Abstract
We report a single-cell bisulfite sequencing method (scBS-Seq) capable of accurately measuring DNA methylation at up to 48.4% of CpGs. We observed that ESCs grown in serum or 2i both display epigenetic heterogeneity, with “2i-like” cells present in serum cultures. In silico integration of 12 individual mouse oocyte datasets largely recapitulates the whole DNA methylome, making scBS-Seq a versatile tool to explore DNA methylation in rare cells and heterogeneous populations.
DNA methylation (5mC) is an epigenetic mark with critical roles in regulation and maintenance of cell-type-specific transcriptional programs1,2. Our understanding of 5mC functionality has been revolutionized by the development of bisulfite sequencing (BS-Seq), which offers single cytosine resolution and absolute quantification of 5mC levels genome-wide. Recent advances have demonstrated the power of single-cell sequencing analyses for the deconvolution of mixed cell populations3-5. Incorporation of epigenetic information into this single-cell arsenal will transform our understanding of gene regulation and reveal new insights into epigenetic heteogeneity6. Here, we report an accurate and reproducible method for single-cell genome-wide bisulfite sequencing (scBS-Seq) that allows assessment of 5mC heterogeneity within cell populations across the entire genome.
In commonly used BS-Seq protocols, sequencing adapters are first ligated to fragmented DNA and bisulfite conversion is performed, resulting in loss of information due to DNA degradation by bisulfite treatment. For scBS-Seq we used a modification of Post-Bisulfite Adaptor Tagging (PBAT)7, where bisulfite treatment is performed first, resulting in simultaneous DNA fragmentation and conversion of unmethylated cytosines (Fig. 1a). Then, complementary strand synthesis is primed using custom oligos containing Illumina adapter sequences and a 3′ stretch of nine random nucleotides. This step is performed five times to ensure that maximum numbers of DNA strands are tagged and to generate multiple copies of each fragment. After capture of the tagged strands, the second adapter is similarly integrated, and PCR amplification is performed with indexed primers allowing multiple single-cell libraries to be sequenced together.
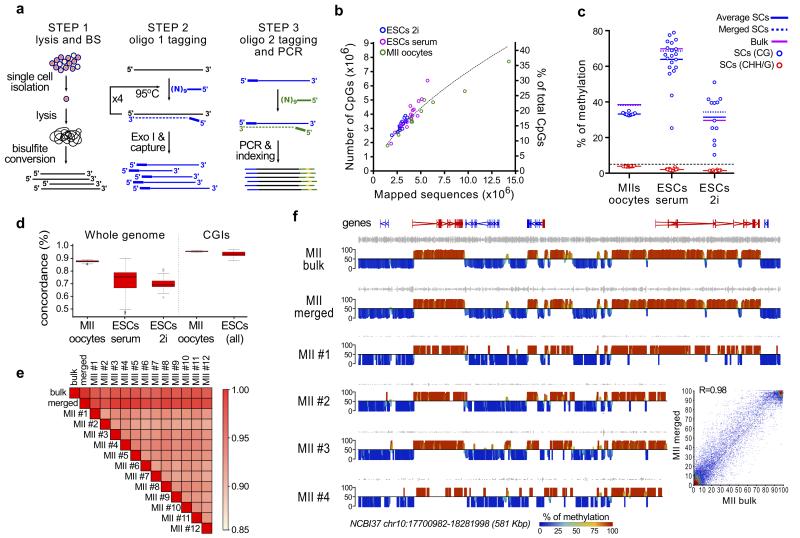
Figure 1. scBS-Seq is an accurate and reproducible method for genome-wide methylation analysis.
(a) scBS-Seq library preparation is performed in three stages: (1) single cells are isolated and lysed before bisulfite conversion is performed; (2) five rounds of random priming and extension are performed using oligo1 (which carries the first sequencing adaptor) and newly synthesized fragments are purified; (3) a second random priming and extension step is performed using oligo2 (which carries the second sequencing adaptor) and the resulting fragments are amplified by PCR. (b) Number of CpGs obtained by scBS-Seq correlates with the number of mapped sequences. (c) Global level of DNA methylation in a CpG and non-CpG context for single cells, in silico merged, and bulk samples. (d) Boxplot representation of the pairwise analysis of CpG concordance genome-wide and in unmethylated CGIs. Boxplots (plotted using the R package) represent the interquartile range, with the median. (e) Pairwise correlation matrix (Pearson’s; 2kb windows) for MII bulk, individual MIIs, and in silico merged MII scBSSeq datasets. (f) Screenshots showing CpG methylation (%) quantified over 2kb windows, with red indicating high methylation and blue low methylation. Data are displayed for four single MII libraries and the in silico merged dataset from all 12 MIIs (MII merged), which closely resemble the methylation landscape of the bulk MII sample. The inset shows the correlation between MII bulk and MII merged.
We performed scBS-Seq on metaphase-II (ovulated) oocytes (MIIs) and mouse embryonic stem cells (ESCs) cultured either in 2i (2i ESCs) or serum (serum ESCs) conditions. MIIs are an excellent model for technical assessment as they: i) can be individually handpicked ensuring only one cell is processed; ii) represent a highly homogeneous population allowing discrimination between technical and biological variability; and iii) present a distinct DNA methylome comprising large-scale hyper- and hypomethylated domains8. ESCs grown in serum conditions exist in a state of dynamic equilibrium characterized by transcriptional heterogeneity9-12, and emerging evidence from immunofluorescence and locus-specific studies has provided hints of 5mC heterogeneity in ESCs13. Recent studies have also demonstrated the remarkable plasticity of the ESC methylome, with genome-wide hypomethylation induced by inhibition of FGF signaling using two kinase inhibitors (2i)13,14. We use serum and 2i ESCs as a model to determine whether scBS-Seq can reveal DNA methylation heterogeneity at the single-cell level.
12 MII, 12 2i ESC, 20 serum ESC scBS-Seq libraries (and seven negative controls) and their bulk counterparts (i.e., pools of cells) were sequenced on the Illumina HiSeq platform (100bp paired-end), at a relatively low sequencing depth (average 19.4 million reads). On average, 3.9 million reads were mapped (1.5M-14.3M range), with an average efficiency of 24.6% (compared to 2.1% in negative controls) (Supplementary Fig. 1 and Supplementary Table 1). This relatively low mapping efficiency is mostly due to the presence of low-complexity sequences (Supplementary Fig. 2). We obtained methylation scores on an average of 3.7 million CpG dinucleotides (CpGs; 1.8M-7.7M range) corresponding to 17.7% of all CpGs (8.5-36.2% range) (Fig. 1b). Of importance, more CpGs can be obtained with deeper sequencing, as the limiting duplication plateau was not reached at this sequencing depth (Supplementary Fig. 3). To validate this, we sequenced two MII libraries close to saturation and with longer sequencing reads (150bp), resulting in a 1.5- and 1.9-fold increase in the number of CpGs measured (Supplementary Table 1). In addition, because of the broad size distribution of fragments in scBS-Seq libraries (Supplementary Fig. 1b), longer reads also resulted in an increase in CpGs covered (9% at saturating sequencing depth, 16% for low sequencing depth). Integrating this additional sequencing revealed that up to 10.1M CpGs (48.4% of all CpGs) can be obtained by scBS-Seq.
Next, we investigated the reproducibility and accuracy of our scBS-Seq approach. Low levels of non-CpG methylation across all samples revealed a minimum bisulfite conversion efficiency of 97.7% (or 98.5% by examining the mitochondrial chromosome in ESCs) (Fig. 1c and Supplementary Table 1). CpG sites in MIIs were overwhelmingly called methylated or unmethylated, consistent with a highly digitized output from single cells (Supplementary Fig.4). As expected, global methylation of MIIs was highly homogeneous (33.1±0.8%) and 2i ESCs were hypomethylated compared to serum ESCs13. Yet, strikingly, both 2i and serum ESCs exhibited 5mC heterogeneity (serum: 63.9±12.4%, 2i: 31.3±12.6%) (Fig. 1c). Global 5mC levels measured in individual MIIs were slightly lower than bulk (39.0%), but merging all MII datasets resulted in 38.8% global methylation. To assess scBS-Seq accuracy at CpG resolution, we calculated the pairwise concordance across single oocyte libraries and found an average of 87.6% genome-wide (85.3-88.9% range) and 95.7% in unmethylated CpG islands (CGIs), a highly homogeneous genomic feature, demonstrating the technical reproducibly of scBS-Seq (Fig. 1d). Of note, CpG concordance in ESCs was lower (serum: 72.7%, 2i: 69.8%), reflecting the heterogeneity of these cells (Fig. 1d and Supplementary Fig. 5). At lower genomic resolution (2kb windows), we observed high correlation between individual MIIs (on average R=0.92), and between individual MIIs and bulk (on average R=0.95) (Fig. 1e). In addition, for each MII, we obtained methylation information on an average of 61.5% of all CGIs (46.3-82.7% range); of 1,615 CGIs identified as methylated from bulk and informative in individual MIIs, ≥92% were called methylated by scBS-Seq, with ≤0.3% incorrectly called unmethylated (Supplementary Fig. 6).
While scBS-Seq mapped reads were distributed homogeneously across the genome, the enrichment towards exons, promoters and CGIs observed in bulk libraries was exaggerated in scBS-Seq libraries (Supplementary Fig. 7). Thus, scBS-Seq provides information on all genomic contexts, including regulatory regions (Supplementary Table 2). Yet, obtaining ~20% coverage of CpGs per cell means that recurrent information across samples is dependent on the nature of analytic units; conversely, in silico merging of individual datasets rapidly increases the number of CpGs with information (Supplementary Fig. 8). Strikingly, we were able to largely reproduce the entire 5mC landscape of oocytes using only 12 single cells (Fig. 1e,f and Supplementary Fig. 9). This capability is particularly beneficial for homogeneous cell populations, and makes our scBS-Seq approach an important tool to investigate the 5mC landscape from very rare material.
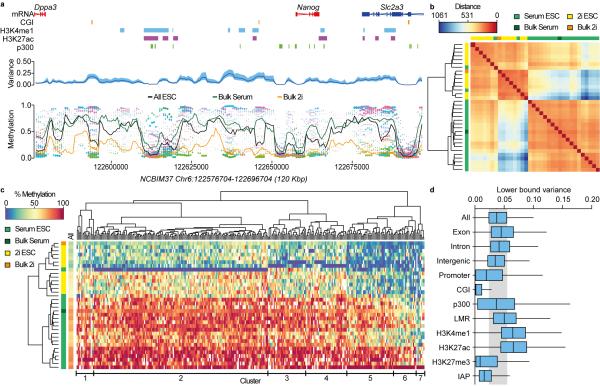
We next used scBS-Seq to explore 5mC heterogeneity in ESCs. A 3kb sliding window was used to estimate the methylation rate across the genome of each ESC, as well as the mean methylation rate and variance across all ESCs (Fig. 2a). Cells were clustered based on estimated methylation rates, while penalizing uncertainty in estimates due to low read counts. Two distinct clusters could be identified, representing the majority of 2i and serum ESCs (Fig. 2b). Intriguingly, outlier cells from the serum condition clustered with 2i ESCs, implying that serum cultures contain “2i-like” ESCs. This demonstrates the ability of scBS-Seq to identify rare cell-types within cell populations. To examine ESC heterogeneity in greater detail, we ranked sites by the estimated cell-to-cell variance and repeated the cluster analysis for the 300 most variable sites (Fig. 2c). The structure of the resulting clusters was grossly similar to that of the genome-wide analysis, and all 300 variable sites followed the global trend of being more highly methylated in serum than 2i ESCs with high similarity between sites (Fig. 1c, Fig. 2b,c, Supplementary Fig. 10 and Supplementary Fig. 11). This observation is consistent with the genomewide hypomethylation observed in 2i-grown ESCs13, and indicates that a major determinant of ESC heterogeneity is the global methylation level. Importantly, detailed analysis by scBS-Seq was also able to identify sites whose methylation varied more than the genome average, including sites with marked heterogeneity even among cells from the same growth condition (e.g. Clusters 5 and 6 in serum ESCs) (Fig. 2c). Regions containing H3K4me1 and H3K27ac, marks associated with active enhancers, had the greatest variance in 5mC, whereas CGIs and IAP elements had lower variance than the genome average (Fig. 2d and Supplementary Fig. 12). These findings are consistent with observations that distal regulatory elements are differentially methylated between tissues and throughout development15-17. Undoubtedly, further analysis will lead to the discovery of new genomic features with dynamic DNA methylation and regulatory function.
Figure 2. scBS-Seq reveals DNA methylation heterogeneity in ESCs.
(a) DNA methylation rates were estimated for each ESC using a sliding window across the genome (each cell is represented by a different color in the bottom panel, size of dot is the inverse of estimation error). The mean methylation rate across cells (black line in bottom panel) and the cell-to-cell variance (blue line in middle panel, 95% confidence interval shaded in light blue) were also estimated. The methylation rates for Bulk serum (green line) and Bulk 2i (orange line) are superimposed in the bottom panel. The region shown as an example includes the Nanog locus with some annotated features. (b) Genome-wide cluster dendrogram and distance matrix for all ESCs and Bulk samples based on the estimated methylation rates. Distance refers to the weighted Euclidean norm between estimated methylation rates. (c) Heatmap for methylation rates of the top 300 most variable sites among single-cell ESC samples. Cluster dendrograms for samples (left) and sites (top) are shown. The genome-wide average methylation rate is displayed in the left track (‘All’). The main clusters of variable sites are indicated at the bottom. (d) Variance of sites located in different genomic contexts. Boxplots represent the interquartile range, with the median. The upper (lower) whiskers correspond to 1.5 times the interquartile range. The shaded gray region indicates the interquartile range for all genome-wide sites.
While this manuscript was in preparation, a single-cell reduced-representation bisulfite sequencing (scRRBS) method was reported18, based on the single-tube RRBS strategy we previously developed19. While scRRBS and scBS-Seq could be seen as complementary, currently our methodology provides, at equivalent sequencing depth, information on ~5-fold more CpGs and ~1.5-fold more CGIs (Supplementary Fig. 13). Future technological developments will undoubtedly allow information to be recovered from most genomic CpGs, the key being the ability to amplify DNA prior to bisulfite conversion. The ability to capture the DNA methylome from individual cells will be critical for a full understanding of early embryonic development, cancer progression and induced pluripotent stem cell (iPSC) generation.
In summary, our work provides a proof-of-principle that large-scale single-cell epigenetic analysis is achievable, and demonstrates that scBS-Seq is a unique and powerful approach to accurately measure 5mC across the genome of single cells and to reveal 5mC heterogeneity within cell populations.
ONLINE METHODS
Sample Collection
MII oocytes were collected from superovulated 4–5-week-old C57BL/6Babr mice, under a stereomicroscope, by mouth pipetting, and stored at −80°C. Prior to scBS-Seq, 2× oocyte lysis buffer (10mM Tris-Cl pH7.4, 2% SDS) and 0.5μl proteinase K were added (final volume 12μl) followed by incubation at 37°C for 1h. E14 ESCs were cultured in serum plus LIF or 2i plus LIF conditions as described previously13. The 2i ESCs had been maintained in this medium for 24 days and matched serum ESCs were cultured in parallel. Single ESCs were collected by FACS in 12μl of ESC lysis buffer (10mM Tris-Cl pH7.4, 0.6% SDS, 0.5μl proteinase K) using a BD Influx instrument in single cell 1 drop mode. ToPro-3 and Hoechst 33342 staining were used to select for live cells with low DNA content (i.e. in G0/G1). ESCs were incubated at 37°C for 1h and stored at −20°C until required for library preparation. Negative controls were either lysis buffer alone (“empty” tubes) or sorted BD Accudrop Beads, and were prepared and processed concomitantly with all single cell samples.
Single-Cell Library Preparation
Bisulfite conversion was performed on cell lysates using the Imprint DNA Modification Kit (Sigma) with the following modifications: all volumes were halved, and chemical denaturation was followed by incubation at 65°C for 90min, 95°C for 3min and 65°C for 20min. Purification was performed as described previously7, and DNA eluted in 10mM Tris-Cl (pH 8.5) and combined with 0.4mM dNTPs, 0.4μM oligo1 ([Btn]CTACACGACGCTCTTCCGATCTNNNNNNNNN) and 1× Blue Buffer (Sigma) (24μl final) before incubation at 65°C for 3min followed by 4°C pause. 50U of Klenow exo- (Sigma) were added and the samples incubated at 4°C for 5min, +1°C/15s to 37°C, 37°C for 30min. Samples were incubated at 95°C for 1min and transferred immediately to ice before addition of fresh oligo1 (10pmol), Klenow exo- (25U), and dNTPs (1nmol) in 2.5μl total. The samples were incubated at 4°C for 5min, +1°C/15s to 37°C, 37°C for 30min. This random priming and extension was repeated a further 3 times (5 rounds in total). Samples were then incubated with 40U exonuclease I (NEB) for 1h at 37°C before DNA was purified using 0.8× Agencourt Ampure XP beads (Beckman Coulter) according to the manufacturer’s guidelines. Samples were eluted in 10mM Tris-Cl (pH 8.5) and incubated with washed M-280 Streptavidin Dynabeads (Life Technologies) for 20min with rotation at room temperature. Beads were washed twice with 0.1N NaOH, and twice with 10mM Tris-Cl (pH 8.5) and re-suspended in 48μl of 0.4mM dNTPs, 0.4μM oligo2 (TGCTGAACCGCTCTTCCGATCTNNNNNNNNN) and 1× Blue Buffer. Samples were incubated at 95°C for 45s and transferred immediately to ice before addition of 100U Klenow exo- (Sigma) and incubation at 4°C for 5min, +1°C/15s to 37°C, 37°C for 90min. Beads were washed with 10mM Tris-Cl (pH 8.5) and resuspended in 50μl of 0.4mM dNTPs, 0.4μM PE1.0 forward primer (AATGATACGGCGACCACCGAGATCTACACTCTTTC-CCTACACGACGCTCTTCCGATCT), 0.4μM indexed iPCRTag reverse primer20, 1U KAPA HiFi HotStart DNA Polymerase (KAPA Biosystems) in 1× HiFi Fidelity Buffer. Libraries were then amplified by PCR as follows: 95°C 2min, 12-13 repeats of (94°C 80s, 65°C 30s, 72°C 30s), 72°C 3min, 4°C hold. Amplified libraries were purified using 0.8× Agencourt Ampure XP beads, according to the manufacturer’s guidelines, and were assessed for quality and quantity using High-Sensitivity DNA chips on the Agilent Bioanalyser, and the KAPA Library Quantification Kit for Illumina (KAPA Biosystems). Pools of 12-14 single cell libraries were prepared for 100bp paired-end sequencing on a HiSeq2500 in rapid-run mode (2 lanes/run).
Bulk Sample Library Preparation
Samples from bulk cell populations were prepared according to the protocol above, with some modifications. For the bulk oocyte sample, 120 MII oocytes were collected and lysed as described above. For ESC bulk cell samples, DNA was purified from cell pellets using the QIAamp micro kit (QIAGEN), according to the manufacturer’s instructions, and 50ng of purified DNA was used in the library preparation. One round of first strand synthesis was performed using 0.8mM dNTPs and 4μM oligo1, and second strand synthesis also used 0.8mM dNTPs and 4μM oligo2. Bulk cell libraries were amplified as above with 9-12 cycles of PCR.
Sequencing Data Processing and Data Analysis
Raw sequence reads were trimmed to remove the first 9 base pairs, adapter contamination and poor quality reads using Trim Galore (v0.3.5, www.bioinformatics.babraham.ac.uk/projects/trim_galore/, parameters: --clip_r1 9 --clip_r2 9 --paired). Due to the multiple rounds of random priming performed with oligo1, scBS-seq libraries are non-directional. Trimmed sequences were first mapped to the human genome (build GRCh37) using Bismark21 (v0.10.1; parameters: --pe, --bowtie2, --non_directional, -- unmapped), resulting in 1.4% mapping efficiency (0.2-13.2% range). Remaining sequences were mapped to the mouse genome (build NCBI37) in single-end mode (Bismark parameters: --bowtie2 -- non_directional). Methylation calls were extracted after duplicate sequences had been excluded. For oocyte bulk analysis, our MII bulk dataset was merged in silico with previously published datasets8 (DDBJ/GenBank/EMBL accession number DRA000570). Data visualization and analysis were performed using SeqMonk, custom R and Java scripts. For Figure 1c, CG methylation was calculated as the average of methylation for each CpG position, and non-CpG methylation was extracted from the Bismark reports. Trend line in Figure 1b was calculated using polynomial regression. Percentage of concordance was calculated as the percentage of CpGs presenting the same methylation call at the same genomic position across two cells. For correlation analysis (Pearson’s), 2kb windows were defined informative if at least 8 CpGs per window were sequenced. CGI annotation used is from CAP-Seq experiments22. Informative CGIs were defined if at least 10 CpGs per CGI were sequenced. Hyper-methylated and hypo-methylated CGIs were defined as ≥80% and ≤20% methylation respectively. Annotation for comparison of genomic contexts (Fig. 2d, Supplementary Fig. 12 and Supplementary Table 2) were extracted from previously published datasets15,23.
Statistical Analyses
Statistical analysis for estimating sample-specific methylation rates, estimating mean methylation rates and for clustering are detailed in Supplementary Note 1.
Supplementary Material
ACKNOWLEDGEMENTS
We thank K. Tabbada and the Welcome Trust Sanger Institute sequencing pipeline team for assistance with Illumina sequencing, R. Walker for assistance with FACS and T. Hore (Babraham Institute, Cambridge, UK) for provision of ESCs maintained in 2i and serum conditions. We thank T. Hore, J. Huang, I. Macaulay, S. Lorenz, M. Quail, T. Voet and H. Swerdlow for helpful discussions. This work was supported by the UK Biotechnology and Biological Sciences Research Council grant BB/J004499/1, UK Medical Research Council grant MR/K011332/1, Wellcome Trust award 095645/Z/11/Z, and EU FP7 EpiGeneSys and BLUEPRINT.
Footnotes
ACCESSION CODES Gene Expression Omnibus (GEO): GSE56879.
COMPETING FINANCIAL INTERESTS Wolf Reik is a consultant to Cambridge Epigenetix Ltd; the authors declare no other competing interests.
REFERENCES
- 1.Jones PA. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 2.Smith ZD, Meissner A. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 3.Jaitin DA, et al. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng Q, et al. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 5.Macaulay IC, Voet T. PLoS Genet. 2014;10:e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee HJ, et al. Cell Stem Cell. 2014;14:710–719. doi: 10.1016/j.stem.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miura F, et al. Nucleic Acids Res. 2012;40:e136. doi: 10.1093/nar/gks454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shirane K, et al. PLoS Genet. 2013;9:e1003439. doi: 10.1371/journal.pgen.1003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers I, et al. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 10.Islam S, et al. Nat. Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi K, et al. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torres-Padilla ME, Chambers I. Development. 2014;141:2173–2181. doi: 10.1242/dev.102624. [DOI] [PubMed] [Google Scholar]
- 13.Ficz G, et al. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Habibi E, et al. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Stadler MB, et al. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 16.Ziller MJ, et al. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hon GC, et al. Nat. Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo H, et al. Genome Research. 2013;23:2126–2135. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smallwood SA, et al. Nat. Genet. 2011;43:811–814. doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quail MA, et al. Nat. Methods. 2012;9:10–11. [Google Scholar]
- 21.Krueger F, Andrews SR. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Illingworth RS, et al. PLoS Genet. 2010;6:e1001134. doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Creyghton MP, et al. P.N.A.S. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, et al. PLoS Biol. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bock C, et al. Molecular Cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.