Abstract
Oncogenic K-Ras proteins, such as K-RasG12D, accumulate in the active, guanosine triphosphate (GTP)–bound conformation and stimulate signaling through effector kinases. The presence of the K-RasG12D oncoprotein at a similar abundance to that of endogenous wild-type K-Ras results in only minimal phosphorylation and activation of the canonical Raf–mitogen-activated or extracellular signal–regulated protein kinase kinase (MEK)–extracellular signal–regulated kinase (ERK) and phosphoinositide-3 kinase (PI3K)–Akt–mammalian target of rapamycin (mTOR) signaling cascades in primary hematopoietic cells, and these pathways remain dependent on growth factors for efficient activation. Here, we show that phospholipase C γ (PLC-γ), PI3K, and their generated second messengers link activated cytokine receptors to Ras and ERK signaling in differentiated bone marrow cells and in a cell population highly enriched for leukemia stem cells. Cells expressing endogenous oncogenic K-RasG12D remained dependent on the second messenger diacylglycerol for the efficient activation of Ras-ERK signaling. These data raise the unexpected possibility of therapeutically targeting proteins that function upstream of oncogenic Ras in cancer.
Introduction
Ras proteins are signal switch molecules that regulate cell fate by cycling between active, guanosine triphosphate (GTP)–bound (Ras-GTP) and inactive, guanosine diphosphate (GDP)–bound (Ras-GDP) conformations (13, 55). Cancer-associated mutant RAS alleles encode oncogenic proteins that accumulate in the GTP-bound conformation because of a defective intrinsic guanosine triphosphatase (GTPase) activity and their resistance to GTPase-activating proteins (GAPs)(6, 13, 55). Based on the high prevalence of somatic RAS mutations in many lethal human malignancies, reversing the biochemical consequences of oncogenic Ras signaling is of fundamental importance for reducing the worldwide burden of cancer. However, the Ras GTPase switch poses extraordinary problems for anti-cancer drug development, because an “ideal” agent must restore normal GTPase activity and responsiveness to GAPs (that is, it must repair a “broken” enzyme) in the context of a highly constrained domain of Ras in which the γ phosphate of GTP interacts with the “arginine finger” of GAPs (6, 13, 55). Based on the assumption that oncogenic Ras-GTP makes cancer cells less reliant on growth factors for survival and proliferation by constitutively activating downstream signaling pathways, intensive efforts are focusing on developing and evaluating small-molecule inhibitors of Ras effectors, particularly components of the phosphoinositide-3 kinase (PI3K)–Akt–mammalian target of rapamycin (mTOR) and Raf–mitogen-activated or extracellular signal–regulated protein kinase kinase (MEK)–extracellular signal–regulated kinase (ERK) pathways (14). Recent studies also raise the possibilites of therapeutically targeting other domains of Ras oncoproteins (39) or interfering with their post-translational processing (56, 59).
Juvenile myelomonocytic leukemia (JMML) and chronic myelomonocytic leukemia (CMML) are myeloproliferative neoplasms (MPNs) that frequently contain “driver” mutations in genes encoding components of Ras signaling networks such as NRAS, KRAS, PTPN11, CBL, and NF1 (35, 53). Germline PTPN11, CBL, and NF1 mutations confer an increased risk of JMML, which implicated hyperactive Ras as initiating this aggressive leukemia. Bone marrow cells from JMML patients form granulocyte macrophage colony–forming unit (CFU-GM) colonies in the absence of cytokine growth factors and at very low concentrations of granulocyte macrophage colony-stimulating factor (GM-CSF). This cellular hallmark of JMML is also observed in bone marrow cells from Mx1-Cre, KrasG12D mice (7, 16). These mice express oncogenic K-RasG12D from its endogenous locus in hematopoietic cells and develop a fatal MPN that recapitulates many features of CMML and JMML (7, 9, 16).
Although Ras-GTP abundance is constitutively increased in bone marrow cells from Mx1-Cre, KrasG12D mice compared to that in cells from wild-type mice, the amounts of phosphorylated Akt and ERK (pAkt and pERK) in cells from these mice are not changed or only minimally increased compared to those in wild-type mice. Bone marrow cells from both wild-type and Mx1-Cre, KrasG12D mice markedly increase pAkt and pERK abundance in response to GM-CSF stimulation (7). Consistent with these biochemical data, CFU-GM colony growth is greatly enhanced by GM-CSF (7, 9). Similarly, mouse embryonic fibroblasts (MEFs) from Mx1-Cre, KrasG12D mice show little or no basal activation of canonical effector pathways despite enhanced abundance of Ras-GTP, and they exhibit marked increases in pERK and pAkt abundances in response to epidermal growth factor (EGF)(21, 52). Administering PD0325901, a potent and selective MEK inhibitor, to Mx1-Cre, KrasG12D mice with MPN results in substantial hematologic improvement, characterized by a restoration of normal white blood cell counts, improvement in anemia, and reduction in splenic enlargement (38). This observation provides direct evidence that aberrant Raf-MEK-ERK signaling underlies the aberrant proliferation of hematopoietic cells in vivo in this model of human MPN. Understanding the biochemical mechanisms required for the full activation of oncogenic Ras in response to growth factor stimulation might therefore reveal new therapeutic targets.
Based on the extensive cell biologic, genetic, and preclinical data implicating aberrant GM-CSF signaling in the pathogenesis of JMML, we combined phospho-flow cytometric analysis and pharmacological pathway mapping (33) to clarify the molecular mechanisms linking the stimulation of primary hematopoietic cells with GM-CSF to the activation of ERK (2, 16, 29, 31, 38). Here, we show that phospholipase C γ (PLC-γ) and PI3K were essential for efficient Ras and ERK activation, and that analogs of diacylglycerol (DAG), the second messenger generated by PLC-γ, by-passed this requirement. Despite having substantially increased amounts of Ras-GTP compared to hematopoietic cells from wild-type mice, hematopoietic cells from Mx1-Cre, KrasG12D mice remained dependent on PLC-γ and PI3K for efficient activation of ERK. Inhibitors of PLC-γ and PI3K also interfered with ERK phosphorylation and activation in response to stem cell factor (SCF) in immature populations of bone marrow cells from wild-type and Mx1-Cre, KrasG12D mice that were highly enriched for hematopoietic stem cells (HSC) and leukemia-initiating cells. Studies of hematopoietic progenitor colony growth and of cytokine signaling after treatment of mice with a PI3K inhibitor confirmed the physiological relevance of this pathway. Together, these data raise the possibility that targeting proteins that function upstream of oncogenic Ras – alone or in combination with other therapies – may represent a viable strategy for treating cancers, particularly those that are highly responsive to growth factors. In addition, these data suggest that PI3K inhibitors could contribute to myelosuppression by interfering with cytokine signaling networks in normal hematopoietic stem and progenitor cells.
Results
PLC-γ and PI3K stimulate ERK activation in hematopoietic cells from wild-type and Mx1-Cre, KrasG12D mice
Compared to those of wild-type mice, the bone marrows of Mx1-Cre, KrasG12D mice contain a preponderance of myeloid cells with a corresponding loss of erythroid and lymphoid cell populations (7, 16). These changes in cellular composition might increase the apparent response to a stimulus that acts on myeloid cells alone. Phospho-flow cytometry avoids this problem by analyzing signaling at the single-cell level in a heterogeneous tissue such as bone marrow (28, 54). We therefore used this methodology to characterize cytokine responses in bone marrow cells from wild-type and Mx1-Cre, KrasG12D mice. Our analysis focused on differentiated myeloid lineage cells (mostly mature granulocytes and monocytes) that expressed the Mac1 and Gr1 cell-surface markers (M+G+ cells), as well as the c-kit+Mac1–Gr1– (K+M–G–) population, which was enriched for stem and progenitor cells and constituted 2 to 5% of nucleated bone marrow cells (fig. S1). We found that basal amounts of pERK were comparable in wild-type and KrasG12D M+G+ and K+M–G– bone marrow cells (Fig. 1A and fig. S2A). Although the bone marrow of Mx1-Cre, KrasG12D mice contained more GM-CSF–responsive M+G+ cells than did bone marrow from wild-type mice (fig. S1), the extent of ERK phosphorylation within responding cells was comparable in wild-type and KrasG12D cells (Fig. 1A and fig. S2A, green curves). Importantly, KrasG12D cells were dependent on GM-CSF stimulation to fully activate ERK (Fig. 1A and fig. S2A).
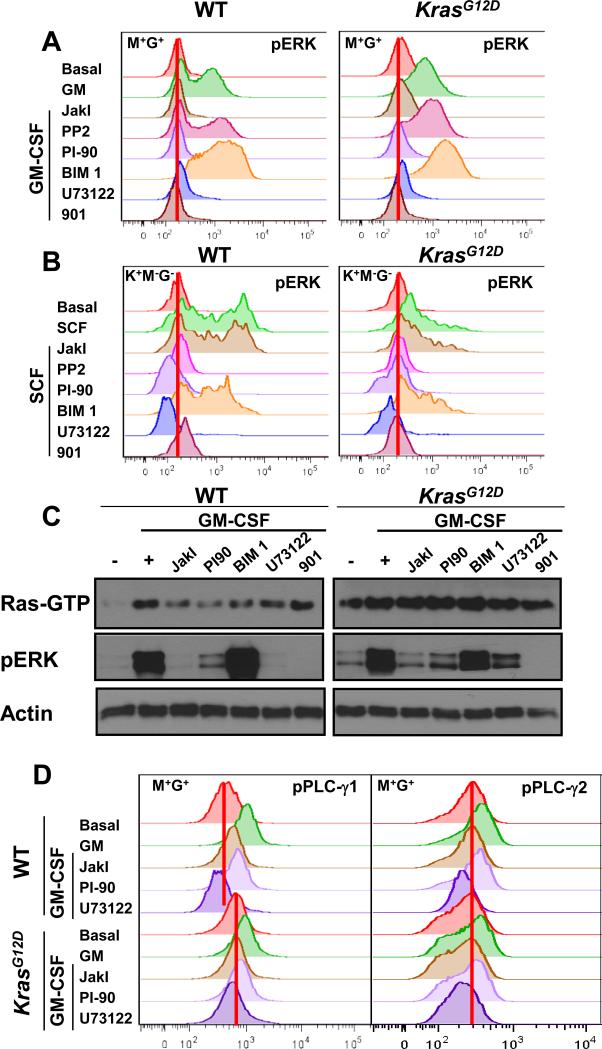
Fig. 1. PI-90 and U73122 impair Ras and ERK activation in wild-type and Mx1-Cre, KrasG12D hematopoietic cells and reduce PLC-γ phosphorylation.
(A) M+G+ cells from wild-type (WT) and Mx1-Cre, KrasG12D (KrasG12D) mice were incubated for 30 min with the indicated inhibitors (each at final concentration of 5 μM), stimulated with GM-CSF for 10 min, and then analyzed by phospho-flow cytometry to determine the abundance of pERK. Basal indicates cells that were treated neither with inhibitor nor cytokine. The red vertical lines indicate basal median fluorescence intensity (MFI) values. (B) K+M−G− cells from WT and KrasG12D mice were incubated for 30 min with the same inhibitors used in (A) before they were stimulated with SCF for 15 min and then analyzed by phospho-flow cytometry to determine pERK abundance. (C) Ras-GTP and pERK abundances were measured in bone marrow cells from WT and KrasG12D mice (as described in the Methods) that were treated with the indicated inhibitors before being stimulated with GM-CSF. Actin was used as a loading control. (D) The amounts of phosphorylated PLC-γ1 (pPLC-γ1) and PLC-γ2 (pPLC-γ2) were determined by flow cytometric analysis of WT and KrasG12D mouse bone marrow M+G+ cells that were pretreated with the indicated inhibitors before being stimulated with GM-CSF. Data from additional independent experiments are shown in figs. S2 B and C, and S3.
Upon ligand binding, the common β chain of the GM-CSF receptor is phosphorylated by Janus-activated kinase 2 (JAK2), generating docking sites for signaling molecules and adaptor proteins (1). To investigate the molecular mechanism linking GM-CSF to ERK activation, we exposed bone marrow cells from wild-type and Mx1-Cre, KrasG12D mice to a panel of kinase inhibitors. Cells incubated with the MEK inhibitor PD0325901 (8) served as a positive control for measuring inhibition of ERK phosphorylation (Figs. 1A and fig. S2A). As expected, JakI, an inhibitor of JAK proteins, abrogated GM-CSF–dependent ERK phosphorylation in wild-type and Kras mutant bone marrow cells. In contrast, GM-CSF–dependent ERK activation was unimpaired by the Src family tyrosine kinase inhibitor PP2, despite there being a marked reduction in the total abundance of tyrosine-phosphorylated proteins (Fig. 1A and figs. S2, A and B). GM-CSF-induced ERK phosphorylation was also unimpaired in cells that were exposed to the protein kinase C (PKC) inhibitor bisindolylmaleimide I (BIM 1)(Fig. 1A). Pre-treatment of cells with chemical inhibitors of PLC-γ (U73122)(4, 50) or PI3K (PI-90)(32) reduced the ability of GM-CSF to increase pERK abundance in differentiated M+G+ and immature K+M−G− cells from wild-type and Mx1-Cre, KrasG12D mice (Fig. 1A and fig. S2A).
The responses of M+G+ and K+M−G− cells to GM-CSF and chemical inhibitors differed in some respects (Fig. 1A and fig. S2A). Wild-type and Mx1-Cre, KrasG12D M+G+ cells had low basal amounts of pERK compared to those of K+M−G− cells, and inhibition of PLC-γ or PI3K almost completely abrogated ERK activation by GM-CSF (Fig. 1A). GM-CSF elicited a more heterogeneous pattern of ERK phosphorylation in K+M−G− cells, which is consistent with the expectation that only the myeloid subset of this mixed population will respond to GM-CSF (fig. S2A). U73122 completely blocked this response, while K+M−G− cells that were exposed to PI-90 showed some residual ERK activation upon GM-CSF stimulation (fig. S2A).
SCF stimulates the proliferation of hematopoietic stem cells and progenitor cells through binding to the receptor tyrosine kinase (RTK) c-Kit. Somatic KIT mutations that constitutively activate the intrinsic kinase activity of c-Kit are found in acute myeloid leukemia (20, 34). We investigated the effects of small-molecule inhibitors on SCF-stimulated ERK activation in K+M−G− cells, which uniformly express c-Kit. As reported previously (54), the phosphorylation of ERK in response to SCF was impaired in Mx1-Cre, KrasG12D cells compared to that in wild-type cells (Fig. 1B). As expected, JakI had no effect on SCF-induced ERK activation, whereas inhibitors of Src family tyrosine kinases (PP2 and SU6656) greatly reduced SCF-stimulated ERK phosphorylation. Similar to our findings from experiments with cells stimulated with GM-CSF, PKC inhibitors had no effect on the ability of SCF to stimulate ERK activation. We found that U73122 and PI-90 markedly reduced the ability of SCF to induce ERK phosphorylation in K+M−G− cells from wild-type and Mx1-Cre, KrasG12D mice (Fig. 1B). Together, these data place PLC-γ and PI3K upstream of ERK activation in GM-CSF and SCF signaling pathways in differentiated and immature myeloid lineage cells from both wild-type and Mx1-Cre, KrasG12D mice.
Inhibitors of PLC-γ and PI3K have effects on Ras-GTP abundance in bone marrow from wild-type and Mx1-Cre, KrasG12D mice
To clarify the level at which PLC-γ and PI3K regulated Ras-Raf-MEK-ERK signaling, we measured Ras-GTP abundance in bone marrow cells from wild-type and Mx1-Cre, KrasG12D mice before and after stimulation with GM-CSF. As expected, GM-CSF induced GTP-loading on Ras in wild-type bone marrow cells. This response was reduced by chemical inhibitors of JAKs, PLC-γ, or PI3K, all of which also abrogated or markedly reduced the extent of ERK phosphorylation (Fig. 1C and figs. S3, A, C, and D). While the PKC inhibitor BIM-I also lowered GM-CSF-dependent generation of Ras-GTP, ERK activation was normal. As expected, PD0325901 completely eliminated ERK phosphorylation in response to GM-CSF, but had no effect on Ras-GTP abundance (Fig. 1C and figs. S3C). PI-90 and U73122 also markedly reduced Akt phosphorylation (fig. S3D).
Consistent with our phospho-flow cytometry data (Fig. 1, A and B) and a previous report (7), ERK is only minimally phosphorylated in unstimulated bone marrow cells from Mx1-Cre, KrasG12D mice despite a constitutive increase in Ras-GTP abundance (Fig. 1C). We detected equivalent abundances of Ras, phospholipase C (PLC)-γ1, PLC-γ2, Akt, MEK, and ERK in the bone marrow cells of wild-type and Mx1-Cre, KrasG12D mice, and therefore used Actin as a general loading control for subsequent Western blot experiments (fig. S3A). Upon exposure to GM-CSF, these cells robustly generated pERK without further increasing Ras-GTP abundance. Whereas JAKI, PI-90, and U73122 did not alter the high basal abundance of Ras-GTP in Mx1-Cre, KrasG12D mouse bone marrow, each inhibitor markedly impaired ERK activation in response to GM-CSF in these cells (Fig. 1A and fig. S3, C and D). Together, these data place PLC-γ and PI3K upstream of Ras in the Raf-MEK-ERK signaling pathway in primary bone marrow cells. We also showed that the constitutively increased abundance of Ras-GTP that resulted from endogenous oncogenic K-RasG12D was insufficient for the efficient phosphorylation of ERK, which required cytokine and was sensitive to inhibition of PLC-γ or PI3K.
To expand on our observations of the effects of PI3K inhibition, we treated M+G+ cells with PI-103, a dual-specificity inhibitor of PI3K and mTOR that is structurally unrelated to PI-90 (32), and with isoform-selective PI3K inhibitors (figs. S4-S6). We found that PI-103 reduced GM-CSF–stimulated ERK activation as did PI-90, and that an inhibitor selective for p110α and p110γ was more potent than inhibitors of p110β and p110δ in altering ERK activation in cells that were stimulated with GM-CSF (figs. S5 and S6).
To further characterize the role of PLC-γ in GM-CSF signaling in bone marrow cells from wild-type and Mx1-Cre, KrasG12D mice, we examined the effects of GM-CSF on the phosphorylation of PLC-γ1 and PLC-γ2 (pPLC-γ1 and pPLC-γ2, respectively) in M+G+ cells in the absence and presence of chemical inhibitors. The basal amount of pPLC-γ1 was increased in Mx1-Cre, KrasG12D cells compared to that in wild-type cells, and GM-CSF stimulated the phosphorylation of both isoforms of PLC-γ in cells of either genotype (Fig. 1D and fig. S7). Pretreatment with U73122 abrogated this response, and JakI also potently inhibited the phosphorylation of both PLC-γ1 and PLC-γ2. Cells treated with PI-90 or PI-103 showed a more modest reduction in GM-CSF–dependent pPLC-γ1 or PLC-γ2 generation than was observed in cells treated with JakI or U73122 (Fig. 1D and S7).
PLC-γ mediates the activation of PI3K and MAPKs
To functionally study signaling networks in primary myeloid cells, we cultured mouse bone marrow cells in macrophage medium that selectively enabled the growth of bone marrow macrophage progenitor cells (BMMPCs), a relatively homogeneous population that exhibits activated Ras signaling in response to GM-CSF (10, 48). The abundances of Ras and of other key signaling molecules were equivalent in BMMPC grown from wild-type and Mx1-Cre, KrasG12D mice (fig. S3B). Western blotting analysis showed that JakI, PI-90, and U73122 inhibited GM-CSF–dependent ERK activation in BMMPCs, whereas the PKC inhibitor BIM I had no effect (fig. S8). These data were consistent with the observed effects of these inhibitors on bone marrow M+G+ cells (Fig. 1A and fig. S2A).
The ability to culture BMMPCs from wild-type and Mx1-Cre, KrasG12D bone marrow provided us with a robust primary cell system in which to exploit RNA interference (RNAi) to interrogate the interaction of PLC-γ and PI3K in ERK activation. We constructed retroviral vectors encoding PLC-γ–specific or control short hairpin RNAs (shRNAs), a neomycin resistance gene, and a fluorescent marker [green fluorescent protein (GFP) for shPLC-γ1 and mCherry for shPLC-γ2]. After their transduction, bone marrow cells were grown in myeloid medium that selectively enabled the growth of BMMPCs in the presence of the antibiotic G418 to increase the number of cells expressing the shRNA constructs (fig. S9A). Flow cytometric analysis confirmed that most of the BMMPCs expressed GFP or mCherry (58 to 79%, fig. S9A). Knockdown of PLC-γ2 reduced the extent of ERK and Akt activation in response to GM-CSF (Fig. 2A), whereas knockdown of PLC-γ1 with either shRNA only had modest effects on pAkt induction (fig. S9B).
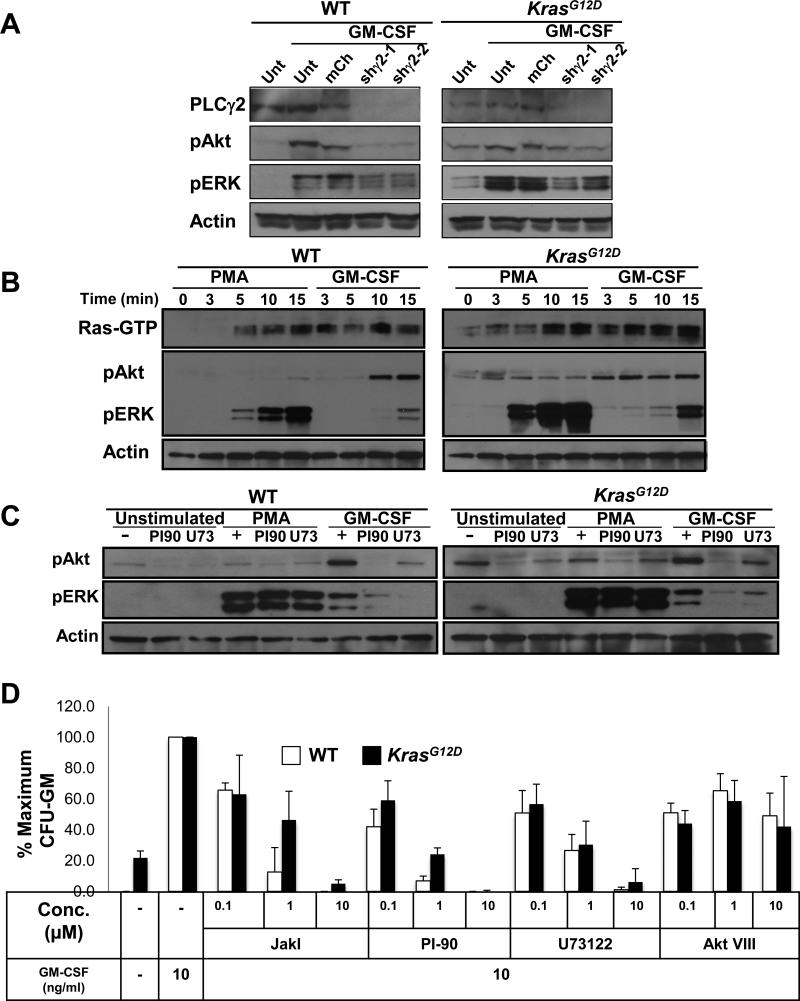
Fig. 2. Effects of inhibiting PLC-γ and PI3K on signaling in BMMPCs.
(A) BMMPCs that were not transduced (Unt) or were transduced with a retroviral vector expressing mCherry (mCh) or a vector encoding mCherry and one of two independent shRNAs specific for PLC-γ2 (shγ2-1 or shγ2-2) were lysed, and the abundances of PLC-γ2, pAkt, and pERK were measured by Western blotting analysis, with actin used as a loading control. (B) Effects of PMA and GM-CSF on Ras-GTP, pERK, and pAkt abundances in BMMPCs from WT and KrasG12D mice. BMMPCs from the indicated mice were treated with PMA or GM-CSF for the indicated times and lysed. Ras-GTP abundance was assessed by a Ras-RBD pull down assay and abundance of pAkt, and pERK was analyzed by Western blotting on the same lysates. Actin was used as a loading control. (C) BMMPCs were pretreated with PI-90 or U73122 (U73) and then were left unstimulated or were stimulated with GM-CSF or PMA for 15 min. Cells were then analyzed by Western blotting for the abundances of pAkt and pERK, with actin used as a loading control. (D) Analysis of CFU-GM growth from bone marrow cells from WT and KrasG12D mice that were cultured in a saturating concentration of GM-CSF (10 ng/ml) in the absence or presence of the inhibitors JakI, PI-90, U73122, or AKT VIII. Data from independent experiments corresponding the panels (B) and (C) are shown in figs S8 – S10. Data in (D) are mean values ± SD from three independent experiments.
DAG analogs by-pass chemical inhibitors to activate Ras-Raf-MEK-ERK signaling in BMMPCs
Activated PLC-γ hydrolyzes phosphatidylinositol 4,5 biphosphate (PIP2) to generate DAG and inositol 1,4,5 trisphosphate (IP3), the latter of which mobilizes intracellular calcium ions (Ca2+). These second messengers, in turn, activate PKC and guanine nucleotide exchange factors (GEFs) of the Ras guanine nucleotide-releasing protein (RasGRP) family (11). We compared Ras-GTP abundance, as well as the extents of ERK and Akt phosphorylation, in wild-type and Mx1-Cre, KrasG12D BMMPCs that were exposed to GM-CSF or to the DAG mimetic phorbol 12-myristate 13-acetate (PMA). We found that PMA increased Ras-GTP and pERK abundances in BMMPCs of both genotypes, but that it had little or no effect on pAkt abundance (Fig. 2B and fig. S10A).
GM-CSF also stimulated an increase in Ras-GTP abundance and ERK phosphorylation in BMMPCs; however, the time courses of these responses differed from those of PMA (Fig. 2B and fig. S10B). In contrast to PMA, GM-CSF also stimulated an increase in pAkt abundance (Fig. 2B, figs. S3D and S10B). The extent and duration of Akt activation differed between wild-type and Mx1-Cre, KrasG12D BMMPCs. In particular, wild-type cells showed a marked response 10 to 15 min after exposure to GM-CSF, whereas Kras mutant cells had increased amounts of basal pAkt but responded less to stimulation (Fig. 2B and fig. S10B). U73122 and PI-90 blocked the GM-CSF–dependent increase in ERK phosphorylation in wild-type and Mx1-Cre, KrasG12D BMMPCs; however, neither inhibitor altered PMA-dependent activation of ERK (Fig. 2C and fig. S11, A and B). Furthermore, U73122 blocked the GM-CSF–dependent phosphorylation of Akt in wild-type BMMPCs, reduced basal amounts of pAkt in Mx1-Cre, KrasG12D cells, and inhibited the activation of Akt by GM-CSF in these cells (Fig. 2C and figs. S3D and S11B). Together with the results of the phospho-flow cytometry and shRNA experiments presented earlier, these data place PLC-γ upstream of PI3K in GM-CSF signaling in wild-type and Mx1-Cre, KrasG12D mutant hematopoietic cells.
We next asked whether the PLC-γ effector Ca2+ and calmodulin–dependent protein kinase II (CaMKII) was required for efficient ERK phosphorylation in Mx1-Cre, KrasG12D BMMPCs by assessing the inhibitory effects of 2-APB (an IP3 receptor antagonist) and KN-62 (a CaMKII inhibitor)(36). Like U73122, 2-APB and KN-62 blocked GM-CSF–dependent ERK phosphorylation, whereas BIM I did not (fig. S12, A and B). These data are consistent with a study showing that 2-APB and KN-62 blocked GM-CSF–dependent ERK activation in wild-type hematopoietic cells (36).
To assess the functional requirements for PLC-γ and PI3K in myeloid progenitors, we grew CFU-GM colonies in methylcellulose medium containing a saturating concentration of GM-CSF together with JakI, PI-90, U73122, or the Akt inhibitor Akt VIII These studies revealed dose-dependent inhibition of CFU-GM growth of both wild-type and Mx1-Cre, KrasG12D bone marrow cells in the presence of 0.1 to 10 μM JakI, U73122, or PI-90 (Fig. 2D). We observed substantial inhibition of CFU-GM growth in cells exposed to 1 to 10 μM of each inhibitor, which correlated with biochemical inhibition of GM-CSF–dependent ERK phosphorylation in BMMPCs (Fig. 2D and fig. S8). In contrast, Akt VIII had modest effects on CFU-GM growth at concentrations that blocked Akt phosphorylation in cultured BMMPCs (Fig. 2D and fig. S8B).
Together, these data suggest that phosphorylation of the GM-CSF receptor by JAK2 creates docking sites for PLC-γ and PI3K binding. These proteins, in turn, generate DAG, Ca2+, and PIP3, which function upstream of Ras in the activation of the Raf-MEK-ERK pathway in wild-type and Mx1-Cre, KrasG12D cells. Furthermore, multiple lines of evidence place PLC-γ upstream of PI3K in modulating the response of myeloid cells to GM-CSF. First, U73122 inhibited Akt phosphorylation, whereas PI-90 only partially reduced PLC-γ activation (Figs. 1D and 2C and fig. S7). Second, knockdown of PLC-γ2 markedly impaired the GM-CSF–dependent phosphorylation of Akt and ERK (Fig. 2A). Third, U73122 inhibited CFU-GM progenitor growth, whereas Akt VIII had only modest effects (Fig. 2D).
RasGRP proteins potentiate the phosphorylation of ERK
We sought DAG effectors that might contribute to activation of Ras-ERK signaling in BMMPCs. PKC isoforms and RasGRP proteins contain DAG-binding domains and regulate the generation of Ras-GTP; however, the PKC inhibitor BIM I did not inhibit cytokine-induced ERK activation in M+G+ or K+M−G− bone marrow cells (Fig. 1, A to C, and fig. S2A) or in cultured BMMPCs (fig. S8). Furthermore, GM-CSF stimulation did not alter the phosphoylation states of multiple PKC isoforms in BMMPCs (fig. S13).
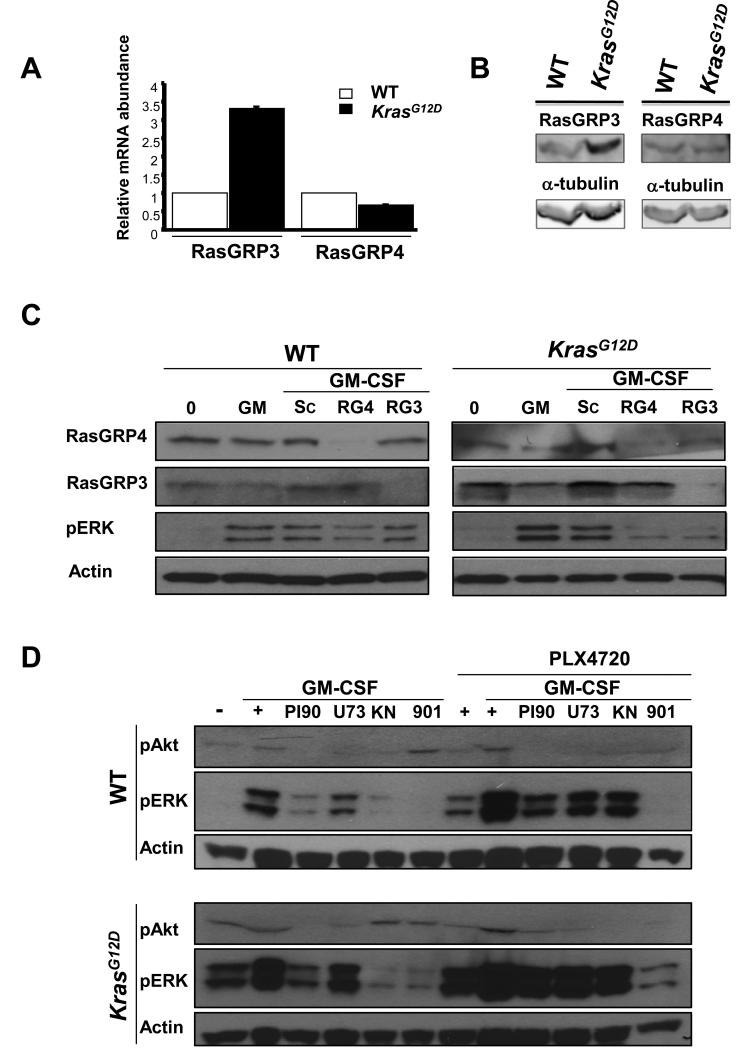
We next asked whether members of the RasGRP family of GEFs might link cytokine receptor activation to the stimulation of Ras and ERK signaling. Quantitative polymerase chain reaction (PCR) analysis showed that BMMPCs express RasGrp3 and RasGrp4, but not RasGrp1. Interestingly, Mx1-Cre, KrasG12D cells expressed substantially increased amounts of Rasgrp3 mRNAs and Western blotting analysis showed increased amounts of RasGRP3 protein compared to wild-type cells (Fig. 3, A and B and fig. S14). We used RNAi to reduce RasGRP3 and RasGRP4 abundances in BMMPCs, and assessed the effects on GM-CSF–dependent ERK phosphorylation (Fig. 3C and fig. S14). Knocking down RasGRP4 modestly reduced the extent of ERK phosphorylation in wild-type BMMPCs, whereas knockdown of RasGRP3 had minimal effects (Fig. 3C and fig. S14). In contrast, knockdown of either RasGRP3 or RasGRP4 inhibited GM-CSF–dependent ERK phosphorylation in Mx1-Cre, KrasG12D cells (Fig. 3C and fig. S14).
Fig. 3. Effects of the knockdown of RasGRP isoforms on ERK phosphorylation in BMMPCs from WT and Mx1-Cre, KrasG12D mice.
(A) Rasgrp3 and Rasgrp4 mRNA and RasGRP3 and RasGRP4 in BMMPCs from WT and KrasG12D mice were determined by quantitative RT-PCR. (B) Protein abundances were assessed by Western blotting (see also fig. S14) (C) BMMPCs that were incubated with or without siRNAs (10 nM) specific for Rasgrp3 (RG3) or RasGrp4 (RG4) or with a control scrambled siRNA (Sc) were stimulated with GM-CSF for 15 min. The cells were lysed and Western blotting analysis was performed to determine the amounts of RasGRP3, RasGRP4, and pERK, with actin used as a loading control. (D) BMMPCs were starved of serum and cytokines overnight, incubated with PI-90, U73122, KN-62, or PD0325901 for 30 min, and then stimulated for 15 min with GM-CSF alone or in the presence of 10 μM PLX4720. Cells were then lysed and analyzed by Western blotting to determine the abundances of pAkt and pERK, with actin used as a loading control. Data from additional independent experiments are presented in fig. S15.
PLX4720 partially reverses the inhibitory effects of U73122 and PI-90 in bone marrow cells
The potent Raf inhibitor PLX4032 (also known as vemurafenib) is a highly promising treatment for human cancers with somatic BRAF mutations (5, 19). PLX4032 and the related compound PLX4720 selectively reduce pERK abundance in cells expressing oncogenic B-Raf proteins, but paradoxically activate signaling in the context of wild-type B-Raf through allosteric effects on homo- and heterodimers of c-Raf and B-Raf (24, 25, 41). Consistent with studies in other cell types, we found that incubating bone marrow cells from wild-type or Mx1-Cre, KrasG12D mice with PLX4720 increased the abundance of pERK in response to GM-CSF compared to that in untreated cells (Fig. 3D and fig. S15). PLX4720 also partially antagonized the inhibitory effects of PI-90 and U73122 on GM-CSF–dependent ERK phosphorylation, but, as expected, did not overcome inhibition by PD0325901 (Fig. 3D and fig. S15). Together with data presented earlier, these findings further support a model in which PLC-γ and PI3K link the activated GM-CSF receptor to ERK activation through Ras and Raf.
Inhibitors of PLC-γ and PI3K perturb SCF signaling in immature hematopoietic cells
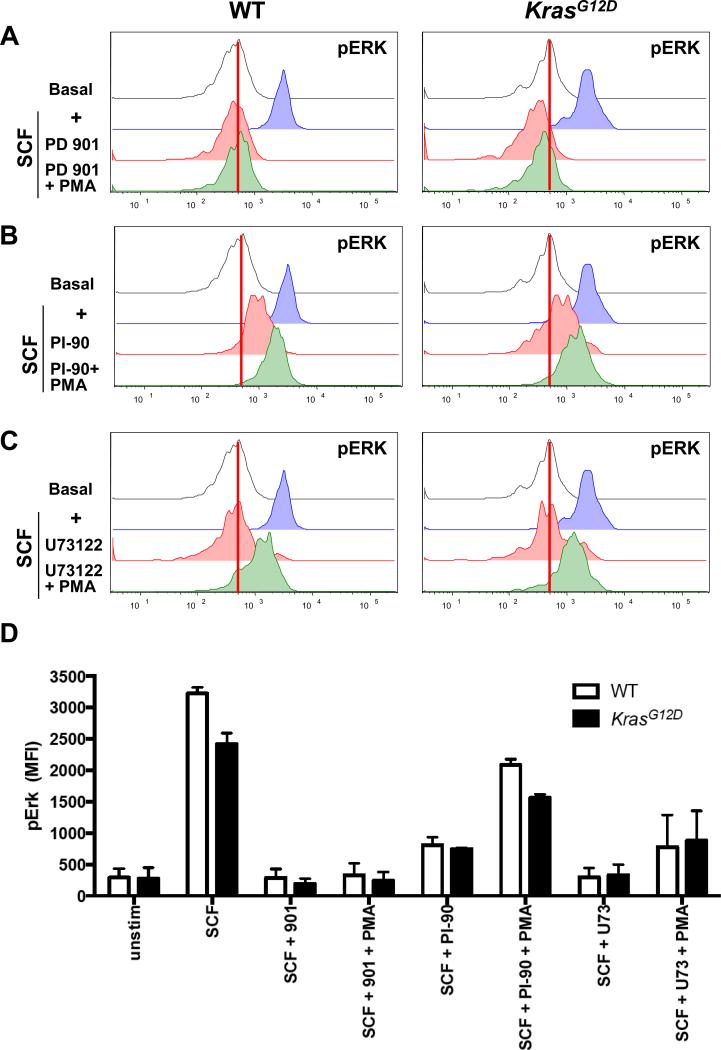
The transplantation of purified populations of Mx1-Cre, KrasG12D bone marrow cells into recipient mice showed that HSCs initiate and maintain leukemic growth in vivo (46, 60). To determine the extent to which PLC-γ and PI3K were essential for efficient ERK activation in a population of bone marrow cells that contains a high percentage of HSC, we performed phospho-flow cytometry to analyze the kit+lindim/−Sca1+CD48− (KLS 48−) subset of bone marrow stimulated with SCF in the presence of the MEK inhibitor PD0325901, the PI3K inhibitor PI-90, or the PLC-γ inhibitor U73122. We found that KLS 48− cells from wild-type and Mx1-Cre, KrasG12D mice showed robust ERK activation in response to SCF (Fig. 4, A to C). As expected, PD0325901 abrogated ERK phosphorylation in response to SCF or the combination of SCF and PMA (Fig. 4A). SCF-induced ERK activation in KLS 48− cells was also sensitive to inhibition by PI-90 and U73122; however, KLS 48− cells that were also stimulated with PMA had increased pERK abundance despite the presence of either inhibitor (Fig. 4, B and C). U73343, a structural analog of U73122 that does not inhibit PLC-γ activity, had no effect on SCF-dependent ERK phosphorylation in KLS 48− cells (fig. S16). These results are consistent with the data from experiments with BMMPCs showing that PLC-γ and PI3K function upstream of DAG in ERK activation.
Fig. 4. Inhibition of PLC-γ and PI3K reduces SCF-induced ERK phosphorylation in WT and Mx1-Cre, KrasG12D cells highly enriched for stem cells.
(A to C) Bone marrow cells from WT and Mx1-Cre, KrasG12D mice were left unpretreated or were pretreated with (A) PD0325901, (B) PI-90, or (C) U73122 before being stimulated with SCF in the absence or presence of PMA. Cells were analyzed by phospho-flow cytometry to determine the abundance of pERK in KLS 48- cells. (D) Quantitation of two independent experiments.
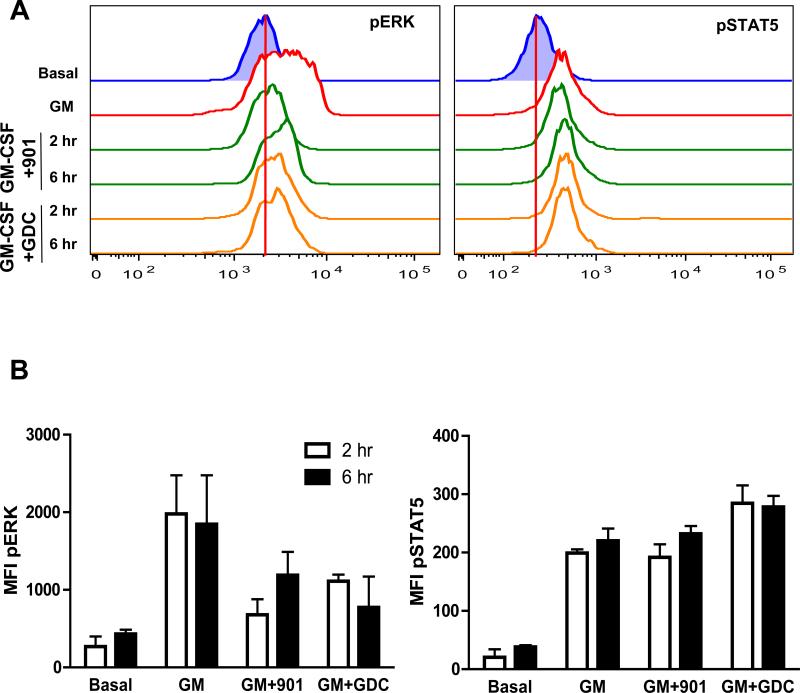
In vivo treatment of mice with a PI3K inhibitor reduces GM-CSF–dependent ERK activation in primary bone marrow cells
GDC-0941 is a PI3K inhibitor that is being evaluated as an anti-cancer agent in humans (42). To investigate whether in vivo treatment with a clinical PI3K inhibitor might affect Raf-MEK-ERK signaling in bone marrow, we administered GDC-0941 to wild-type mice at an oral dose (100 mg/kg) that was efficacious in preclinical models (n= 3 per time point) and euthanized the mice 2 or 6 hours later. Mice that received PD0325901 (5 mg/kg) served as a positive control in this experiment. Treatment with a single dose of either GDC-0941 or PD0325901 markedly reduced the extent of ERK phosphorylation in cells that were stimulated with GM-CSF ex vivo, but had no effect on STAT5 phosphorylation (Fig. 5, A and B). These pharmacodynamic data demonstrate that administering a therapeutically relevant dose of a PI3K inhibitor to mice reduces the ability of bone marrow cells to activate ERK in response to cytokines.
Fig. 5. Treatment of mice with inhibitors of MEK or PI3K impairs cytokine-induced ERK activation in bone marrow cells.
(A and B) WT mice were left untreated or received either PD0325901 (901, 5 mg/kg) or GDC-0941 (GDC, 100 mg/kg) by oral gavage, and were euthanized 2 or 6 hours later. Bone marrow cells were harvested and left untreated or were stimulated with GM-CSF for 10 min, and the abundances of pERK (A, left panel) and pSTAT5 (A, right panel) were determined by flow cytometric analysis. Data are representative of three mice for each time point. The red vertical lines display basal MFI values. (B) MFI plots of the data shown in (A) depict the effects of GM-CSF on pERK and pSTAT5 abundances, as well as the extent of inhibition observed in mice that were treated with the MEK inhibitor PD032501 or the PI3K inhibitor GDC-0941. Data are medians ± SD from three mice for each time point.
Discussion
Here, we demonstrated cooperativity between PLC-γ and PI3K in linking GM-CSF receptor activation to Ras and ERK signaling in primary hematopoietic cells (Fig. 6). This module regulates growth factor responses in immature and differentiated cells and is engaged by cytokine receptors with and without intrinsic RTK activity, suggesting that it may represent a general mechanism for activating the Raf-MEK-ERK cascade. Inhibitors of PLC-γ and PI3K interfered with ERK phosphorylation in a highly enriched population of normal and Mx1-Cre, KrasG12D bone marrow cells that included leukemia-initiating cells. Despite having markedly increased basal amounts of Ras-GTP, cells expressing oncogenic K-RasG12D from the endogenous genetic locus remained dependent on PLC-γ and PI3K for efficient ERK activation in response to GM-CSF and SCF. Studies of hematopoietic progenitor colony growth, and of cytokine signaling after treatment with a PI3K inhibitor in vivo, confirmed the physiological relevance of our in vitro observations. Together, these data have implications for clinical trials that combine Raf-MEK-ERK and PI3K-Akt-mTOR or PLC-γ inhibitors, as well as for nascent efforts to develop small molecules that directly target oncogenic Ras proteins.
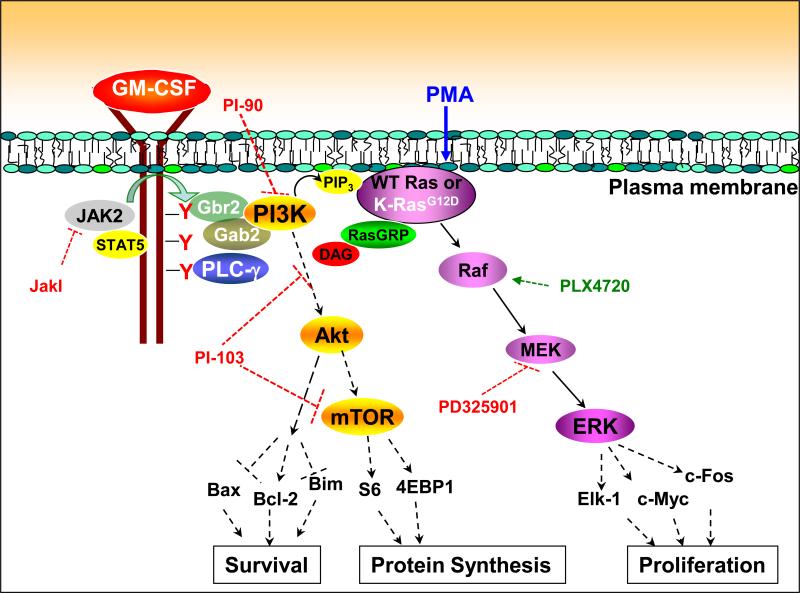
Fig. 6. Proposed mechanism of cytokine-induced Ras and ERK activation in hematopoietic cells expressing WT and oncogenic K-Ras.
In response the binding of GM-CSF to its receptor, JAK2 trans-phosphorylates multiple tyrosine residues on the β common chain of the GM-CSF receptor, creating docking sites for adaptor proteins and signaling molecules. PLC-γ1 and PLC-γ2 are recruited to the receptor complex and becomes activated, generating DAG. The p85 regulatory subunit of PI3K binds to the receptor through adaptor proteins, which results in PI3K activation and PIP3 production. Second messengers generated by PI3K and PLC-γ stimulate nucleotide exchange on Ras, likely by both localizing Ras to the plasma membrane and activating RasGRPs. The DAG mimetic PMA activates Ras and ERK independently of PLC-γ and PI3K. This scheme does not directly address the interaction between PLC-γ and PI3K; however, multiple lines of evidence suggested that PLC-γ is upstream of PI3K. The levels at which the different inhibitors used in this study exerted their effects are indicated by dashed red bars. Raf activation by PLX4720 is indicated as a dashed green arrow. Studies using many of these inhibitors support a similar role for PLC-γ and PI3K in linking the activated receptor tyrosine kinase c-Kit to Ras and ERK signaling.
We addressed additional questions regarding the role of PLC-γ in the activation of the Raf-MEK-ERK pathway by GM-CSF. First, our data showing that PMA overcame the inhibitory effects of U73122 supports an essential role for DAG in the efficient activation of ERK. We also implicated RasGRP proteins as potential effectors of DAG. Second, the ability of U73122 to reduce Ras-GTP loading in wild-type bone marrow cells and of PLX4720 to rescue ERK phosphorylation in the presence of this inhibitor identified Ras and Raf as linking PLC-γ proteins to the activation of MEK and ERK. Third, we showed that PI3K was also required to activate Ras and ERK in response to cytokines. Finally, we showed that primary hematopoietic cells with endogenous amounts of oncogenic K-RasG12D remained dependent on PLC-γ and PI3K signaling for efficient ERK activation despite having increased basal amounts of Ras-GTP compared to those in wild-type cells.
Ras-GTP binds to and activates class I and II PI3Ks, which, in turn, are crucial for Ras-induced transformation of many cell types. For example, engineering oncogenic Ras proteins with “second site” amino acid substitutions that eliminate or severely impair PI3K binding results in defective transforming activity(37, 44, 58), and strains of mice lacking the regulatory p85 subunit of PI3K or carrying a “knock in” mutation in the catalytic p110α subunit that abrogates Ras binding are resistant to tumorigenesis driven by expression of oncogenic K-RasG12D (17, 22). These and other data establish PI3Ks as critical effectors of oncogenic Ras proteins in cancer pathogenesis.
However, PI3K inhibitors can also block the activation of Ras and ERK in some contexts(15, 27, 45, 47, 57). Three independent studies of cultured epithelial cell lines found that PI3K was essential for efficient ERK activation only at low concentrations of growth factor, which likely reflects physiologic conditions in most normal tissues (15, 47, 57). Duckworth and Cantley(15) also identified a PI3K-independent PLC-γ–PKC pathway in 3T3 cells that was engaged at high concentrations of platelet-derived growth factor and overcame the inhibitory effects of wortmannin (a PI3K inhibitor). In contrast, our data demonstrate that PLC-γ and PI3K cooperatively activate ERK in primary hematopoietic cells through a mechanism that is very likely independent of PKCs. These observations suggest that the architecture of signaling networks downstream of activated growth factor receptors varies in a tissue-specific manner. Our data showing that U73122 and PLC-γ2 knockdown greatly reduced GM-CSF–dependent Akt phosphorylation in BMMPCs place PLC-γ upstream of PI3K; however, PI-90 also modestly reduced the extent of PLC-γ phosphorylation, which raises the possibility that PI3K functions in a “feed forward” manner to amplify the signal from PLC-γ (Fig. 6). We speculate that lipid second messengers generated by PI3K and PLC-γ after recruitment to growth factor receptors, specifically PIP3 and DAG, cooperate to localize and assemble signaling complexes that efficiently activate exchange factors, Ras, and the Raf-MEK-ERK pathway. In agreement with the GM-CSF network that we described here for myeloid cells, we also showed that the increased abundance of RasGRP1 links cytokine receptors to Ras in T cell leukemia (23). These data suggest that different RasGRP family members regulate Ras activation in response to cytokines in distinct hematopoietic lineages.
It is unclear why constitutively increased amounts of Ras-GTP in Mx1-Cre, KrasG12D hematopoietic cells only minimally activate Raf-MEK-ERK signaling. One provocative idea is that flux through the pathway in increased, but cells use negative feedback mechanisms that reduce basal amounts of pERK. Alternatively, we speculate that efficient activation of Raf-MEK-ERK signaling requires that Ras both bind to GTP and be localized at signaling complexes on activated cytokine receptors. Lipid second messengers, in particular PIP3, can recruit and retain K-Ras and other polybasic proteins to specific domains within the plasma membrane (26). PIP3 might also colocalize PH-domain containing proteins, including SOS and RasGRFs, within membrane nanoclusters to promote the assembly of signaling complexes on activated receptors(40, 51). It is likely that wild-type Ras localizes to activated cytokine receptors at which GEFs mediate GTP loading. In contrast, whereas oncogenic Ras accumulates in its GTP-bound conformation in the absence of growth factors, our data are consistent with the idea that it still requires a localization signal that is dependent on PLC-γ and PI3K to fully activate the Raf-MEK-ERK pathway.
RasGRP4, a Ras-specific GEF that is highly abundant in myeloid cells, is mutated in some cases of acute myeloid leukemia, and modulates Ras-GTP amounts in response to PMA (43). Our data support a role for RasGRP4 in activating Ras and ERK in the physiologic setting of GM-CSF signaling. Whereas RasGRP4 played a dominant role in wild-type cells, we found that Mx1-Cre, KrasG12D BMMPCs had increased amounts of RasGRP3 and become dependent on this isoform, which suggests that K-RasG12D remodels signaling networks in unanticipated ways. The observation that PLC-γ engaged Ras though RasGRP proteins in myeloid cells is consistent with previous studies in T and B lymphocytes in which the PLC-γ–dependent product DAG plays a critical role in recruiting RasGRP1 and RasGRP3 to the plasma membrane (3).
These unexpected results also raise the intriguing possibility that inhibiting the biochemical output of proteins upstream of oncogenic Ras may represent a viable approach for treating some cancers, particularly those that are highly responsive to growth factors. Maurer et al. (39) used fragment-based lead discovery to develop chemical inhibitors to a previously uncharacterized binding pocket on the surface of Ras. DCA1, a compound from this screen, inhibits Ras activation by interfering with SOS-mediated guanine nucleotide exchange, and also likely has activity against other GEFs (39). The results of our study support testing the ability of DCA1 and related compounds to block GM-CSF– and SCF-mediated ERK activation in wild-type and Mx1-Cre, KrasG12D cells to further evaluate the potential of this general therapeutic strategy.
Preclinical data supporting the simultaneous targeting of the Raf-MEK-ERK and PI3K-Akt-mTOR signaling pathways in KRAS mutant cancers (17) are currently being translated clinically. Although this approach may enhance tumor killing, our data suggest that suppression of cytokine signaling in normal hematopoietic stem and progenitor cells will emerge as a clinically relevant adverse effect of PI3K inhibition. This potential toxicity may become especially problematic when PI3K inhibitors are used in combination with other anti-cancer drugs that suppress blood cell production. If this proves true, Akt or mTOR inhibitors may have a better therapeutic index in certain clinical settings.
Finally, because some primary cells that have amounts of oncogenic Ras similar to that of wild-type Ras remain dependent on extracellular stimuli to fully activate downstream effector pathways, inhibitors of proteins upstream of Ras may be effective in some cancers. For example, PLC-γ and PI3K are attractive therapeutic targets in JMML, CMML, and other myeloproliferative neoplasms that have a high incidence of RAS mutations in which hematopoietic growth factors play a central role in driving aberrant growth (53). This general paradigm may also apply to non-hematologic cancers that remain dependent on growth factors and other extracellular stimuli that contribute to fully activate oncogenic signaling pathways.
Materials and Methods
Antibodies
Antibodies specific for pERK (9101), RasGRP3 (3334), pPKCα (pT638/641; 9375), pPKCβ (pS660; 9371), pPKCδ (pT505; 9374), pPKCθ (pT538; 9377), and β-actin (4967) were purchased from Cell Signaling Technology; antibody against pAkt (pS473; 44-621G) was from Invitrogen; antibody against Ras (clone Ras10) (05-516) was from Millipore; antibody specific for RasGRP4 (pab0554-1) was from Covalab; antibody against PLC-γ1 (sc-7290) was from Santa Cruz; and antibody against PLC-γ2 (bs-3532R) was from Bioss. Horseradish peroxidase (HRP)-conjugated polyclonal goat anti-rabbit immunoglobulin G (IgG, P0448) was from DakoCytomation, and HRP-conjugated sheep anti-mouse IgG (NA931V) was from GE Healthcare (UK). In flow cytometry experiments, primary antibodies were detected with fluorescein isothiocyanate (FITC)–conjugated anti-rabbit IgG (711-096-152) or allophycocyanin (APC)-conjugated anti-rabbit IgG (711-136-152), both of which were purchased from Jackson Immunoresearch. Phosphorylated STAT5 (pY694) was detected with an antibody directly conjugated to Alexa Fluor 647 (BD Biosciences #612599), and antibodies specific for pPLC-γ1 (bs-3343R) and pPLC-γ2 (bs-3339R) were purchased from Bioss. Cell-surface proteins were detected with directly conjugated antibodies from BD Pharmingen [for CD11b/Mac1 (553311) and Gr1 (553128)] and eBioscience [for CD117/c-kit (15-1171-320)]. Antibody against CD16 and CD32 (BD Biosciences 553142) was used to block Fc receptors.
Chemical inhibitors and ligands
The PI3K inhibitors PI-90, PI-103, AS605240 (for PI3Kα andPI3Kγ), TGX 115 (PI3Kβ), IC87114 (PI3Kδ), and SW13 (PI3Kδ) (all used at a final concentration of 5 μM) were synthesized in the Shokat laboratory. JakI (catalog #420099), SU6656 (#572635), PP2 (#529576), bisindolylmaleimide I (BIM I, #203291), bryostatin (#203811), U73122 (#662035) and U73343 (#662041) were purchased from Calbiochem; PMA (catalog #P8139), 2-APB (#D9754), and KN-62 (#I2142) were obtained from Sigma-Aldrich; PD0325901 was provided by Pfizer Inc.; GDC-0941 was provided by Genentech, Inc.; and PLX4720 was obtained from Plexxikon, Inc.
siRNA oligonucleotides and reagents
Specific siRNAs for RasGRP3 (MSS216067), RasGRP4 (MSS239172), the Negative Universal Control low GC (12935-110), and lipofectamine RNAiMAX (13778-150) were purchased from Invitrogen.
shRNA oligonucleotides and reagents
The shRNAs specific for PLC-γ1 and PLC-γ2 were generated according to published rules(18). The sequences of the PLC-γ hairpins used to generate the data shown in Fig. 2A and fig. S6 are provided in table S1.
Mice
KrasLSL-G12D mice were described previously (30). Mx1-Cre, KrasLSL-G12D mice (7) received a single 250-μg injection of polyinosinic-polycytidilic acid [poly(I:C), Sigma] at 21 days of age to induce KrasG12D expression. All animals were maintained in the rodent barrier facility at the University of California, San Francisco (UCSF), and the UCSF Committee on Animal Research approved the experimental procedures.
Harvesting and stimulation of bone marrow cells
Bone marrow (BM) cells were harvested from mouse femurs into Iscove's Modified Dulbecco's Medium (IMDM) containing 1% bovine serum albumin (BSA, Sigma-Aldrich). Erythrocytes were lysed and mononuclear cells were incubated at 37°C for 2 to 2.5 hours. Where indicated, the cells were stimulated with the following murine cytokines (all purchased from Peprotech) at the indicated concentrations: GM-CSF (10 or 1 ng/ml), SCF (100 ng/ml), and M-CSF (10 ng/ml). PMA and bryostatin were used at concentrations of 200 ng/ml and 500 nM, respectively. For in vitro experiments, BM cells were harvested as described earlier, starved of cytokines and serum for 90 min, and incubated for 30 min with each inhibitor before being stimulated with GM-CSF (10 ng/ml) or SCF (100 ng/ml) for 10 min (for primary bone marrow cells) or 15 min [for cultured bone marrow macrophage progenitor cells (BMMPCs)].
Flow cytometric analysis and myeloid progenitor cell proliferation
For flow cytometry, cells were fixed, permeabilized, and analyzed as described previously, with staining of Sca1 and CD48 after formaldehyde fixation and prior to methanol permeabilization (54){REF: Lyubynska}. CFU-GM colonies were grown from bone marrow cells in methylcellulose medium M3231 (Stem Cell Technologies) and scored at day 8 by indirect microscopy.
Culture and stimulation of BMMPCs
BMMPCs were grown in macrophage medium as described previously (10, 49). After reaching confluence, the cells were passaged once and grown for an additional 2 to 3 days. Before being stimulated, BMMPC culture medium was replaced with minimal medium (IMDM supplemented with 1% BSA) for 4 hours. Next, supernatants were removed, 3 ml of phosphate-buffered saline (PBS)-based dissociation buffer (Gibco) was added, and the plates were placed at 37°C for 5 min. Cells were then gently scrapped off the plates with a rubber scrapper, washed once with PBS, and resuspended in IMDM, 1% BSA for stimulation.
Western blotting analysis and Ras-GTP assays
After stimulation, cells were pelleted and lysed in 1% NP-40 buffer containing 30 mM NaF, 30 mM β-glycerophosphate, 20 mM Na4P2O7, 1 mM Na3VO4, aprotinin (10 μg/ml), leupeptin (10 μg/ml), and 1 mM phenylmethylsulfonyl fluoride (PMSF). Protein concentrations were determined with the BCA Protein Assay Kit (Pierce). Samples were resolved on precast Criterion polyacrylamide gels (BioRad) and transferred to Immobilon-P membranes (Millipore). After incubation with the appropriate antibodies and washing, the blots were treated with ECL (Amersham Biosciences) or ECL plus (GE Healthcare) to visualize target proteins. Ras-GTP abundance was measured with a Raf-Ras binding domain (RBD) pulldown assay, as described previously (12, 49).
Analysis of the effects of PLX4720 on ERK activation
BMMPCs were starved of serum and cytokines overnight, resuspended in IMDM, 1% BSA medium, incubated with 5 μM PI-90 or 5 μM U73122 for 30 min, and then stimulated with GM-CSF in the absence or presence of 5 μM PLX4720 for 15 min. Cells were then lysed and subjected to Western blotting analysis to determine ERK activation.
Analysis of RasGRP expression by quantitative PCR
Total RNA was extracted with an RNeasy kit (Qiagen) according to the manufacturer's instructions. After quantifying the RNA with a Nanodrop spectrophotometer, we performed reverse-transcription polymerase chain reaction (RT-PCR) assays with standard procedures and random primers (Invitrogen). Real-time (Taqman) RT-PCR analysis was performed with an ABI Taqman 7300 (Applied Biosystems) with specific TAMRA-labeled probes. All steps were performed in Amplitaq Gold mix (Applied Biosystems). During the exponential phase of the PCR reaction, the crossing threshold (Ct) was determined for each amplification curve. DNA from cells not stimulated were used as calibrators. All results were normalized to those of the gene encoding β-actin and were expressed as ratios related to the expression of the gene of interest in the calibrator. The relative quantification ratio was evaluated with quantification based on the ΔΔCt method(23). RasGRP1-specific primers and probe (Operon) were designed as follows: Forward: aagctccaccaactacagaact; Reverse: agggagatgaggtccttgagat; probe: [6-FAM]ccacatgaaatcaataaggttctcggtgag[TAMRA-6-FAM]. Expression of the genes encoding β-actin and RasGRP3, and RasGRP4, was determined with the following primer-probe sets purchased from Applied Biosystems: Mm01205647_g1 (β-actin), Mm01233143_m1 (Ras-GRP3), and Mm00460898_m1 (Ras-GRP4).
RasGRP knockdown by RNAi
For RNAi experiments, BMMPCs (1 × 106) were replated in 60 × 15-mm petri dishes (BD Biosciences #351007) in 4 ml of macrophage culture medium containing M-CSF one day before the experiment. After 24 hours, the medium was aspirated and replaced with 2.5 ml of IMDM, 1% BSA. After 3 hours, a mixture containing 5 to 20 pmol (0.75 to 3 μl) of each siRNA duplex together with 12 μl of Lipofectamine RNAiMAX solution suspended in 500 μl of IMDM, 1% BSA was added to each plate. Cells were mixed gently and incubated for 3 hours at 37°C in 5% CO2 before serum was added to a final concentration of 10%. After 36 hours, cells were harvested with cell dissociation buffer as described earlier, resuspended in 1 ml of IMDM, 1% BSA medium, and transferred to microfuge tubes for stimulation with GM-CSF for 15 min. Cells were then spun down at 1130 g, lysed, and subjected to SDS-PAGE, transfer, and Western blotting analysis as described earlier.
Retrovirus production and knockdown of PLC-γ1 and PLC-γ2 by shRNA
HEK 293T cells were transfected with the calcium phosphate transfection protocol as described previously (59) to produce retrovirus containing LMN vectors (61) expressing both GFP and shRNA constructs specific for PLC-γ1 or both mCherry and shRNA constructs specific for PLC-γ2. Cells were cultured for 72 hours to produce a high viral titer, and the supernatant was collected. Bone marrow cells from wild-type mice were then freshly harvested in IMDM, 10% FBS and spin-infected with the virus supernatant at 1000 g for 2 hours at room temperature on 6-well plates containing 4 × 106 cells per construct. After spin infection, cells were incubated for 5 hours at 37°C and were then transferred to 15-ml tubes. After centrifugation at 1300 rpm for 5 min, the supernatant was removed and replaced with fresh macrophage culture medium [IMDM, 20% bovine growth serum, containing M-CSF (30 ng/ml)], and the cells were plated again in 6-well plates and cultured for 24 hours. The next day, more virus was collected and the spin infection protocol was repeated. After changing the culture medium, cells were grown for 5 days until macrophages were generated. G418 (0.5 mg/ml) was added to the cells for 5 days to select the transduced cells. G418-containing medium was replaced by macrophage medium containing M-CSF (30 ng/ml), and cells were cultured for an additional 7 to 8 days until the macrophages reached 80% confluencey. Cells were then cultured in starving medium (IMDM, 1% BSA) overnight and then were collected before being stimulated with GM-CSF (10 ng/ml) for 15 min and then pelleted for Western blotting analysis, as described earlier.
Supplementary Material
Acknowledgments
We thank D. Tuveson and T. Jacks for Kras mutant mice; Johannes Zuber and Scott Lowe for the LMN vector; Genentech, Inc. for GDC-0941, Pfizer Inc. for PD0325901, Plexxikon, Inc. for PLX4970; and L. Cantley, F. McCormick, and D. Stokoe for advice and helpful suggestions. Funding: This work was supported by NIH grants U01 CA84221, R37 CA72614, K01 CA118425, and U54 CA143874; by a Specialized Center of Research Award from the Leukemia and Lymphoma Society (LLS 7019-04); by the V Foundation for Cancer Research; by a Young Investigator Award from the Children's Tumor Foundation; by the St. Baldrick's Foundation; and by the Frank A. Campini Foundation. Kevan Shokat is an Investigator of the Howard Hughes Medical Institute, Kevin Shannon is an American Cancer Society Research Professor, Benjamin Braun is a St. Baldrick's Foundation Scholar, and Jeroen Roose is a Kimmel Foundation Scholar.
Footnotes
Author contributions: E.D-F., P.D., J.P.R., and B.S.B. generated reagents, designed and performed the experiments, interpreted the data, and wrote the manuscript with technical assistance from H.G., V.N., J.A., K.K., and M.C; M.R.B, O.W., B.H., K. Shokat, D.S., and G.B. generated essential reagents and assisted with experimental design and data analysis; and K. Shannon conceived the work, interpreted experimental data, wrote the paper, and supervised all aspects of the project.
Competing interests: Deepak Sampath is a full time employee of Genentech, Inc. and Gidoen Bollag is a full time employee of Plexxikon, Inc.
References
- 1.Bagley CJ, Woodcock JM, Stomski FC, Lopez AF. The structural and functional basis of cytokine receptor activation: lessons from the common beta subunit of the granulocyte-macrophage colony-stimulating factor, interleukin-3 (IL-3), and IL-5 receptors. Blood. 1997;89:1471–1482. [PubMed] [Google Scholar]
- 2.Birnbaum RA, O'Marcaigh A, Wardak Z, Zhang YY, Dranoff G, Jacks T, Clapp DW, Shannon KM. Nf1 and Gmcsf interact in myeloid leukemogenesis. Mol Cell. 2000;5:189–195. doi: 10.1016/s1097-2765(00)80415-3. [DOI] [PubMed] [Google Scholar]
- 3.Bivona TG, Perez De Castro I, Ahearn IM, Grana TM, Chiu VK, Lockyer PJ, Cullen PJ, Pellicer A, Cox AD, Philips MR. Phospholipase Cgamma activates Ras on the Golgi apparatus by means of RasGRP1. Nature. 2003;424:694–698. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- 4.Bleasdale JE, Thakur NR, Gremban RS, Bundy GL, Fitzpatrick FA, Smith RJ, Bunting S. Selective inhibition of receptor-coupled phospholipase C-dependent processes in human platelets and polymorphonuclear neutrophils. The Journal of pharmacology and experimental therapeutics. 1990;255:756–768. [PubMed] [Google Scholar]
- 5.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, Burton EA, Wong B, Tsang G, West BL, Powell B, Shellooe R, Marimuthu A, Nguyen H, Zhang KY, Artis DR, Schlessinger J, Su F, Higgins B, Iyer R, D'Andrea K, Koehler A, Stumm M, Lin PS, Lee RJ, Grippo J, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, Chapman PB, Flaherty KT, Xu X, Nathanson KL, Nolop K. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 7.Braun BS, Tuveson DA, Kong N, Le DT, Kogan SC, Rozmus J, Le Beau MM, Jacks TE, Shannon KM. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004;101:597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown AP, Carlson TC, Loi CM, Graziano MJ. Pharmacodynamic and toxicokinetic evaluation of the novel MEK inhibitor, PD0325901, in the rat following oral and intravenous administration. Cancer Chemother Pharmacol. 2007;59:671–679. doi: 10.1007/s00280-006-0323-5. [DOI] [PubMed] [Google Scholar]
- 9.Chan IT, Kutok JL, Williams IR, Cohen S, Kelly L, Shigematsu H, Johnson L, Akashi K, Tuveson DA, Jacks T, Gilliland DG. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest. 2004;113:528–538. doi: 10.1172/JCI20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan RJ, Leedy MB, Munugalavadla V, Voorhorst CS, Li Y, Yu M, Kapur R. Human somatic PTPN11 mutations induce hematopoietic-cell hypersensitivity to granulocyte-macrophage colony-stimulating factor. Blood. 2005;105:3737–3742. doi: 10.1182/blood-2004-10-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen PJ, Lockyer PJ. Integration of calcium and Ras signalling. Nat Rev Mol Cell Biol. 2002;3:339–348. doi: 10.1038/nrm808. [DOI] [PubMed] [Google Scholar]
- 12.Donovan S, See W, Bonifas J, Stokoe D, Shannon KM. Hyperactivation of protein kinase B and ERK have discrete effects on survival, proliferation, and cytokine expression in Nf1-deficient myeloid cells. Cancer Cell. 2002;2:507–514. doi: 10.1016/s1535-6108(02)00214-3. [DOI] [PubMed] [Google Scholar]
- 13.Donovan S, Shannon KM, Bollag G. GTPase activating poteins: critical regulators of intracellular signaling. BBA Rev Cancer. 2002;1602:23–45. doi: 10.1016/s0304-419x(01)00041-5. [DOI] [PubMed] [Google Scholar]
- 14.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 15.Duckworth BC, Cantley LC. Conditional inhibition of the mitogen-activated protein kinase cascade by wortmannin. Dependence on signal strength. J Biol Chem. 1997;272:27665–27670. doi: 10.1074/jbc.272.44.27665. [DOI] [PubMed] [Google Scholar]
- 16.Emanuel PD, Bates LJ, Castleberry RP, Gualtieri RJ, Zuckerman KS. Seletive hypersensitivity to granulocyte-macrophage colony stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood. 1991;77:925–929. [PubMed] [Google Scholar]
- 17.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, Chirieac LR, Kaur R, Lightbown A, Simendinger J, Li T, Padera RF, Garcia-Echeverria C, Weissleder R, Mahmood U, Cantley LC, Wong KK. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, Elledge SJ, Hannon GJ, Lowe SW. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol Cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goemans BF, Zwaan CM, Miller M, Zimmermann M, Harlow A, Meshinchi S, Loonen AH, Hahlen K, Reinhardt D, Creutzig U, Kaspers GJ, Heinrich MC. Mutations in KIT and RAS are frequent events in pediatric core-binding factor acute myeloid leukemia. Leukemia. 2005;19:1536–1542. doi: 10.1038/sj.leu.2403870. [DOI] [PubMed] [Google Scholar]
- 21.Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, Campuzano V, Barbacid M. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 22.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 23.Hartzell C, Ksionda O, Lemmens E, Coakley K, Yang M, Dail M, Harvey RC, Govern C, Bakker J, Lenstra TL, Ammon K, Boeter A, Winter SS, Loh M, Shannon K, Chakraborty AK, Wabl M, Roose JP. Dysregulated RasGRP1 responds to cytokine receptor input in T cell leukemogenesis. Sci Signal. 2013;6:ra21. doi: 10.1126/scisignal.2003848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, Morales T, Aliagas I, Liu B, Sideris S, Hoeflich KP, Jaiswal BS, Seshagiri S, Koeppen H, Belvin M, Friedman LS, Malek S. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 25.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, Marais R. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Q, Klippel A, Muslin AJ, Fantl WJ, Williams LT. Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol-3 kinase. Science. 1995;268:100–102. doi: 10.1126/science.7701328. [DOI] [PubMed] [Google Scholar]
- 28.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, Nolan GP. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 29.Iversen PO, Lewis ID, Turczynowicz S, Hasle H, Niemeyer C, Schmiegelow K, Bastiras S, Biondi A, Hughes TP, Lopez AF. Inhibition of granulocyte-macrophage colony-stimulating factor prevents dissemination and induces remission of juvenile myelomonocytic leukemia in engrafted immunodeficient mice. Blood. 1997;90:4910–4917. [PubMed] [Google Scholar]
- 30.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim A, Morgan K, Hasz DE, Wiesner SM, Lauchle JO, Geurts JL, Diers MD, Le DT, Kogan SC, Parada LF, Shannon K, Largaespada DA. Beta common receptor inactivation attenuates myeloproliferative disease in Nf1 mutant mice. Blood. 2007;109:1687–1691. doi: 10.1182/blood-2006-05-025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B, Balla T, Weiss WA, Williams RL, Shokat KM. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–430. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 34.Kohl TM, Schnittger S, Ellwart JW, Hiddemann W, Spiekermann K. KIT exon 8 mutations associated with core-binding factor (CBF)-acute myeloid leukemia (AML) cause hyperactivation of the receptor in response to stem cell factor. Blood. 2005;105:3319–3321. doi: 10.1182/blood-2004-06-2068. [DOI] [PubMed] [Google Scholar]
- 35.Lauchle JO, Braun BS, Loh ML, Shannon K. Inherited predispositions and hyperactive Ras in myeloid leukemogenesis. Pediatr Blood Cancer. 2006;46:579–585. doi: 10.1002/pbc.20644. [DOI] [PubMed] [Google Scholar]
- 36.Leon CM, Barbosa CM, Justo GZ, Borelli P, Resende JD, Jr., de Oliveira JS, Ferreira AT, Paredes-Gamero EJ. Requirement for PLCgamma2 in IL-3 and GM-CSF-stimulated MEK/ERK phosphorylation in murine and human hematopoietic stem/progenitor cells. Journal of cellular physiology. 2011;226:1780–1792. doi: 10.1002/jcp.22507. [DOI] [PubMed] [Google Scholar]
- 37.Lim KH, Counter CM. Reduction in the requirement of oncogenic Ras signaling to activation of PI3K/AKT pathway during tumor maintenance. Cancer Cell. 2005;8:381–392. doi: 10.1016/j.ccr.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 38.Lyubynska N, Gorman MF, Lauchle JO, Hong WX, Akutagawa JK, Shannon K, Braun BS. A MEK inhibitor abrogates myeloproliferative disease in Kras mutant mice. Sci Transl Med. 2011;3:76ra27. doi: 10.1126/scitranslmed.3001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, Ludlam MJ, Bowman KK, Wu J, Giannetti AM, Starovasnik MA, Mellman I, Jackson PK, Rudolph J, Wang W, Fang G. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1116510109. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plowman SJ, Muncke C, Parton RG, Hancock JF. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sci U S A. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raynaud FI, Eccles SA, Patel S, Alix S, Box G, Chuckowree I, Folkes A, Gowan S, De Haven Brandon A, Di Stefano F, Hayes A, Henley AT, Lensun L, Pergl-Wilson G, Robson A, Saghir N, Zhyvoloup A, McDonald E, Sheldrake P, Shuttleworth S, Valenti M, Wan NC, Clarke PA, Workman P. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol Cancer Ther. 2009;8:1725–1738. doi: 10.1158/1535-7163.MCT-08-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reuther GW, Lambert QT, Rebhun JF, Caligiuri MA, Quilliam LA, Der CJ. RasGRP4 is a novel Ras activator isolated from acute myeloid leukemia. J Biol Chem. 2002;277:30508–30514. doi: 10.1074/jbc.M111330200. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 45.Rubio I, Wetzker R. A permissive function of phosphoinositide 3-kinase in Ras activation mediated by inhibition of GTPase-activating proteins. Curr Biol. 2000;10:1225–1228. doi: 10.1016/s0960-9822(00)00731-4. [DOI] [PubMed] [Google Scholar]
- 46.Sabnis AJ, Cheung LS, Dail M, Kang HC, Santaguida M, Hermiston ML, Passegu, Emmanuelle, Shannon K, Braun BS. Oncogenic Kras Initiates leukemia in hematopoietic stem cells. PLoS Biology. 2009;7:e59. doi: 10.1371/journal.pbio.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sampaio C, Dance M, Montagner A, Edouard T, Malet N, Perret B, Yart A, Salles JP, Raynal P. Signal strength dictates phosphoinositide 3-kinase contribution to Ras/extracellular signal-regulated kinase 1 and 2 activation via differential Gab1/Shp2 recruitment: consequences for resistance to epidermal growth factor receptor inhibition. Mol Cell Biol. 2008;28:587–600. doi: 10.1128/MCB.01318-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schubbert S, Lieuw K, Rowe SL, Lee CM, Li X, Loh ML, Clapp DW, Shannon KM. Functional analysis of leukemia-associated PTPN11 mutations in primary hematopoietic cells. Blood. 2005;106:311–317. doi: 10.1182/blood-2004-11-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schubbert S, Zenker M, Rowe SL, Boll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner LE, Nguyen H, West B, Zhang KY, Sistermans E, Rauch A, Niemeyer CM, Shannon K, Kratz CP. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 50.Smith RJ, Sam LM, Justen JM, Bundy GL, Bala GA, Bleasdale JE. Receptor-coupled signal transduction in human polymorphonuclear neutrophils: effects of a novel inhibitor of phospholipase C-dependent processes on cell responsiveness. The Journal of pharmacology and experimental therapeutics. 1990;253:688–697. [PubMed] [Google Scholar]
- 51.Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol. 2007;9:905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- 52.Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, Hingorani SR, Zaks T, King C, Jacobetz MA, Wang L, Bronson RT, Orkin SH, DePinho RA, Jacks T. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 53.Van Etten RA, Shannon KM. Focus on myeloproliferative diseases and myelodysplastic syndromes. Cancer Cell. 2004;6:547–552. doi: 10.1016/j.ccr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Van Meter ME, Diaz-Flores E, Archard JA, Passegue E, Irish JM, Kotecha N, Nolan GP, Shannon K, Braun BS. K-RasG12D expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood. 2007;109:3945–3952. doi: 10.1182/blood-2006-09-047530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 56.Wahlstrom AM, Cutts BA, Liu M, Lindskog A, Karlsson C, Sjogren AK, Andersson KM, Young SG, Bergo MO. Inactivating Icmt ameliorates K-RAS-induced myeloproliferative disease. Blood. 2008;112:1357–1365. doi: 10.1182/blood-2007-06-094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wennstrom S, Downward J. Role of phosphoinositide 3-kinase in activation of ras and mitogen-activated protein kinase by epidermal growth factor. Mol Cell Biol. 1999;19:4279–4288. doi: 10.1128/mcb.19.6.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White MA, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler MH. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 59.Xu J, Hedberg C, Dekker FJ, Li Q, Haigis KM, Hwang E, Waldmann H, Shannon K. Inhibiting the palmitoylation/depalmitoylation cycle selectively reduces the growth of hematopoietic cells expressing oncogenic Nras. Blood. 2012;119:1032–1035. doi: 10.1182/blood-2011-06-358960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J, Wang J, Liu Y, Sidik H, Young KH, Lodish HF, Fleming MD. Oncogenic Kras-induced leukemogeneis: hematopoietic stem cells as the initial target and lineage-specific progenitors as the potential targets for final leukemic transformation. Blood. 2009;113:1304–1314. doi: 10.1182/blood-2008-01-134262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.