Abstract
Lyme disease spirochetes demonstrate strain- and species-specific differences in tissue tropism. For example, the three major Lyme disease spirochete species, Borrelia burgdorferi sensu stricto, B. garinii, and B. afzelii, are each most commonly associated with overlapping but distinct spectra of clinical manifestations. Borrelia burgdorferi sensu stricto, the most common Lyme spirochete in the U.S., is closely associated with arthritis. The attachment of microbial pathogens to cells or to the extracellular matrix of target tissues may promote colonization and disease, and the Lyme disease spirochete encodes several surface proteins, including the decorin- and dermatan sulfate-binding adhesin DbpA, which vary among strains and have been postulated to contribute to strain-specific differences in tissue tropism. DbpA variants differ in their ability to bind to its host ligands and to cultured mammalian cells. To directly test whether variation in dbpA influences tissue tropism, we analyzed murine infection by isogenic B. burgdorferi strains that encode different dbpA alleles. Compared to dbpA alleles of B. afzelii strain VS461 or B. burgdorferi strain N40-D10/E9, dbpA of B. garinii strain PBr conferred the greatest decorin- and dermatan sulfate-binding activity, promoted the greatest colonization at the inoculation site and heart, and caused the most severe carditis. The dbpA of strain N40-D10/E9 conferred the weakest decorin- and GAG-binding activity, but the most robust joint colonization and was the only dbpA allele capable of conferring significant joint disease. Thus, dbpA mediates colonization and disease by the Lyme disease spirochete in an allele-dependent manner and may contribute to the etiology of distinct clinical manifestations associated with different Lyme disease strains. This study provides important support for the long-postulated model that strain-specific variations of Borrelia surface proteins influence tissue tropism.
Author Summary
Lyme disease, the most common vector-borne disease in the United States, is caused by a bacterium, Borrelia burgdorferi. This bacterium infects the skin at the site of the tick bite and then can spread to other tissues, such as the heart, joints or nervous system, causing carditis, arthritis or neurologic disease. To colonize human tissues, the pathogen produces surface proteins that promote bacterial attachment to these sites. For example, DbpA binds to decorin, a component of human tissue. Different Lyme disease strains differ in the particular tissues they colonize and the disease they cause, but we do not understand why. Different strains also make distinct versions of DbpA that bind decorin differently, so variation of DbpA might contribute to strain-to-strain variation in clinical manifestations. To test this, we infected mice with Lyme disease strains that were identical except for the particular DbpA variant they produced. We found that the strains colonized different tissues and caused different diseases, such as arthritis or carditis. These results provide the first solid evidence that variation of an outer surface protein, in this case DbpA, influences what tissues are most affected during Lyme disease.
Introduction
Lyme disease is distributed worldwide and is the most common arthropod-borne infectious disease in the United States [1]–[3]. The causative agent is the spirochete Borrelia burgdorferi sensu lato, which includes B. burgdorferi sensu stricto, B. garinii, and B. afzelii [4] [5]. Following the bite of an infected Ixodes tick, the Lyme disease spirochete produces a local infection, resulting in the characteristic skin lesion erythema migrans. In the absence of antibiotic treatment, spirochetes may disseminate to multiple organs, including joints, the central nervous system, and the heart, resulting in diverse manifestations such as arthritis, neurological abnormalities, and carditis [2], [6]–[9].
Lyme disease spirochetes demonstrate strain- and species-specific differences in tissue tropism. For example, B. burgdorferi sensu stricto, most prevalent in the United States, B. garinii and B. afzelii, each more common in Europe [1], [5], are genetically distinct and are associated with different typical chronic manifestations: B. burgdorferi with arthritis, B. garinii with neuroborreliosis, and B. afzelii with the chronic skin lesion acrodermatitis [10]. In addition, the severity of human symptoms and the dissemination activities of different strains within a single Lyme disease species may also differ significantly [9], [11], [12]. Strain-to-strain variation in dissemination and disease manifestation has also been observed in animal studies [13], [14].
The basis for differences in tissue tropism and/or disease severity is not well understood. Several documented or putative virulence factors encoded by Lyme disease spirochete vary in a strain-specific manner [3], [15]–[17]. In some instances this variation is associated with differences in the postulated biological activity of the factor, e.g. binding of complement regulators by CspZ and other CRASPs (complement regulator-acquiring surface proteins) or binding of plasminogen by the outer surface protein OspC [16]–[19]. Moreover, in a set of three Lyme disease strains, invasiveness correlated with the ability of OspC to bind plasminogen [18], [20], giving rise to the hypothesis that allelic variation of B. burgdorferi surface proteins have the capacity to contribute to tissue tropism of different Lyme disease spirochete strains [9], [12], [18], [21]. However, to date rigorous demonstration that isogenic strains harboring allelic variants of virulence genes indeed behave differently during animal infection has been lacking.
Adhesion of bacterial pathogens to host cells or extracellular matrix (ECM) of target tissues, often mediated by outer surface protein adhesins, is thought to be an important early step in tissue colonization [22]. In fact, Borrelia sp. encode a plethora of adhesins that have been found to recognize different ECM components and/or to promote binding to diverse mammalian cell types [23]–[25]. Two related Borrelia adhesins, decorin binding proteins A and B (DbpA and DbpB, respectively), encoded by a bicistronic operon [26], bind to both decorin and to the glycosaminoglycan (GAG) dermatan sulfate [27], [28]. Whereas the DbpB sequence is highly conserved in different strains of B. burgdorferi sensu lato, the DbpA sequence is highly polymorphic, with sequence similarities as low as 58% between variants [29].
Spirochetes disseminate less efficiently in decorin-deficient compared to wild type mice, suggesting an important function for decorin binding in spirochete tissue spread. [30]. B. burgdorferi lacking DbpA and DbpB in fact exhibited both reduced colonization and dissemination activity and a three- to four-log increase in ID50, indicating that these adhesins play a significant role in infection [31]–[35]. Consistent with this role, dbpA and dbpB are expressed efficiently in culture conditions that may reflect the host environment, such as at mammalian body temperature or in the presence of atmospheric CO2 [36]–[38].
The ability of DbpA to bind to decorin and/or dermatan sulfate requires an intact C-terminus, and DbpA variants demonstrate differences in decorin- and/or dermatan sulfate-binding activities [21], [39]. Given the abovementioned strain- and species-specific differences in tissue tropism among Lyme disease spirochetes, an attractive hypothesis is that the decorin and/or GAG-binding activities of DbpA (and DbpB) are critical for promoting colonization, and that allelic variation of dbpA might influence the tissue tropism of Lyme disease spirochetes. In the current study, we infected mice with various isogenic B. burgdorferi strains encoding DbpA variants, or a non-binding mutant. These studies indicate that decorin- and/or GAG-binding activity of DbpA is required for colonization functions. Importantly we also found that allelic variation of dbpA contributes to differences in tissue tropism.
Results
Recombinant DbpA protein variants show differences in binding to decorin and dermatan sulfate
We previously tested the ability of DbpA mutants or variants to mediate binding of a non-adhesive and non- infectious B. burgdorferi strain to decorin, dermatan sulfate or mammalian cells [39]. DbpAVS461ΔC11, which lacks the 11 C-terminal residues of DbpAVS461, was shown to be unable to promote spirochetal binding to decorin or dermatan sulfate. In addition, a set of variants that included DbpA from B. burgdorferi strains B31 (DbpAB31), 297 (DbpA297), N40-D10/E9 (DbpAN40-D10/E9), B356 (DbpAB356), B. afzelii VS461 (DbpAVS461), and B. garinii PBr (DbpAPBr), showed variant-specific differences in the ability to promote bacterial adhesion to the two substrates. By using semi-quantitative ELISA, this study also analyzed the binding of recombinant versions of DbpA variants except DbpA297 and DbpAB356, which display 90% and 99% similarities to DbpAB31 and DbpAN40-D10/E9, respectively. To measure the decorin- and dermatan sulfate-binding affinities of DbpA variants more precisely, here we utilized quantitative ELISA and surface plasmon resonance (SPR; Fig. S2A and Table 1). The two independent methods for assessing binding gave results entirely consistent with each other and revealed dissociation constants indicating (1) robust decorin-binding by DbpAPBr (KD = 0.06–0.09 µM); (2) moderate decorin-binding by DbpAB31, DbpA297, and DbpAVS461 (KD = 0.14–0.30 µM); (3) less efficient decorin-binding by DbpAN40-D10/E9 and DbpAB356 (KD = 0.71-0.95 µM). Interestingly, a BXBB motif (residues 64 to 67) that has been proposed to form a positively charged pocket that binds to decorin and/or dermatan sulfate [40], is not found in DbpAPBr (Fig. S6), suggesting that BXBB is not essential for decorin- or dermatan sulfate-binding.
Table 1. DbpA variants differ in binding to decorin and dermatan sulfate.
| ELISA | ----- Surface Plasmon Resonance ----- | ||||
| DbpA variant | Ligand | KD (µM) | KD (µM) | kon (105 s−1M−1) | koff (s−1) |
| B. burgdorferi | |||||
| DbpAB31 | Decorin | 0.21±0.03 | 0.28±0.03 | 4.17±0.70 | 0.11±0.01 |
| Derm SO4 | 0.91±0.02 | 0.63±0.04 | 0.36±0.01 | 0.02±0.01 | |
| DbpA297 | Decorin | 0.14±0.06 | 0.30±0.08 | 3.51±0.86 | 0.10±0.01 |
| Derm SO4 | 0.78±0.18 | 0.50±0.01 | 0.52±0.02 | 0.02±0.01 | |
| DbpAB356 | Decorin | 0.95±0.12 | 0.84±0.04 | 0.66±0.33 | 0.06±0.01 |
| Derm SO4 | 3.68±0.21 | 1.91±0.87 | 0.44±0.04 | 0.07±0.03 | |
| DbpAN40-D10/E9 | Decorin | 0.85±0.12 | 0.71±0.01 | 0.97±0.42 | 0.07±0.01 |
| Derm SO4 | 3.10±0.26 | 2.58±0.17 | 0.13±0.06 | 0.03±0.01 | |
| B. garinii | |||||
| DbpAPBr | Decorin | 0.06±0.01 | 0.09±0.06 | 18.3±1.10 | 0.13±0.02 |
| Derm SO4 | 0.21±0.03 | 0.16±0.02 | 0.56±0.02 | 0.009±0.001 | |
| B. afzelii | |||||
| DbpAVS461 | Decorin | 0.29±0.07 | 0.26±0.02 | 1.52±0.23 | 0.04±0.01 |
| Derm SO4 | 1.55±0.63 | 1.61±0.48 | 0.43±0.06 | 0.07±0.03 | |
| DbpAVS461ΔC11 | Decorin | n.b a | n.b. | n.b. | n.b. |
| Derm SO4 | n.b. | n.b. | n.b. | n.b. | |
All values represent the mean ± SEM of three experiments.
No binding activity was detected.
With the exception of DbpAVS461, the calculated KD for dermatan sulfate binding of each DbpA variant was approximately two- to four-fold higher than its KD for decorin binding; DbpAVS461 bound to dermatan sulfate approximately six-fold less efficiently than to decorin (Fig. S2 and Table 1). Finally, recombinant protein DbpAVS461ΔC11, which was found by far-UV CD analysis (Fig. S1) to retain the secondary structure of wild-type DbpAVS461, was unable to bind to decorin or dermatan sulfate. These findings were entirely consistent with previous results determined with less quantitative methods [39], and likely reflect the fact that sequence lacking in DbpAVS461ΔC11 includes conserved K170, a lysine residue previously shown to be critical for decorin-binding activity [41] (Fig. S6).
DbpA variants alter the ability of an infectious strain of B. burgdorferi to bind decorin and dermatan sulfate
DbpAPBr, DbpAVS461, and DbpAN40-D10/E9 each represent one of the three binding profiles described above, as well as collectively encompass the three major genospecies of Lyme disease spirochetes, each of which has been associated with different human clinical manifestations. To focus on how variations in DbpA binding to decorin and dermatan sulfate may influence the infectious process and avoid potential functional redundancy associated with the production of another decorin- and dermatan sulfate-binding adhesin, we generated DbpA-producing strains that did not produce DbpB. We generated a set of plasmids that encode the bbe22 gene, which is required for spirochete survival in a mammalian host [42], and the coding region of dbpAPBr, dbpAVS461, or dbpAN40-D10/E9, or dbpAVS461ΔC11 (as a non-binding control) under the control of the dbpBA promoter of B. burgdorferi strain B31. The plasmids encoding different dbpA alleles were then individually introduced into a dbpBA deletion mutant of the highly transformable infectious strain B. burgdorferi ML23, a derivative of B. burgdorferi B31 that lacks bbe22 and therefore cannot survive in the mouse in the absence of a complementing bbe22-encoding plasmid [43]. We verified by flow cytometry analysis that the DbpA variants produced in B. burgdorferi ML23ΔdbpBA were located on the surface of the recombinant spirochetes, and at levels indistinguishable from that of their DbpA-proficient parental strain ML23 (Fig. S3).
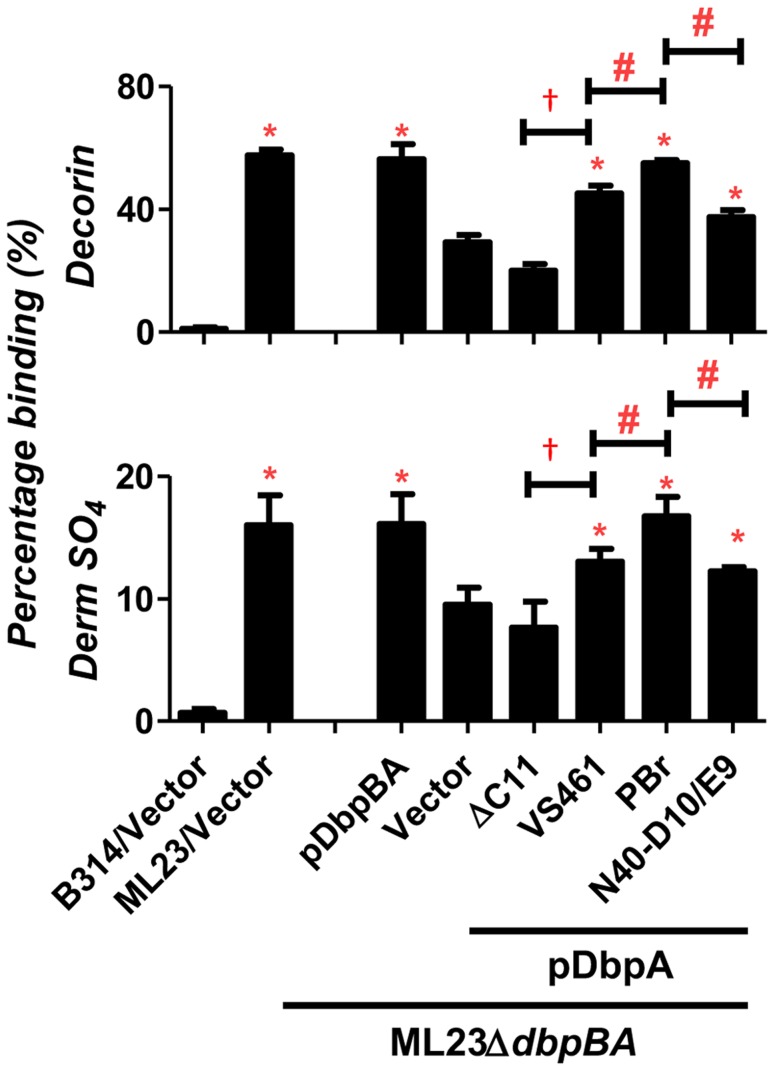
We next investigated the distinct decorin- and dermatan sulfate-binding activities specifically conferred to infectious strain ML23 by the various dbpA alleles. We measured binding of radiolabeled ML23ΔdbpBA strains producing DbpA variants to microtiter wells coated with decorin or dermatan sulfate. Chondroitin-6-sulfate, included as a negative control, mediated binding of less than 5% of inoculum (data not shown). Strain ML23 harboring the vector alone, a positive control that expresses both DbpA and DbpB, bound to decorin or dermatan sulfate with an efficiency of approximately 55% or 15%, respectively (Fig. 1). This level of binding was significantly greater than binding by strain ML23ΔdbpBA harboring vector alone, i.e. approximately 30% or 10% for binding to decorin or dermatan sulfate, respectively (Fig. 1). This “background” (i.e., DbpB- and DbpA-independent) decorin- and dermatan sulfate-binding activity of strain ML23ΔdbpBA is considerably greater than that of the high-passage strain B. burgdorferi B314 (i.e., less than 2% for either substrate), suggesting that decorin- and dermatan sulfate-binding adhesins other than DbpA and DbpB are expressed by strain ML23ΔdbpBA. As expected, the production of both DbpA and DbpB in strain ML23ΔdbpBA (Fig 1, “pDbpBA”) restored binding to the levels of strain ML23.
Figure 1. DbpA variants produced in B. burgdorferi promote distinct binding activities to decorin and dermatan sulfate.
Binding of radiolabeled B. burgdorferi ML23/pBBE22, ML23ΔdbpBA/pBBE22 (“Vector”), and the deletion strain bearing a plasmid encoding DbpA and DbpB (“pDbpBA”), DbpAVS461ΔC11 (“ΔC11”), DbpAVS461 (“VS461”), DbpAPBr (“PBr”) or DbpAN40-D10/E9 (“N40-D10/E9”), to decorin, dermatan sulfate (Derm SO4), or the negative control chondroitin-6-sulfate (see Materials and Methods). The non-adherent B. burgdorferi strain B314 harboring the empty vector pJF21 (“B314/Vector”) was also included as negative control. The percentage of bound bacteria was determined by radioactive counts in bound bacteria normalized to the counts in the inoculum.Each bar represents the mean of four independent determinations ± SEM. Statistical significance was determined using the one-way ANOVA test. Significant (P<0.05) differences in binding relative to the dbpBA deletion strain (“*”), between two strains relative to each other (“#”), or relative to the dbpAVS461ΔC11-complemented strain (“†”) are indicated.
The production of DbpAVS461, DbpAPBr, or DbpAN40-D10/E9 in strain ML23ΔdbpBA resulted in decorin- and dermatan sulfate-binding significantly greater than strain ML23ΔdbpBA harboring vector alone, indicating that these DbpA variants provide significant adhesive function to this strain (Fig. 1). DbpAVS461ΔC11 conferred no detectable increase in binding, indicating, as predicted, that the 11 C-terminal amino acids of DbpAVS461 are essential for binding to decorin and dermatan sulfate [39]. DbpAPBr promoted significantly greater spirochete binding to decorin and dermatan sulfate than did DbpAVS461 or DbpAN40-D10/E9 (Fig. 1). Thus, the degree of decorin- and dermatan sulfate-binding conferred to strain ML23ΔdbpBA by each DbpA variant was consistent with both the quantitative binding analysis of purified recombinant DbpA proteins described above (Fig. S2) and with our previous study of these variants expressed in a non-adherent, non-infectious strain B314 [39].
DbpA lacking the decorin and GAG-binding activities fails to facilitate colonization
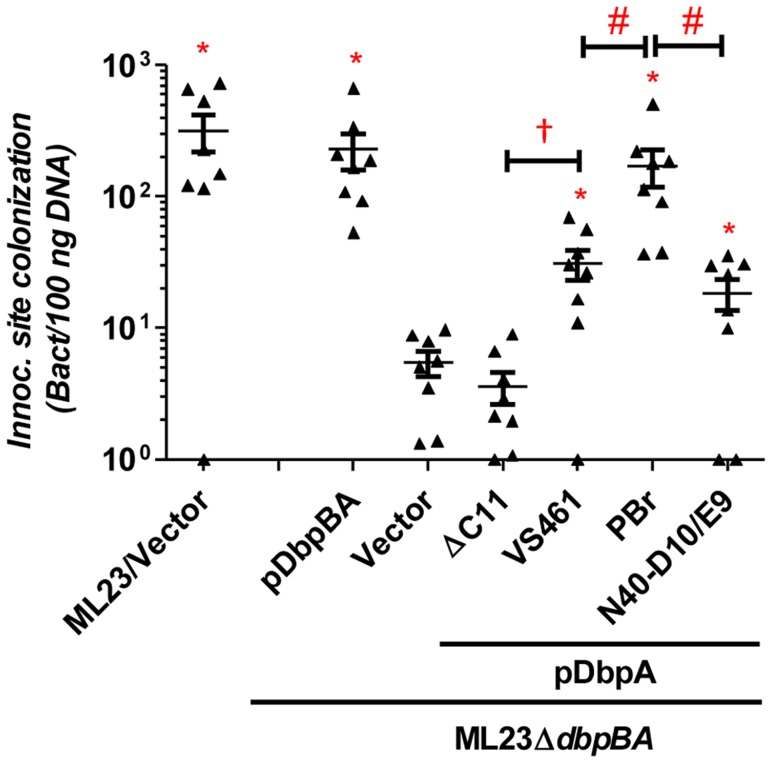
The defect in decorin- and/or dermatan sulfate-binding by DbpAVS461ΔC11 provided an opportunity to determine if these activities of DbpA are essential to promote B. burgdorferi colonization. C3H/HeN mice were infected with ML23ΔdbpBA producing DbpAVS461 or DbpAVS461ΔC11 and the bacterial load at the inoculation site was assessed at 3 days post-infection. Strains ML23 and ML23ΔdbpBA/pDbpBA were included as positive controls and colonized the site efficiently (∼300 bacteria per 100 ng of DNA), 60-fold higher than that of ML23ΔdbpBA harboring vector alone (Fig. 2). ML23ΔdbpBA producing DbpAVS461 promoted significant colonization (∼30 bacteria per 100 ng DNA, or ∼six-fold more than ML23ΔdbpBA) at the inoculation site. This finding indicated that production of DbpA alone could partially complement the defect of a B. burgdorferi ΔdbpBA mutant, consistent with previous studies [32], [33]. In contrast, ML23ΔdbpBA producing DbpAVS461ΔC11 did not mediate colonization at a level any greater than ML23ΔdbpBA carrying the empty vector.
Figure 2. DbpA variants promote distinct B. burgdorferi inoculation site colonization during early infection.
C3H/HeN mice infected with 104 B. burgdorferi strain ML23/pBBE22 (“ML23/Vector”), dbpBA deletion strain ML23ΔdbpBA/pBBE22(“Vector”), or the deletion strain bearing a plasmid encoding the indicated variants were sacrificed at 3 days post-infection. Bacterial loads at the inoculation site were determined by qPCR. Data shown are the mean bacterial loads ± SEM of 10 mice per group. Statistical significance was determined using a one-way ANOVA test. Significant (P<0.05) differences in spirochete number relative to the dbpBA deletion strain (“*”), between two strains relative to each other (“#”), or relative to the dbpAVS461ΔC11-complemented strain (“†”) are indicated. (n.d.): not determined.
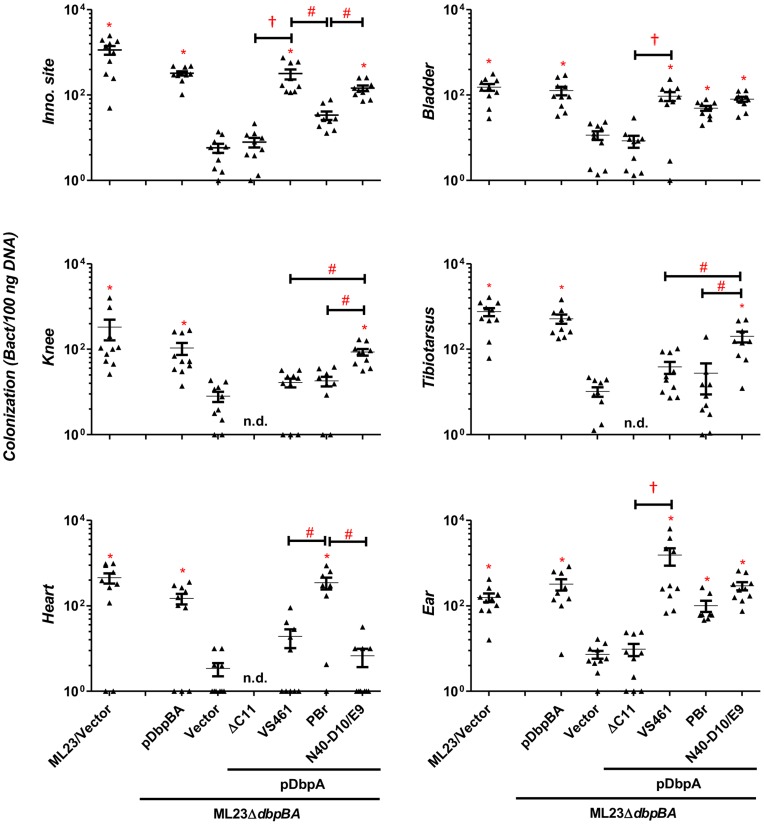
To determine if DbpAVS461ΔC11 might promote colonization at a later time point, we also assessed infected mice at 28 days post-infection. The positive control strains B. burgdorferi ML23 and B. burgdorferi ML23ΔdbpBA/pDbpBA displayed efficient colonization at all sites tested (Fig. 3). In particular, colonization of the inoculation site, bladder and ear was 30-200-fold higher than that of ML23ΔdbpBA harboring vector alone. The production of DbpAVS461 by ML23ΔdbpBA producing DbpAVS461 did not promote colonization of the joints or heart at this time point but did promote colonization of the inoculation site, bladder, and ear at levels indistinguishable from the positive control strains. In contrast, ML23ΔdbpBA/pDbpAVS461ΔC11 did not mediate colonization at a level any greater than ML23ΔdbpBA carrying the empty vector at any of the sites tested.
Figure 3. DbpA variants promote distinct B. burgdorferi tissue colonization profiles at 28 days post-infection.
C3H/HeN mice infected with 104 B. burgdorferi strain ML23/pBBE22 (“ML23/Vector”), dbpBA deletion strain ML23ΔdbpBA/pBBE22 (“Vector”), or the deletion strain bearing a plasmid encoding the indicated DbpA variants were sacrificed at 28 days post-infection. The bacterial loads at the inoculation site, ear, bladder, heart, knee, and tibiotarsus joint were determined by qPCR. Data shown are the mean bacterial loads ± SEM of 10 mice per group. Statistical significance was determined using a one-way ANOVA test. Significant (P<0.05) differences in spirochete number relative to the dbpBA deletion strain (“*”), between two strains relative to each other (“#”), or relative to the dbpAVS461ΔC11-complemented strain (“†”), are indicated. (n.d.): not determined. These data are described comprehensively with other post-infection time points in Table S1.
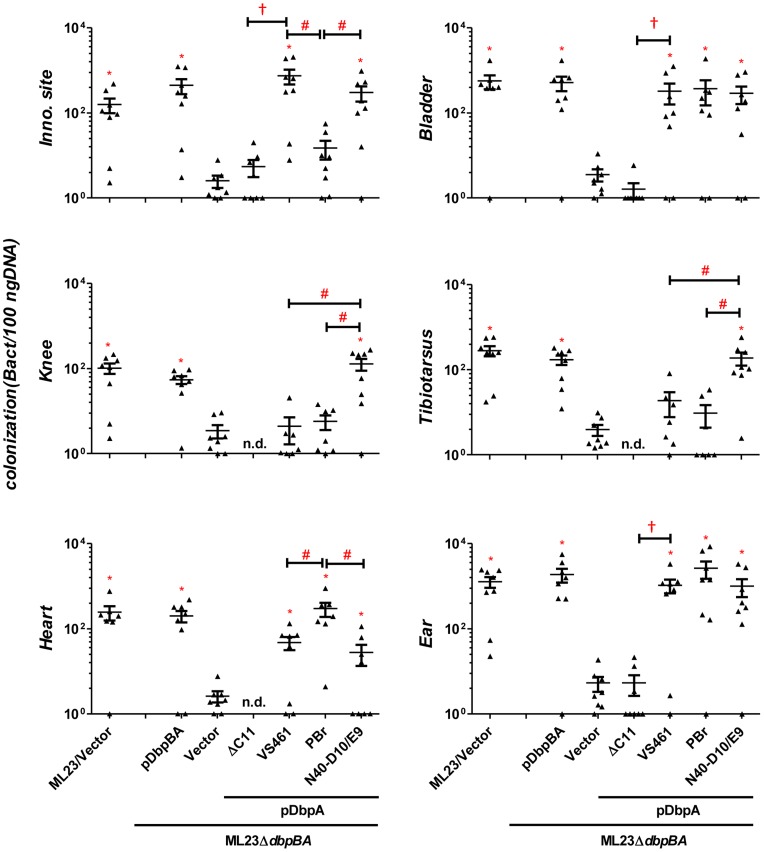
The bacterial load in a particular tissue may be in part a reflection of the rate of immune clearance. To determine if the colonization defect of ML23ΔdbpBA/pDbpAVS461ΔC11 might be due to the induction of a particularly robust immune response, at 28 days post-infection we measured B. burgdorferi-specific IgG or IgM in the sera of mice inoculated with this strain. No B. burgdorferi-specific antibodies were detected (Fig. S4), suggesting that ML23ΔdbpBA/pDbpAVS461ΔC11 is incapable of establishing a productive infection that triggers an adaptive immune response. In addition, the results suggest that the colonization defect of this strain was independent of an adaptive immune response. Consistent with this, a 28-day infection of the mice strain deficient for adaptive immune response (SCID mice) revealed that the production of DbpAVS461ΔC11 was unable to enhance the ability of ML23ΔdbpBA to colonize any of the tissues tested, in contrast to the production of DbpAVS461 (Fig. 4). Together, these results strongly suggest that the decorin- and/or dermatan sulfate-binding activity of DbpA is required for its ability to facilitate spirochetal colonization.
Figure 4. Differences in tissue tropism promoted by DbpA variants are not a function of an adaptive immune response.
C3H/HeN-SCID mice infected with 103 B. burgdorferi ML23/pBBE22 (“ML23/Vector”), ML23ΔdbpBA/pBBE22 (“Vector”), or ML23ΔdbpBA bearing a plasmid encoding the indicated DbpA variants/mutant were sacrificed at 28 days post-infection. Bacterial loads at the inoculation site, ear, bladder, heart, knee, and tibiotarsus joint were determined by qPCR. Data shown are the mean bacterial loads ± SEM of 10 mice per group. Statistical significance was determined using a one-way ANOVA test. Significant differences (P<0.05) in spirochete number relative to the ML23ΔdbpBA (“*”), between two strains relative to each other (“#”), or relative to the pdbpAVS461ΔC11-complemented strain (“†”), are indicated.
Differences in the decorin- and dermatan sulfate-binding activities of DbpA variants influence colonization
To test whether variation in the decorin or dermatan sulfate binding capabilities by DbpA correlates with differences in colonization and/or disease, we chose to analyze the colonization promoting abilities of three DbpA variants that display distinct decorin- and dermatan sulfate-binding activities. C3H/HeN mice were infected with ML23ΔdbpBA producing DbpAPBr, DbpAN40-D10/E9, or DbpAVS461, and differences in colonization at the inoculation site, heart, joints, bladder and ear were assessed at 3, 7, 14, 21, or 28 days post-infection. Strains ML23 and ML23ΔdbpBA/pDbpBA were included as positive controls, and ML23ΔdbpBA harboring vector alone served as a negative control. As previously observed [31], [34], [44], the kinetics of colonization by B. burgdorferi producing DbpA and DbpB varied with tissue: the bladder and joints were colonized by day 7 post-infection whereas the heart and ear were detectably colonized only at the 14 and 21-day time point, respectively (Figs. 3, 5 and Fig. S5; for comprehensive summary of bacterial loads at all times points, see Table S1). ML23ΔdbpBA harboring vector alone was defective for colonization at all time points.
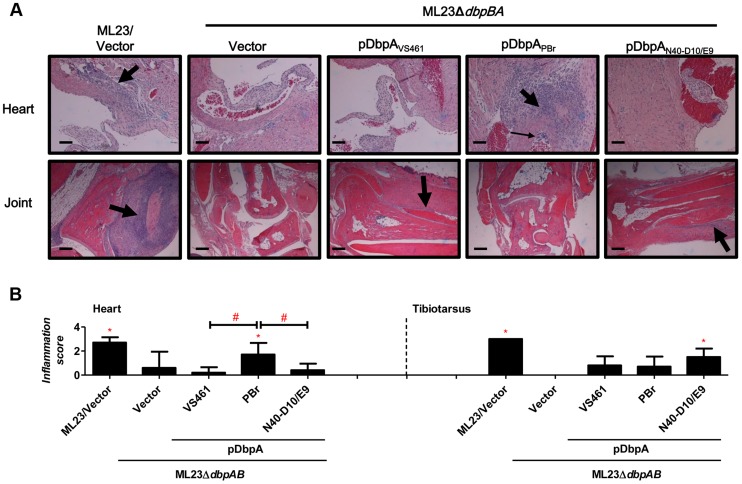
Figure 5. DbpA variants produced by B. burgdorferi lead to differences in tissue inflammation in C3H/HeN-infected mice.
(A) C3H/HeN mice infected with 104 B. burgdorferi strain ML23/pBBE22 (“ML23/Vector”), dbpBA deletion strain ML23ΔdbpBA/pBBE22 (“Vector”), or the deletion strain bearing a plasmid encoding the indicated DbpA variants were sacrificed at 28 days post-infection. Inflammation of the heart (upper panel) and joints (lower panel) were assessed using Hematoxylin and Eosin staining histopathology. The upper panel and lower panel indicate the higher-resolution (10×, bar = 58 µm) and lower-resolution (4×, bar = 141 µm) images. (B) To quantitate inflammation of heart and joint tissues, at least ten random sections from each infection group were scored on a scale of 0–3 for the severity of carditis and arthritis, as indicated in Material and Methods. Statistical significance was determined using a one-way ANOVA test. Data shown are the mean inflammation score ± SD of 5 mice per group. Statistical significance was determined using a one-way ANOVA test. Significant (P<0.05) differences in the inflammation score relative to the dbpBA deletion strain (“*”) or between two strains relative to each other (“#”), are indicated.
Upon infection with ML23ΔdbpBA producing DbpAPBr, DbpAN40-D10/E9, or DbpAVS461, we found that at 21 days post-infection, each of the DbpA variants tested was capable of fully replacing the colonization function of the endogenous (strain B31) DbpA and DbpB in the inoculation site, bladder, knee, and tibiotarsus (Table S1). On the other hand, in the ear or heart, production of these DbpA variants was associated with delayed colonization (28- vs. 14-day colonization in the ear; 21- vs. 14-day in the heart) compared to these DbpA- and DbpB-proficient strains (Figs. 3, 5 and Fig. S5), which is consistent with the findings reported previously [44].
Importantly, in several tissues, the different DbpA variants conferred significant differences in the efficiency or kinetics of colonization. At the inoculation site at three days post-infection, production of DbpAPBr, which displayed greater decorin- and dermatan sulfate-binding activity than DbpAVS461 or DbpAN40-D10/E9, conferred approximately six- to nine-fold greater colonization (P<0.05; Fig. 2). The production of DbpAPBr was also associated with diminished late colonization of the inoculation site, because by 28 days post-infection, ML23ΔdbpBA producing DbpAPBr was present at this site at levels approximately 20- to 50-fold lower than ML23ΔdbpBA producing DbpAVS461 or DbpAN40-D10/E9 (Fig. 3).
The production of the high affinity binding variant DbpAPBr also resulted in enhanced colonization of the heart at 21 and particularly 28 days post-infection compared to production of the other two DbpA variants (Figs. 3 and 5). At the later time point, ML23ΔdbpBA producing DbpAPBr was present at levels 15- to 50-fold higher than ML23ΔdbpBA producing DbpAVS461 or DbpAN40-D10/E9 (P<0.05), which were not present at levels significantly higher than the negative control strain ML23ΔdbpBA (Fig. 3).
Interestingly, although as mentioned above, no dbpA allele-specific differences were observed in joint colonization at 21 days post-infection (Table S1), DbpAN40-D10/E9, which binds to decorin and dermatan sulfate with the lowest affinity among the variants analyzed, promoted the greatest level of colonization of the tibiotarsus and knee at 28 days post-infection (Fig. 3). Whereas by this time ML23ΔdbpBA producing DbpAVS461 or DbpAPBr were no longer present in the knee or tibiotarsus at levels significantly greater than the DbpA- and DbpB-deficient ML23ΔdbpBA harboring vector alone, ML23ΔdbpBA producing DbpAN40-D10/E9 was present at levels 18 to 32-fold higher than strains producing either DbpAVS461 or DbpAPBr (Fig. 3) (P<0.04).
An adaptive immune response does not account for the differences in B. burgdorferi tissue tropism promoted by distinct DbpA variants
To test whether the distinct colonization levels in the inoculation site, heart, knee or tibiotarsus late in infection might be due to differences in the humoral immune response triggered by different DbpA variants, we quantitated serum IgG and IgM titers against each DbpA variant. We found no significant difference in anti-DbpA antibody production among animals infected with strains producing the different variants (Fig. S4). To determine if the ability to generate an adaptive immune response was required to elicit the differences in apparent tissue tropism among strains, we infected SCID mice with strains encoding each of the three alleles. Pilot experiments revealed that a dose of 103 bacteria, i.e., ten-fold lower than that inoculated into wild type mice, was optimal for discerning colonization differences between strains ML23 and ML23ΔdbpBA (data not shown; see Materials and Methods). We assessed tissue burden at 28 days post-infection and found that compared to ML23ΔdbpBA producing DbpAVS461 or DbpAN40-D10/E9, an isogenic strain expressing DbpAPBr was present at approximately five- to ten-fold lower levels at the inoculation site (P<0.02) and 15 to 87-fold higher levels in the heart (P<0.02; Fig. 4). ML23ΔdbpBA producing DbpAN40-D10/E9 colonized the knee and tibiotarsus six- to eight-fold more efficiently than ML23ΔdbpBA expressing DbpAPBr or DbpAVS461 (P<0.04). Thus, the strain-specific colonization pattern for the heart and joints were identical to those observed upon infection of immunocompetent mice.
Strain-specific DbpA variation influences carditis and arthritis
To determine whether the observed DbpA strain-specific differences in tissue tropism resulted in corresponding differences in disease severity, C3H/HeN mice infected with isogenic ML23ΔdbpBA derivatives producing DbpAVS461, DbpAPBr or DbpAN40-D10/E9 were subjected to histopathological analysis. We evaluated carditis at the heart base at 28 days post-infection, at which time ML23ΔdbpBA producing DbpAPBr colonized the heart at levels six to nine-fold greater than the negative control strain ML23ΔdbpBA or strains producing DbpAVS461 or DbpAN40-D10/E9 (Fig. 3). When coded samples were scored blindly for carditis on a scale of 0 to 3 depending on the number and intensity of inflammatory foci at the heart base (see Materials and Methods), the positive control strain ML23 induced robust (grade 3) carditis (Fig. 5B, left panel). Focal subendocardial mononuclear cell infiltrates were present upon infection by this strain (thick arrow in Fig. 5A, top row), whereas strain ML23ΔdbpBA harboring vector alone or producing DbpAVS461 or DbpAN40-D10/E9 showed, at most, a very mild mononuclear cell infiltrate (carditis score near grade 0; Fig. 5, left panel), reflecting their relative levels of cardiac colonization at this time point (Fig. 3). Importantly, consistent with the persistent cardiac colonization by ML23ΔdbpBA producing DbpAPBr, the mice infected with this strain exhibited significant (grade ∼2) carditis (Fig. 5B, left panel). Focal interstitial subacute myocarditis (thick arrow at Fig. 5A, top row) and vasculitis (thin arrow), involving mostly mononuclear cells was present.
Histopathological analyses were also performed on the tibiotarsus joint at 28 days post-infection, a time point at which ML23ΔdbpBA producing DbpAN40-D10/E9 colonized the joints at levels 18- to 32-fold greater than isogenic strains producing DbpAVS461 or DbpAPBr or the negative control strain ML23ΔdbpBA (Fig. 3). When coded H&E-stained tibiotarsus joint samples were scored blindly for inflammatory infiltrates, severe (grade 3) arthritis was triggered by the positive control strain ML23 (Fig. 5B, right panel). Severe inflammation surrounding some tendons and of the synovial membrane was observed, with periostitis and some proliferation of new bone (thick arrow at Fig. 5A, bottom row). In contrast, the negative control strain ML23ΔdbpBA carrying vector alone or producing DbpAPBr triggered no arthritis (grade 0; Fig. 5A, bottom row and Fig. 5B, right panel). The strain producing DbpAVS461 appeared to induce mild (grade ∼1; Fig. 5B, right) arthritis evidenced by mild and focal inflammation (thick arrow at Fig. 5A, bottom row). These findings are consistent with the low levels of joint colonization by those strains at this time point (Fig. 3). Importantly, reflecting the persistent joint colonization by ML23ΔdbpBA producing DbpAN40-D10/E9, mice infected with this strain exhibited higher levels of arthritis (grade ∼2; Fig. 5B right panel), with moderate mononuclear infiltrates commonly near connective tissue (thick arrow at Fig. 5A, bottom row). We conclude that the higher levels of heart or joint colonization associated with the production of DbpAPBr or DbpAN40-D10/E9, respectively, resulted in greater levels of pathology.
Discussion
Although it has long been known that different genospecies or strains of Borrelia burgdorferi sensu lato cause infections with different clinical manifestations in humans and distinct pathogenicity and/or tissues tropism in animal infection models, the reasons for these differences have remained obscure [9]–[12]. An attractive hypothesis put forth has been is that variation in spirochetal factors that control spread to or survival in different tissues contribute to the disparate behavior during mammalian infection [9], [12], [18], [21]. The ospC gene, which encodes a surface lipoprotein required for infection, is allelic variable, and a sampling of recombinant OspC variants from three invasive or noninvasive strains demonstrated a correlation between plasminogen binding and invasiveness in mice [18]. CRASP's (complement regulator acquiring surface proteins) variants, which promote serum resistance, differ in their ability to bind to the complement regulatory proteins factor H and factor H like protein (FHL-1) [16], [17], [19]. Rigorous demonstration that allelic variation of genes encoding documented or putative virulence factors influences tissue tropism and/or disease manifestation requires experimental infection using isogenic strains, and has thus far been lacking. The dbpA gene, which encodes a Lyme disease spirochete adhesin required for full infectivity, is allelic variable, and DbpA variants differ in their ability to promote spirochetal attachment to decorin, dermatan sulfate, or mammalian cells [21], [39]. DbpAVS461ΔC11, a DbpA truncation that lacks 11 C-terminal amino acids was previously shown in semi-quantitative binding assays to be unable to bind dermatan sulfate or decorin [39]. We confirmed this finding by quantitative ELISA and SPR. The C-terminal 11 amino acids lacking in DbpAVS461ΔC11 are not generally well conserved among DbpA variants but do encompass the universally conserved residue K170, which has been shown to be required for decorin/dermatan sulfate-binding [30], [40], [45]. To test whether the adhesive activity of DbpA is specifically required for colonization, mice were infected with a B. burgdorferi dbpBA deletion mutant that ectopically produced wild type DbpAVS461 or DbpAVS461ΔC11. DbpAVS461ΔC11 was, in fact, also unable to facilitate colonization at the inoculation site, bladder, or ear, indicating that this binding activity of DbpA is likely required for tissue colonization.
The requirement for DbpA adhesive activity for efficient mammalian colonization raised the possibility that the variability of ligand binding among DbpA variants found among Lyme disease spirochetes contributes to the observed strain-to-strain differences in tissue tropism and disease severity [21], [29], [39]. Thus, we quantitatively characterized the decorin- and dermatan sulfate-binding activities of three DbpA variants, i.e. DbpAPBr, DbpAVS461 and DbpAN40-D10/E9, which together represent the three major Lyme disease spirochete genospecies, and generated a set of isogenic B. burgdorferi strains derived from a B. burgdorferi ΔdbpBA mutant that expressed each of these variants. These DbpA-producing strains exhibited the predicted differences in their ability to bind to decorin and dermatan sulfate, with DbpAPBr promoting the most efficient spirochetal binding to purified decorin and dermatan sulfate and DbpAN40-D10/E9 promoting the least.
When mice were infected with these strains, the B. burgdorferi strain producing DbpAPBr promoted better early (i.e., three days post-infection) colonization at the inoculation site. This result is consistent with reports that decorin is enriched in the skin [46] and that spirochetal overproduction of DbpA enhanced colonization of the inoculation site [47]. Importantly, the strain producing DbpAPBr infected the heart at levels one to two orders of magnitude greater than strains producing DbpAVS461 or DbpAN40-D10/E9, and this more intense infection of the heart was associated with enhanced carditis. B. burgdorferi selectively colonizes decorin-rich heart microenvironments such as the tunica adventitia [35], and upon infection, decorin-deficient mice harbor fewer B. burgdorferi in the heart than do littermate control mice [30]. We found that the relative tropism of the DbpAPBr-producing strain for skin and heart was also observed in SCID mice, indicating that the higher level of colonization of these sites by this strain is not accounted for by an adaptive immune response that might be generated more efficiently against one DbpA variant than another. Rather, the tissues that are more efficiently colonized by B. burgdorferi producing a variant of DbpA that binds tightly to decorin corresponds to what is currently understood about the relative enrichment of decorin in these tissues.
Nevertheless, DbpA-mediated colonization is not a simple reflection of its ability to bind decorin because DbpAN40-D10/E9, which displayed the weakest decorin and dermatan sulfate binding, promoted the most robust colonization of the joints late (28 days) after inoculation. Upon scoring of coded histological samples, DbpAN40-D10/E9 was the only variant that promoted arthritis significantly more severe than the non-DbpA-producing control strain. Our in vitro assays indicate that compared to DbpAVS461 or DbpAPBr, DbpAN40-D10/E9 poorly recognizes human recombinant decorin and the (commercially available porcine skin) dermatan sulfate utilized in this study. However, it is possible that DbpAN40-D10/E9 binds to a host ligand present in the murine joint better than these other DbpA variants. Dermatan sulfate, like other GAGs, is heterogeneous with respect to epimerization and modification, raising the possibility that murine joint decorin may be well recognized by DbpAN40-D10/E9. In addition, biglycan, which like the other class I proteoglycan decorin contains ten leucine-rich repeats and two dermatan sulfate GAGs, is present in the joint at higher levels than decorin and could be an additional (well recognized) ligand for DbpAN40-D10/E9 [48]–[50]. Finally, the tibiotarsus joint apparently presents B. burgdorferi with functionally distinct microenvironments, because B. burgdorferi colonizes both synovial and adjacent connective tissues in the joints of untreated SCID mice, but are cleared specifically from synovium by administration of anti-DbpA serum [51]. In our study, the tropism of DbpAN40-D10/E9 for joints was not a simple function of adaptive immunity because it was recapitulated in SCID mice, but it is possible that the different DbpA variants, by recognizing host ligands differently, promote distinct distributions of spirochetes among joint microenvironments. A technical challenge to experimental validation of this hypothesis is the relative paucity of spirochetes in the joints of infected animals.
One interesting observation is that an allele of dbpA from a B. burgdorferi sensu stricto strain (i.e. strain N40-D10/E9) promoted joint colonization and disease in the mouse, an apparent tropism that correlates with the common manifestation of Lyme arthritis upon infection by this genospecies of Lyme disease spirochete [10]. This is not to imply, however, that the production of a particular DbpA variant fully explains the tissue tropism of a given strain. Tissue tropism is undoubtedly multifactorial, so any approach that addresses the contribution of a single allelic variable gene, in this case dbpA, is inherently limited due to its concomitant inability to assess the role of other potential determinants. In addition, here we assessed strains that did not produce DbpB, which may have partially redundant function, and to what degree allelic variation of dbpA contributes to the etiology of distinct symptoms associated with different Lyme disease strains in otherwise wild-type strains will require further study. Nevertheless, the demonstration that dbpA influences colonization and disease by the Lyme disease spirochete in an allele-specific manner provides important support for the long-postulated model that allelic variation of a Borrelia surface protein influences tissue tropism.
Materials and Methods
Ethics statement
All mouse experiments were performed in strict accordance with all provisions of the Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals, and the PHS Policy on Humane Care and Use of Laboratory Animals. The protocol was approved by the Tufts University School of Medicine Institutional Animal Care and Use Committee (IACUC), protocol docket number 2011–140. All efforts were made to minimize animal suffering.
Bacterial strains and growth conditions
The Borrelia and E. coli strains used in this study are described in Table S3. Escherichia coli strains DH5α, BL21 and derivatives were grown in Luria-Bertani (BD Bioscience, Franklin lakes, NJ) broth or agar, supplemented with kanamycin (50 µg/ml) or ampicillin (100 µg/ml) where appropriate. All B. burgdorferi strains were grown in BSK-II completed medium supplemented with kanamycin (200 µg/ml) or Gentamycin (50 µg/ml).
Generation of recombinant DbpA proteins and antisera
To generate recombinant histidine-tagged DbpA proteins, the dbpA open reading frames lacking the putative signal sequences from B. burgdorferi strains B31 and N40-D10/E9, B. garinii strain PBr, and B. afzelii strain VS461 were inserted into pET15b (Novagen, Madison, WI) as previously described [39] (see Table S3). In addition, dbpA open reading frames (lacking the putative signal sequence) from B. burgdorferi strain 297 (encoding residues 30 to 187), B356 (encoding residues 33 to 194), and an altered open reading frame encoding DbpAVS461ΔC11 (residues 22 to 158, lacking the 11 C-terminal amino acids, from B. afzelii strain VS461), were amplified using the primers described in Table S3. Amplified fragments were engineered to encode a BamHI site at the 5′ end and a stop codon followed by a SalI site at the 3′ end. PCR products were sequentially digested with BamHI and SalI and then inserted into the BamHI and SalI sites of pQE30 (Qiagen, Valencia, CA). The resulting plasmids were transformed into E. coli strain M15 (for dbpA297, dbpAB356, and dbpAVS461ΔC11) or BL21 (for all other dbpA alleles) and the plasmid inserts were sequenced (Tufts core sequencing facility). The histidine-tagged DbpA variants were produced and purified by nickel affinity chromatography according to the manufacturer's instructions (Qiagen, Valencia, CA). Antisera against DbpAN40-D10/E9, DbpAPBr, or DbpAVS461 were generated by immunizing five-week-old BALB/C mice with each of the DbpA proteins as described previously [51].
Purification of human decorin
Recombinant human decorin, a generous gift from David Mann (MedImmune, Inc.), was purified from stably transfected Chinese hamster ovary cells (ATCC CCL 61) as described previously [52].
Circular dichroism (CD) spectroscopy
CD analysis was performed on a Jasco 810 spectropolarimeter (Jasco Analytical Instrument, Easton, MD) under N2. CD spectra were measured at RT (25°C) in a 1 mm path length quartz cell. Spectra of DbpAVS461 (10 µM) and DbpAVS461ΔC11 (10 µM) were recorded in Tris buffer at 25°C, and three far-UV CD spectra were recorded from 190 to 250 nm for far-UV CD in 1 nm increments. The background spectrum of buffer without protein was subtracted from the protein spectra. CD spectra were initially analyzed by the software Spectra Manager Program. Analysis of spectra to extrapolate secondary structures was performed by Dichroweb (http://dichroweb.cryst.bbk.ac.uk/html/home.shtml) using the K2D and Selcon 3 analysis programs [53].
GAG and decorin binding assays
Quantitative ELISA for decorin and dermatan sulfate binding by DbpA proteins was performed similarly to that previously described [54]. One µg of decorin, dermatan sulfate, chondroitin 6 sulfate, or BSA was coated onto microtiter plate wells. One hundred microliters of increasing concentrations (0.03125, 0.0625, 0.125, 0.25, 0.5, 1, 2 µM) of histidine-tagged RevA (negative control) or a DbpA variant, including DbpAB31, DbpA297, DbpAN40-D10/E9, DbpAB356, DbpAVS461, DbpAPBr, or DbpAVS461ΔC11, were then added to the wells. To detect the binding of histidine-tagged proteins, mouse anti-histidine tag (Sigma-Aldrich, St. Louis, MO; 1∶200) and HRP-conjugated goat anti-mouse IgG (Promega, Fitchburg, WI; 1∶1,000) were used as primary and secondary antibodies. The plates were washed three times with PBST (0.05% Tween20 in PBS buffer), and 100 µl of tetramethyl benzidine (TMB) solution (Kirkegaard and Perry Laboratories, Gaithersburg, MD) were added to each well and incubated for five minutes. The reaction was stopped by adding 100 µl of 0.5% hydro sulfuric acid to each well. Plates were read at 405 nm using a Synergy HT ELISA plate reader (BioTek, Winooski, VT). To determine the dissociation constant (KD), the data were fitted by the following equation using KaleidaGraph software (Version 4.1.1 Abekbecj Software, Reading, PA).
| (1) |
Surface Plasmon Resonance (SPR)
Interactions of DbpA with decorin or dermatan sulfate were analyzed by a SPR technique using a Biacore 3000 (GE Healthcare, Piscataway, NJ). Ten µg of biotinylated decorin or dermatan sulfate was conjugated to an SA chip (GE Healthcare, Piscataway, NJ). A control flow cell was injected with PBS buffer without decorin or dermatan sulfate. For quantitative SPR experiments to determine decorin- or dermatan sulfate–binding, ten µl of increasing concentrations (0, 15.625, 31.25, 62.5, 125, 250, 500 nM) of a DbpA variant, including DbpAB31, DbpA297, DbpAN40-D10/E9, DbpAB356, DbpAVS461, DbpAPBr, or DbpAVS461ΔC11, were injected into the control cell and flow cell immobilized with decorin or dermatan sulfate at 10 µL/min, 25°C. To obtain the kinetic parameters of the interaction, sensogram data were fitted by means of BIAevaluation software version 3.0 (GE Healthcare, Piscataway, NJ), using the one step biomolecular association reaction model (1∶1 Langmuir model), resulting in optimum mathematical fit with the lowest Chi values.
Shuttle plasmid construction
To generate the plasmids encoding dbpA alleles, genes dbpAN40-D10/E9, dbpAVS461, dbpAPBr, or dbpAVS461ΔC11 were first PCR amplified with the addition of a SalI site and a BamH1 site at the 5′ and 3′ ends, respectively, using the primers listed in Table S1. Amplified DNA fragments were inserted into TA cloning vector pCR2.1-TOPO (Invitrogen, Houston, TX; see Table S3), to generate the plasmids pCR2.1- dbpAN40-D10/E9, pCR2.1-dbpAVS461, pCR2.1-dbpAPBr, and pCR2.1-dbpAVS461ΔC11. The plasmids were then digested with SalI and BamHI to release the dbpA alleles, which were then inserted into the SalI and BamHI sites of pBBE22 (see Table S3). The promoter region of dbpBA from B. burgdorferi B31, 289 bp upstream from the start codon of dbpB, was also PCR amplified, adding, SphI and SalI sites at the 5′ and 3′ ends, respectively, using primers pdbpBAfp and pdbpBArp (Table S3). Promoter fragments were then inserted into the SphI and SalI sites of pBBE22 to drive the expression of dbpAN40-D10/E9, dbpAVS461, dbpAPBr, and dbpAVS461ΔC11.
Plasmid transformation into B. burgdorferi
Electrocompetent B. burgdorferi ML23ΔdbpBA was transformed separately with 80 µg of each of the shuttle plasmids encoding dbpAN40-D10/E9, dbpAVS461, dbpAPBr, or dbpAVS461ΔC11 (see Table S3) and cultured in BSK II medium at 33°C for 24 hours. Aliquots of the culture were mixed with 1.8% analytical grade agarose (BioRad; Hercules, CA) and plated on a solidified BSK II/agarose layer in sterilized 100×20 mm tissue culture dishes (Corning Incorporated, Corning, NY). Plates were incubated at 33°C in 5% CO2 for two weeks. Kanamycin- and gentamycin-resistant colonies of dbpA-complemented B. burgdorferi were obtained and expanded at 33°C in liquid BSK II medium containing kanamycin and gentamycin, followed by genomic DNA preparation as previously described [55]. PCR was performed with primers (Fig. S1) specific for kan (encoding the kanamycin resistance gene), to verify its presence in the transformants. The plasmid profiles of the dbpBA deficient mutant complemented with dbpA alleles were examined as described previously [56] and found to be identical to those of this strain harboring the empty vector (data not shown).
Flow cytometry
To determine the production and the surface localization of DbpA variants and of OspC in B. burgdorferi, 1×108 B. burgdorferi cells were washed thrice with HBSC buffer containing DB (25 mM Hepes acid, 150 mM sodium chloride, 1 mM MnCl2, 1 mM MgCl2, 0.25 mM CaCl2, 0.1% glucose, and 0.2% BSA, final concentration) and then resuspended into 500 µL of the same buffer. A mixture of mouse antisera raised against DbpAB31, DbpAN40-D10/E9, DbpAVS461, and DbpAPBr [39] and rabbit anti-OspC (Rockland, Gilbertsville, PA) was used as a primary antibody, and Alexa488-conjugated goat anti-mouse IgG (Invitrogen; 1∶250×) and Alexa 635-conjugated goat anti-rabbit IgG (Invitrogen; 1∶250×) were used as secondary antibodies. 300 µL of formalin (0.1%) was then added for fixing. Surface production of DbpA and OspC was measured by flow cytometry using a Becton-Dickinson FACSCalibur (BD Bioscience, Franklin Lakes, NJ). All flow cytometry experiments were performed within two days of collection of B. burgdorferi samples. Spirochetes in the suspension were distinguished on the basis of their distinct light scattering properties in a Becton Dickinson FACSCalibur flow cytometer equipped with a 15 mW, 488 nm air-cooled argon laser, a standard three-color filter arrangement, and CELLQuest Software (BD Bioscience, Franklin Lakes, NJ). The mean fluorescence index (MFI) of each sample was obtained from FlowJo software (Three star Inc, Ashland, OR) representing the surface production of the indicated proteins. To compare the surface production of DbpA and OspC proteins in different strains, results in Fig. S3 are shown as relative production, the MFI normalized to that of B. burgdorferi strain ML23. The results shown in Fig. S3 represent the mean of twelve independent determinations ± the standard deviation. Each standard deviation value was no more than 7 percent of its mean value.
Binding of radiolabeled B. burgdorferi to purified decorin or GAG
Binding of B. burgdorferi to purified decorin or dermatan sulfate was determined as previously described [39]. Briefly, spirochetes were radiolabeled with [35S] methionine, and 1×108 radiolabeled bacteria were added to break-apart microtiter plate wells previously incubated with 250 µg/mL decorin, dermatan sulfate or chondroitin 6 sulfate (as a negative control). After 16 hours at 4°C, unbound bacteria were removed by washing with PBS containing 0.2% BSA. Plates were air-dried, and percent binding was determined by liquid scintillation counting. The percentage of bound bacteria was determined by radioactive counts in bound bacteria normalized to the counts in the inoculum.
Mouse infection experiments
Four-week-old female C3H/HeN mice (Charles River, Wilmington, MA) were used for all experiments. Mice were infected by intradermal injection as previously described [31] with ∼104 B. burgdorferi ML23ΔdbpBA/vector, or derivatives expressing dbpAN40-D10/E9, dbpAVS461, dbpAPBr, or dbpAVS461ΔC11. For the mice sacrificed at 3 days post-infection, the skin at the inoculation site was collected. For the mice sacrificed at 7, 14, 21, or 28 days post-infection, skin at the inoculation site, the tibiotarsal joint, knee joint, bladder, heart, and ear were collected. For infections of mice defective in adaptive immunity, four-week-old C3H-SCID mice (Jackson Lab, Bar Harbor, ME) were infected as described above for C3H/HeN mice. In C3H-SCID mice, a dose of 103 resulted in a 30- to 236-fold difference in bacterial load of B. burgdorferi strain ML23 and ML23ΔdbpBA/vector, whereas a dose of 104 resulted in indistinguishable colonization by two strains. Hence to maximize the chances of revealing differences in colonization due to the production of the DbpA variants, the lower (i.e. 103) dose was used in this study. All SCID mice were sacrificed on 28 days post-infection, and skin at the inoculation site, the tibiotarsal joint, knee joint, bladder, heart, and ear were collected.
Quantification of B. burgdorferi in infected tissues and blood samples
DNA was extracted from tissue using the DNeasy Blood & Tissue kit (Qiagen). The quantity and quality of DNA for each tissue sample have been assessed by measuring the concentration of DNA and the ratio of the UV absorption at 280 to 260. The amount of DNA used in this study was 100 ng for each sample, and the 280∶260 ratio was between 1.75 to 1.85, indicating the lack of contaminating RNA or proteins. qPCR was then performed to quantitate bacterial load, using 100 ng of DNA per reaction. B. burgdorferi genomic equivalents were calculated using an CFX Connect Real-Time PCR detection system (BioRad, Hercules, CA) in conjunction with SYBR green PCR Mastermix (BioRad), based on amplification of the B. burgdorferi recA gene using primers BBRecAfp and BBRecArp (Table S2), as described previously [57]. The number of recA copies was calculated by establishing a threshold cycle (Ct) standard curve of a known number of recA gene extracted from B. burgdorferi strain B31, then comparing the Ct values of the experimental samples. To assure the low signals were not simply a function of the presence of PCR inhibitors in the DNA preparation, we subjected 5 samples from tibiotarsal joint, bladder, and heart of the mice infected by B. burgdorferi strain ML23/vector, ML23ΔdbpBA/vector (i.e. the dbpBA mutant), or dbpBA mutant complemented with dbpAN40, dbpAVS461, dbpAPBr, or dbpAVS461ΔC11 to qPCR using mouse nidogen primers mNidfp and mNidrp (Table S3) as an internal standard [58]. As predicted, we detected 107 copies of the nidogen gene from 100 ng of each DNA sample, ruling out the presence of PCR inhibitors in these samples.
Antibody titer determinations
Nunc maxisorp flat-bottom 96-well plates were coated with 1 µg of recombinant DbpAB31, DbpAN40-D10/E9, DbpAPBr, or DbpAVS461 protein in 100 µl of coating buffer (0.05 M Na2CO3, pH 9.0) overnight in 4°C. The next day, plates were washed three times with wash buffer (0.05% PBS Tween 20) and blocked for 1 hour in blocking buffer (0.05% PBS Tween 20 with 1% BSA). Plates were then washed three times, and incubated for 1 hour with serum (diluted 1∶100, 1∶300 and 1∶900) at room temperature. Then, after washing plates three times, a 1∶10,000 dilution of HRP-conjugated goat anti-mouse IgM or IgG antibodies (Bethyl Lab, Montgomery, TX) was added to each well for one hour at room temperature. Subsequently, plates were washed and 100 µl of SureBlue Reserve TMB 2-Component Microwell Peroxidase Substrate system (Kirkegaard and Perry Laboratories) were added to each well. Plates were then read at OD650 using a Synergy HT ELISA plate reader (BioTek). For kinetic ELISA experiments, readings were taken every minute for 10 minutes. Vmax (milli-optical density unit per minute) based on the slope of the continuous readings were calculated using the Gen5 Software (Version 2.00.18, BioTek, Winooski, VT).
Controls included three dilutions (1∶100, 1∶300 and 1∶900) of purified IgG or IgM (125 µg/mL; Bethyl Lab) coated on microtiter plates, and uninfected (“naïve”) serum run in parallel with sample sera. The product of Vmax × inverse serum dilution factor was largely independent of serum dilution factor. Arbitrary units of a given serum sample were chosen as the largest Vmax × inverse serum dilution factor product within the dilution range, and were expressed relative to the arbitrary units of control pooled sera, set to 100 (Marty-Roix, R. and Maung, N., unpublished data). Antibody units of sample sera were normalized by subtracting the antibody unit “background” of naïve mice, and expressed relative to the control wells coated with purified IgG and IgM.
Histological evaluation of arthritis and carditis
At least 10 tibiotarsus joints and 5 hearts were collected from each group of mice (5 animal/group) infected with the different B. burgdorferei isolates. For histology, joints and hearts were fixed in 10% formalin and processed for Hematoxylin and Eosin staining. Sections were evaluated for signs of arthritis using histological parameters for B. burgdorferi-induced inflammation [51], [59], such as exudation of inflammatory cells into joints, altered thickness of tendons or ligament sheaths, and hypertrophy of the synovium. Signs of carditis [51], [60] were evaluated based on cardiac inflammatory infiltrate, including transmural infiltration of neutrophils in the blood vessels and infiltration by macrophages into the surrounding connective tissue. Inflammation was scored as 0 (no inflammation), 1 (mild inflammation with less than two small foci of infiltration), 2 (moderate inflammation with two or more foci of infiltration), or 3 (severe inflammation with focal and diffuse infiltration covering a large area).
Statistical analysis
Significant differences between samples were determined using the one-way ANOVA test following logarithmic transformation of the data. P-values were determined for each sample.
Supporting Information
The 11 C-terminal amino acids of DbpAVS461 do not affect its structure. Far-UV CD analysis of DbpAVS461 and DbpAVS461ΔC11. The molar ellipticity, Φ, was measured from 190 to 250 nm for 10 µM of each protein in PBS buffer.
(TIF)
Recombinant DbpA variants exhibit distinct decorin- and dermatan sulfate-binding activities. Left panel: The indicated concentrations of various recombinant histidine-tagged DbpA variants, including DbpAB31 (“B31”), DbpAN40-D10/E9 (“N40-D10/E9”), DbpA297 (“297”), DbpAB356 (“B356”), DbpAPBr (“PBr), DbpAVS461 (“VS461”), DbpAVS461ΔC11 (“VS461ΔC11”), or RevA (negative control), were added to quadruplicate wells coated with (A) decorin or (B) dermatan sulfate (Derm SO4)., and protein binding was quantitated by ELISA (Y axis). Numbers represent the mean ± standard deviation. Binding of DbpAVS461ΔC11 to decorin and dermatan sulfate was not statistically different than RevA (p>0.05 by Student's t test). (The other DbpA variants bound to these substrates significantly better than RevA, and their KD was obtained from the average of three independent experiments is shown on Table 1. Shown is a representative of three independently performed experiments. Right panel: 15.625 to 500 nM of histidine-tagged DbpA protein was flowed over a surface coated with 10 µg (A) decorin or (B) dermatan sulfate (Derm SO4). Binding was measured in response units (RU) by SPR (see Materials and Methods). Shown is a representative of six experiments performed on three different occasions and in Table 1 are the kon, koff, and KD values obtained from average of these six experiments.
(TIF)
DbpA variants are localized at the surface of B. burgdorferi . Flow cytometry analysis of DbpA localized on the surface of B. burgdorferi. (A) Flow cytometry analysis of DbpA localized to the surface of parental strain B. burgdorferi ML23/pBBE22 (“ML23/Vector”), dbpBA deletion strain ML23ΔdbpBA/pBBE22 (“ML23ΔdbpBA/Vector”), and the dbpBA deletion strain bearing a plasmid encoding DbpBA (“ML23ΔdbpBA/pDbpBA”). (B) The production of OspC (control) and DbpA on the surface of B. burgdorferi strain ML23/pBBE22 (“ML23/vector”), dbpBA deletion strain ML23ΔdbpBA/pBBE22 (“Vector”), and the deletion strain bearing a plasmid encoding the indicated DbpA variants were detected by flow cytometry (see Materials and Methods). Non-adherent B. burgdorferi strain B314 carrying the shuttle vector (“B314/Vector”) was included as a negative control. Values are shown relative to the production levels of DbpA on the surface of B. burgdorferi strain ML23/pBBE22 (“ML23/vector”). Each bar represents the mean of twelve independent determinations ± the standard deviation. Each standard deviation value is no more than 7 percent of its mean value. (*): indicates that surface production of the indicated proteins was significantly lower (P<0.05) than surface production of DbpA by B. burgdorferi strain ML23/pBBE22.
(TIF)
DbpA variants produced in B. burgdorferi trigger similar adaptive immune responses. C3H/HeN mice infected with B. burgdorferi strain ML23/pBBE22 (“ML23/Vector”), dbpBA deletion strain ML23ΔdbpBA/pBBE22 (“Vector”), or the deletion strain bearing a plasmid encoding DbpA (“pDbpBA”), DbpAVS461 (“VS461”), DbpAPBr (“PBr”), DbpAN40-D10/E9 (“N40-D10/E9”), or DbpAVS461ΔC11 (“ΔC11”) at doses of 104 spirochetes, were sacrificed at 28 days post-infection. Serum titers of IgG (top panel) and IgM (bottom panel) in mice infected with the indicated strains. Statistical significance was determined using a one-way ANOVA test. Data shown are the mean bacterial loads ± SEM of 8 mice per group. Statistical significance was determined using a one-way ANOVA test. Significant (P<0.05) differences in antibody titers relative to the dbpBA deletion strain (“*”) or relative to the dbpAVS461ΔC11-complemented strain (“†”), are indicated.
(TIF)
The kinetics of B. burgdorferi dissemination in C3H/HeN mice. C3H/HeN mice infected (104 spirochetes) with B. burgdorferi strain ML23/pBBE22 (“ML23/Vector”), dbpBA deletion strain ML23ΔdbpBA/pBBE22 (“Vector”), or the deletion strain bearing a plasmid encoding the indicated DbpA variants were sacrificed at 7 and 14 days post-infection. Bacterial loads at the (A) inoculation site, (B) knee joint, (C) heart, (D) bladder, (E) tibiotarsus joint, and (F) earwere determined by qPCR. Data shown are the mean bacterial loads ± SEM of 10 mice per group. Statistical significance was determined using a one-way ANOVA test. Significant (P<0.05) differences in spirochete number relative to the dbpBA deletion strain (“*”), or between two strains relative to each other (“#”) are indicated. These data are described comprehensively with other post-infection time points in Table S1.
(TIF)
Sequence alignment of DbpA variants found in B31, 297, N40-D10/E9, PBr and VS461 strains for Borrelia . DbpAB356 is not shown because it has 99% sequence identity with strain DbpAN40-D10/E9 [39]. Black shaded residues are the critical residues for decorin- and dermatan sulfate-binding [41]. Gray shaded residues are the residues in BXBB motif previously suggested to be important for decorin- and dermatan sulfate-binding [40], [45].
(TIF)
Summary of tissue colonization promoted by diverse dbpA alleles.
(DOCX)
Primers used in this study.
(DOCX)
Bacterial strains and plasmids used in this study.
(DOCX)
Supplemental references.
(DOCX)
Acknowledgments
We thank Roderick Bronson and Vairamuthu Subbiah for histopathological analysis, Nikhat Parveen, George Chaconas, Linden Hu, Tanja Petnicki-Ocwieja, Stacie Clark, Nang Maung and Jenifer Coburn for valuable advice, and Emily Mallick and Marcia Osburne for critical reading of the manuscript. Jon Skare generously provided B. burgdorferi strain ML23, the dbpBA mutant strain JF105 (ΔdbpBA), and the dbpBA-complemented strain JF105/pJBF17 (ML23/ΔdbpBA/pDbpAB). We thank David Mann and Nancy Ulbrandt for recombinant decorin, Patti Rosa and Philip Stewart for pBSV2, Jorge Benach for α-FlaB CB1 monoclonal antibody, James Baleja for help with circular dichroism, Allen Parmelee and Stephen Kwok of Flow Cytometry Core in Pathology at Tufts University School of Medicine for assisting with flow cytometry, and Lawrence Stern for kindly allowing us access to his surface plasmon resonance equipment. We also thank Jon Blevins, Jemiina Salo and Jukka Hytonen for the communication of unpublished results.
Funding Statement
This work was supported by grants from NIH R01AI37601 and R01AI093104 (to JML), R01AI080615 (to UP), R03AI090389 (to XY), and American Heart Association Postdoctoral Fellowship 12POST11660031 (to YPL). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Steere AC, Coburn J, Glickstein L (2004) The emergence of Lyme disease. J Clin Invest 113: 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Radolf JD, Caimano MJ, Stevenson B, Hu LT (2012) Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol 10: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kenedy MR, Lenhart TR, Akins DR (2012) The role of Borrelia burgdorferi outer surface proteins. FEMS Immunol Med Microbiol 66: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Steere AC (2001) Lyme disease. N Engl J Med 345: 115–125. [DOI] [PubMed] [Google Scholar]
- 5. Brisson D, Drecktrah D, Eggers CH, Samuels DS (2012) Genetics of Borrelia burgdorferi. Annu Rev Genet 46: 515–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hildenbrand P, Craven DE, Jones R, Nemeskal P (2009) Lyme neuroborreliosis: manifestations of a rapidly emerging zoonosis. AJNR Am J Neuroradiol 30: 1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lelovas P, Dontas I, Bassiakou E, Xanthos T (2008) Cardiac implications of Lyme disease, diagnosis and therapeutic approach. Int J Cardiol 129: 15–21. [DOI] [PubMed] [Google Scholar]
- 8.Puius YA, Kalish RA (2008) Lyme arthritis: pathogenesis, clinical presentation, and management. Infect Dis Clin North Am 22: : 289–300, vi–vii. [DOI] [PubMed] [Google Scholar]
- 9. Wang G, Ojaimi C, Wu H, Saksenberg V, Iyer R, et al. (2002) Disease severity in a murine model of lyme borreliosis is associated with the genotype of the infecting Borrelia burgdorferi sensu stricto strain. J Infect Dis 186: 782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang G, van Dam AP, Schwartz I, Dankert J (1999) Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin Microbiol Rev 12: 633–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang G, Ojaimi C, Iyer R, Saksenberg V, McClain SA, et al. (2001) Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect Immun 69: 4303–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones KL, Glickstein LJ, Damle N, Sikand VK, McHugh G, et al. (2006) Borrelia burgdorferi genetic markers and disseminated disease in patients with early Lyme disease. J Clin Microbiol 44: 4407–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coburn J, Barthold SW, Leong JM (1994) Diverse Lyme disease spirochetes bind integrin alpha IIb beta 3 on human platelets. Infect Immun 62: 5559–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Craig-Mylius KA, Lee M, Jones KL, Glickstein LJ (2009) Arthritogenicity of Borrelia burgdorferi and Borrelia garinii: comparison of infection in mice. Am J Trop Med Hyg 80: 252–258. [PubMed] [Google Scholar]
- 15. Wilske B, Preac-Mursic V, Jauris S, Hofmann A, Pradel I, et al. (1993) Immunological and molecular polymorphisms of OspC, an immunodominant major outer surface protein of Borrelia burgdorferi. Infect Immun 61: 2182–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kraiczy P, Skerka C, Brade V, Zipfel PF (2001) Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect Immun 69: 7800–7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rogers EA, Marconi RT (2007) Delineation of species-specific binding properties of the CspZ protein (BBH06) of Lyme disease spirochetes: evidence for new contributions to the pathogenesis of Borrelia spp. Infect Immun 75: 5272–5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lagal V, Portnoi D, Faure G, Postic D, Baranton G (2006) Borrelia burgdorferi sensu stricto invasiveness is correlated with OspC-plasminogen affinity. Microbes Infect 8: 645–652. [DOI] [PubMed] [Google Scholar]
- 19. Wallich R, Pattathu J, Kitiratschky V, Brenner C, Zipfel PF, et al. (2005) Identification and functional characterization of complement regulator-acquiring surface protein 1 of the Lyme disease spirochetes Borrelia afzelii and Borrelia garinii. Infect Immun 73: 2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seinost G, Dykhuizen DE, Dattwyler RJ, Golde WT, Dunn JJ, et al. (1999) Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect Immun 67: 3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salo J, Loimaranta V, Lahdenne P, Viljanen MK, Hytonen J (2011) Decorin binding by DbpA and B of Borrelia garinii, Borrelia afzelii, and Borrelia burgdorferi sensu Stricto. J Infect Dis 204: 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patti JM, Allen BL, McGavin MJ, Hook M (1994) MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol 48: 585–617. [DOI] [PubMed] [Google Scholar]
- 23. Antonara S, Ristow L, Coburn J (2011) Adhesion mechanisms of Borrelia burgdorferi. Adv Exp Med Biol 715: 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coburn J, Fischer JR, Leong JM (2005) Solving a sticky problem: new genetic approaches to host cell adhesion by the Lyme disease spirochete. Mol Microbiol 57: 1182–1195. [DOI] [PubMed] [Google Scholar]
- 25. Coburn J, Leong J, Chaconas G (2013) Illuminating the roles of the Borrelia burgdorferi adhesins. Trends Microbiol 21: 372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hagman KE, Lahdenne P, Popova TG, Porcella SF, Akins DR, et al. (1998) Decorin-binding protein of Borrelia burgdorferi is encoded within a two-gene operon and is protective in the murine model of Lyme borreliosis. Infect Immun 66: 2674–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo BP, Brown EL, Dorward DW, Rosenberg LC, Hook M (1998) Decorin-binding adhesins from Borrelia burgdorferi. Mol Microbiol 30: 711–723. [DOI] [PubMed] [Google Scholar]
- 28. Parveen N, Caimano M, Radolf JD, Leong JM (2003) Adaptation of the Lyme disease spirochaete to the mammalian host environment results in enhanced glycosaminoglycan and host cell binding. Mol Microbiol 47: 1433–1444. [DOI] [PubMed] [Google Scholar]
- 29. Roberts WC, Mullikin BA, Lathigra R, Hanson MS (1998) Molecular analysis of sequence heterogeneity among genes encoding decorin binding proteins A and B of Borrelia burgdorferi sensu lato. Infect Immun 66: 5275–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown EL, Wooten RM, Johnson BJ, Iozzo RV, Smith A, et al. (2001) Resistance to Lyme disease in decorin-deficient mice. J Clin Invest 107: 845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weening EH, Parveen N, Trzeciakowski JP, Leong JM, Hook M, et al. (2008) Borrelia burgdorferi lacking DbpBA exhibits an early survival defect during experimental infection. Infect Immun 76: 5694–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi Y, Xu Q, McShan K, Liang FT (2008) Both decorin-binding proteins A and B are critical for the overall virulence of Borrelia burgdorferi. Infect Immun 76: 1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blevins JS, Hagman KE, Norgard MV (2008) Assessment of decorin-binding protein A to the infectivity of Borrelia burgdorferi in the murine models of needle and tick infection. BMC Microbiol 8: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hyde JA, Weening EH, Chang M, Trzeciakowski JP, Hook M, et al. (2011) Bioluminescent imaging of Borrelia burgdorferi in vivo demonstrates that the fibronectin-binding protein BBK32 is required for optimal infectivity. Mol Microbiol 82: 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Imai DM, Feng S, Hodzic E, Barthold SW (2013) Dynamics of connective-tissue localization during chronic Borrelia burgdorferi infection. Lab Invest 93: 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hyde JA, Trzeciakowski JP, Skare JT (2007) Borrelia burgdorferi alters its gene expression and antigenic profile in response to CO2 levels. J Bacteriol 189: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cassatt DR, Patel NK, Ulbrandt ND, Hanson MS (1998) DbpA, but not OspA, is expressed by Borrelia burgdorferi during spirochetemia and is a target for protective antibodies. Infect Immun 66: 5379–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, et al. (2003) Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect Immun 71: 1689–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benoit VM, Fischer JR, Lin YP, Parveen N, Leong JM (2011) Allelic variation of the Lyme disease spirochete adhesin DbpA influences spirochetal binding to decorin, dermatan sulfate, and mammalian cells. Infect Immun 79: 3501–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morgan A, Wang X (2013) The Novel Heparin-Binding Motif in Decorin-Binding Protein A from Strain B31 of Borrelia burgdorferi Explains the Higher Binding Affinity. Biochemistry 52: 8237–8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brown EL, Guo BP, O'Neal P, Hook M (1999) Adherence of Borrelia burgdorferi. Identification of critical lysine residues in DbpA required for decorin binding. J Biol Chem 274: 26272–26278. [DOI] [PubMed] [Google Scholar]
- 42. Purser JE, Lawrenz MB, Caimano MJ, Howell JK, Radolf JD, et al. (2003) A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol Microbiol 48: 753–764. [DOI] [PubMed] [Google Scholar]
- 43. Seshu J, Esteve-Gassent MD, Labandeira-Rey M, Kim JH, Trzeciakowski JP, et al. (2006) Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol Microbiol 59: 1591–1601. [DOI] [PubMed] [Google Scholar]
- 44. Shi Y, Xu Q, Seemanaplli SV, McShan K, Liang FT (2008) Common and unique contributions of decorin-binding proteins A and B to the overall virulence of Borrelia burgdorferi. PLoS One 3: e3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang X (2012) Solution structure of decorin-binding protein A from Borrelia burgdorferi. Biochemistry 51: 8353–8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liang FT, Brown EL, Wang T, Iozzo RV, Fikrig E (2004) Protective niche for Borrelia burgdorferi to evade humoral immunity. Am J Pathol 165: 977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu Q, Seemanaplli SV, McShan K, Liang FT (2007) Increasing the interaction of Borrelia burgdorferi with decorin significantly reduces the 50 percent infectious dose and severely impairs dissemination. Infect Immun 75: 4272–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Iozzo RV (1998) Matrix proteoglycans: from molecular design to cellular function. Annu Rev Biochem 67: 609–652. [DOI] [PubMed] [Google Scholar]
- 49. Poole AR, Rosenberg LC, Reiner A, Ionescu M, Bogoch E, et al. (1996) Contents and distributions of the proteoglycans decorin and biglycan in normal and osteoarthritic human articular cartilage. J Orthop Res 14: 681–689. [DOI] [PubMed] [Google Scholar]
- 50. Scott PG, Nakano T, Dodd CM (1997) Isolation and characterization of small proteoglycans from different zones of the porcine knee meniscus. Biochim Biophys Acta 1336: 254–262. [DOI] [PubMed] [Google Scholar]
- 51. Barthold SW, Hodzic E, Tunev S, Feng S (2006) Antibody-mediated disease remission in the mouse model of lyme borreliosis. Infect Immun 74: 4817–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McBain AL, Mann DM (2001) Purification of recombinant human decorin and its subdomains. Methods Mol Biol 171: 221–229. [DOI] [PubMed] [Google Scholar]
- 53. Lin YP, Greenwood A, Nicholson LK, Sharma Y, McDonough SP, et al. (2009) Fibronectin binds to and induces conformational change in a disordered region of leptospiral immunoglobulin-like protein B. J Biol Chem 284: 23547–23557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lin YP, Lee DW, McDonough SP, Nicholson LK, Sharma Y, et al. (2009) Repeated domains of leptospira immunoglobulin-like proteins interact with elastin and tropoelastin. J Biol Chem 284: 19380–19391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Parveen N, Leong JM (2000) Identification of a candidate glycosaminoglycan-binding adhesin of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 35: 1220–1234. [DOI] [PubMed] [Google Scholar]
- 56. Labandeira-Rey M, Skare JT (2001) Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect Immun 69: 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liveris D, Wang G, Girao G, Byrne DW, Nowakowski J, et al. (2002) Quantitative detection of Borrelia burgdorferi in 2-millimeter skin samples of erythema migrans lesions: correlation of results with clinical and laboratory findings. J Clin Microbiol 40: 1249–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Petnicki-Ocwieja T, DeFrancesco AS, Chung E, Darcy CT, Bronson RT, et al. (2011) Nod2 suppresses Borrelia burgdorferi mediated murine Lyme arthritis and carditis through the induction of tolerance. PLoS One 6: e17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang X, Ma Y, Weis JH, Zachary JF, Kirschning CJ, et al. (2005) Relative contributions of innate and acquired host responses to bacterial control and arthritis development in Lyme disease. Infect Immun 73: 657–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yang X, Coleman AS, Anguita J, Pal U (2009) A chromosomally encoded virulence factor protects the Lyme disease pathogen against host-adaptive immunity. PLoS Pathog 5: e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 11 C-terminal amino acids of DbpAVS461 do not affect its structure. Far-UV CD analysis of DbpAVS461 and DbpAVS461ΔC11. The molar ellipticity, Φ, was measured from 190 to 250 nm for 10 µM of each protein in PBS buffer.
(TIF)
Recombinant DbpA variants exhibit distinct decorin- and dermatan sulfate-binding activities. Left panel: The indicated concentrations of various recombinant histidine-tagged DbpA variants, including DbpAB31 (“B31”), DbpAN40-D10/E9 (“N40-D10/E9”), DbpA297 (“297”), DbpAB356 (“B356”), DbpAPBr (“PBr), DbpAVS461 (“VS461”), DbpAVS461ΔC11 (“VS461ΔC11”), or RevA (negative control), were added to quadruplicate wells coated with (A) decorin or (B) dermatan sulfate (Derm SO4)., and protein binding was quantitated by ELISA (Y axis). Numbers represent the mean ± standard deviation. Binding of DbpAVS461ΔC11 to decorin and dermatan sulfate was not statistically different than RevA (p>0.05 by Student's t test). (The other DbpA variants bound to these substrates significantly better than RevA, and their KD was obtained from the average of three independent experiments is shown on Table 1. Shown is a representative of three independently performed experiments. Right panel: 15.625 to 500 nM of histidine-tagged DbpA protein was flowed over a surface coated with 10 µg (A) decorin or (B) dermatan sulfate (Derm SO4). Binding was measured in response units (RU) by SPR (see Materials and Methods). Shown is a representative of six experiments performed on three different occasions and in Table 1 are the kon, koff, and KD values obtained from average of these six experiments.
(TIF)
DbpA variants are localized at the surface of B. burgdorferi . Flow cytometry analysis of DbpA localized on the surface of B. burgdorferi. (A) Flow cytometry analysis of DbpA localized to the surface of parental strain B. burgdorferi ML23/pBBE22 (“ML23/Vector”), dbpBA deletion strain ML23ΔdbpBA/pBBE22 (“ML23ΔdbpBA/Vector”), and the dbpBA deletion strain bearing a plasmid encoding DbpBA (“ML23ΔdbpBA/pDbpBA”). (B) The production of OspC (control) and DbpA on the surface of B. burgdorferi strain ML23/pBBE22 (“ML23/vector”), dbpBA deletion strain ML23ΔdbpBA/pBBE22 (“Vector”), and the deletion strain bearing a plasmid encoding the indicated DbpA variants were detected by flow cytometry (see Materials and Methods). Non-adherent B. burgdorferi strain B314 carrying the shuttle vector (“B314/Vector”) was included as a negative control. Values are shown relative to the production levels of DbpA on the surface of B. burgdorferi strain ML23/pBBE22 (“ML23/vector”). Each bar represents the mean of twelve independent determinations ± the standard deviation. Each standard deviation value is no more than 7 percent of its mean value. (*): indicates that surface production of the indicated proteins was significantly lower (P<0.05) than surface production of DbpA by B. burgdorferi strain ML23/pBBE22.
(TIF)
DbpA variants produced in B. burgdorferi trigger similar adaptive immune responses. C3H/HeN mice infected with B. burgdorferi strain ML23/pBBE22 (“ML23/Vector”), dbpBA deletion strain ML23ΔdbpBA/pBBE22 (“Vector”), or the deletion strain bearing a plasmid encoding DbpA (“pDbpBA”), DbpAVS461 (“VS461”), DbpAPBr (“PBr”), DbpAN40-D10/E9 (“N40-D10/E9”), or DbpAVS461ΔC11 (“ΔC11”) at doses of 104 spirochetes, were sacrificed at 28 days post-infection. Serum titers of IgG (top panel) and IgM (bottom panel) in mice infected with the indicated strains. Statistical significance was determined using a one-way ANOVA test. Data shown are the mean bacterial loads ± SEM of 8 mice per group. Statistical significance was determined using a one-way ANOVA test. Significant (P<0.05) differences in antibody titers relative to the dbpBA deletion strain (“*”) or relative to the dbpAVS461ΔC11-complemented strain (“†”), are indicated.
(TIF)
The kinetics of B. burgdorferi dissemination in C3H/HeN mice. C3H/HeN mice infected (104 spirochetes) with B. burgdorferi strain ML23/pBBE22 (“ML23/Vector”), dbpBA deletion strain ML23ΔdbpBA/pBBE22 (“Vector”), or the deletion strain bearing a plasmid encoding the indicated DbpA variants were sacrificed at 7 and 14 days post-infection. Bacterial loads at the (A) inoculation site, (B) knee joint, (C) heart, (D) bladder, (E) tibiotarsus joint, and (F) earwere determined by qPCR. Data shown are the mean bacterial loads ± SEM of 10 mice per group. Statistical significance was determined using a one-way ANOVA test. Significant (P<0.05) differences in spirochete number relative to the dbpBA deletion strain (“*”), or between two strains relative to each other (“#”) are indicated. These data are described comprehensively with other post-infection time points in Table S1.
(TIF)
Sequence alignment of DbpA variants found in B31, 297, N40-D10/E9, PBr and VS461 strains for Borrelia . DbpAB356 is not shown because it has 99% sequence identity with strain DbpAN40-D10/E9 [39]. Black shaded residues are the critical residues for decorin- and dermatan sulfate-binding [41]. Gray shaded residues are the residues in BXBB motif previously suggested to be important for decorin- and dermatan sulfate-binding [40], [45].
(TIF)
Summary of tissue colonization promoted by diverse dbpA alleles.
(DOCX)
Primers used in this study.
(DOCX)
Bacterial strains and plasmids used in this study.
(DOCX)
Supplemental references.
(DOCX)