transit/extreme temperature exposure during transportation, labeling error, hemolysed specimen, or low cell count; 4) Processing of the sample was inadequate which usually refers to incorrect tissue culture conditions or only one culture set up; 5) For other reasons.
Statistical Methods
Associations between continuous variables were assessed using the Wilcoxon rank-sum test. Associations between categorical variables were evaluated using Fisher's exact test. Survival curves were estimated using the Kaplan-Meier method. Log-rank tests were used to assess differences between survival curves. Cox regression models were used to evaluate covariate associations with OS. (Cox DR. 1972) The significance level for analyses was ≤.05. For regression analyses, age, hemoglobin, platelets, marrow blast percentage, and white blood cell count at diagnosis were measured as continuous variables. Performance status (PS) was divided according to groups with PS 3 and 4 being combined in a single group due to the small number of patients. Secondary AML was not included in the regression analysis as it represented an exclusion criterion for most clinical trials included in this analysis or was not captured in the database.
RESULTS
Baseline characteristics of patients with UC studies
Among the 1,623 patients with centrally reviewed cytogenetic studies, 220 cytogenetic reports were rejected during central review (14%). Reasons for rejection included (samples could have been rejected for more than one reason or for reasons not listed): inadequate banding (n = 34), too few metaphases were available for analysis (n = 86), inadequate specimen handling (n = 22), and inadequate processing (n = 95).
Initially, we determined if there were differences in outcomes between patients with inadequate banding or too few metaphases available for analysis (n - 94) and those with inadequate specimen handling or processing (pre-analytical) (n – 126). As some patients had more than one reason for rejection and since inadequate processing can lead to suboptimal sample quality, all patients with inappropriate sampling were included on the pre-analytical cohort irrespective of whether they also had inadequate banding or too few metaphases. No significant differences between CR rates (43% vs. 52%), 4-week mortality (13% vs. 12%), or OS (HR=1.3 95% confidence interval [CI] 0.97-1.74, p=0.08) were noted between these two large groups. These differences remained non-significant after adjusting for age, PS, and white blood cell (WBC) count (CR adjusted OR=0.94 95% CI 0.82-1.07, p=0.33; for ED adjusted OR=0.98 95% CI 0.90-1.07, p=0.60; and for OS adjusted HR=1.16 95% CI 0.86, 1.57, p=0.33). Given that unique biological properties are unlikely to be associated with inadequate specimen handling or processing, we limited our subsequent analyses to patients only with inadequate banding or metaphases (n -94 [6%]) (Hereafter UC). Table 1 summarizes the baseline characteristics and cytogenetic classification of the cohorts. Patients with UC were older at diagnosis (median 60 years vs. 54 years, p= 0.08) and we noted an increased incidence with advancing age (p=0.08). No differences in treatment were observed between patients with accepted cytogenetic studies, UC and pre-analytical cohorts (p-value comparing the three cohorts = 0.55) (data not shown).
TABLE 1.
Baseline characteristics between patients with approved and failed cytogenetic studies
| Approved Cytogenetics (n=1,403) |
Unsuccessful Cytogenetics (n=94) |
p-value | |
|---|---|---|---|
| Age (years) | 54 | 60 | 0.08 |
| Incidence | 0.08 | ||
| < 40 (%) | 95 | 5 | |
| 40-49 (%) | 95 | 5 | |
| 50-59 (%) | 95 | 5 | |
| 60-69 (%) | 91 | 9 | |
| >70 (%) | 92 | 8 | |
| WBC (109/L) | 19 | 17 | 0.94 |
| Marrow Blasts (%) | 65 | 70 | 0.15 |
| Platelets (109/L) | 51 | 59 | 0.34 |
| Female (%) | 45 | 50 | 0.57 |
| ECOG PS (%) | 0.55 | ||
| 0 | 33 | 29 | |
| 1 | 48 | 46 | |
| 2 | 14 | 16 | |
| Karyotype (%) | |||
| Favorable | 155 (10) | ||
| Intermediate | 668 (41) | ||
| Unknown | 125 (8) | ||
| Unfavorable * | 272 (17) | ||
| Very Unfavorable (MK) | 183 (11) | ||
| Unsuccessful | 94 (6) | ||
Legend:
- Either N (%) or medians are presented. Some columns may not add to total n due to missing data.
- Abbreviations: WBC- White blood cells count at diagnosis, PS- Performance Status, MK- Monosomal Karyotype.
- Excludes patients with monosomal karyotype
Impact of UC on complete remission and early death rates
Next, we investigated the significance of UC on early treatment outcomes in patients with AML. (Table 2) The CR rate for UC patients (43%) was similar when compared to patients with unfavorable karyotype (excluding monosomal karyotype [MK+ AML]) (p-0.72), but significantly better than patients with MK+ AML (hereafter “very unfavorable”) (p-0.04), and inferior to patients with favorable (p<0.001), or intermediate risk cytogenetics (p-0.01). After adjusting for age, PS, gender, treatment, WBC count, hemoglobin (Hb) level, platelets, and marrow blasts percentage at diagnosis, UC remained an independent factor for lower CR rate similar to those observed in unfavorable karyotypes (Table 2).
TABLE 2.
Impact of Unsuccessful Cytogenetic Analysis on complete remission, 4-week mortality and 5-year overall survival rates
| UC | Favorable | Intermediate | Unfavorable | Very Unfavorable | |||||
|---|---|---|---|---|---|---|---|---|---|
| p- value |
p- value |
p- value |
p- value |
||||||
| Remission | |||||||||
| CR Rate (%) |
43 | 80 | <0.01 | 58 | 0.01 | 46 | 0.72 | 30 | 0.04 |
| OR (95% CI) |
5.02 (2.68,9.4) |
<0.01 | 1.60 (1.00,2.58) |
0.05 | 0.93 (0.55,1.57) |
0.79 | 0.57 (0.33,1.0) |
0.05 | |
| Early Death | |||||||||
| 4-week (%) |
12 | 3 | >0.3 | 8 | >0.3 | 12 | >0.3 | 21 | 0.07 |
| OR (95% CI) |
0.41 (0.12,1.44) |
0.17 | 0.80 (0.37,1.71) |
0.56 | 1.26 (0.54,2.95) |
0.59 | 2.20 (0.98,4.96) |
0.06 | |
| Survival | |||||||||
| 5-year (%) |
16 | 55 | <0.01 | 26 | <0.01 | 14 | 0.79 | 2 | <0.01 |
| HR (95% CI) |
0.38 (0.27, 0.54) |
<0.01 | 0.71 (0.55, 0.91) |
0.01 | 1.20 (0.91, 1.57) |
0.19 | 2.14 (1.62, 2.84) |
<0.01 | |
Abbreviations: UC- Unsuccessful cytogenetics, CR- complete remission; OR- Odds Ratio, CI- Confidence Interval, HR- Hazard Ratio.
Early death was similar for patients with UC compared to patients with successful cytogenetic analysis (12% vs. 9%, p=0.47). Unadjusted and adjusted Cox analyses demonstrated a trend towards lower ED rate only when compared to MK+ AML (p=0.07 and p=0.06, respectively).
Association of unsuccessful cytogenetic studies and survival in patients with AML
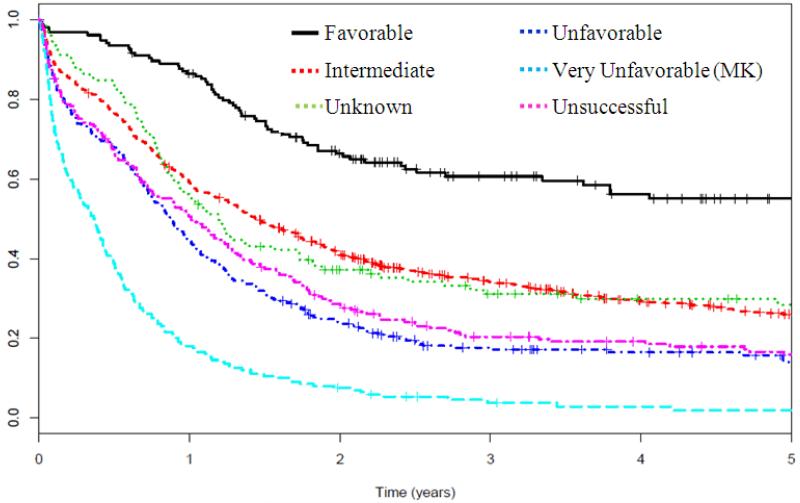
Compared to patients with successful cytogenetic analysis, patients with UC had worse survival (HR=1.42 95% CI 1.29, 1.55), p=0.002). As shown in Figure 1 and Table 2, survival was similar to patients with unfavorable risk cytogenetics (excluding MK+ AML). An unadjusted Cox model demonstrated that UC patients had worse OS compared to favorable or intermediate risk karyotype (p<0.01 for both comparisons), improved survival compared to MK+ AML (p<0.01) and similar survival to unfavorable karyotype (p=0.79). Multivariate analysis demonstrated that UC remained independently associated with worse OS compared to favorable (p<0.001) or intermediate risk karyotype (p=0.01) and better OS than patients with MK+ AML (p<0.01). The 5-year OS of UC patients decreased with advancing age, where patients 60 and older had dismal survival rates of less than 5% at 5 years (data not shown).
Figure 1.
Five-year overall survival Kaplan-Meier plots according to revised cytogenetic risk category definition
Legend: The cytogenetic risk classification has been previously described by SWOG.5 In brief, it compares the survival outcomes of patients with unsuccessful cytogenetics (magenta dashed) with those in previously described subgroups, including favorable (black solid), intermediate (orange dashed), unknown (green dashed), unfavorable (navy dashed), very unfavorable (light blue dashed).
DISCUSSION
Given the risk-adapted nature of AML treatment and the prognostic implication of cytogenetic risk stratification, it is disappointing and frustrating when diagnostic cytogenetic studies fail to produce analyzable metaphases. The data presented here should help physicians with this dilemma. Our results demonstrate that UC studies occur more commonly in older patients, predict for poor response to chemotherapy and should be considered a high-risk feature.
The significance of UC on the outcome of AML patients has been scarcely reported. Schiffer et al described the median age (50 years) and CR rate (60%) of patients with “no mitoses”. (Schiffer CA et al, 1989) The United Kingdom Medical Research Council (MRC) only concisely reported an improved 10-year survival in UC patients compared to those with successful studies or no sample submitted for analysis (46% vs. 40% vs. 34%, p<0.001). (Grimwade D et al, 2010) The HOVON reported that between 5% and 12% of their patients had “no cytogenetic testing results available”. (Löwenberg B et al, 2009 and Löwenberg B et al, 2011) In contrast to the MRC data, older patients (>60 years) with UC had similar outcomes (CR and 2-year survival rates) to patients with unfavorable karyotype (excluding MK+ AML). (Löwenberg B et al, 2011) Our findings support this observation that patients with UC should be considered high-risk patients, however, they do not provide a clear explanation for the discordant results observed in the MRC trials. Similarly, in myelodysplasia, the presence of UC at diagnosis was associated with worse survival when compared to normal karyotype. (Cervera J et al, 2009)
Our results confirm previous reports that metaphase analysis is unsuccessful in ~10% of AML patients. (Grimwade D et al, 2010, Fischer K et al, 1996 and Löwenberg B et al, 2011) What remains unclear is how and why UC results into poor response to chemotherapy. The routine use of targeted fluorescence in situ hybridization (FISH) studies has been proposed as a strategy to overcome UC. (Tiu RV et al, 2009) This method can be very sensitive for the detection of specific AML-related recurrent structural and numerical abnormalities. However, detection of genomic abnormalities by FISH requires multiple assays and is limited by the number of DNA probe sets available, therefore limiting its applicability to uncommon cytogenetic aberrations. Single nucleotide polymorphism microarrays (SNP-Array) could be considered an alternative karyotyping tool in patients with unsuccessful cytogenetic studies. SNP-Array has excellent resolution, requires small amounts of DNA and detects copy number neutral loss of heterozygosity. In AML, SNP-Arrays enhance the detection of chromosomal abnormalities compared to conventional cytogenetic analysis, although it cannot detect balanced translocations. (Tiu RV et al, 2009) In a recent study, SNP-arrays obtained karyotypic information in all patients with UC, including 60% of abnormal karyotypes. (Dougherty MJ et al, 2011) Unfortunately the commercial implementation of these SNP-arrays is not widespread.
Clear limitations to this work exist. The data were collected from patients enrolled in multiple sequential trials testing different treatment regimens and post-remission strategies. Thus, these results, especially those regarding survival, need to be interpreted with caution. The impact of alloHSCT cannot be determined as only ~10% of patients participated in studies testing the impact of alloHSCT on outcome .(Slovak ML et al, 2000 and List AF et al, 2001) Retrospective determination of important prognostic genomic abnormalities routinely used in current clinical practice, such as FLT3-ITD, NPM1 and CEBPA mutations, or FISH for AML specific recurrent cytogenetic abnormalities (inv(16), t(8;21), −7/del(7q), −5/del(5q)), was not possible for lack of archived material.
Nevertheless, our results demonstrate, in a large cohort of patients with AML, that UC studies are relatively common in AML and are associated with increasing age and poor outcomes. Although new techniques may overcome the technical challenges associated with UC, these findings should be considered an unfavorable prognostic factor in untreated AML patients.
Acknowledgements
Support: This work was supported in part by the following Public Health Service Cooperative Agreement grant numbers awarded by the National Cancer Institute, National Institutes of Health, Department of Health and Human Services: CA32102 and CA38926
Footnotes
Clinical Trials Registration Numbers: ClinicalTrials.gov Identifier: NCT014343329; NCT01338974; NCT00899171; NCT1059734; NCT01059734; NCT00899743; NCT0143329, NCT00023777; NCT00085709; NCT01360125; NCT00004217
Conflict of Interest Disclosures: The authors indicated no potential conflicts of interest.
REFERENCES
- Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS, Koduru PR, Moore JO, Stone RM, Mayer RJ, Feldman EJ, Davey FR, Schiffer CA, Larson RA, Bloomfield CD, Cancer and Leukemia Group B (CALGB 8461) Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- Cervera J, Solé F, Haase D. Prognostic impact on survival of an unsuccessful conventional cytogenetic study in patients with myelodysplastic syndromes (MDS) Leuk Res. 2009;33(S1):S75–76. [Google Scholar]
- Cox DR. Regression models and life-tables (with discussion) J R Stat Soc Series B. 1972;34:187–220. [Google Scholar]
- Dougherty MJ, Wilmoth DM, Tooke LS, Shaikh TH, Gai X, Hakonarson H, Biegel JA. Implementation of high resolution single nucleotide polymorphism array analysis as a clinical test for patients with hematologic malignancies. Cancer Genet. 2011;204:26–38. doi: 10.1016/j.cancergencyto.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Fischer K, Scholl C, Sàlat J, Fröhling S, Schlenk R, Bentz M, Stilgenbauer S, Lichter P, Döhner H. Design and validation of DNA probe sets for a comprehensive interphase cytogenetic analysis of acute myeloid leukemia. Blood. 1996;88:3962–71. [PubMed] [Google Scholar]
- Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, Wheatley K, Harrison CJ, Burnett AK, National Cancer Research Institute Adult Leukaemia Working Group Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- International System for Human Cytogenetic Nomenclature . An International System for Human Cytogenetic Nomenclature. S. Karger; Basel, Switzerland: 2005. [Google Scholar]
- List AF, Kopecky KJ, Willman CL, Head DR, Persons DL, Slovak ML, Dorr R, Karanes C, Hynes HE, Doroshow JH, Shurafa M, Appelbaum FR. Benefit of cyclosporine modulation of drug resistance in patients with poor-risk acute myeloid leukemia: a Southwest Oncology Group study. Blood. 2001;98:3212–20. doi: 10.1182/blood.v98.12.3212. [DOI] [PubMed] [Google Scholar]
- Löwenberg B, Pabst T, Vellenga E, van Putten W, Schouten HC, Graux C, Ferrant A, Sonneveld P, Biemond BJ, Gratwohl A, de Greef GE, Verdonck LF, Schaafsma MR, Gregor M, Theobald M, Schanz U, Maertens J, Ossenkoppele GJ, Dutch-Belgian Cooperative Trial Group for Hemato-Oncology (HOVON) and Swiss Group for Clinical Cancer Research (SAKK) Collaborative Group Cytarabine dose for acute myeloid leukemia. N Engl J Med. 2011;364:1027–36. doi: 10.1056/NEJMoa1010222. [DOI] [PubMed] [Google Scholar]
- Löwenberg B, Ossenkoppele GJ, van Putten W, Schouten HC, Graux C, Ferrant A, Sonneveld P, Maertens J, Jongen-Lavrencic M, von Lilienfeld-Toal M, Biemond BJ, Vellenga E, van Marwijk Kooy M, Verdonck LF, Beck J, Döhner H, Gratwohl A, Pabst T, Verhoef G, Dutch-Belgian Cooperative Trial Group for Hemato-Oncology (HOVON) German AML Study Group (AMLSG) Swiss Group for Clinical Cancer Research (SAKK) Collaborative Group High-dose daunorubicin in older patients with acute myeloid leukemia. N Engl J Med. 2009;361:1235–48. doi: 10.1056/NEJMoa0901409. [DOI] [PubMed] [Google Scholar]
- Medeiros BC, Othus M, Fang M, Roulston D, Appelbaum FR. Prognostic impact of monosomal karyotype in young adult and elderly acute myeloid leukemia: the Southwest Oncology Group (SWOG) experience. Blood. 2010;116:2224–8. doi: 10.1182/blood-2010-02-270330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer CA, Lee EJ, Tomiyasu T, Wiernik PH, Testa JR. Prognostic impact of cytogenetic abnormalities in patients with de novo acute nonlymphocytic leukemia. Blood. 1989;73:263–70. [PubMed] [Google Scholar]
- Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, Paietta E, Willman CL, Head DR, Rowe JM, Forman SJ, Appelbaum FR. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 200;96:4075–83. [PubMed] [Google Scholar]
- Tiu RV, Gondek LP, O'Keefe CL, Huh J, Sekeres MA, Elson P, McDevitt MA, Wang XF, Levis MJ, Karp JE, Advani AS, Maciejewski JP. New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia. J Clin Oncol. 2009;27:5219–26. doi: 10.1200/JCO.2009.21.9840. [DOI] [PMC free article] [PubMed] [Google Scholar]