Abstract
The ‘tubulin-code’ hypothesis proposes that different tubulin genes or posttranslational modifications (PTMs), which mainly confer variation in the carboxy-terminal tail (CTT), result in unique interactions with microtubule-associated proteins for specific cellular functions. However, the inability to isolate distinct and homogenous tubulin species has hindered biochemical testing of this hypothesis. Here, we have engineered 25 α/β tubulin heterodimers with distinct CTTs and PTMs and tested their interactions with four different molecular motors using single molecule assays. Our results show that tubulin isotypes and PTMs can govern motor velocity, processivity and microtubule depolymerization rates, with substantial changes conferred by even single amino acid variation. Revealing the importance and specificity of PTMs, we show that kinesin-1 motility on neuronal β-tubulin (TUBB3) is increased by polyglutamylation and that robust kinesin-2 motility requires detyrosination of α-tubulin. Our results also show that different molecular motors recognize distinctive tubulin “signatures”, which supports the premise of tubulin-code hypothesis.
Microtubules (MTs), polymers of α/β-tubulin heterodimers, interact with many MT-associated proteins to carry out a variety of cellular functions, including intracellular transport and cell division1,2. Reflecting its essential role, tubulin is among the most highly conserved of all eukaryotic genes. Lower eukaryotes have relatively few tubulin genes (e.g. 2 α- and 1 β-tubulin in budding yeast). However, the tubulin gene family expanded considerably in vertebrates (7 α- and 8 β-tubulins in humans)3. Almost all of the amino acid and length variation is confined to the unstructured C-terminal tail, which protrudes out from the polymer. In contrast, the ~400 amino acid structural core is highly conserved (97% and 95% identity among α and β vertebrate tubulin genes respectively). β-tubulin isotypes have the most diverse CTTs and many of these tubulin isotypes are enriched in certain cell or tissue types; for example, βII (TUBB2A/B) is enriched in brain and epithelial cells4, βIII (TUBB3) in specific neuronal cells5, βIV (TUBB4) in ciliary and flagellar structures6, and βVI (TUBB1) in platelets and hematopoietic cells7,8. Mutations in the TUBB2B, TUBB3 and TUBB5 also have been linked to diseases of the human nervous system9–13. However, the roles of these different tubulin genes remains poorly understood.
In addition to the genetic variation, tubulins in higher eukaryotes are substrates for several post-translational modifications (PTMs). With the exception of acetylation of a lysine residue that occurs within the lumen of the microtubule14, the other PTMs take place on the CTTs15,16. The very C-terminal Tyr residue of α-tubulin can undergo a regulated cycle in which it is cleaved in the polymer state by an unknown carboxy-peptidase to produce ‘Glu-tubulin’ (or de-tyrosinated tubulin, ΔY) and then added back by tubulin tyrosine ligases (TTLs)15,16. The presence or absence of the C-terminal Tyr has been implicated in the regulation of motor proteins17–20 and CAP-Gly domain containing microtubule plus-end binding proteins21. Another well-known tubulin PTM is the addition of glutamate residues by TTL-like family enzymes to the γ-carboxyl side chain of one or more glutamate residues in the CTT of both α- and β-tubulin22–26,15. Polyglutamylation is abundant in neurons and axonemal strucutres, the number of gultamate residues attached as branches varies from 1–6 in neuronal cells and even longer in the cilia and flagella structures22,27–29. Polyglutamylation has been shown to enhance the MT severing activity of spastin and katanin30 and MAP binding31, but their contribution towards molecular motors is unknown.
The numerous tubulin gene products and PTMs have the potential of encoding information that might specify the localization or activity for the numerous microtubule-associated proteins in the cell12,15,32,33. However, the molecular details of this proposed ‘tubulin-code’ or ‘multi-tubulin hypothesis’ are still poorly understood. Biochemical dissection of this problem has been hampered by the inhomogeneity of tubulin purified from native vertebrate sources (usually brain), which contains a complex mixture of tubulin gene products and PTMs3,15,28. Furthermore, the roles of CTTs, the most divergent element in tubulin, have been studied through subtilisin digestion, which removes both CTTs and also cleaves tubulins at a variety of sites including within the most C-terminal secondary structural element, helix-1234–36.
To circumvent these limitations, we devised an approach to produce homgenous tubulin that contains a specific human tubulin isotype CTTs with or without a specific PTM. We tested these engineered microtubules against molecular motors, which are prime candidates among microtubule-associated proteins for interpreters of a tubulin code15,33. We find that genetic and PTM variations of tubulin result in different motor activity, as reflected by changes in velocity or processivity. Furthermore, by comparing a panel of isotypes and PTMs, we show that kinesin-1 and kinesin-2 display very different responses to distinct tubulin signatures. Overall, these in vitro results support the premise of the tubulin code hypothesis.
Results
Strategy for engineering tubulin isotype and PTM variations
Using a yeast expression system37, we attempted to express human tubulin, choosing the most abundant human α/β-tubulin heterodimer combination of TUBA1A/TUBB2A. However, we found that human tubulin was insoluble in yeast, possibly because it could not be folded properly by yeast chaperones. Given that the yeast and human tubulin structural cores are nearly identical, particularly at the binding interface with motor proteins38–40 (Supplementary Figure 1–3), we prepared a chimeric tubulin in which the yeast tubulin cores were fused to the human CTTs (Fig. 1a). The yeast core-TUBA1A/TUBB2A CTT chimera could be purified using a His-6 tag in an internal loop and polymerized into microtubules (Supplementary Figure 4 and Methods).
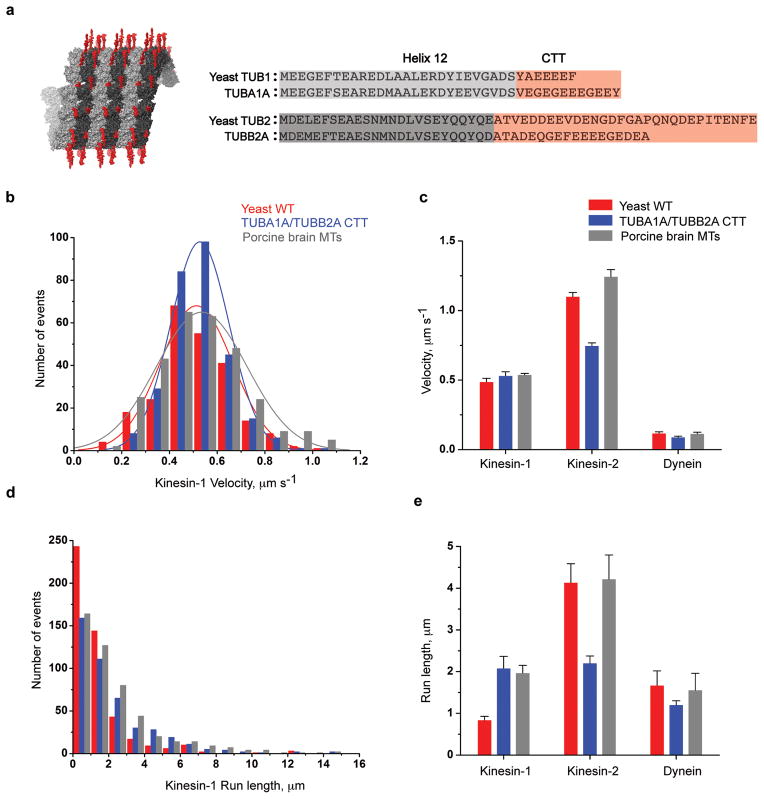
Figure 1. Recombinant tubulin for testing the role of CTTs in motor function.
a. Structure and sequence of the α (light grey) and β (dark grey) tubulin cores (ending with helix 12; pdb 4FFB) and the C-terminal tails (CTTs; red), which are disordered and extend from the polymer. Chimeric tubulins were created with the yeast core fused to the CTTs of TUBA1A/TUBB2A human tubulin at the junction shown.
b & d. Histograms of kinesin-1 motor velocity and run lengths on yeast MTs (red), yeast core with TUBA1A/TUBB2A CTTs (blue) and porcine brain microtubules (grey).
c & e. Single molecule motility of human kinesin-1, -2 and yeast dynein motor on yeast WT, yeast core-TUBA1A/TUBB2A CTT chimeric and porcine brain microtubules (mean and s.e.m, n = 3 independent experiments, each containing >100 motor measurements, for statistics source data see Supplementary Table 1), for absolute values see Table 1; mean velocity and run length were determined as described in Methods.
We first tested how molecular motors interact with microtubules composed of either TUBA1A/TUBB2A CTT chimeric tubulin (herein referred to as TUBA1A/TUBB2A), wildtype yeast tubulin or porcine brain tubulin. Three different cargo-transporting motors (human kinesin-1, human kinesin-2 and yeast cytoplasmic dynein) were chosen for analysis based upon their well-characterized single molecule motility properties. Truncated dimeric versions of all of these motors were used (see Methods). Kinesin-1 (human KIF5B41) is a ubiquitously-expressed motor that transports mitochondria, small vesicles, mRNA, intermediate filaments and other cargos42. Kinesin-2 (human KIF17) is a homodimeric kinesin-2 motor that powers intraflagellar transport of cargos to the tips of cilia and flagella along axonemal microtubules and axons42. Yeast cytoplasmic dynein moves the mitotic spindle into the bud43. A yeast recombinant dynein was used44, since purified mammalian cytoplasmic dynein does not show robust unidirectional processive motion45.
The velocity and processivity for yeast dynein were similar on wildtype yeast, porcine brain and recombinant TUBA1A/TUBB2A MTs (Fig. 1c & e); however, the kinesin motors showed differences. Kinesin-1 velocities were very similar on all three different microtubule substrates (Fig. 1b & c). However, the average run length for kinesin-1 (a measure of processivity) on yeast microtubules was two-fold lower than on TUBA1A/TUBB2A and porcine brain MTs (Fig. 1d, e and Table 1). Interestingly, a different relationship was observed for kinesin-2; both its velocity and processivity were over two-fold lower on TUBA1A/TUBB2A than on yeast and porcine brain MTs (Fig. 1c & e and Table 1). These results hint that the CTTs from human and yeast tubulin confer distinct motor phenotypes.
Table 1. Motor parameters on microtubules composed of different recombinant tubulins.
Velocities shown are the mean ± s.d. (n, the total number of measurements). The mean run length was calculated using a cumulative probability function as described earlier41. The inverse cumulative probability distribution was fit to a simple exponential decay function; the errors reported here are the standard errors of the fit. For kinesin-1, -2 and dynein ‘n’ represents number of motile motors analyzed, in the case of kinesin-13 ‘n’ represents number of microtubule ends analyzed. The values are combined from 2–5 different experiments. Kinesin-13 – Chinese Hamster MCAK, PMTs – Porcine brain microtubules, YMTs – Yeast microtubules, α1a – TUBA1A, βII – TUBB2A, βIII – TUBB3, βIV – TUBB4A, βV – TUBB6, βVI – TUBB1, βVII – TUBB7, βVIII – TUBB8, ND – not determined.
| Human Kinesin-1 | Yeast Dynein | Human Kinesin-2 | C. elegans Kinesin-2 | Kinesin-13 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Velocity, μm s−1 | Processivity, μm | Velocity, μm s−1 | Processivity, μm | Velocity, μm s−1 | Processivity, μm | Velocity, μm s−1 | Processivity, μm | Rate, μm min−1 | |
| pMTs | 0.53 ± 0.19 | 1.99 ± 0.02 (n=340) | 0.11 ± 0.04 | 2.1 ± 0.01 (n=312) | 1.26 ± 0.33 | 4.0 ± 0.04 (n=297) | 0.75 ± 0.17 | 4.0 ± 0.04 (n=423) | 0.16 ± 0.05 (n=140) |
| yMTs | 0.51 ± 0.14 | 0.95 ± 0.01 (n=363) | 0.13 ± 0.03 | 1.5 ± 0.02 (n=284) | 1.08 ± 0.24 | 3.95 ± 0.04 (n=497) | 0.3 ± 0.1 | 4.6 ± 0.04 (n=200) | 0.37 ± 0.13 (n=30) |
| αΔCTT/βΔCTT | 0.22 ± 0.10 | 0.61 ± 0.01 (n=531) | 0.1 ± 0.04 | 0.56 ± 0.01 (n=375) | 1.1 ± 0.32 | 2.2 ± 0.03 (n=508) | 0.3 ± 0.1 | 4.2 ± 0.03 (n=665) | 0.3 ± 0.12 (n=30) |
| α1a/βΔCTT | 0.22 ± 0.10 | 0.98 ± 0.03 (n=388) | 0.1 ± 0.03 | 0.61 ± 0.01 (n=472) | 0.39 ± 0.18 | 0.8 ± 0.01 (n=472) | 0.27 ± 0.16 | 1.8 ± 0.04 (n=374) | 0.7 ± 0.3 (n=138) |
| α1aΔY/βΔCTT | 0.25 ± 0.12 | 0.64 ± 0.01 (n=296) | 0.1 ± 0.05 | 0.91 ± 0.01 (n=386) | 1.16 ± 0.27 | 1.91± 0.02 (n=266) | 0.29 ± 0.14 | 4.1 ± 0.02 (n=553) | 0.32 ± 0.18 (n=54) |
| αΔCTT/βII | 0.64 ± 0.15 | 1.52 ± 0.01 (n=431) | 0.09 ± 0.03 | 1.30 ± 0.02 (n=527) | 1.36 ± 0.3 | 4.8 ± 0.05 (n=318) | 0.24 ± 0.12 | 3.7 ± 0.05 (n=366) | 0.49 ± 0.2 (n=68) |
| α1a/βII | 0.52 ± 0.12 | 1.84 ± 0.01 (n=329) | 0.1 ± 0.03 | 1.22 ± 0.01 (n=424) | 0.74 ± 0.2 | 2.2 ± 0.02 (n=354) | 0.26 ± 0.13 | 2.4 ± 0.02 (n=342) | 1.52 ± 0.5 (n=130) |
| α1aΔY/βII | 0.50 ± 0.14 | 1.34 ± 0.01 (n=471) | 0.1 ± 0.03 | 1.13 ± 0.02 (n=466) | 1.32 ± 0.29 | 5.6 ± 0.08 (n=445) | 0.27 ± 0.12 | 4.4 ± 0.03 (n=465) | 0.51 ± 0.2 (n=92) |
| α1aE452C/βIIE435C | 0.58 ± 0.16 | 1.12 ± 0.01 (n=137) | 0.09 ± 0.03 | 1.28 ± 0.02 (n=343) | 0.8 ± 0.22 | 2.0 ± 0.02 (n=477) | 0.22 ± 0.1 | 2.5 ± 0.04 (n=367) | 0.9 ± 0.27 (n=60) |
| α1a-10E/βII-3E* | 0.52 ± 0.22 | 1.0 ± 0.01 (n=708) | ND | ND | 1.26 ± 0.25 | 3.7 ± 0.04 (n=298) | ND | ND | ND |
| α1a-10E/βII-10E* | 0.54 ± 0.17 | 1.61 ± 0.02 (n=199) | 0.1 ± 0.03 | 1.32 ± 0.01 (n=358) | 1.21 ± 0.27 | 3.7 ± 0.05 (n=392) | 0.4 ± 0.15 | 4.3 ± 0.05 (n=411) | 0.9 ± 0.4 (n=84) |
| αΔCTT/βIII | 0.62 + 0.18 | 0.60 ± 0.01 (n=263) | 0.1 ± 0.03 | 2.1 ± 0.03 (n=215) | 1.31 ± 0.26 | 3.2 ± 0.05 (n=190) | ND | ND | ND |
| αΔCTT/βIIIΔK | 0.64 ± 0.17 | 1.35 ± 0.02 (n=170) | ND | ND | ND | ND | ND | ND | ND |
| α1aE452C/βIII | 0.63 ± 0.18 | 0.54 ± 0.02 (n=154) | ND | ND | ND | ND | ND | ND | ND |
| α1a-3E/βIII* | 0.48 ± 0.19 | 1.04 ± 0.01 (n=199) | ND | ND | ND | ND | ND | ND | ND |
| α1a-10E/βIII* | 0.60 ± 0.20 | 1.50 ± 0.02 (n=153) | ND | ND | ND | ND | ND | ND | ND |
| αΔCTT/βIV | 0.65 ± 0.17 | 1.26 ± 0.02 (n=185) | 0.08 ± 0.02 | 1.31 ± 0.03 (n=215) | 1.33 ± 0.25 | 3.3 ± 0.041 (n=247) | 0.2 ± 0.07 | 4.7 ± 0.1 (n=162) | 0.52 ± 0.15 (n=54) |
| αΔCTT/βV | 0.58 ± 0.15 | 1.17 ± 0.02 (n=107) | 0.07 ± 0.02 | 1.53 ± 0.02 (n=228) | ND | ND | ND | ND | ND |
| αΔCTT/βVI | 0.53 ± 0.18 | 0.71 ± 0.02 (n=145) | 0.08 ± 0.02 | 1.31 ± 0.02 (n=108) | ND | ND | ND | ND | ND |
| αΔCTT/βVII | 0.25 ± 0.11 | 0.91 ± 0.01 (n=214) | 0.09 ± 0.03 | 1.02 ± 0.03 (n=141) | ND | ND | ND | ND | ND |
| αΔCTT/βVIII | 0.64 ± 0.16 | 1.01 ± 0.01 (n=353) | 0.08 ± 0.03 | 1.35 ± 0.02 (n=214) | ND | ND | ND | ND | ND |
| α1a/βIV | ND | ND | ND | ND | ND | ND | ND | ND | 1.6 ± 0.5 (n=104) |
| α1aΔY/βIV | ND | ND | ND | ND | ND | ND | ND | ND | 0.53 ± 0.18 (n=72) |
| α1aE452C/βIVE435C | ND | ND | ND | ND | ND | ND | ND | ND | 0.91 ± 0.3 (n=54) |
| α1a-10E/βIV-10E* | ND | ND | ND | ND | ND | ND | ND | ND | 0.89 ± 0.3 (n=74) |
3 or 10 glutamate peptide crosslinked to cysteine light tubulin mutants as indicated.
CTT requirements for normal motor function
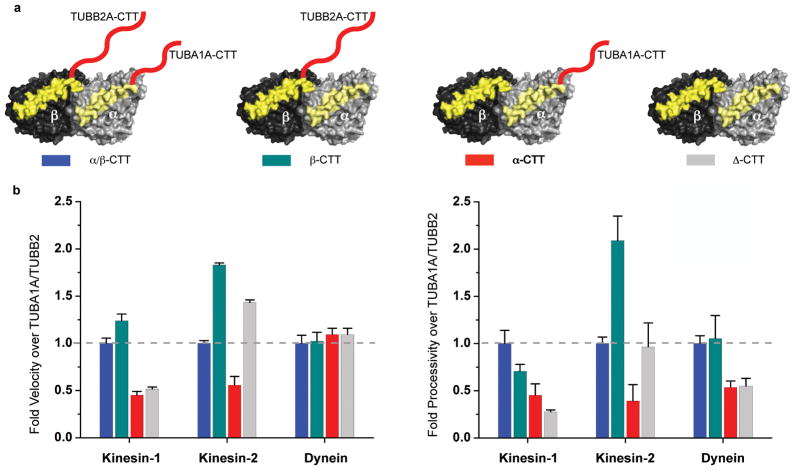
We next examined whether the CTTs on α, β tubulin or both are required for motor function (Fig. 2a), a question that cannot be addressed by subtilisin treatment, which cleaves both CTTs and also cleaves at heterogeneous sites35,36. Deletion of both CTTs (Δ-CTT microtubules) reduced the velocity and processivity of kinesin-1 movement by ~50% and ~75% respectively, which is consistent with prior results with subtilisin-treated porcine brain MTs46. Interestingly, the β-CTT alone restored the velocity and most of the processivity of kinesin-1 (100% and ~75% of α/β-CTT respectively), while α-CTT alone had little effect (Fig. 2b). The velocity of yeast dynein was unaffected by deletion of the CTTs, while processivity was reduced by 50%, in agreement with early results of mammalian dynein bead motility assays46. Yeast dynein processivity could be restored by β- but not the α-CTT. Interestingly, for human kinesin-2, removal of the α-CTT (creating MTs with only β-CTT) substantially increased both the velocity and processivity (2-fold and 2.5-fold respectively; Fig. 2b) to values similar to those observed with porcine brain microtubules (Fig. 1 and Table 1). Collectively, these results show that α- and β-CTTs exhibit distinct effects on motors. Kinesin-1 and yeast dynein require only the β-CTT for full activity. However, kinesin-2 motility appears to be regulated by both of the CTTs, with an inhibitory element being present within the α-CTT.
Figure 2. Minimal CTT requirement for motor function.
a. Illustration of recombinant tubulins with TUBA1A/TUBB2A CTTs (blue), TUBB2A CTT alone (green), TUBA1A CTT alone (red) or no (Δ) CTTs (grey). Helix 12 highlighted in yellow and CTTs in red.
b. Velocity and processivity of human kinesin-1, -2 and dynein was compared as a ratio to TUBA1A/TUBB2A CTT chimeric microtubule (mean and s.e.m, n = 3 independent experiments; see Supplementary Table 1 for individual experiment values and Table 1 for absolute values).
Effects of α-tubulin terminal tyrosine residue on motors
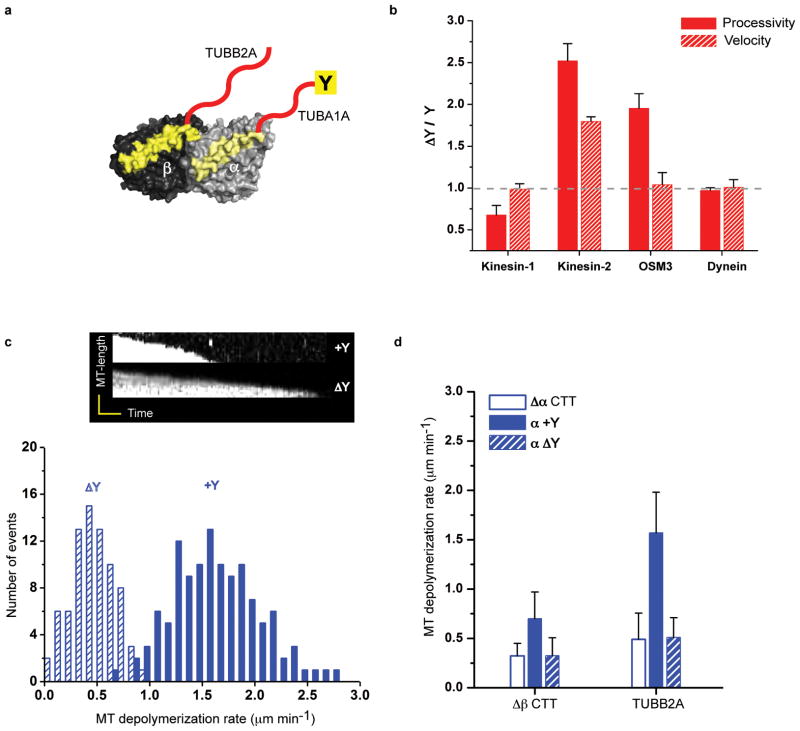
We next tested the role of the C-terminal tyrosine in α-tubulin (Fig. 3a), which is subject to a de-tyrosination/tyrosination cycle in metazoans15,16. The presence or absence of the C-terminal tyrosine had little effect on yeast dynein motility (Fig. 3b). When tested on kinesin-1, removal of the C-terminal tyrosine decreased processivity by ~25% (TUBA1A-ΔY compared to TUBA1A; Fig. 3b). Strikingly for kinesin-2, the removal of the tyrosine in α-CTT (TUBA1A-ΔY) caused the opposite effect, increasing velocity and processivity by ~2 and 2.5-fold respectively (Fig. 3b, shown as the ratio of values for ΔY/Y microtubules; absolute values found in Table 1 and Supplementary Table 1). We also tested whether this phenomenon is conserved in kinesin-2 motors from a distant metazoan species. We found that the processivity, but not the velocity, of C. elegans Osm-3 motors could be increased by ~2-fold upon de-tyrosination of α-CTT (Fig. 3b). Our previously described CTT truncation results suggested that α-CTT possesses an inhibitory element for kinesin-2. These results suggest that this inhibitory element is the C-terminal tyrosine.
Figure 3. The effects of the α-tubulin C-terminal tyrosine on motor performance.
a. Microtubules were prepared from tubulin with (TUBA1A/TUBB2A) or without (TUBA1A-ΔY/TUBB2A) the C-terminal tyrosine (Y) on α-tubulin.
b. The fold velocity and processivity of de-tyrosinated (ΔY) over tyrosinated (Y) microtubules is shown (mean and s.e.m, n = 3 independent experiments; see Supplementary Table 1 for individual experiment values and Table 1 for absolute values).
c. The effect of the C-terminal α-tubulin tyrosine on microtubule depolymerization by mammalian kinesin-13; the kymographs (time versus microtubule end position) show the decrease in the length of the microtubule polymer over time. Histograms of kinesin-13 microtubule depolymerization rates of tyrosinated (TUBA1A/TUBB2A) and de-tyrosinated (TUBA1A-ΔY/TUBB2A) microtubules. The mean and s.d. were 1.5 ± 0.5 (n = 130) and 0.5 ± 0.2 (n = 92) respectively; n represents the number of microtubule ends analyzed.
d. Kinesin-13 depolymerization activity with no α-CTT (Δα), the complete TUBA1A-CTT with its genetically encoded C-terminal tyrosine (α+Y), or the TUBA1A-CTT lacking this tyrosine (αΔY); experiments were performed with a tubulin heterodimer lacking β-CTT (Δβ) or containing the TUBB2A-CTT. Similar results were obtained with the βIV-CTT from two independent experiments (for absolute values and sample number see Table 1).
We also tested the effect of the C-terminal tyrosine on Chinese Hamster MCAK, a kinesin-13 motor that uses ATP energy to depolymerize microtubules rather than to move along them. We choose Chinese Hamster MCAK, as it is a very well-characterized mammalian kinesin-13 family motor17,47, The motor domains of human and Chinese Hamster kinesin-13 are extremely similar to one another (~96% amino acid identity, Supplementary Figure 5) and the TUBA1A and TUBB2A isotypes are 100% identical between human and hamster. We found a 2.5-fold faster depolymerization rate of genetically pure tyrosinated versus de-tyrosinated α-tubulin; the latter depolymerized at a comparable rate to Δ-CTT (Fig. 3c, d). These results are in agreement with previous studies showing that brain microtubule depolymerization is inhibited by detyrosination17 and CTT removal by subtilisin48. Unlike the requirements for motile kinesins, we also found that both α- and β-CTTs were required for maximal activity of kinesin-13 (Fig. 3d; Table 1).
In summary, the presence or absence of a single C-terminal tyrosine residue on α-tubulin influences the processivity of kinesin-1 and -2 (in opposite directions), the velocity of kinesin-2, and the depolymerizing activity of kinesin-13.
Polyglutamylation increases kinesin processivity
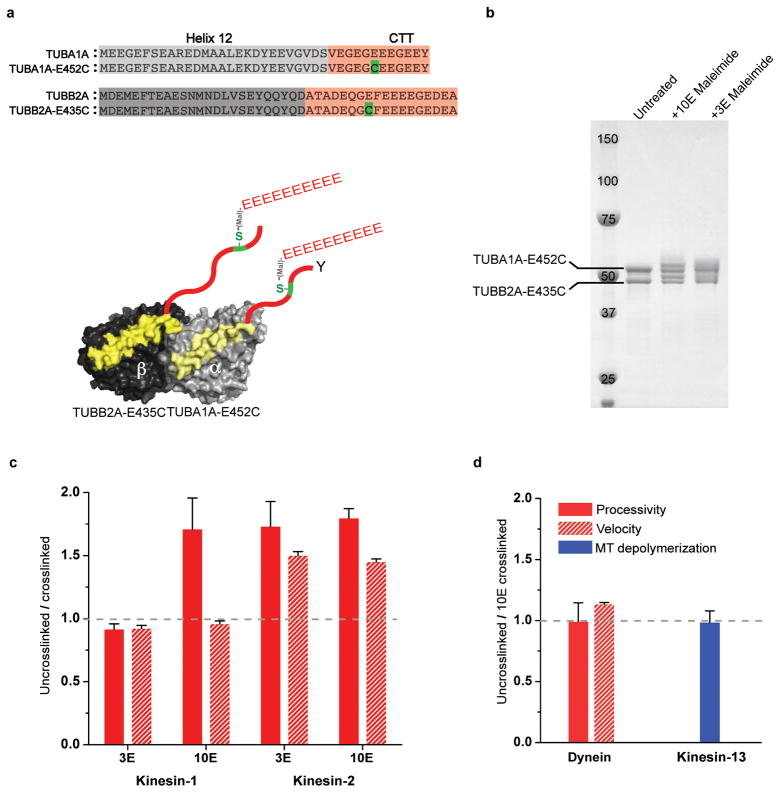
We next tested the effects of tubulin polyglutamylation, which involves the enzymatic addition of glutamate residues to the γ-carboxyl side chain of a glutamate residue in the CTT of both α- and β-tubulin15,22,23. Glutamic acid residue (E445 in α- and E435 in β-tubulin) were identified as the most common sites of polyglutamylation22,49,50, although other nearby glutamic acids are also modified by polyglutamylases25,28. Cell biology22,23,27, mass spectrometry28 and biochemical studies with purified enzymes26 also have shown that the glutamate chain length can vary considerably. In neurons, the chain lengths vary between 1–6 glutamates22,28; in cilia, >10 glutamates are common28. To create a biochemically homogenous population of polyglutamylated tubulin, we substituted a cysteine for α-E452 (E445 equivalent in chimeric α-tubulin) and β-E435 and used maleimide chemistry to crosslink glutamate peptides to this cysteine residue using a cys-light tubulin (see Methods) (Fig. 4a & b). To examine the effects of chain length, we prepared tubulins modified with either a short (3E) or a long (10E) glutamate chain. The addition of the branching glutamate peptide on α- and β-CTTs (3E- and 10E-CTTs) did not affect the activity of yeast dynein or kinesin-13 (Fig. 4d, Table 1 and Supplementary Table 1). However, both the 3E- and 10E-CTTs increased the velocity (1.2 μm s−1) and processivity (~3.5 μm run length) of kinesin-2 by ~1.5-fold (Fig. 4c), similar to values observed with porcine brain microtubules and detyrosinated recombinant microtubules (Fig. 1c, 1e, 3b, and Table 1). Unlike kinesin-2 where both 3E and 10E showed marked increases in motility, only the 10E-CTT increased the processivity of kinesin-1 (by ~1.5-fold) (Fig. 4c). Thus, in summary, the yeast cytoplasmic dynein and kinesin-13 appear to be insensitive to polyglutamylation, while the two cargo-transporting kinesins (kinesin-1 and kinesin-2) show gain of functions by the addition of glutamate chains to the CTT.
Figure 4. Effects of polyglutamylation on motor performance.
a. To study how polyglutamylation affects motor function, peptide of either 3 or 10 glutamates (3E or 10E) was crosslinked using maleimide chemistry to a single cysteine in the CTT (see Methods). A 10E peptide is shown in this illustration.
b. The crosslinked reaction product is shown by SDS PAGE as reflected by an upward gel shift of the indicated α- and β-tubulin bands (estimated ~60% crosslinked product). In a control without the introduced cysteine in the CTT, this gel shift did not occur, indicating the crosslinking occurred at the correct position of the CTT (Supplementary Figure 6).
c and d. The ratio of the motor velocity (red hatched), processivity (solid red) or microtubule depolymerization rate (solid blue) on uncrosslinked versus crosslinked microtubules is shown (mean and s.e.m; n = 3 independent experiments, see Supplementary Table 1). Absolute values are shown in Table 1.
Specificity of motors towards human β-tubulin isotypes
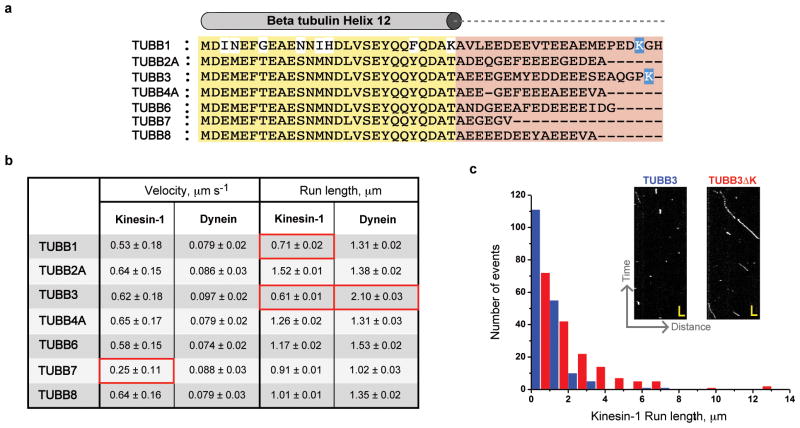
Next, we investigated whether the genetic sequence diversity in human β-tubulin CTTs influences motor activity (Fig. 5a). Chimeric tubulins with CTTs from human TUBB2-8 were prepared in a Δα-CTT background, so that the influence of β-CTTs could be unambiguously evaluated. The velocity of dynein was the same for all β-CTTs. On the other hand, kinesin-1 moved ~50% slower on the TUBB7-CTT isotype. Since this velocity is comparable to Δ-CTT microtubules (Fig. 2a) and TUBB7 is the shortest among the human isotypes (Fig. 5b), it is possible that the short length of the TUBB7 CTT accounts for the reduction in velocity of kinesin-1. Interestingly, the run length of kinesin-1 on TUBB3 was significantly lower (3-fold reduction) compared with TUBB2. This TUBB3 inhibitory effect on kinesin-1 was specific, since yeast dynein exhibited a modest increase (1.5-fold; Fig. 5b) and kinesin-2 showed no significant change in run length (Table 1). The addition of the TUBA1A CTT (to produce microtubules composed of TUBA1A/TUBB3 heterodimers) did not increase the run length of kinesin (Table 1). The run length of kinesin-1 on TUBB1 also was reduced by ~3-fold compared with TUBB2, while yeast dynein processivity was unaffected (Fig. 5b).
Figure 5. Motility regulation by β-tubulin isotypes.
a. Sequence alignment of helix 12 (yellow) and CTTs (red) of human β-tubulin isotypes. The basic residues in TUBB1 and TUBB3 CTTs are highlighted in blue.
b. Kinesin-1 and dynein motors velocity and processivity values measured against various β-tubulin isotypes as indicated (in Δα-CTT background). Mean velocity and run length were determined as described in Fig 1.
c. Run length histograms of kinesin-1 motors moving on TUBB3 (blue) and TUBB3ΔK (red) microtubules (mean run length of 0.6 and 1.35 μm respectively). Examples of raw kymographs (inset panel; scale, 2 sec and 1.5 μm).
TUBB1 and TUBB3 are the only β-tubulin isotypes that possess a positively charged lysine residue in the CTTs (Fig. 5a). To test whether this positive charge affects processivity, we removed the terminal lysine residue of TUBB3-CTT (TUBB3ΔK). Single molecule experiments showed that the removal of this single lysine fully restored the run length of kinesin-1 to the same level as TUBB2 (Fig. 5c & Supplementary Table 1). Thus a single basic residue within the β-CTT can dramatically alter the processivity of kinesin-1.
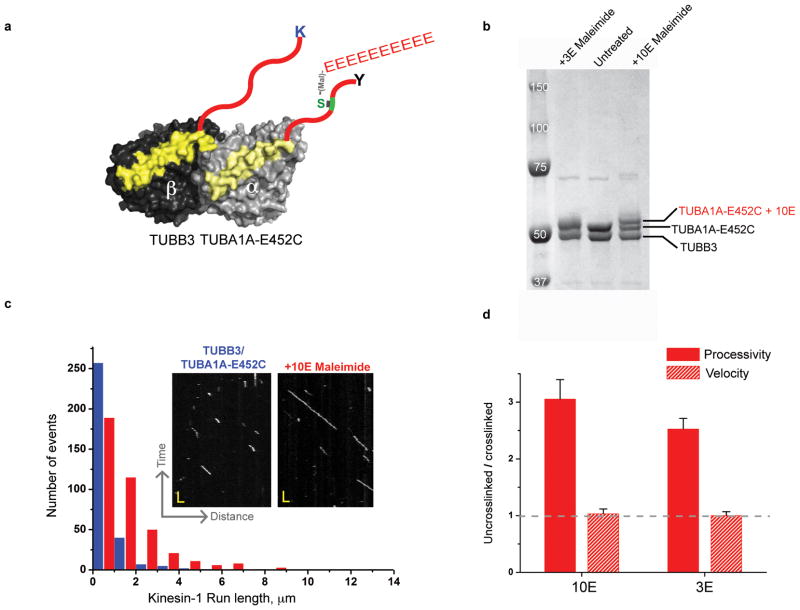
Polyglutamylation rescues kinesin-1 processivity against TUBB3
Since the lysine residue in TUBB3 decreased kinesin-1 processivity by ~3-fold, we wanted to determine if addition of negative charge in the form of polyglutamylation might overcome this effect. We utilized the same strategy for crosslinking a polyglutamate peptide to the CTT, as described earlier. However, in this experiment, the 3E or 10E peptide was crosslinked only to the TUBA1A-CTT so as to avoid any potential cis-interaction with lysine on TUBB3 CTT (Fig. 6a & b). Crosslinking of either the 3E or 10E peptides to these TUBB3 microtubules dramatically increased the processivity of kinesin-1 motors (Fig. 6c, 6d). These results reveal that polyglutamylation on the TUBA1A-CTT can overcome the inhibitory effect of the lysine on TUBB3.
Figure 6. Cross-regulation of isotype specificity by polyglutamylation.
a. Illustration of TUBA1A-E452C/TUBB3 heterodimer crosslinking strategy with 10E.
b. SDS-PAGE of crosslinked product of 10 and 3 glutamic acid peptides (10E and 3E) as indicated in Fig. 4.
c. Kinesin-1 motility kymographs (inlet panel) and run length histograms of TUBB3/TUBA1A- E452C and +10E (red) microtubule (mean run lengths of 0.5 and 1.5 μm respectively). Scale bars and bin size are similar to 4c.
c. The ratio of the motor velocity (red hatched), processivity (solid red) on uncrosslinked versus 10E and 3E crosslinked TUBA1A- E452C/TUBB3 microtubules is shown (mean and s.e.m; n = 3 independent experiments, see Supplementary Table 1). Absolute values are shown in Table 1.
Discussion
In this study, we show that CTTs can regulate motor proteins in distinct ways. For example, kinesin-1 and dynein motility are regulated primarily by the β–CTT, while kinesin-2 and kinesin-13 show more complex regulation involving both the CTTs. Moreover, kinesin-2 and -13 show different responses to the C-terminal tyrosine on the α–chain, being negatively and positively regulated respectively (Fig. 7). Kinesin-1 and kinesin-2 motility are also strongly enhanced by polyglutamylation, while kinesin-13 and dynein are insensitive to this modification (Fig. 7). Since kinesin-2 motility and yeast dynein motility is not affected by TUBB3 microtubules (Table 1), the influence of terminal lysine in TUBB3 isotype seems to be a kinesin-1 specific effect.
Figure 7. Summary of CTT mediated effects on different motors.
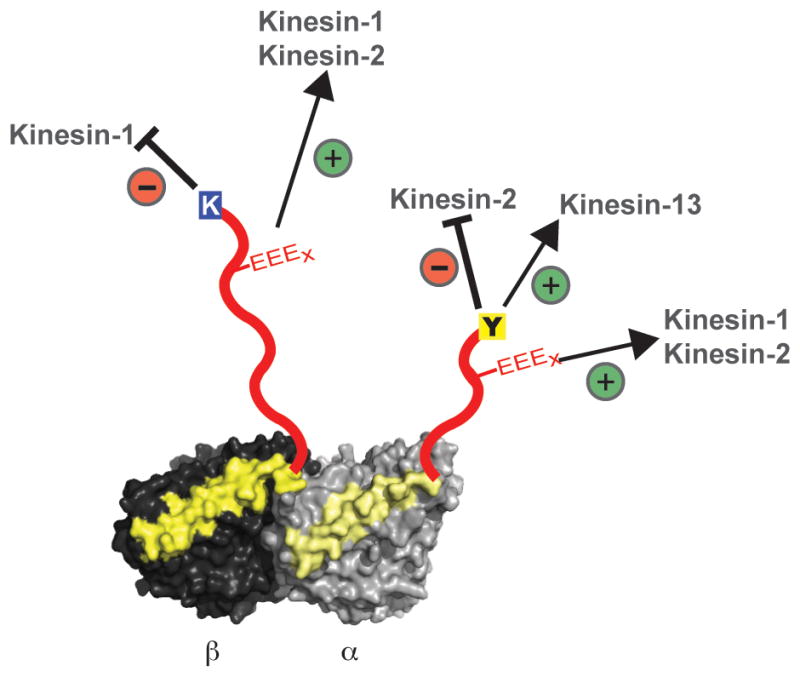
α/β-tubulin heterodimer in light and dark grey respectively with helix 12 colored in yellow and unstructured C-terminal tails (CTTs) in red. K (blue), Y (yellow) and the EEEx (red) represent the TUBB3 β-isotype, tyrosination in α-tubulin and polyglutamylation respectively. The specific inhibitory and enhancing effects of tubulin variations on different motors are as indicated.
The CTT effects described above are likely to be mediated by different mechanisms. It has been suggested that the processivity of kinesin-1 is aided by weak electrostatic interactions between positively-charged patches on the motor and the negatively charged CTTs41,51,52. This electrostatic interaction probably serves as a weak tether to retain kinesin near the microtubule surface under circumstances in which both motor domains detach. Previous work has shown that altering the charge of the kinesin-1 neck region can increase or decrease the processivity of the motor; this effect was largely abolished using subtilisin-cleaved tubulin that lacks CTTs41. Our current finding that a positive charge in the CTTs (lysine in the case of TUBB3) can interfere with motor processivity and can be rescued through polyglutamylation further supports the notion that electrostatic interaction regulates kinesin-1 processivity. Similarly, the increase in kinesin-2 processivity by polyglutamate chains (in the tyrosinated α-tubulin background) might be explained due to the electrostatic effects. However, the pronounced inhibitory effect of the C-terminal α-tubulin tyrosine on kinesin-2 velocity as well as the enhancing effect of this same residue of kinesin-13 depolymerization might be mediated by a more specific docking interaction between this residue and these motor domains. Similarly, the requirement of β-CTT for optimal kinesin-1 velocity could involve a site-specific interaction between the CTT and the motor domain. The current structures of motors bound to tubulin or microtubules do not identify interactions between motors and CTTs38–40,53, perhaps due to either insufficient resolution or poor occupancy as a result of weak affinity or the heterogeneity in the native tubulin used in these studies. Therefore, future structural and mutagenesis studies with our recombinant tubulin system might provide further mechanistic information on how tubulin CTTs regulates motor proteins.
Our in vitro results also raise interesting new questions on the roles of tubulin isotypes and PTMs in cells. Kinesin-1 has been reported to move preferentially along detyrosinated microtubules in cells18–20, but this preference was not manifest on recombinant detyrosinated microtubules. This result suggests that a factor associated with detyrosinated MT, rather than detyrosination per se, may underlie this effect. Another intriguing observation for kinesin-1 is its low processivity on TUBB3 microtubules and that polyglutamylation (both the 3E and 10E peptides) can restore normal movement. The TUBB3 isotype is expressed highly in neurons3,5 and is linked to a spectrum of neurological syndromes10,12. Polyglutamylation of tubulin, particularly shorter glutamate chains (<6), is also prevalent in brain22,23,27,28. The combination of negative (TUBB3) and positive (polyglutamylation) regulators may enable subtle regulation of motor protein function, such as tilting the balance of movement between kinesin-dynein motor54,55 or favoring a subset of microtubule tracks for transport activity. The effects in vivo, however, could be even more complex than what we report here, since microtubules in neurons are likely to be composites of different isotypes and PTMs rather than the homogeneous tubulin prepared in this study.
Our study also raise the possibility that anterograde intraflagellar transport by homodimeric kinesin-242 may be activated by detyrosination. The tubulin in the axoneme is heavily de-tyrosinated and polyglutamylated15,25,29. We also discovered that even short glutamate chain (3E) could overcome the inhibitory effects of tyrosinated α-tubulin on kinesin-2. A recent study has shown that knockdown of the deglutamylase enzyme CCP1 (cytosolic carboxypeptidase-1) leads to decrease in velocity of OSM3/KIF17 mediated anterograde intraflagellar transport56. In addition to its side chain deglutamylase activity, CCP1 is known to remove genetically-encoded glutamate residues of α-tubulin, generating an irreversible de-tyrosinated tubulin (Δ2-tubulin)57. Thus, in addition to the proposed auto-inhibition by the C-terminal cargo binding domain58,59, our study reveals an additional tier of regulating kinesin-2 mediated transport through a preference for de-tyrosinated or polyglutamylated tracks.
Taken together, our in vitro results provide support for the ‘tubulin code’ hypothesis that specific tubulin isotypes or PMTs confer unique biochemical activities. We also provide evidence for previous speculations15,33 that different PTMs and isotypes can influence one another in a combinatorial manner, similar to the histone code. Future experiments with defined stoichiometry of different tubulin isotypes and PTMs should reveal more information on the intricate balances, interplay and dominating roles of various tubulin species in the cell. In addition to molecular motors, numerous microtubule-associated proteins interact and influence the stability and dynamics of microtubules. The recombinant tubulins described here can be used to dissect how different PTMs and isotypes affect these diverse regulatory activities, which will provide a deeper understanding of the tubulin code and how it may be used to control microtubule function in cells.
Methods
Expression, purification and assembly of recombinant tubulin
The yeast expression system used in this study for purifying recombinant tubulin was adapted from Johnson et al37. This method overexpresses the recombinant yeast α/β-tubulin from two yeast 2 micron galactose-inducible expression plasmids. To separate the recombinant from the native tubulin (which is expressed at low levels), a metal-affinity chromatography was used as described by Johnson et al37. However, we found that the hexa-histidine tag (His-6) at C-terminus of either α- or β-tubulin affected the velocity of kinesin-1 compared to porcine microtubule (50% reduced for His-6 C-terminal β-tubulin and 25% increased for His-6 C-terminal α-tubulin). To overcome this problem and also not to add non-native residues to the human CTTs, a hexa-histidine (His-6) tag was placed in less conserved luminal loop in α-tubulin (Supplementary Figure 4). Since the His-6tag is in the luminal surface, it will not be exposed to motor proteins. In addition, a previous study has shown that up to 17 amino acids can be inserted into this luminal loop without disrupting tubulin function60. Using this approach, we were able to purify the recombinant tubulin (yeast α-int-His/β-tubulin) and assemble it into microtubules (Supplementary Figure 4). The velocity of kinesin-1 movement on recombinant, His-6 tagged yeast microtubules (Fig. 1b & c) were similar to that described with yeast microtubules61. Since we did not observe any discernable effects of an internal His-6 tag on microtubule polymerization and motor behavior, all engineered tubulin heterodimers were purified in the α-int-His background.
To express recombinant tubulin, an initial culture was scaled to 6 liters and grown in bioreactor with continuous airflow and stirring as described before62. The expression was induced by addition of galactose to the growth media and the tubulin from the expressed yeast cells was purified as described earlier37. To make chimeric yeast core-human CTT tubulin heterodimer, the end of helix-12 of yeast tubulin was chosen as a junction point (Fig. 1a) and the human CTT (codon optimized for yeast) was fused to the gene. The purified tubulin heterodimer (~20 μM) was stored in BRB80 buffer (80 mM Pipes pH 6.8, 2 mM MgCl2, 1 mM EGTA and 200 μM GTP) at −80° C. Using quantitative western blots with DM1a antibody (Sigma), we estimated that our preparations contain <3% percentage of endogenous yeast β-tubulin.
Purification of motor proteins
Kinesin-1 (human K560)41 and C. elegans homodimeric kinesin-2 (OSM3-ΔH2)58 motors with GFP-6xHis at the C-terminus were purified using Ni-NTA beads as described earlier. A truncated, dimeric yeast dynein (GST-D6-dynein) was purified using IgG beads as previously reported44. In order to remove inactive molecules, motors were subjected to additional purification step involving microtubule binding in the absence of ATP and their release from microtubules with ATP63. The Chinese Hamster MCAK (kinesin-13 motor) was purified from insect cells using baculovirus expression system as described earlier17. Human homodimeric kinesin-2 (KIF17), which could not be expressed in soluble form in bacteria, was cloned into a lentiviral expression vector (pHR) with in frame super-folded GFP (sfGFP) and Strep-tag at the 5′ end. Because the full length KIF17 undergoes auto-inhibition and is inactive, a truncated KIF17 (1–738 amino acids) based on previous study was chosen here59. The pHR-KIF17-sfGFP-Strep plasmid was initially transfected into the HEK293 cells for producing the virus. The lentivirus was harvested after two days of infection and used to make stable HEK293 cells expressing KIF17-sfGFP-Strep. After 18 hr of infection the HEK293 cells were resuspended in 50 mM PIPES pH 6.8, 100 mM KCl, 2 mM MgCl2 buffer and the cells were lysed using 0.5% Triton X-100 and protease inhibitors. The lysed cells were centrifuged at high-speed 30,000xg at 4° C for 30 min. The supernatant was incubated using Strep-Tactin superflow beads (Qiagen) for 60 min at 4° C. The beads were washed with 50 mM PIPES pH 6.8, 100 mM KCl, 2 mM MgCl2 and eluted using 2.5 mM d-Desthiobiotin (Sigma). The eluted fractions were flash frozen and stored in −80° C until further use.
Single molecule assays with motor proteins
For each day of assays, an aliquot of tubulin was thawed and 2 mM GTP with 5 μM Epothilone-B (Sigma) was added and polymerized overnight at 30° C, yielding typical microtubule lengths of >20 μm. ~1:250 ratio of biotinylated porcine brain tubulin was added to the mixture to enable attachment of the microtubules to streptavidin adsorbed onto acid-washed glass coverslips in flow chambers (~5 μL). A motility mixture containing the desired motor (typically 100–200 pM), 2 mM ATP, and oxygen-scavenging reagents (catalase, glucose oxidase, and glucose)64 were added to buffer containing 25 mM Pipes (pH 6.8), 2 mM MgCl2, 1 mM EGTA and 1 mg/ml casein (BRB25 + casein). The motility mix was flowed in and the chamber was sealed. Single molecule motility was imaged using a Nikon Ti microscope (1.49 N.A., 100x objective) using total internal reflection microscopy (TIRF); images were acquired with an Andor EM-CCD camera and MicroManager software65 (acquisition rates of 5 and 1 Hz for kinesins and dynein respectively). Velocities and run lengths were calculated using kymograph analysis in ImageJ software (minimum distance of 0.25 μm). The mean run length was calculated using a cumulative probability distribution function as described earlier41. The inverse cumulative probability distribution was fit to a simple exponential decay function using Origin 8; errors reported for run lengths in Table 1 and Fig. 5b are the standard errors of the fit. All measurements are in agreement with at least two experiments with independently polymerized microtubules.
Microtubule depolymerization by MCAK
Recombinant tubulin was polymerized with Cy5-labelled pig brain tubulin (1:100 ratio) and biotinylated tubulin (1:250 ratio) with 1 mM GMPPCP (slow hydrolyzing GTP analogue) overnight at 30° C. The Cy5-labeled microtubules were attached using biotin streptavidin methods in a flow chamber as described earlier. The MCAK assay buffer contained BRB80, 1 mg/ml casein, oxygen-scavenging reagents, 20 nM chMCAK, and 1 mM ATP. MT depolymerization events were recorded using TIRF microscopy at 2 Hz and the rates were calculated from kymographs using ImageJ software.
Crosslinking of a 10E peptide to tubulin CTTs
A total of 11 and 6 cysteine residues are present in yeast α- and β-tubulin respectively, out of which only α-C130 and β-C127 is exposed on the surface of polymerized microtubules. Both cysteine residues are located in the grove between adjacent protofilaments, but do not participate in lateral protofilament interactions. In order to generate a cysteine-light tubulin, both the α-C130 and β-C127 were mutated to serine. This cysteine-light tubulin was used to introduce crosslinking cysteines at the polyglutamylation site of α-tubulin (TUBA1A-E452C) and β-tubulin (TUBB2-E435C). Although polyglutamylation can occur in multiple glutamic acid residues25, we choose the most commonly reported modified glutamic acid residue of E445 and E435 equivalent residues in our chimeric α- and β-tubulin chains respectively22,49,50. The TUBA1A-E452C/TUBB2A-E435C and TUBA1A-E452C/TUBB3 heterodimer was purified as described for other recombinant tubulins and stored with additional 1 mM TCEP (reducing agent). A linear chain of three-glutamate and deca-glutamate peptides were synthesized with a reactive maleimide group at the N-terminus (3E and 10E maleimide, with 95% purity) was obtained from Elim Biopharm (Hayward, California, USA). The 3E represents a median length of glutamate chain reported in brain22,27,28 and 10E represents a longer glutamate chains, which is predominant in cilia and flagella28,29. Both 3E and 10E maleimide peptides were stored as 5 mM aliquots in BRB80, 1 mM TCEP buffer at −20° C. For crosslinking reactions, the microtubules with cysteine at the CTTs was incubated with a ~50X molar excess maleimide-10E for 15 min at room temperature. After 15 min, the reaction was quenched using 5 mM DTT. The crosslinking product was analyzed by SDS-PAGE (4–12% gradient gel with NuPAGE - MOPS buffer, Invitrogen) and a typical reaction yields >75% 3E crosslinked tubulin and ~60% 10E crosslinked tubulin (Fig. 4b and 6b). Cysteine-light tubulin without an introduced cysteine in the CTT was not crosslinked by maleimide-10E (Fig. 6b and Supplement Fig 6).
Tubulin and microtubule model generation
For illustrative purpose a linear polypeptide chain of TUBA1A and TUBB2A CTTs were generated using Pymol program and fused to the last visible residue of alpha and beta chains in the microtubule model (generated using PDB:1JFF) shown in Fig. 1a. The tubulin model shown in Figure 2a, 3a, 4a and 5a was generated from PDBID:4FFB using Pymol, CTTs were added using Adobe Illustrator.
Supplementary Material
Acknowledgments
The authors thank Nico Stuurman for microscopy assistance, Marvin Tanenbaum for help in lentiviral expression and members of the Vale lab for comments on the manuscript. R.D.V is a Howard Hughes Medical Institute investigator and M.S is a Human Frontiers Science Program - Long-term fellow (LT-000120/2009). L.M.R is supported by NSF MCB-1054947. This work received support from an NIH grant (38499).
Footnotes
Author Contributions:
M.S and R.D.V conceived the project. M.S performed the experiments, analyzed the data and wrote the paper. L.M.R developed and tested the internal His-tagged tubulin and advised in this study. R.D.V supervised the work and wrote the paper. All authors discussed and commented on the manuscript.
The authors declare no competing financial interests.
References
- 1.Nogales E. Structural insight into microtubule function. Annu Rev Biophys Biomol Struct. 2001;30:397–420. doi: 10.1146/annurev.biophys.30.1.397. [DOI] [PubMed] [Google Scholar]
- 2.Amos LA, Schlieper D. Microtubules and maps. Adv Protein Chem. 2005;71:257–298. doi: 10.1016/S0065-3233(04)71007-4. [DOI] [PubMed] [Google Scholar]
- 3.Ludueña RF, Banerjee A. Role Microtubules Cell Biol Neurobiol Oncol. Humana Press; 2008. pp. 123–175. (MD, T. F.) at < http://link.springer.com/chapter/10.1007/978-1-59745-336-3_6>. [Google Scholar]
- 4.Banerjee A, et al. A monoclonal antibody against the type II isotype of beta-tubulin. Preparation of isotypically altered tubulin. J Biol Chem. 1988;263:3029–3034. [PubMed] [Google Scholar]
- 5.Burgoyne RD, Cambray-Deakin MA, Lewis SA, Sarkar S, Cowan NJ. Differential distribution of beta-tubulin isotypes in cerebellum. EMBO J. 1988;7:2311–2319. doi: 10.1002/j.1460-2075.1988.tb03074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raff EC, Fackenthal JD, Hutchens JA, Hoyle HD, Turner FR. Microtubule architecture specified by a beta-tubulin isoform. Science. 1997;275:70–73. doi: 10.1126/science.275.5296.70. [DOI] [PubMed] [Google Scholar]
- 7.Leandro-García LJ, et al. Hematologic β-tubulin VI isoform exhibits genetic variability that influences paclitaxel toxicity. Cancer Res. 2012;72:4744–4752. doi: 10.1158/0008-5472.CAN-11-2861. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Villasante A, Lewis SA, Cowan NJ. The mammalian beta-tubulin repertoire: hematopoietic expression of a novel, heterologous beta-tubulin isotype. J Cell Biol. 1986;103:1903–1910. doi: 10.1083/jcb.103.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cederquist GY, et al. An inherited TUBB2B mutation alters a kinesin-binding site and causes polymicrogyria, CFEOM and axon dysinnervation. Hum Mol Genet. 2012;21:5484–5499. doi: 10.1093/hmg/dds393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tischfield MA, et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tischfield MA, Cederquist GY, Gupta ML, Jr, Engle EC. Phenotypic spectrum of the tubulin-related disorders and functional implications of disease-causing mutations. Curr Opin Genet Dev. 2011;21:286–294. doi: 10.1016/j.gde.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tischfield MA, Engle EC. Distinct alpha- and beta-tubulin isotypes are required for the positioning, differentiation and survival of neurons: new support for the ‘multi-tubulin’ hypothesis. Biosci Rep. 2010;30:319–330. doi: 10.1042/BSR20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breuss M, et al. Mutations in the β-tubulin gene TUBB5 cause microcephaly with structural brain abnormalities. Cell Rep. 2012;2:1554–1562. doi: 10.1016/j.celrep.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.L’Hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry (Mosc) 1985;24:473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- 15.Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol. 2011;12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 16.Garnham CP, Roll-Mecak A. The chemical complexity of cellular microtubules: tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskelet Hoboken NJ. 2012;69:442–463. doi: 10.1002/cm.21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peris L, et al. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J Cell Biol. 2009;185:1159–1166. doi: 10.1083/jcb.200902142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat Neurosci. 2009;12:559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- 19.Dunn S, et al. Differential trafficking of Kif5c on tyrosinated and detyrosinated microtubules in live cells. J Cell Sci. 2008;121:1085–1095. doi: 10.1242/jcs.026492. [DOI] [PubMed] [Google Scholar]
- 20.Liao G, Gundersen GG. Kinesin is a candidate for cross-bridging microtubules and intermediate filaments. Selective binding of kinesin to detyrosinated tubulin and vimentin. J Biol Chem. 1998;273:9797–9803. doi: 10.1074/jbc.273.16.9797. [DOI] [PubMed] [Google Scholar]
- 21.Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 22.Eddé B, et al. Posttranslational glutamylation of alpha-tubulin. Science. 1990;247:83–85. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- 23.Audebert S, et al. Reversible polyglutamylation of alpha- and beta-tubulin and microtubule dynamics in mouse brain neurons. Mol Biol Cell. 1993;4:615–626. doi: 10.1091/mbc.4.6.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janke C, et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- 25.Wloga D, Gaertig J. Post-translational modifications of microtubules. J Cell Sci. 2010;123:3447–3455. doi: 10.1242/jcs.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Dijk J, et al. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol Cell. 2007;26:437–448. doi: 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 27.Audebert S, et al. Developmental regulation of polyglutamylated alpha- and beta-tubulin in mouse brain neurons. J Cell Sci. 1994;107 (Pt 8):2313–2322. doi: 10.1242/jcs.107.8.2313. [DOI] [PubMed] [Google Scholar]
- 28.Redeker V. Mass spectrometry analysis of C-terminal posttranslational modifications of tubulins. Methods Cell Biol. 2010;95:77–103. doi: 10.1016/S0091-679X(10)95006-1. [DOI] [PubMed] [Google Scholar]
- 29.Gaertig J, Wloga D. Ciliary tubulin and its post-translational modifications. Curr Top Dev Biol. 2008;85:83–113. doi: 10.1016/S0070-2153(08)00804-1. [DOI] [PubMed] [Google Scholar]
- 30.Lacroix B, et al. Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J Cell Biol. 2010;189:945–954. doi: 10.1083/jcb.201001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonnet C, et al. Differential binding regulation of microtubule-associated proteins MAP1A, MAP1B, and MAP2 by tubulin polyglutamylation. J Biol Chem. 2001;276:12839–12848. doi: 10.1074/jbc.M011380200. [DOI] [PubMed] [Google Scholar]
- 32.Fulton C, Simpson PA. Cell Motil. Vol. 3. Cold Spring Harbor: 1976. pp. 987–1005. [Google Scholar]
- 33.Verhey KJ, Gaertig J. The tubulin code. Cell Cycle Georget Tex. 2007;6:2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- 34.Serrano L, Avila J, Maccioni RB. Controlled proteolysis of tubulin by subtilisin: localization of the site for MAP2 interaction. Biochemistry (Mosc) 1984;23:4675–4681. doi: 10.1021/bi00315a024. [DOI] [PubMed] [Google Scholar]
- 35.Redeker V, Melki R, Promé D, Le Caer JP, Rossier J. Structure of tubulin C-terminal domain obtained by subtilisin treatment. The major alpha and beta tubulin isotypes from pig brain are glutamylated. FEBS Lett. 1992;313:185–192. doi: 10.1016/0014-5793(92)81441-n. [DOI] [PubMed] [Google Scholar]
- 36.Lobert S, Hennington BS, Correia JJ. Multiple sites for subtilisin cleavage of tubulin: effects of divalent cations. Cell Motil Cytoskeleton. 1993;25:282–297. doi: 10.1002/cm.970250308. [DOI] [PubMed] [Google Scholar]
- 37.Johnson V, Ayaz P, Huddleston P, Rice LM. Design, overexpression, and purification of polymerization-blocked yeast αβ-tubulin mutants. Biochemistry (Mosc) 2011;50:8636–8644. doi: 10.1021/bi2005174. [DOI] [PubMed] [Google Scholar]
- 38.Sindelar CV, Downing KH. An atomic-level mechanism for activation of the kinesin molecular motors. Proc Natl Acad Sci U S A. 2010;107:4111–4116. doi: 10.1073/pnas.0911208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Redwine WB, et al. Structural basis for microtubule binding and release by dynein. Science. 2012;337:1532–1536. doi: 10.1126/science.1224151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gigant B, et al. Structure of a kinesin-tubulin complex and implications for kinesin motility. Nat Struct Mol Biol. 2013;20:1001–1007. doi: 10.1038/nsmb.2624. [DOI] [PubMed] [Google Scholar]
- 41.Thorn KS, Ubersax JA, Vale RD. Engineering the processive run length of the kinesin motor. J Cell Biol. 2000;151:1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 43.Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reck-Peterson SL, et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trokter M, Mücke N, Surrey T. Reconstitution of the human cytoplasmic dynein complex. Proc Natl Acad Sci U S A. 2012;109:20895–20900. doi: 10.1073/pnas.1210573110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Sheetz MP. The C-terminus of tubulin increases cytoplasmic dynein and kinesin processivity. Biophys J. 2000;78:1955–1964. doi: 10.1016/S0006-3495(00)76743-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooper JR, Wagenbach M, Asbury CL, Wordeman L. Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat Struct Mol Biol. 2010;17:77–82. doi: 10.1038/nsmb.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hertzer KM, Walczak CE. The C-termini of tubulin and the specific geometry of tubulin substrates influence the depolymerization activity of MCAK. Cell Cycle Georget Tex. 2008;7:2727–2737. doi: 10.4161/cc.7.17.6590. [DOI] [PubMed] [Google Scholar]
- 49.Rüdiger M, Plessman U, Klöppel KD, Wehland J, Weber K. Class II tubulin, the major brain beta tubulin isotype is polyglutamylated on glutamic acid residue 435. FEBS Lett. 1992;308:101–105. doi: 10.1016/0014-5793(92)81061-p. [DOI] [PubMed] [Google Scholar]
- 50.Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- 51.Okada Y, Hirokawa N. Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc Natl Acad Sci U S A. 2000;97:640–645. doi: 10.1073/pnas.97.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lakämper S, Meyhöfer E. The E-hook of tubulin interacts with kinesin’s head to increase processivity and speed. Biophys J. 2005;89:3223–3234. doi: 10.1529/biophysj.104.057505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skiniotis G, et al. Modulation of kinesin binding by the C-termini of tubulin. EMBO J. 2004;23:989–999. doi: 10.1038/sj.emboj.7600118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Derr ND, et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science. 2012;338:662–665. doi: 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rai AK, Rai A, Ramaiya AJ, Jha R, Mallik R. Molecular adaptations allow dynein to generate large collective forces inside cells. Cell. 2013;152:172–182. doi: 10.1016/j.cell.2012.11.044. [DOI] [PubMed] [Google Scholar]
- 56.O’Hagan R, et al. The tubulin deglutamylase CCPP-1 regulates the function and stability of sensory cilia in C. elegans. Curr Biol CB. 2011;21:1685–1694. doi: 10.1016/j.cub.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rogowski K, et al. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 2010;143:564–578. doi: 10.1016/j.cell.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 58.Imanishi M, Endres NF, Gennerich A, Vale RD. Autoinhibition regulates the motility of the C. elegans intraflagellar transport motor OSM-3. J Cell Biol. 2006;174:931–937. doi: 10.1083/jcb.200605179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J Cell Biol. 2010;189:1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schatz PJ, Georges GE, Solomon F, Botstein D. Insertions of up to 17 amino acids into a region of alpha-tubulin do not disrupt function in vivo. Mol Cell Biol. 1987;7:3799–3805. doi: 10.1128/mcb.7.10.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uchimura S, et al. Identification of a strong binding site for kinesin on the microtubule using mutant analysis of tubulin. EMBO J. 2006;25:5932–5941. doi: 10.1038/sj.emboj.7601442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomishige M, Vale RD. Controlling kinesin by reversible disulfide cross-linking. Identifying the motility-producing conformational change. J Cell Biol. 2000;151:1081–1092. doi: 10.1083/jcb.151.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 65.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μManager. Curr Protoc Mol Biol Ed Frederick M Ausubel Al. 2010;Chapter 14(Unit14.20) doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.