Abstract
Genes on the mammalian X chromosome are present in one copy in males and two copies in females. The complex mechanisms that regulate the X chromosome lead to evolutionary and physiological variability in gene expression between species, the sexes, individuals, developmental stages, tissues and cell types. In early development, delayed and incomplete X chromosome inactivation (XCI) in some species causes variability in gene expression. Additional diversity stems from escape from XCI and from mosaicism or XCI skewing in females. This causes sex-specific differences that manifest as differential gene expression and associated phenotypes. Furthermore, the complexity and diversity of X dosage regulation affect the severity of diseases caused by X-linked mutations.
Regulation of the X chromosome involves several mechanisms of dosage compensation that ensure balanced gene expression levels between the X chromosome and autosomes, and similar expression levels between the sexes. Such mechanisms have evolved in many organisms owing to the differentiation between the sex chromosomes. In many diploid species such as mammals, sex is determined by the sex chromosome complement: XY in males and XX in females. The sex chromosomes differentially evolved into a relatively large, gene-rich X chromosome and a small, gene-poor Y chromosome that degenerated owing to suppressed recombination to avoid abnormal transfer of the male determinant. In mammals, dosage compensation is achieved by increasing expression levels of dosage-sensitive X-linked genes (that is, X upregulation) in both sexes and by random silencing of one X chromosome (that is, X chromosome inactivation (XCI)) in females. These mechanisms are not absolute, and substantial variability in gene expression is thus observed between species, the sexes, individuals, developmental stages, tissues and cell types. Furthermore, the X chromosome contains a unique set of genes with sex-specific expression. A better definition of the functional content and patterns of regulation of the sex chromosomes will help to understand sex biases in gene expression.
This Review is based on surprising new findings about the gene content and regulation of the X chromosome, as well as new information on the onset and distribution of XCI. Here, we address the causes and consequences of variability in expression of X-linked genes. Recent advances in this research field have been possible owing to new methodologies to obtain complete DNA sequence data, to access early embryos from humans and other mammalian species for XCI analyses, and to develop systems to evaluate allele-specific gene expression and manipulate XCI in somatic cells and tissues.
We first briefly review our knowledge of the function of genes that are located on the mammalian X chromosome and focus on genes that are involved in reproduction. Second, we discuss dosage regulation of the active X chromosome, with an emphasis on regulatory mechanisms that affect subsets of dosage-sensitive genes. Third, we report the substantial diversity in the timing of XCI in mammalian species. Most of the molecular details of XCI onset have been elucidated in mice to facilitate studies in embryos. However, analyses in humans and other mammalian species show that, at the onset of XCI, the X chromosome is not as tightly silenced as it is in rodents, which causes differences between the sexes during development. Fourth, we show how next-generation sequencing approaches have helped to obtain complete profiles of allele-specific X-linked gene expression in both engineered mouse models and human systems. Such systems have generated maps of XCI and catalogues of genes that escape XCI (hereafter termed ‘escape genes’), which differ between species, tissues and individuals. The expression of these genes results in sex biases, which are only beginning to be recognized. Finally, we discuss the importance of considering the peculiar modes of X chromosome regulation in the interpretation of phenotypic effects caused by X-linked mutations. In particular, the role of mosaicism and XCI skewing in the manifestation of specific diseases is only starting to be investigated. Our goal is to clarify the salient features of the X chromosome that influence sex biases in health and disease.
Functional diversity of X-linked genes
To understand X chromosome regulation and function, it is important to first define the gene content of this chromosome. Indeed, the function of X-linked genes determines the constraints and consequences of dosage regulation. The sex chromosomes independently evolved from ordinary autosomes in diverse lineages. At least two adaptive and opportunistic features evolved in convergence: the emergence of dosage compensation and the functional specialization of the gene content of the sex chromosomes1,2. The well-known accumulation of male-beneficial genes on the Y chromosome is probably due to its sole transmission in males3. Intriguingly, the X chromosome also has a special complement of genes.
Male-specific genes
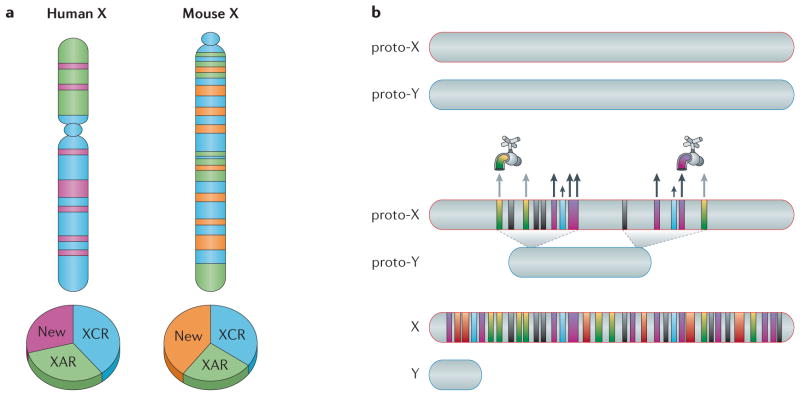
The accumulation of male-advantageous genes on the mammalian X chromosome can be explained by several theories, including sexual antagonism, X hemizygosity in males and female-biased X transmission (that is, a twofold transmission time in females relative to males)4. Most male-specific X-linked genes are predominantly or exclusively expressed in the testes at both premeiotic and postmeiotic stages5,6, which supports the sexual antagonism hypothesis7. In between these two stages, meiotic sex chromosome inactivation (MSCI) silences X-linked genes. To compensate for MSCI, functional pseudogene copies of essential housekeeping genes have been retrotransposed on autosomes8. Both protein-coding genes and microRNAs (miRNAs) that are expressed in the testes are enriched on the X chromosome9. Among these genes there is a marked preponderance of ampliconic genes (BOX 1; TABLE 1), which represent ~13% of X-linked genes in humans and 17% in mice6,10,11 (FIG. 1a; TABLE 1). The prevailing theory to explain this special feature is that the increase in copy number helps to regain normal gene expression levels in round spermatids, in which only a low level of postmeiotic X reactivation occurs6. Surprisingly, most ampliconic X-linked genes have been independently acquired in humans and mice11. By contrast, >94% of single-copy X-linked genes are conserved between these species11, and there are only few examples of single-copy genes (such as chloride channel 4-2 (Clcn4-2)) that are located on the X chromosome in some species but on an autosome in others12. As germ cell development and testicular function have similar features between humans and mice, the ampliconic X-linked genes probably have convergent and overlapping functions despite their diverse origin. Conversely, phenotypic differences in testicular function between species may be partly determined by independent acquisition of distinct gene families. Functional studies will help to characterize the new male-biased ampliconic gene sets. However, owing to the lack of conservation of these genes between species, it will be difficult to use model systems for such studies.
Box 1. Summary of different types of X-linked genes.
X-linked genes can be categorized on the basis of their copy number, evolutionary history, mode of X upregulation and X chromosome inactivation (XCI) status.
Copy number
X-linked genes are classified as single-copy, multicopy (that is, genes with ≥2 copies but that are not present in ampliconic regions) or ampliconic (that is, genes present in segmental duplications of >10 kb in length that share >99% nucleotide identity). Similar to autosomal genes, both single-copy and multicopy X-linked genes do not show a testis-biased expression pattern. By contrast, ampliconic X-linked genes are expressed predominantly in testicular germ cells11 (TABLE 1).
Evolutionary history
‘Old’ X-linked genes are those found on chicken orthologous autosomes 1 and 4, whereas ‘new’ X-linked genes are those that have been acquired during the evolution of the mammalian X chromosome. Genes within the X conserved region (XCR) have orthologues on the marsupial X chromosome, and genes within the X added region (XAR) have been added since the divergence between eutherian mammals and marsupials13. Most single-copy and multicopy X-linked genes are shared between humans and mice, whereas most ampliconic X-linked genes have been acquired independently in these mammalian species11 (FIG. 1a; TABLE 1).
Mode of X upregulation
Two main mechanisms of X upregulation have so far been described: a histone acetyltransferase KAT8-mediated mechanism to enhance transcription initiation and a mechanism that increases RNA half-life. Some genes are more sensitive to KAT8 regulation and some to regulation of RNA stability27. Genes implicated in networks and large protein complexes are dosage sensitive30 (FIG. 1b; TABLE 1).
XCI status
X-linked genes either undergo or escape XCI to a variable degree. Usually, a gene that escapes XCI has been defined as one that shows ≥10% expression from the inactive X allele compared with the active X allele76. Escape genes can be exclusively expressed from the inactive X chromosome, for example, X inactive specific transcript (XIST). Some escape genes are members of X–Y gene pairs with a paralogue on the Y chromosome, which may have the same function as the X paralogue. Other escape genes have lost their Y paralogue, or their Y paralogue has evolved a separate, often testis-specific, function66. Genes located in the pseudoautosomal regions have equivalent copies on the sex chromosomes (FIG. 3; TABLE 1).
Table 1.
Classification of X-linked protein-coding genes in relation to copy number, evolution, X upregulation and escape from XCI
| X-linked protein-coding genes | Copy number*
|
Evolutionary origin‡
|
Human–mouse comparison§
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Single | Multicopy | Ampliconic | ‘Old’ | ‘New’ | XCR | XAR | Shared | Independently acquired | |
| Human X-linked genes (n = 800) | 72% | 15% | 13% | 71% | 29% | 39% | 31% | 82% | 10% |
|
| |||||||||
| Mouse X-linked genes (n = 853) | 69% | 14% | 17% | 60% | 40% | 35% | 25% | 77% | 16% |
|
| |||||||||
| Percentage of genes per category | |||||||||
|
| |||||||||
| Human testis-specific genes|| | 11% | 38% | 84% | Low | High | Low | Low | 16% | 89% |
|
| |||||||||
| Mouse testis-specific genes|| | 16% | 23% | 69% | Low | High | Low | Low | 18% | 67% |
|
| |||||||||
| Genes upregulated by transcription initiation and/or RNA stability¶ | Yes | Yes | ND | Yes | Yes | Yes | Yes | Yes | ND |
|
| |||||||||
| Human escape genes (n = 518) | 15%# | ND | ND | 15%** | 12% | 5% | 27% | ND | 0% |
|
| |||||||||
| Mouse escape genes (n = 393) | 3%# | ND | ND | ND | ND | 2%** | 8% | ND | 0% |
ND, not determined; XAR, X added region; XCI, X chromosome inactivation; XCR, X conserved region.
Percentages of protein-coding X-linked genes that are classified as single-copy, multicopy or ampliconic in humans and in mice11.
Percentages of X-linked genes in each category based on evolutionary origin: old or new genes are those present or absent, respectively, on orthologous autosomes in chickens; genes in the XCR are subsets of old genes that are orthologous to genes on chicken autosome 1, whereas genes in the XAR are subsets of old genes that are orthologous to genes on chicken autosome 4. Percentages for human genes are obtained from REF. 13, and percentages for mouse genes are estimates on the basis of their human orthologues.
Percentages of X-linked genes that are shared between humans and mice or that are independently acquired after the human–mouse divergence11.
Percentages of genes predominantly expressed in the testes in each category of human and mouse X-linked genes11. If percentages were not available, ‘high’ and ‘low’ indicate the trend.
‘Yes’ indicates that the gene category is regulated by transcription initiation and/or RNA stability. Note that new genes are less affected by histone acetyltransferase KAT8 knockdown than old X-linked genes. However, transcripts of new genes have enhanced half-lives compared with transcripts of old genes27.
Percentage of X-linked genes that consistently escape XCI in human–rodent hybrid cell lines and in a mouse cell line69,76. The total number of genes assayed (n), excluding genes in the pseudoautosomal regions, is indicated in parentheses.
Percentage of escape genes in each category. Not all escape genes could be categorized as old, new, XCR, XAR or shared, as some escape genes were lost in mice. In humans, 336 genes could be categorized on the basis of REF. 13.
Figure 1. Variable gene content and dosage upregulation of the mammalian X chromosome.
a | Schematics of the human and mouse X chromosomes1,11 are shown. The X conserved region (XCR; blue), which occupies the long arm and a small region of the short arm of the human X chromosome, and the X added region (XAR; green) are indicated. In mice, the XAR and XCR are rearranged. Many of the newly acquired ampliconic genes differ between humans (pink) and mice (orange). Note that the position of these regions does not necessarily represent the position of the ampliconic genes. b | The proto-X and proto-Y chromosomes originally had similar sizes, and their expression level was balanced with that of autosomal genes (which are present in two copies). After the Y chromosome degenerated, hemizygous X-linked genes became upregulated. There is evidence of at least two feedforward mechanisms, which are schematically represented by the different colour faucets for upregulation of X-linked genes in mammals, including increased transcription (yellow and green; large grey arrows) and extended RNA half-life (pink and purple; large black arrows). Additional feedback and buffering mechanisms that respond to any type of aneuploidy may also be implicated in X upregulation (blue; small black arrows). Genes that represent dosage-insensitive genes (black) would not be upregulated. Note that some genes may be regulated by more than one mechanism (red and orange). Part b is adapted from REF. 128.
Female-specific genes
Mammalian X-linked genes have been grouped into two categories: ‘old’ genes that are conserved on chicken orthologous autosomes and ‘new’ genes that are absent on chicken orthologous autosomes13 (BOX 1; TABLE 1). In humans, the old group of X-linked genes is often female biased and has high expression levels in the ovaries compared with a control group of oldest autosomal genes14. Analyses of old conserved genes indicate that the mouse X chromosome is also enriched in female-biased genes that are expressed in the ovaries15. In female germ cells, a double dose of X-linked gene expression is observed following X reactivation that is mediated by the germ cell factor PR domain zinc-finger protein 14 (PRDM14)16,17. Increased expression of X-linked genes is essential for normal ovarian function. This is supported by the fact that individuals with Turner syndrome, who have a single X chromosome, show ovarian dysgenesis.
Brain-specific genes
In addition to genes that are important in reproduction, the mammalian X chromosome is also enriched in genes related to neurological function18. Many of the same genes that are expressed in the testes are also expressed in the brain, which suggests that sexual selection has a role in this enrichment19. Overall, X-linked genes are highly expressed in the brain20, and the proportion of X-linked genes expressed in the brain is significantly higher than that in other somatic tissues21. Consistently, X-linked forms of intellectual disability are 3.5 times more common than autosomal forms, and nearly 100 human X-linked genes have mutations in individuals with intellectual disability22,23. The effects of these mutations are variable in females owing to XCI skewing or escape from XCI (see below).
Diverse mechanisms of X upregulation
X upregulation consists of increased expression levels of genes on the single active X chromosome to balance expression with the autosomes (which are present in two copies). This phenomenon was demonstrated by analyses of several somatic tissues and embryonic stem cells (ESCs)21,24,25. Molecular studies have shown enrichment in RNA polymerase II and in histone marks that are indicative of active transcription, such as acetylation of histone H4 at lysine 16 (H4K16ac), at the 5′ promoter-proximal region of X-linked genes relative to autosomal genes, which would help to open chromatin structure and increase transcription levels26,27. Another mechanism of X upregulation apparently operates by enhancement of RNA stability, but the molecular causes of increased RNA half-life are not yet understood27,28. Interestingly, both old and new X-linked genes — as defined by their conservation in chickens (see above) — are regulated by these mechanisms but to different extents27 (BOX 1; TABLE 1). Whether the ampliconic X-linked genes that are exclusively expressed in the testes are also upregulated is not entirely clear owing to limited analyses of gene expression in this organ27. Evolutionary comparisons between mammalian X-linked genes and their autosomal homologues in chickens did not show evidence of global X upregulation, which may be limited to subsets of dosage-sensitive genes29. The highly specialized nature of the X chromosome gene content should be taken into account when evaluating dosage regulation. Products of X-linked genes that interact with those from autosomal genes in large protein complexes and networks would be expected to be more tightly regulated in terms of dosage30. Tissue specificity also has an important role: genes that are strictly expressed in male germ cells are silent in somatic tissues in which they are not upregulated27.
X-linked genes may be classifiable on the basis of the mechanisms that evolved to upregulate them. Indeed, a subset of 69 X-linked genes seem to be more sensitive to knockdown of histone acetyltransferase KAT8 (also known as MOF, which acetylates H4K16) in mouse ESCs. Among the KAT8-sensitive genes only 60% also show enhanced RNA half-life, which suggests that increased transcription levels and RNA stability have independently evolved to upregulate individual X-linked genes after they lost their Y copy27 (FIG. 1b; TABLE 1). The existence of differentially regulated subsets of X-linked genes suggests that X upregulation may have evolved gene by gene. In addition to feedforward mechanisms, X upregulation may also depend on basal feedback or buffering mechanisms that dampen the effects of any aneuploidy31,32 (FIG. 1b). Furthermore, it has been proposed that an inverse dosage effect similar to that described in Drosophila melanogaster33 may downregulate autosomal genes in mammals to balance their expression with X-linked genes29. Thus, several strategies may have evolved to balance expression of the genome, and some genes may be regulated by more than one mechanism (FIG. 1b). The finding of diverse mechanisms to achieve a balanced expression dosage raises the question: does variability in the mechanisms and effectiveness of dosage regulation of the X chromosome in relation to the rest of the genome have any effect on phenotypes in health or disease? Dysregulation of specific regulatory pathways may differentially affect specific subsets of X-linked genes, as exemplified by KAT8 knockdown in mouse ESCs27.
Variable XCI initiation and mosaic X expression
Most studies of XCI in early mammalian development have been carried out in mice, in which detailed analyses can be done. Surprisingly, XCI initiation in rodents turned out to be different from that in humans, rabbits and cows34. Although XCI initiation is tightly controlled throughout mouse development from the two-cell stage, this is not the case in other mammalian species. Thus, previously unsuspected levels of variability in X-linked gene expression occur in embryonic development before gonadal development. These early sex biases are poorly understood but slowly becoming recognized35.
Another important level of variability in X-linked gene expression relates to the randomness of XCI at its onset. A limited number of cells are present at the time of random XCI, which leads to measurable levels of unequal distribution of cells that have either the paternal or the maternal X chromosome inactivated in somatic tissues. Thus, female tissues are composed of patches of cells with different allele-specific expression patterns of X-linked genes36.
XCI initiation in mice
In mice, two different forms of XCI — imprinted and random — occur through regulation of the long non-coding RNAs (lncRNAs) inactive X specific transcripts (Xist) (FIG. 2). Extensive reviews of this process have recently been published37–39. XCI is initially paternally imprinted in preimplantation mouse embryos following upregulation of the paternal Xist allele at the two-cell stage by the maternally deposited X-linked activator E3 ubiquitin protein ligase RLIM, whereas the maternal X chromosome is protected by a repressive Xist imprint that is established in the female germ line. The antisense lncRNA Tsix also has an important role in Xist repression40,41. The paternal X chromosome remains inactive in extra-embryonic tissues and is reactivated in the inner cell mass (ICM) of the mouse mid-stage blastocyst42. XCI is then reinitiated by upregulating Xist from one randomly chosen X chromosome. Random XCI can be recapitulated in vitro upon differentiation of female mouse ESCs in systems that have greatly advanced our understanding of the molecular mechanisms of XCI, but such findings need to be confirmed in vivo. Interestingly, increased X dosage blocks differentiation of female ESCs43. Following XCI initiation Xist RNA spreads in cis44 and recruits repressive complexes to implement silencing through specific histone modifications, DNA methylation at CpG islands and changes in replication timing37. Additional proteins such as SMCHD1 — a member of the structural maintenance of chromosomes (SMC) family of proteins that is important in chromatin condensation — and repeat elements such as long interspersed nuclear elements (LINEs) help the formation of the condensed inactive X compartment39,45,46. The timing of onset of these modifications and of their associated silencing varies between genes, which would cause gene-specific sex bias in early development47.
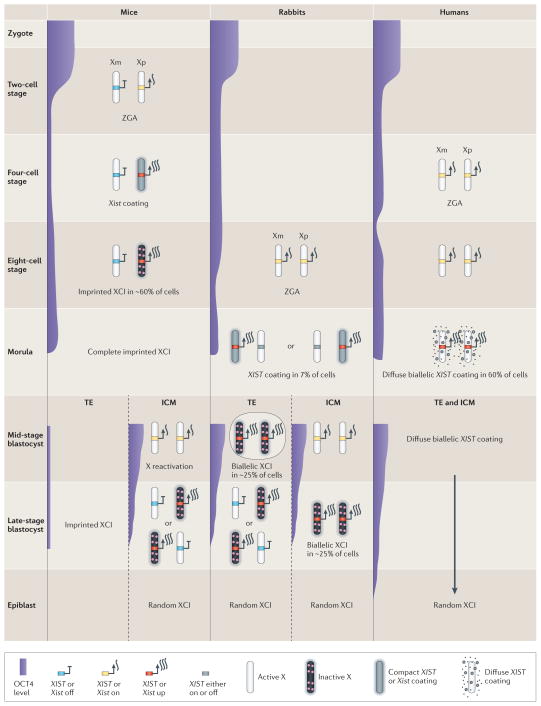
Figure 2. XCI initiation varies in mammals.
A comparison of X chromosome inactivation (XCI) in early embryonic development (that is, from zygote to epiblast) in three eutherian species (mice, rabbits and humans) is shown. XCI is mainly mediated by regulation of X inactive specific transcript (XIST) in eutherians; high expression levels of XIST start at zygotic genome activation (ZGA), and mice have the earliest onset of Xist expression. In mice, XCI is initially imprinted and triggered by exclusive Xist expression and coating of the paternal X chromosome (Xp) at the 2–4-cell stage. At the morula stage, imprinted XCI is completed. The Xp remains inactive in the trophectoderm (TE) that develops into extra-embryonic tissues such as the placenta, whereas Xp is reactivated in the inner cell mass (ICM) of the mid-stage blastocyst. XCI is then reinitiated by upregulating Xist from one randomly chosen X chromosome (that is, either the maternal X chromosome (Xm) or Xp) at the late-stage blastocyst and epiblast stage. In rabbits, XIST is not imprinted; thus, XIST upregulation and coating can occur on one X chromosome or both X chromosomes in some cells at the morula and early blastocyst stages followed by silencing. In late-stage blastocysts, cells with random monoallelic XCI in both the ICM and TE are selected by an unknown mechanism. In humans, XIST is also not imprinted, and diffuse XIST coating is visible on both X chromosomes but without initiation of silencing. Random XCI is initiated much later, probably after the blastocyst stage. Pluripotency regulatory factors such as octamer-binding protein 4 (OCT4) can repress XIST. OCT4 expression is very high in zygotes, decreases sharply at the two-cell stage and then gradually declines until ZGA, followed by an increase in preimplantation embryos. OCT4 levels remain high in both the ICM and TE in rabbit and human blastocysts but only in the ICM in mice. Data from REF. 34.
XCI initiation in other mammals
The idea that imprinted XCI is the primitive state of X silencing in mammals has been challenged by studies in rabbits and humans48 (FIG. 2). In these species XIST is not imprinted but is expressed from all X chromosomes in males and females at the 4–8-cell stage48,49. Surprisingly, some ICM cells show XIST RNA coating on both X chromosomes in early rabbit blastocysts, which is quickly resolved because most cells display only one inactive X chromosome in late blastocysts. In humans, the two X chromosomes remain active in trophectoderm and ICM cells even if there is XIST RNA coating, which suggests a late onset of XCI48. Lack of imprinted XCI is supported by studies in rabbit parthenogenotes, and in human and horse placenta34,50. By contrast, early X silencing seems to be essential in mice to quickly ensure correct dosage given fast embryonic development51. Comparisons between mice and humans with sex chromosome aneuploidy also highlight the importance of early paternal X silencing in mice: only XmXpY (where Xm denotes the maternal X chromosome and Xp the paternal X chromosome), but not XmXmY, karyotypes survive in rodents52, whereas equal percentages of these karyotypes are observed in humans53. Species-specific differences in expression patterns of the pluripotency factor octamer-binding protein 4 (OCT4), which acts as a XIST repressor, could mediate such adaptation. Whereas species with delayed implantation timing (for example, rabbits, pigs, rhesus monkeys and humans) show OCT4 expression in both the ICM and the trophectoderm to allow expansion of these layers, species with rapid implantation (for example, mice) lack OCT4 expression in trophoblasts, which are end cells adapted to immediate implantation54 (FIG. 2).
Random XCI in rabbits and humans may represent a stochastic process that is initiated by X-encoded activators such as RLIM34, followed by selection of cells with one active X chromosome against those with no or two active X chromosomes during development43,48. Interestingly, a stochastic XCI process can also be observed in mouse cells55, but it may be too slow to be adapted in rapidly developing extra-embryonic tissues. In the mouse ICM, random XCI is tightly controlled by monoallelic regulation of Xist mainly through the antisense Tsix, and such a mechanism may be lacking in other mammals56. Recently, another lncRNA — X active specific transcript (XACT) — was reported to coat the active X chromosome in human ESCs but not in mouse ESCs, which suggests a species-specific role in XCI57. Incomplete stochastic XCI could be an ancient compensation mechanism adapted from random monoallelic expression1,34. In marsupials, in which the X chromosome is homologous to the X conserved region (XCR) in eutherian mammals but lacks the X added region (XAR) (FIG. 1a), XCI is paternally imprinted58–61. Most strikingly, the XIST gene does not exist in marsupials62, but the marsupial-specific lncRNA RNA-on-the-silent X (RSX) has been shown to have XIST-like properties60. This finding indicates the extensive diversity of silencing mechanisms in mammals.
In conclusion, diverse regulatory mechanisms involved in XCI can quickly adapt to the species-specific modalities of embryonic development. Differences between the sexes in early stages of development will need to be clarified in view of XCI diversity between species. Even though XCI patterns in eutherian species apparently vary in early development, they eventually resolve to the silencing of one X chromosome in female somatic cells; nevertheless, XCI is ultimately less complete in humans than in mice (see below).
Mosaic X expression in female somatic tissues
Following random XCI initiation, tissues develop from variable numbers of progenitor cells, which leads to diverse patterns of mosaicism. This female-specific mosaicism has been comprehensively reviewed elsewhere36. The extent of patches of cells with a specific X allele expressed depends on whether gene expression is cell autonomous, on the number of progenitor cells at the time of silencing and on potential growth rate advantage of cells with a given X allele inactivated. The stochastic nature of the process can lead to marked differences in the distribution of expressed alleles even between the two halves of the brain63. Mosaicism for X allele expression has important consequences for the expression of phenotypes owing to X-linked mutations (see below).
Sex bias in gene expression
Most X-linked genes are subject to XCI in mammals, which results in similar expression levels between the sexes. However, a subset of X-linked genes escape silencing and thus have higher expression levels in females than in males. XIST is unusual, as it is completely female biased owing to exclusive expression from the inactive X chromosome in adult tissues. For other escape genes, expression from the inactive X allele is usually lower than that from the active X allele, which leads to a female bias of less than twofold in expression levels64. Some genes are present on both X and Y chromosomes: among these, genes located within the pseudoautosomal regions (PARs) have near equal expression levels between the sexes except for some male bias, which is probably due to partial spreading of XCI in females65. By contrast, X–Y gene pairs located outside the PARs often show a female bias for the X-linked paralogue owing to a lack of compensation by the Y-linked paralogue that has often acquired a testicular function66. Rare imprinted X-linked genes also display sexual dimorphisms, as males do not express maternally imprinted X-linked genes67.
Tissue-specific escape from XCI
Allele-specific expression analyses in mice indicate that 3–6% of X-linked genes consistently escape XCI in cell lines68,69 and in the brain63. Escape genes are dispersed along the mouse X chromosome, but most are in the XAR (FIG. 3; TABLE 1). These genes lack both Xist RNA coating and repressive chromatin marks44,69–71. Chromatin conformation analyses show that these genes apparently adopt a specific chromatin configuration with different domains interacting with each other, which suggests that escape genes may occupy a specific nuclear compartment71. Domains of silencing may be protected by insulator elements from encroachment by escape domains72,73. Furthermore, lncRNAs (the genes of which are often located adjacent to protein-coding genes) may facilitate their escape74,75.
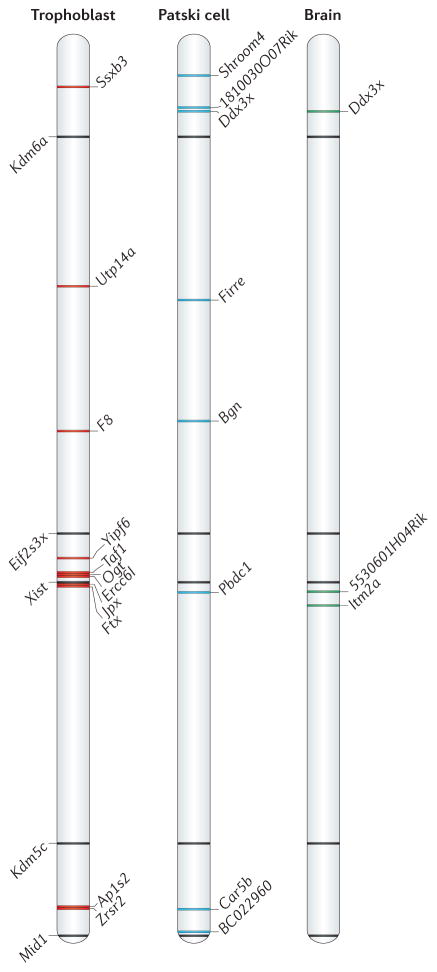
Figure 3. Escape from XCI varies between cell types and tissue in mice.
Schematics show the location of genes that escape X chromosome inactivation (XCI) along the mouse X chromosome on the basis of complete surveys of allele-specific gene expression by RNA sequencing in three systems: trophoblasts, Patski cells (which are derived from the embryonic kidney) and the brain63,68,69. The positions of five escape genes on the mouse X chromosome in all three cell types and tissue (black) are indicated, with their names shown at the left. Genes that escape XCI only in trophoblasts are shown in red, those only in Patski cells in blue and those only in the mouse brain in green; the gene names are indicated at the right of each chromosome. Only genes expressed from the inactive X chromosome at a level ≥10% of the active X chromosome in Patski cells (13 genes) and in the brain (8 genes), and genes with P < 0.05 in trophoblasts (16 genes; common in both reciprocal crosses) are included. Escape gene location is based on coordinates from University of California Santa Cruz (UCSC) genome browser build NCBI37/mm9.
In humans, escape genes are more abundant than those in mice and are concentrated within large domains in the evolutionarily recent XAR (TABLE 1). About 15% of human X-linked genes consistently escape XCI, and an additional 10% escape silencing in a variable manner in cell lines76. Whether this is representative of expression patterns in human tissues is still unclear, as only a few escape genes have been confirmed in vivo owing to technical difficulties in measuring allele-specific expression. However, indirect determination of the XCI status of genes on the basis of their epigenetic features suggests diversity between tissues. For example, on the basis of the finding that escape genes have markedly higher levels of intragenic non-CpG methylation in females than in males, new escape genes were identified specifically in neuronal cells77. Similarly, a lack of methylation at CpG islands has been exploited to identify putative escape genes in one tissue but not in others78. Variability in the degree of escape has been confirmed for some genes in vivo at the cell and whole-organism levels; for example, the TIMP metallopeptidase inhibitor 1 (TIMP1) gene has a propensity to escape silencing in some women owing to acetylation of histones that are associated with the gene79.
Escape from XCI in development
The XCI status of specific genes can change during development. Some genes that escape XCI in adult mouse tissues are initially silenced in embryos followed by reactivation during development80, whereas other genes escape XCI from the onset81. Whether specific human genes escape XCI at a given developmental stage is not well studied. Such information would be helpful to understand the diversity of developmental defects observed in patients with Turner syndrome. One study reports that in placental and extra-embryonic tissues, there is a wide range of individual differences for genes that are expressed either monoallelically or biallellically82. Further studies of the developmental plasticity of the domains of escape and XCI will help to better understand their role in health and disease.
Sex effects of escape from XCI
An extensive analysis of X-linked gene expression in hundreds of human lymphoblastoid cell lines determined that only 5% of X-linked genes have increased expression levels in females relative to males, which suggests a limited role for escape genes in eliciting sex biases in this in vitro system65. However, this may not be the case in vivo, as indicated by new studies in tissues. A recent RNA sequencing analysis showed significant sex-specific dimorphisms in gene expression, the extent of which differed between subregions of the brain83. Sex bias in X-linked gene expression is not limited to protein-coding genes, as some mouse miRNAs show a strong male bias in the brain and testes84. In some instances the sex bias can be due to imprinting, for example, in the case of the maternally imprinted Fthl17 (ferritin, heavy polypeptide-like 17) gene family85.
There is good evidence that transcriptional sex-specific differences exist before gonadal development in several species86. In fact, some sex biases can be detected as early as the blastocyst stage; for example, the X-linked reproductive homeobox genes Rhox6 and Rhox9 show a female bias that is already present in ESCs and that is later maintained in the ovaries87. Interestingly, expression of Rhox6 and Rhox9 is regulated by the histone demethylase KDM6A, which is encoded by an escape gene that has higher expression levels in female than in male mouse embryos and adults87. An early sex bias in gene expression has also been observed in preimplantation cow embryos88. Thus, sex-biased expression of some genes precedes the onset of gonadal differentiation, which indicates that such bias is independent of sex hormones.
Sex chromosome complement and sex bias
Studying the role of the sex chromosomes in eliciting sexual dimorphisms in gene expression in adult tissues is complicated by the potential interference of gonadal hormones. Useful mouse models have been developed to circumvent this problem. One such model — the four core genotype (4CG), in which XY and XX males, and XX and XY females are produced — has clearly shown that both the sex chromosome complement and gender have a role in phenotypic sex-specific differences89. For example, the number of X chromosomes can influence adiposity and cardiovascular sexual dimorphisms90,91. Using the 4CG mouse model, global gene expression differences that originate from the sex chromosomes have been shown to influence the entire genome, which suggests genome-wide effects92. Analyses of brain specimens collected before gonadal development in another mouse model that is based on a rearranged Y chromosome (Y*) have also identified genes with differential expression between mice with one or two X chromosomes93,94. Although the majority of these genes were X-linked, some differentially expressed genes were autosomal. This finding confirms that the sex chromosome complement influences the entire genome, which is also supported by expression analysis of somatic tissues from XX and XO mice95. Some of these genome-wide effects could potentially be attributed to female bias in expression of master regulators, for example, the histone demethylases KDM6A and KDM5C (both of which are encoded by escape genes)87,96,97. KDM6A also has a histone demethylase-independent function that is essential for normal development98. Other escape genes may also have chromatin-wide effects on the genome, which could explain specific sex biases, but these effects remain to be discovered.
X chromosome regulation and disease
The phenotypic manifestations of mutations on the X chromosome are greatly influenced by the modalities of dosage regulation of this particular chromosome (FIG. 4). Males are affected by both recessive and dominant mutations because they only have one allele. Duplications of X-linked genes have severe consequences in males owing to their inability to become inactivated99. In females, the manifestations of mutations in X-linked genes can either be dampened by random XCI that results in a population of cells that express the normal allele or even be obliterated by complete skewing of XCI36. Phenotypic effects also differ depending on whether the mutated gene is subject to XCI or escapes XCI (FIG. 4). Age-related X reactivation could further influence the diversity of phenotypes observed in diseases, for example, in neurological disorders and cancer100. A recent mapping of the topography of XCI in mouse tissues demonstrates important fluctuations in the extent and distribution of mosaicism63. Notably, this elegant study mapped allele-specific expression of X-linked genes using fluorescent markers and suggested that XCI has a role in stochastic diversity of gene expression in females. Females may also be susceptible to aberrant X reactivation in somatic cells. Here, we consider the interaction of dosage compensation with the phenotypic effects of X-linked mutations.
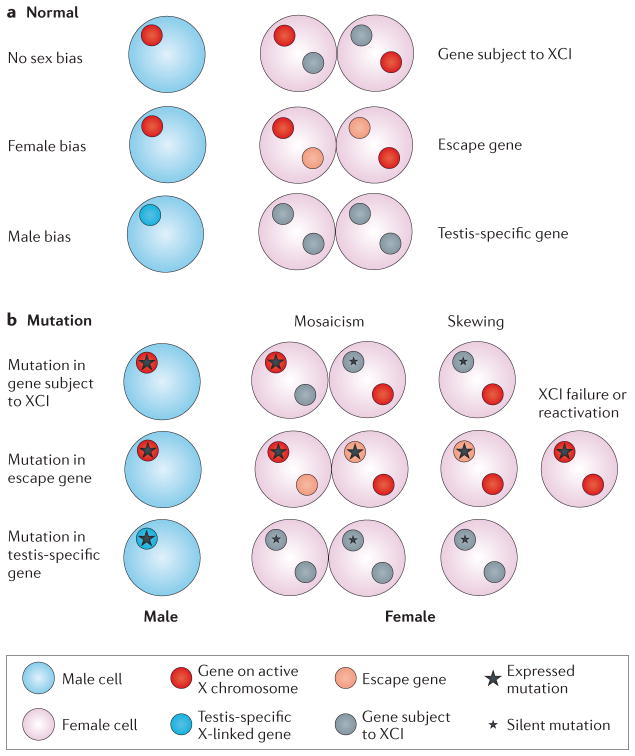
Figure 4. Variability of X-linked gene expression and sex bias.
a | Sex bias in X-linked gene expression in normal individuals is shown. There is no sex bias for genes that are subject to X chromosome inactivation (XCI) or for genes with equivalent X–Y paralogues (not shown). A female bias occurs for genes that escape XCI but that have no equivalent Y paralogue; a male bias occurs for genes that are solely expressed in male organs such as the testes. b | The effects of X-linked gene mutation depend on XCI patterns. For genes that are subject to XCI, a mutation that affects males does not necessarily affect females, who can be rescued either by random XCI (which leads to mosaicism for cells with the mutant allele silenced) or by selective skewing in favour of cells that express the normal allele. For escape genes, males can be partly rescued by expression of a Y paralogue with similar function (if it exists), whereas females will be haploinsufficient even if there is skewing in favour of the normal allele. Severity of the haploinsufficiency in females depends on the expression levels from the inactive X chromosome. Mutations in genes that are essential for XCI (such as the gene that encodes X inactive specific transcript (XIST)) specifically affect females by causing XCI failure. If a mutation is in an X-linked gene that is solely expressed in male organs (such as the testes), only males will be affected. However, as many of the testis-specific genes are ampliconic, the effect of a mutation in one copy will be partly rescued by the normal copies.
X-linked mutations and XCI skewing
XCI skewing in favour of cells in which the normal allele is active is often observed in female carriers of an X-linked mutation, which can alleviate the phenotypic outcome (FIG. 4). If the gene escapes XCI, skewing will only partially restore a normal phenotype36. For genes that are subject to XCI, protection from deleterious effects depends on the degree of skewing that is related to the extent of cell selection in specific tissues. Factors that influence XCI skewing include whether the mutated gene produces a cell autonomous protein or a secreted protein, and whether the protein product is part of a multimeric complex. XCI skewing can selectively occur in cell types in which expression of a normal allele is crucial, and the degree of skewing can also be related to developmental stage or age101. Innate XCI skewing can also occur as observed for mouse X chromosome controlling element (Xce) alleles that directly influence the choice of which X chromosome will be silenced102. In humans, progressive XCI skewing is suspected to occasionally have a deleterious effect by uncovering mutations, as proposed in autoimmune diseases that are prevalent in women103. Interestingly, age-related XCI skewing may be an indicator of clonal expansion in the bone marrow even in normal healthy individuals104. XCI skewing can certainly have a role in cancer; for example, expression of the breast tumour suppressor gene forkhead box P3 (FOXP3) is altered by skewing105.
Mutations in escape genes
Mutations, deletions and duplications of individual escape genes are known to cause a range of abnormal phenotypes, including cancer. Chromatin modifiers are of particular interest, as they can influence global gene expression through epigenetic changes. For example, mutations in the escape gene KDM6A have been implicated in medulloblastoma, prostate cancer and renal carcinoma106–108. Constitutional KDM6A mutations cause Kabuki syndrome, which is a rare congenital syndrome characterized by skeletal abnormalities, growth retardation and mild to severe intellectual disability109,110. Interestingly, heterozygous females are affected to various degrees, probably because of low expression levels from the normal allele in cells in which it is on the inactive X chromosome109,111. As individuals with Turner syndrome do not have Kabuki syndrome, monoallelic KDM6A expression is apparently sufficient to prevent this phenotype. Another escape gene associated with intellectual disability is KDM5C, which is important for neural cell development. In this case, only men are affected, which suggests that haploinsufficiency only has mild consequences in women112,113. Additional studies will help to better define dosage effects that are related to escape from XCI.
Sex chromosome aneuploidy
Abnormal phenotypes in individuals with sex chromosome aneuploidy are partly due to abnormal expression levels of escape genes. Effects on female and male fertility have been extensively discussed114. For example, 45,X females exhibit Turner syndrome, which is associated with severe phenotypes such as frequent death in utero, ovarian dysgenesis, short stature, webbed neck and other physical abnormalities115,116. As the Y chromosome protects men from these deficiencies, the most likely candidate genes for Turner syndrome would have copies on the Y chromosome except for genes that control female-specific phenotypes, such as those regulating ovarian function or XCI itself. Note that germ cells from patients with Turner syndrome have a deficiency in all X-linked genes and not only in escape genes, as the X chromosome is reactivated in normal ovaries114. Variability in the severity of phenotypes of Turner syndrome is thought to result, in large part, from mosaicism for XX or XY cells, which can help survival117.
A common type of sex chromosome aneuploidy can also affect men in the form of an additional X chromosome, for example, in Klinefelter’s syndrome. It is unclear whether the phenotypic features of Klinefelter’s syndrome are caused by altered hormonal dosage due to testicular malfunction or to increased expression levels of escape genes. One hypothesis is that male physiology is not adapted to a female bias in gene dosage, which would contribute to the clinical manifestations. To address this hypothesis a recent study analysed a mouse model of Klinefelter’s syndrome, which confirmed that genes known to escape XCI had significantly higher expression levels in the brain (but not in the kidney and liver) of mice with two X chromosomes (XX and XXY) than male controls118. These brain-specific differences suggest that dosage of escape genes contributes to the cognitive phenotypes of patients with Klinefelter’s syndrome. The variable extent of adverse phenotypes associated with Klinefelter’s syndrome and X aneuploidy in general is partly due to XCI skewing and mosaicism119. In addition, X aneuploidy could indirectly alter gene expression genome wide through epigenetic effects; for example, the presence of an extra heterochromatic chromosome might act as a sink for specific protein complexes120.
Diseases associated with somatic X reactivation
The lncRNA XIST was initially thought to be dispensable for maintenance of silencing121. However, a new study shows that ablation of Xist in the blood compartment causes an aggressive form of cancer in female mice122. The authors hypothesize that upregulation of X-linked genes due to the loss of Xist leads to genome-wide changes in important homeostatic pathways. Mutations in Smchd1, which is also associated with cancer in mice123, lead to X reactivation partly as a result of hypomethylation of the CpG islands of genes that are normally inactivated46. Conversely, reactivation of Xist in immune cells can induce gene silencing of the mouse X chromosome124. In humans, abnormalities of XCI have also been reported in cancer100. Age-related reactivation of specific genes has been occasionally reported, for example, in the case of the ornithine transcarbamylase (Otc) gene125. Furthermore, reactivation of the CD40 ligand (CD40LG) gene by hypomethylation has been proposed as a mechanism that increases protein levels and that contributes to systemic lupus erythematosus126. Interestingly, this disease shows a strong female bias and is also common in patients with Klinefelter’s syndrome. Genes that confer susceptibility to immune disorders in women are only beginning to be identified, and it is expected that X-linked genes will be shown to have an important role in such disordes127.
Conclusions and perspectives
In this Review, we have considered specific aspects of the gene content and regulation of the mammalian X chromosome that lead to substantial differences both between species and between the sexes at the whole organism, tissue and cellular levels. Important questions have been raised, notably regarding the function of the newly identified families of X-linked genes that are expressed in the testes and that are poorly conserved between species. Other unanswered questions include: how early do sex-specific differences in X-linked gene expression manifest themselves, and are these important for phenotypes? Further studies of X-linked gene expression in early human embryos are needed to understand their role in health and disease, for example, in terms of effects associated with sex chromosome aneuploidy. In particular, single-cell analyses using next-generation sequencing hold great promise in clarifying patterns of X-linked gene expression and mosaicism. With the availability of single-cell analyses, systems for skewing XCI or for sorting cells are not necessary, although methods to identify alleles on the basis of frequent single-nucleotide polymorphisms (SNPs) are still needed. Such approaches will provide a better determination of the extent of escape from XCI in specific tissues and developmental stages, which is still lacking to fully understand their role in sexually dimorphic phenotypes. Direct or indirect epigenetic effects mediated by X-linked genes are only beginning to be studied in cancer. Currently, disruptions of XCI but not of X upregulation have been identified. Further molecular analyses of X upregulation will help to understand its potential roles in diseases.
The complexity of dosage regulation of the X chromosome in terms of molecular mechanisms of X upregulation and XCI renders these systems susceptible to a range of genetic or epigenetic mutations with strong sex bias, which can have tremendous advantages for the evolution of new traits, especially those related to sexual reproduction. However, the X chromosome is also vulnerable to mutations that are differentially expressed in males and females. A sizeable sex bias due to differences in the dosage of X-linked genes is becoming recognized as an important contributor to the susceptibility to specific diseases.
Acknowledgments
This work was supported by grants GM046883, GM098039, MH083949, and MH099628 from the US National Institutes of Health.
Glossary
- X upregulation
A process to increase X-linked gene expression levels to balance expression between the X chromosome and autosomes
- X chromosome inactivation
(XCI). A process to silence one X chromosome in females to achieve similar expression levels between the sexes
- Sex biases in gene expression
Differential gene expression between males and females
- Allele-specific gene expression
Specific expression from the maternal or paternal allele that can be assayed by single-nucleotide polymorphisms
- Genes that escape XCI
A subset of X-linked genes that are biallelically expressed in female somatic cells
- XCI skewing
Preferential inactivation of one X chromosome
- Sexual antagonism
Evolution of alleles that are beneficial to one sex but detrimental to the other
- Meiotic sex chromosome inactivation
(MSCI). A process to silence both X-linked and Y-linked genes during male meiosis
- X reactivation
A process to reactivate the silenced X chromosome at different developmental stages (for example, in the mouse inner cell mass and in female germ cells)
- Turner syndrome
Syndrome in females with a single X chromosome (that is, 45,X)
- XCI initiation
Onset of chromosome-wide X-linked gene silencing that is controlled by a master regulatory locus known as the X inactivation centre
- Inactive X specific transcripts
(Xist). Long non-coding RNAs transcribed from the X inactivation centre that function as the master regulator of mouse X chromosome inactivation
- X conserved region
(XCR). A region on the mammalian X chromosome that is found on chicken orthologous autosome 4 and on the marsupial X chromosome
- X added region
(XAR). A region on the mammalian X chromosome that is found on chicken orthologous autosome 1 and that was added since the divergence between eutherian mammals and marsupials
- Mosaicism for X allele expression
A mixture of somatic cells that carry an active paternal or maternal X chromosome owing to random X chromosome inactivation
- Pseudoautosomal regions
(PARs). Regions of homology and pairing on the sex chromosomes
- X–Y gene pairs
Genes with paralogues located on the X and Y chromosomes
- X chromosome controlling element
(Xce). A locus at the X inactivation centre that controls the choice of a particular X chromosome to be inactivated or to remain active
- Haploinsufficiency
Insufficiency due to the presence of a single copy instead of two copies of a gene in a diploid cell
- Klinefelter’s syndrome
A syndrome in males that carry an additional X chromosome (that is, 47,XXY)
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Livernois AM, Graves JA, Waters PD. The origin and evolution of vertebrate sex chromosomes and dosage compensation. Hered (Edinb) 2012;108:50–58. doi: 10.1038/hdy.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Disteche CM. Dosage compensation of the sex chromosomes. Annu Rev Genet. 2012;46:537–560. doi: 10.1146/annurev-genet-110711-155454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachtrog D. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nature Rev Genet. 2013;14:113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurbich TA, Bachtrog D. Gene content evolution on the X chromosome. Curr Opin Genet Dev. 2008;18:493–498. doi: 10.1016/j.gde.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nature Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 6.Mueller JL, et al. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nature Genet. 2008;40:794–799. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice WR. Sexually antagonistic genes: experimental evidence. Science. 1992;256:1436–1439. doi: 10.1126/science.1604317. [DOI] [PubMed] [Google Scholar]
- 8.Adra CN, Ellis NA, McBurney MW. The family of mouse phosphoglycerate kinase genes and pseudogenes. Somat Cell Mol Genet. 1988;14:69–81. doi: 10.1007/BF01535050. [DOI] [PubMed] [Google Scholar]
- 9.Ro S, Park C, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed microRNAs. Dev Biol. 2007;311:592–602. doi: 10.1016/j.ydbio.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross MT, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller JL, et al. Independent specialization of the human and mouse X chromosomes for the male germ line. Nature Genet. 2013;45:1083–1087. doi: 10.1038/ng.2705. This paper shows that many ampliconic X-linked genes are expressed predominantly in testicular germ cells and were independently acquired since the divergence between humans and mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rugarli EI, et al. Different chromosomal localization of the Clcn4 gene in Mus spretus and C57BL/6J mice. Nature Genet. 1995;10:466–471. doi: 10.1038/ng0895-466. [DOI] [PubMed] [Google Scholar]
- 13.Bellott DW, et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature. 2010;466:612–616. doi: 10.1038/nature09172. This study comprehensively compares the chicken Z and human X chromosomes, and shows that they share common features such as acquisition and amplification of testis-specific genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang YE, Vibranovski MD, Landback P, Marais GA, Long M. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 2010;8:e1000494. doi: 10.1371/journal.pbio.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nature Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- 16.Ohhata T, Wutz A. Reactivation of the inactive X chromosome in development and reprogramming. Cell Mol Life Sci. 2012;70:2443–2461. doi: 10.1007/s00018-012-1174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payer B, et al. Tsix RNA and the germline factor, PRDM14, link X reactivation and stem cell reprogramming. Mol Cell. 2013;52:805–818. doi: 10.1016/j.molcel.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zechner U, et al. A high density of X-linked genes for general cognitive ability: a run-away process shaping human evolution? Trends Genet. 2001;17:697–701. doi: 10.1016/s0168-9525(01)02446-5. [DOI] [PubMed] [Google Scholar]
- 19.Marshall Graves JA. The rise and fall of SRY. Trends Genet. 2002;18:259–264. doi: 10.1016/s0168-9525(02)02666-5. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen DK, Disteche CM. High expression of the mammalian X chromosome in brain. Brain Res. 2006;1126:46–49. doi: 10.1016/j.brainres.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 21.Deng X, et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nature Genet. 2011;43:1179–1185. doi: 10.1038/ng.948. This paper confirms that there is dosage compensation of gene expression between the X chromosome and autosomes in both sexes of mammals, nematodes and fruitflies, and proposes the need to take into consideration the skewed gene content and regulation of the X chromosome in the study of dosage compensation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skuse DH. X-linked genes and mental functioning. Hum Mol Genet. 2005;14 (suppl 1):R27–R32. doi: 10.1093/hmg/ddi112. [DOI] [PubMed] [Google Scholar]
- 23.Ropers HH. Genetics of early onset cognitive impairment. Annu Rev Genom Hum Genet. 2010;11:161–187. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nature Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 25.Lin H, et al. Relative overexpression of X-linked genes in mouse embryonic stem cells is consistent with Ohno’s hypothesis. Nature Genet. 2011;43:1169–1170. doi: 10.1038/ng.992. [DOI] [PubMed] [Google Scholar]
- 26.Yildirim E, Sadreyev RI, Pinter SF, Lee JT. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nature Struct Mol Biol. 2011;19:56–61. doi: 10.1038/nsmb.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng X, et al. Mammalian X upregulation is associated with enhanced transcription initiation, RNA half-life, and MOF-mediated H4K16 acetylation. Dev Cell. 2013;25:55–68. doi: 10.1016/j.devcel.2013.01.028. This study shows that subsets of mammalian X-linked genes rely on different X upregulation mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin S, et al. Evidence that the nonsense-mediated mRNA decay pathway participates in X chromosome dosage compensation in mammals. Biochem Biophys Res Commun. 2009;383:378–382. doi: 10.1016/j.bbrc.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 29.Julien P, et al. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 2012;10:e1001328. doi: 10.1371/journal.pbio.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GA. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci USA. 2012;109:5346–5351. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 2010;8:e1000320. doi: 10.1371/journal.pbio.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prestel M, Feller C, Becker PB. Dosage compensation and the global re-balancing of aneuploid genomes. Genome Biol. 2010;11:216. doi: 10.1186/gb-2010-11-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birchler JA, Pal-Bhadra M, Bhadra U. Dosage dependent gene regulation and the compensation of the X chromosome in Drosophila males. Genetica. 2003;117:179–190. doi: 10.1023/a:1022935927763. [DOI] [PubMed] [Google Scholar]
- 34.Escamilla-Del-Arenal M, da Rocha ST, Heard E. Evolutionary diversity and developmental regulation of X-chromosome inactivation. Hum Genet. 2011;130:307–327. doi: 10.1007/s00439-011-1029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold AP. The end of gonad-centric sex determination in mammals. Trends Genet. 2011;28:55–61. doi: 10.1016/j.tig.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Migeon BR. Females are Mosaic: X Inactivation and Sex Differences in Disease. Oxford Univ. Press; 2014. [DOI] [PubMed] [Google Scholar]
- 37.Lessing D, Lee JT. X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu Rev Genom Hum Genet. 2013;14:85–110. doi: 10.1146/annurev-genom-091212-153530. [DOI] [PubMed] [Google Scholar]
- 38.Morey C, Avner P. The demoiselle of X-inactivation: 50 years old and as trendy and mesmerising as ever. PLoS Genet. 2011;7:e1002212. doi: 10.1371/journal.pgen.1002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pollex T, Heard E. Recent advances in X-chromosome inactivation research. Curr Opin Cell Biol. 2013;24:825–832. doi: 10.1016/j.ceb.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 40.Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128:1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- 41.Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103:17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 42.Mak W, et al. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- 43.Schulz EG, et al. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell. 2014;14:203–216. doi: 10.1016/j.stem.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 44.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nozawa RS, et al. Human inactive X chromosome is compacted through a PRC2-independent SMCHD1–HBiX1 pathway. Nature Struct Mol Biol. 2013;20:566–573. doi: 10.1038/nsmb.2532. [DOI] [PubMed] [Google Scholar]
- 46.Gendrel AV, et al. Epigenetic functions of Smchd1 repress gene clusters on the inactive X chromosome and on autosomes. Mol Cell Biol. 2013;33:3150–3165. doi: 10.1128/MCB.00145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gendrel AV, et al. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev Cell. 2012;23:265–279. doi: 10.1016/j.devcel.2012.06.011. This study shows that X-linked genes are silenced at different times during development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okamoto I, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. doi: 10.1038/nature09872. This paper is the first study of XCI in rabbit and human early embryos, and shows that these species have different strategies to initiate XCI compared with mice. [DOI] [PubMed] [Google Scholar]
- 49.van den Berg IM, et al. X chromosome inactivation is initiated in human preimplantation embryos. Am J Hum Genet. 2009;84:771–779. doi: 10.1016/j.ajhg.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Miller DC, Clark AG, Antczak DF. Random X inactivation in the mule and horse placenta. Genome Res. 2012;22:1855–1863. doi: 10.1101/gr.138487.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mugford JW, Yee D, Magnuson T. Failure of extra-embryonic progenitor maintenance in the absence of dosage compensation. Development. 2012;139:2130–2138. doi: 10.1242/dev.076497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russell LB. Genetics of mammalian sex chromosomes. Science. 1961;133:1795–1803. doi: 10.1126/science.133.3467.1795. [DOI] [PubMed] [Google Scholar]
- 53.Thomas NS, Hassold TJ. Aberrant recombination and the origin of Klinefelter syndrome. Hum Reprod Update. 2003;9:309–317. doi: 10.1093/humupd/dmg028. [DOI] [PubMed] [Google Scholar]
- 54.Kirchhof N, et al. Expression pattern of Oct-4 in preimplantation embryos of different species. Biol Reprod. 2000;63:1698–1705. doi: 10.1095/biolreprod63.6.1698. [DOI] [PubMed] [Google Scholar]
- 55.Monkhorst K, Jonkers I, Rentmeester E, Grosveld F, Gribnau J. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell. 2008;132:410–421. doi: 10.1016/j.cell.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 56.Chang SC, Brown CJ. Identification of regulatory elements flanking human XIST reveals species differences. BMC Mol Biol. 2010;11:20. doi: 10.1186/1471-2199-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vallot C, et al. XACT, a long noncoding transcript coating the active X chromosome in human pluripotent cells. Nature Genet. 2013;45:239–241. doi: 10.1038/ng.2530. [DOI] [PubMed] [Google Scholar]
- 58.Graves JA. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 59.Al Nadaf S, et al. Activity map of the tammar X chromosome shows that marsupial X inactivation is incomplete and escape is stochastic. Genome Biol. 2010;11:R122. doi: 10.1186/gb-2010-11-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grant J, et al. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature. 2012;487:254–258. doi: 10.1038/nature11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, Douglas KC, Vandeberg JL, Clark AG, Samollow PB. Chromosome-wide profiling of X-chromosome inactivation and epigenetic states in fetal brain and placenta of the opossum, Monodelphis domestica. Genome Res. 2014;24:70–83. doi: 10.1101/gr.161919.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 63.Wu H, et al. Cellular resolution maps of X chromosome inactivation: implications for neural development, function, and disease. Neuron. 2014;81:103–119. doi: 10.1016/j.neuron.2013.10.051. This study obtains comprehensive maps of XCI in vivo by differentially labelling each X chromosome with fluorescent markers and evaluates escape from XCI in the brain by cell sorting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. Genes that escape from X inactivation. Hum Genet. 2011;130:237–245. doi: 10.1007/s00439-011-1011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnston CM, et al. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet. 2008;4:e9. doi: 10.1371/journal.pgen.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jegalian K, Page DC. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature. 1998;394:776–780. doi: 10.1038/29522. [DOI] [PubMed] [Google Scholar]
- 67.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 68.Calabrese JM, et al. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell. 2012;151:951–963. doi: 10.1016/j.cell.2012.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. This is the first report of a comprehensive XCI profile of the mouse X chromosome based on an allele-specific RNA sequencing analysis, which shows fewer escape genes in mice than in humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Changolkar LN, et al. Genome-wide distribution of macroH2A1 histone variants in mouse liver chromatin. Mol Cell Biol. 2010;30:5473–5483. doi: 10.1128/MCB.00518-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Splinter E, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–1383. doi: 10.1101/gad.633311. This paper reports allele-specific chromosome conformation capture-on-chip assays to produce high-resolution topology maps of the active and inactive X chromosomes, and to show that escape genes are engaged in long-range contacts with each other. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Filippova GN, et al. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. This paper shows that the chromatin insulator CCCTC-binding factor (CTCF) is bound to transition regions between domains of inactivation and escape, which suggests a role in protection of these domains from encroachment of neighbouring domains. [DOI] [PubMed] [Google Scholar]
- 73.Horvath LM, Li N, Carrel L. Deletion of an X-inactivation boundary disrupts adjacent gene silencing. PLoS Genet. 2013;9:e1003952. doi: 10.1371/journal.pgen.1003952. This study shows that deletion of boundary elements between inactivated and escape chromatin leads to the spreading of escape and thus inappropriate reactivation of genes that are normally silenced. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reinius B, et al. Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics. 2010;11:614. doi: 10.1186/1471-2164-11-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lopes AM, Arnold-Croop SE, Amorim A, Carrel L. Clustered transcripts that escape X inactivation at mouse XqD. Mamm Genome. 2011;22:572–582. doi: 10.1007/s00335-011-9350-6. [DOI] [PubMed] [Google Scholar]
- 76.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. This is the first comprehensive report of XCI and escape from XCI in humans that was obtained in interspecific fibroblast-based systems; it shows that a remarkable degree of expression heterogeneity exists among females. [DOI] [PubMed] [Google Scholar]
- 77.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cotton AM, et al. Chromosome-wide DNA methylation analysis predicts human tissue-specific X inactivation. Hum Genet. 2011;130:187–201. doi: 10.1007/s00439-011-1007-8. This study shows tissue-specific differences in escape from XCI on the basis of analyses of DNA methylation patterns in humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anderson CL, Brown CJ. Variability of X chromosome inactivation: effect on levels of TIMP1 RNA and role of DNA methylation. Hum Genet. 2002;110:271–278. doi: 10.1007/s00439-002-0676-8. [DOI] [PubMed] [Google Scholar]
- 80.Lingenfelter PA, et al. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nature Genet. 1998;18:212–213. doi: 10.1038/ng0398-212. [DOI] [PubMed] [Google Scholar]
- 81.Patrat C, et al. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci USA. 2009;106:5198–5203. doi: 10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moreira de Mello JC, et al. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS ONE. 2010;5:e10947. doi: 10.1371/journal.pone.0010947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trabzuni D, et al. Widespread sex differences in gene expression and splicing in the adult human brain. Nature Commun. 2013;4:2771. doi: 10.1038/ncomms3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morgan CP, Bale TL. Sex differences in microRNA regulation of gene expression: no smoke, just miRs. Biol Sex Differ. 2012;3:22. doi: 10.1186/2042-6410-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kobayashi S, et al. The X-linked imprinted gene family Fthl17 shows predominantly female expression following the two-cell stage in mouse embryos. Nucleic Acids Res. 2010;38:3672–3681. doi: 10.1093/nar/gkq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bermejo-Alvarez P, Rizos D, Lonergan P, Gutierrez-Adan A. Transcriptional sexual dimorphism during preimplantation embryo development and its consequences for developmental competence and adult health and disease. Reproduction. 2011;141:563–570. doi: 10.1530/REP-10-0482. [DOI] [PubMed] [Google Scholar]
- 87.Berletch JB, Deng X, Nguyen DK, Disteche CM. Female bias in Rhox6 and 9 regulation by the histone demethylase KDM6A. PLoS Genet. 2013;9:e1003489. doi: 10.1371/journal.pgen.1003489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A. Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc Natl Acad Sci USA. 2010;107:3394–3399. doi: 10.1073/pnas.0913843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arnold AP. Mouse models for evaluating sex chromosome effects that cause sex differences in non-gonadal tissues. J Neuroendocrinol. 2009;21:377–386. doi: 10.1111/j.1365-2826.2009.01831.x. This paper describes the 4CG mouse model to evaluate effects of the sex chromosome complement separately from hormonal effects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Caeiro XE, Mir FR, Vivas LM, Carrer HF, Cambiasso MJ. Sex chromosome complement contributes to sex differences in bradycardic baroreflex response. Hypertension. 2011;58:505–511. doi: 10.1161/HYPERTENSIONAHA.111.175661. [DOI] [PubMed] [Google Scholar]
- 91.Chen X, et al. The number of X chromosomes causes sex differences in adiposity in mice. PLoS Genet. 2012;8:e1002709. doi: 10.1371/journal.pgen.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wijchers PJ, et al. Sexual dimorphism in mammalian autosomal gene regulation is determined not only by Sry but by sex chromosome complement as well. Dev Cell. 2010;19:477–484. doi: 10.1016/j.devcel.2010.08.005. This paper shows genome-wide effects on X-linked and autosomal gene expression that originate from differences in the number of X and Y chromosomes in mice. [DOI] [PubMed] [Google Scholar]
- 93.Eicher EM, et al. The mouse Y* chromosome involves a complex rearrangement, including interstitial positioning of the pseudoautosomal region. Cytogenet Cell Genet. 1991;57:221–230. doi: 10.1159/000133152. [DOI] [PubMed] [Google Scholar]
- 94.Wolstenholme JT, Rissman EF, Bekiranov S. Sexual differentiation in the developing mouse brain: contributions of sex chromosome genes. Genes Brain Behav. 2013;12:166–180. doi: 10.1111/gbb.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lopes AM, et al. Transcriptional changes in response to X chromosome dosage in the mouse: implications for X inactivation and the molecular basis of Turner Syndrome. BMC Genomics. 2010;11:82. doi: 10.1186/1471-2164-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu J, Deng X, Disteche CM. Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS ONE. 2008;3:e2553. doi: 10.1371/journal.pone.0002553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grafodatskaya D, et al. Multilocus loss of DNA methylation in individuals with mutations in the histone H3 lysine 4 demethylase KDM5C. BMC Med Genom. 2013;6:1. doi: 10.1186/1755-8794-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012;8:e1002964. doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leppig KA, Disteche CM. Ring X and other structural X chromosome abnormalities: X inactivation and phenotype. Semin Reprod Med. 2001;19:147–157. doi: 10.1055/s-2001-15395. [DOI] [PubMed] [Google Scholar]
- 100.Spatz A, Borg C, Feunteun J. X-chromosome genetics and human cancer. Nature Rev Cancer. 2004;4:617–629. doi: 10.1038/nrc1413. [DOI] [PubMed] [Google Scholar]
- 101.Gentilini D, et al. Age-dependent skewing of X chromosome inactivation appears delayed in centenarians’ offspring. Is there a role for allelic imbalance in healthy aging and longevity? Aging Cell. 2012;11:277–283. doi: 10.1111/j.1474-9726.2012.00790.x. [DOI] [PubMed] [Google Scholar]
- 102.Clerc P, Avner P. Random X-chromosome inactivation: skewing lessons for mice and men. Curr Opin Genet Dev. 2006;16:246–253. doi: 10.1016/j.gde.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 103.Invernizzi P, Pasini S, Podda M. X chromosome in autoimmune diseases. Expert Rev Clin Immunol. 2008;4:591–597. doi: 10.1586/1744666X.4.5.591. [DOI] [PubMed] [Google Scholar]
- 104.Busque L, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nature Genet. 2012;44:1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Medema RH, Burgering BM. The X factor: skewing X inactivation towards cancer. Cell. 2007;129:1253–1254. doi: 10.1016/j.cell.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 106.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miyake N, et al. KDM6A point mutations cause Kabuki syndrome. Hum Mutat. 2012;34:108–110. doi: 10.1002/humu.22229. [DOI] [PubMed] [Google Scholar]
- 110.Lederer D, et al. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am J Hum Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lindgren AM, et al. Haploinsufficiency of KDM6A is associated with severe psychomotor retardation, global growth restriction, seizures and cleft palate. Hum Genet. 2013;132:537–552. doi: 10.1007/s00439-013-1263-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jensen LR, et al. A distinctive gene expression fingerprint in mentally retarded male patients reflects disease-causing defects in the histone demethylase KDM5C. Pathog. 2010;3:2. doi: 10.1186/1755-8417-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Simensen RJ, et al. Short-term memory deficits in carrier females with KDM5C mutations. Genet Couns. 2013;23:31–40. [PubMed] [Google Scholar]
- 114.Heard E, Turner J. Function of the sex chromosomes in mammalian fertility. Cold Spring Harb Perspect Biol. 2011;3:a002675. doi: 10.1101/cshperspect.a002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sybert VP, McCauley E. Turner’s syndrome. N Engl J Med. 2004;351:1227–1238. doi: 10.1056/NEJMra030360. [DOI] [PubMed] [Google Scholar]
- 116.Zinn AR, Page DC, Fisher EM. Turner syndrome: the case of the missing sex chromosome. Trends Genet. 1993;9:90–93. doi: 10.1016/0168-9525(93)90230-f. [DOI] [PubMed] [Google Scholar]
- 117.Hassold T, Pettay D, Robinson A, Uchida I. Molecular studies of parental origin and mosaicism in 45,X conceptuses. Hum Genet. 1992;89:647–652. doi: 10.1007/BF00221956. [DOI] [PubMed] [Google Scholar]
- 118.Werler S, Poplinski A, Gromoll J, Wistuba J. Expression of selected genes escaping from X inactivation in the 41, XX(Y)* mouse model for Klinefelter’s syndrome. Acta Paediatr. 2011;100:885–891. doi: 10.1111/j.1651-2227.2010.02112.x. [DOI] [PubMed] [Google Scholar]
- 119.Gropman A, Samango-Sprouse CA. Neurocognitive variance and neurological underpinnings of the X and Y chromosomal variations. Am J Med Genet C Semin Med Genet. 2013;163:35–43. doi: 10.1002/ajmg.c.31352. [DOI] [PubMed] [Google Scholar]
- 120.Blewitt ME, et al. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc Natl Acad Sci USA. 2005;102:7629–7634. doi: 10.1073/pnas.0409375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brown CJ, Willard HF. The human X-inactivation centre is not required for maintenance of X chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- 122.Yildirim E, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. This study shows that loss of Xist in the blood compartment of female mice causes an aggressive form of cancer, which suggests that abnormal X reactivation and consequent genome-wide changes can lead to disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Leong HS, et al. Epigenetic regulator Smchd1 functions as a tumor suppressor. Cancer Res. 2012;73:1591–1599. doi: 10.1158/0008-5472.CAN-12-3019. [DOI] [PubMed] [Google Scholar]
- 124.Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol Cell Biol. 2006;26:7167–7177. doi: 10.1128/MCB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wareham KA, Lyon MF, Glenister PH, Williams ED. Age related reactivation of an X-linked gene. Nature. 1987;327:725–727. doi: 10.1038/327725a0. [DOI] [PubMed] [Google Scholar]
- 126.Lu Q, et al. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007;179:6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 127.Zandman-Goddard G, Peeva E. Connective tissue diseases. Unravelling aetiology in male SLE — the X chromosome dose effect. Nature Rev Rheumatol. 2012;8:310–312. doi: 10.1038/nrrheum.2012.44. [DOI] [PubMed] [Google Scholar]
- 128.Deng X, Disteche CM. Genomic responses to abnormal gene dosage: the X chromosome improved on a common strategy. PLoS Biol. 2010;8:e1000318. doi: 10.1371/journal.pbio.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]