Abstract
We conducted data-mining analyses of genome wide association (GWA) studies of the CATIE and MGS-GAIN datasets, and found 13 markers in the two physically linked genes, PTPN21 and EML5, showing nominally significant association with schizophrenia. Linkage disequilibrium (LD) analysis indicated that all 7 markers from PTPN21 shared high LD (r2>0.8), including rs2274736 and rs2401751, the two non-synonymous markers with the most significant association signals (rs2401751, P=1.10×10−3 and rs2274736, P=1.21×10−3). In a meta-analysis of all 13 replication datasets with a total of 13,940 subjects, we found that the two non-synonymous markers are significantly associated with schizophrenia (rs2274736, OR=0.92, 95% CI: 0.86–0.97, P=5.45×10−3 and rs2401751, OR = 0.92, 95% CI: 0.86–0.97, P=5.29×10−3). One SNP (rs7147796) in EML5 is also significantly associated with the disease (OR = 1.08, 95% CI: 1.02-1.14, P=6.43×10−3). These 3 markers remain significant after Bonferroni correction. Furthermore, haplotype conditioned analyses indicated that the association signals observed between rs2274736/rs2401751 and rs7147796 are statistically independent. Given the results that 2 non-synonymous markers in PTPN21 are associated with schizophrenia, further investigation of this locus is warranted.
Keywords: Data-mining, Informatic prioritization, Genetic association study, PTPN21, Non-synonymous SNP
1. Introduction
Schizophrenia, a chronic and devastating psychiatric disorder, is characterized by delusions, hallucinations and cognitive deficits. It has a world-wide prevalence of 0.5–1%. Although family and twin studies have suggested the existence of a strong genetic component in schizophrenia (heritability 80–85%) (Sullivan, 2005), the search for genetic susceptibility factors remains a challenge. In recent genetic studies, advancements have been made on several fronts. First, in a small proportion of schizophrenia cases, rare, large, high-penetrance copy number variations (CNVs) may be causative (Stefansson et al., 2008; The International Schizophrenia Consortium, 2008; Grozeva et al., 2010; Mulle et al., 2010; Raychaudhuri et al., 2010). GWA studies have revealed an enrichment of rare CNVs in cases relative to controls, and in studies with larger samples, specific CNV regions were identified to be associated with schizophrenia (Walsh et al., 2008). Second, GWA studies of common single-nucleotide polymorphisms (SNPs) have identified several genes and a broad region in chromosome 6p22.1 to be associated with schizophrenia (O'Donovan et al., 2008; Purcell et al., 2009; Shi et al., 2009; Stefansson et al., 2009). Third, polygenic model of the disease whereby a common variation in many hundreds or even thousands of genes contributes to schizophrenia is supported from the analyses of the study of International Schizophrenia Consortium (Purcell et al., 2009). In a recent study, data showed that common polygenic variation accounts for roughly one-third of the total variation in schizophrenia (The International Schizophrenia Consortium et al., 2008). Overall, the main conclusion from these studies is that schizophrenia risk is influenced by both rare variants with large effect and common variants with very small effect, a combination that contributes to the genetic heterogeneity of schizophrenia.
While GWA studies provide a promising approach to the genetics of complex diseases, it is clear that GWA studies have not explained most of the underlying genetic risks for schizophrenia. Identifying individual candidate genes/variants with small effects on disease risks remains important. Recently, using an alternative approach that combines data-mining of GWA datasets and bioinformatic prioritization to select promising candidate genes followed by verification and meta-analyses of a large number of independent datasets, we have successfully identified a schizophrenia risk gene, the cardiomyopathy associated 5 (CMYA5) (Chen et al., 2010). Encouraged by this discovery, here we report our study of two other loci: PTPN21 (protein tyrosine phosphatase, non-receptor type 21) and EML5 (echinoderm microtubule associated protein like 5). These two genes were identified by the same procedures we used to identify the CMYA5 gene, and they are close neighbors located on chromosome 14q31.3, occupying a region of about 300 kb genomic distance. From our previous statistical and bioinformatic prioritization procedures, we found that PTPN21 has 2 non-synonymous SNPs (rs2274736 and rs2401751) showing nominally significant association with schizophrenia in 2 GWA datasets and EML5 has multiple markers showing similar associations as well. Therefore, we decided to pursue further investigation of these two genes together. We conducted a replication study with 13 independent datasets including our Irish family (IFAM) and Irish case control study of schizophrenia (ICCSS) samples, and other 11 datasets from GWA studies with both Caucasian and African-American populations.
2. Methods and materials
2.1. Subjects and genotyping
In this study, we used 15 datasets with a total of 17,795 subjects, including 455 families with 1875 subjects, 8003 cases and 7917 controls (Table 1). Detailed description of each individual dataset were reported elsewhere (Chen et al., 2006; Sullivan et al., 2008; Purcell et al., 2009; Shi et al., 2009). In order to maintain the independence of the datasets, the overlapping subjects between the CATIE and MGS-GAIN, CATIE and MGS-nonGAIN, and CATIE-AA and MGS-GAIN-AA were removed. The CATIE and MGS-GAIN datasets were used as our data-mining and hypothesis-generating datasets as described in our previous study (Chen et al., 2010). The other 13 were used as replication datasets. All subjects were of Caucasian ancestry, except CATIE-AA and MGS-GAIN-AA datasets, which were African-American (For details see Table 1). Of these 15 datasets, 13 were used in GWA studies by individual groups and the subjects in these datasets were typed by either the Affymetrix or Perlgen microarray methods. Our two Irish samples, the IFAM and ICCSS, were genotyped by the TaqMan method (Livak, 1999). The quality of genotyping was assessed by individual groups to be satisfactory.
Table 1.
Sample description.
| Samples | Principle investigator | Ethnicity | Sample size (# family, case/ctrl) | Genotyping method | Reference |
|---|---|---|---|---|---|
| CATIE | Patrick Sullivan | European American | 492/523 | Perlgen | Sullivan et al. 2008 |
| MGS-GAIN | Pablo Gejman | European American | 1166/1132 | Affymetrix 6.0 | Shi et al. 2009 |
| MGS-nonGAIN | Pablo Gejman | European American | 1089/1065 | Affymetrix 6.0 | Shi et al. 2009 |
| IFAM | Kenneth Kendler | Irish/English | 455 | TaqMan | Chen et al. 2006 |
| ICCSS | Kenneth Kendler | Irish/English | 792/625 | TaqMan | Chen et al. 2006 |
| ISC-Aberdeen | David St Clair | English | 720/702 | Affymetrix 5.0 | Purcell et al. 2009 |
| ISC-Bulgaria | Michael O'Donovan | Bulgarian | 528/611 | Affymetrix 6.0 | Purcell et al. 2009 |
| ISC-Dublin | Aiden Corvin | Irish/English | 275/866 | Affymetrix 6.0 | Purcell et al. 2009 |
| ISC-Edinburgh | Douglas Blackwood | English | 369/287 | Affymetrix 6.0 | Purcell et al. 2009 |
| ISC-London | Hugh Gurling | English | 523/505 | Affymetrix 5.0 | Purcell et al. 2009 |
| ISC-Portugal | Carlos Pato | Portuguese | 347/216 | Affymetrix 5.0 | Purcell et al. 2009 |
| ISC-Sweden1 | Patrick Sullivan | Sweden | 170/170 | Affymetrix 5.0 | Purcell et al. 2009 |
| ISC-Sweden2 | Patrick Sullivan | Sweden | 390/230 | Affymetrix 6.0 | Purcell et al. 2009 |
| CATIE-AA | Patrick Sullivan | African American | 227/228 | Perlgen | Purcell et al. 2009 |
| MGS-GAIN-AA | Pablo Gejman | African American | 915/757 | Affymetrix 6.0 | Shi et al. 2009 |
|
| |||||
| All samples | 455 families, 8003/7917 | ||||
2.2. Data-mining and bioinformatic prioritization
The GWA analyses were conducted with the quality-control filtered markers from the NIMH (http://nimhgenetics.org/) and GAIN (http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gap) repositories for the CATIE and MGS-GAIN datasets respectively. Association of individual SNPs to schizophrenia was tested by logistic regression (trend test) using the PLINK program (Purcell et al., 2007) for each individual dataset. In these analyses, only Caucasian subjects (CATIE, 492 cases and 523 controls; MGS-GAIN, 1166 cases and 1132 controls) were used and markers with a minor allele frequency < 1% or a Hardy– Weinberg Equilibrium p value < 0.0001 were excluded. For the CATIE dataset, the 7 principle components identified in the previous study (Sullivan et al., 2008) were used as covariates and a total of 446,225 markers were analyzed. For the MGS-GAIN dataset, no covariate was used since there was no significant stratification found in this sample based on previous analyses (Shi et al., 2009). The number of markers analyzed for the MGS-GAIN was 727,905.
For bioinformatic prioritization, we first selected all markers with P values ≤0.05 in the CATIE and MGS-GAIN datasets, and matched them against each other. After the matching, there were 1128 SNPs with unadjusted P values ≤0.05 in both datasets. We then conducted bioinformatic prioritization of these SNPs based on whether they are located in evolutionarily conserved regions, genic regions (exons, introns, untranslated regions (UTRs), or within 2 kb of a gene), transcription factor binding sites, or whether they were on the list of known schizophrenia candidate genes (as listed in the sczgene database http://www.schizophreniaforum.org/res/sczgene/default.asp by June, 2008), or whether the SNPs are non-synonymous. SNPs in each of these categories were assigned an empirical score: 2 for the non-synonymous and known schizophrenia candidate gene categories, 1 for the evolutionary conserved regions, transcription factor binding sites, UTR, and the synonymous SNP category, and 0.5 for the “within 2 kb of a gene” category. SNPs were ranked by the sum of the scores (Sun et al., 2009).
When the PTPN21/EML5 gene regions were identified as one of the leading candidates, we performed LD structure analyses of this region using the HAPLOVIEW program (Barrett et al., 2005). We extracted all markers in the two genes plus 20 kb upstream and downstream sequences for the CATIE and MGS-GAIN datasets, and selected the common markers between the two datasets. We conducted a meta-analysis for the two datasets with all shared markers using the GWAMA program (Magi and Morris, 2010).
2.3. Replication and meta-analyses of 13 independent datasets
Based on our prioritization and LD analyses, we selected 5 SNPs, rs2274736, rs2401751, rs10150311, rs17260415 and rs7147796 in PTPN21/EML5 genes for genotyping in the IFAM and ICCSS samples. Replication was also conducted for the MGS-nonGAIN, MGS-GAIN-AA, CATIE-AA and the 8 ISC datasets using extracted data of these 5 markers.
Meta-analyses for the replication datasets only and all datasets were conducted with the GWAMA software package. The primary reason for using GWAMA was that the methods implemented take genomic control to adjust for potential stratification, and have good estimates for heterogeneity. These features were important for us since we analyzed data from multiple ethnic groups. For association analyses, we used logistic regression from the PLINK program for case control samples and used the UNPHASED program (Dudbridge, 2008) for the family sample (IFAM). For the CATIE-AA dataset, the 7 principle components mentioned above were used as covariates, and for the MGS-GAIN-AA dataset, we fitted a logistic regression model including the first principal component of population stratification as a covariate. Summary statistics (Odds ratio, 95% confident interval, and sample size) were extracted from individual analyses and used in the meta-analyses. The GWAMA package performs both fixed effect and random effect meta-analyses. In our analyses, since the heterogeneity tests (both Cochran's Q statistics and I2) were insignificant (Table 3), we reported the results from the fixed effect analyses. Meta-analysis results were plotted with the R package rmeta (http://www.wessa.net).
Table 3.
Heterogeneity statistics across all studies.
| SNPs | Affected allele | Replication samples | All samples | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Cochran's Q | P value for Q | I2 | Cochran's Q | P value for Q | I2 | ||
| rs2274736 | C | 4.60 | 0.80 | 0.00 | 7.17 | 0.71 | 0.00 |
| rs2401751 | A | 4.43 | 0.82 | 0.00 | 6.96 | 0.73 | 0.00 |
| rs10150311 | G | 11.00 | 0.53 | 0.00 | 17.32 | 0.24 | 0.19 |
| rs17260415 | G | 11.60 | 0.48 | 0.00 | 16.84 | 0.26 | 0.17 |
| rs7147796 | C | 9.88 | 0.54 | 0.00 | 12.47 | 0.49 | 0.00 |
2.4. Testing the independent effects between rs2274736/rs2401751 and rs7147796
As there were three SNPs showing association in the PTPN21/EML5 gene region (the two non-synonymous SNPs from PTPN21 shared very high LD, but they shared relatively low LD with the SNP, rs7147796, from EML5), we evaluated whether the association signals observed at the two genes between rs2274736/rs2401751 and rs7147796 were statistically independent. We took the conditioned analysis approach implemented in the PLINK program that compares the risks between haplotypes with identical alleles in the background locus but different alleles at the locus to be evaluated. Since PLINK is unable to combine family data and case–control data for such haplotype-based analyses, we combined all 12 Caucasian case–control samples in our studies including the two discovery datasets. We first inferred all four haplotypes for rs2274736–rs7147796, and tested the effects of haplotypes with the same allele at rs7147796, but different alleles at rs2274736. A similar approach was applied to rs2401751–rs7147796. Our aim was to evaluate whether the effect of rs2274736/rs2401751 is independent of rs7147796.
3. Results
3.1. GWA studies data-mining and bioinformatic prioritization
In the process of data-mining in the CATIE and MGS-GIAN datasets, the PTPN21 gene was one of our top candidate genes (supplementary Table S1). There were two non-synonymous SNPs showing significant association in each individual dataset and in the combined datasets (Table 2). Another 5 SNPs in the PTPN21 gene also showed similar association in the same direction in both datasets. The P values for the two non-synonymous SNPs in the combined datasets were P=1.10×10−3 for rs2401751 and P=1.21×10−3 for rs2274736. Six SNPs from EML5 gene, which is about 100 kb telomeric to the PTPN21 gene, were also significant in the combined discovery datasets, with the strongest signal at rs7157149 (P=2.52×10−3). rs17260415 was the only SNP among the 6 SNPs from EML5 that were nominally significant in the each individual dataset and the combined datasets (combined P=4.25×10−3).
Table 2.
Association analysis of the CATIE, MGS-GAIN datasets and combined datasets from PTPN21/EML5 gene regions.
| No. | SNP | Chr | Position (bp) | Gene | Function | CATIE | MGS-GAIN | CATIE_MGS-GAIN | ||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| P value | P value | P value | OR(95%CI) | MAF | ||||||
| 1 | rs2295136 | 14 | 88004455 | PTPN21 | Intron | 0.52270 | 0.95880 | 0.76720 | 1.02(0.92–1.12) | 0.40 |
| 2 | rs2274736 | 14 | 88008405 | PTPN21 | Missense | 0.01705 | 0.02076 | 0.00121 | 0.84(0.76–0.93) | 0.33 |
| 3 | rs2401751 | 14 | 88016375 | PTPN21 | Missense | 0.01883 | 0.01769 | 0.00110 | 0.84(0.75–0.93) | 0.33 |
| 4 | rs1864744 | 14 | 88020759 | PTPN21 | Intron | 0.02501 | 0.02016 | 0.00155 | 0.84(0.76–0.94) | 0.33 |
| 5 | rs7160647 | 14 | 88043437 | PTPN21 | Intron | 0.03853 | 0.01391 | 0.00145 | 0.84(0.75–0.93) | 0.30 |
| 6 | rs10138002 | 14 | 88046168 | PTPN21 | Intron | 0.03834 | 0.01258 | 0.00130 | 0.84(0.75–0.93) | 0.30 |
| 7 | rs10150311 | 14 | 88046225 | PTPN21 | Intron | 0.04164 | 0.01711 | 0.00191 | 0.84(0.75–0.94) | 0.30 |
| 8 | rs11847417 | 14 | 88046703 | PTPN21 | Intron | 0.03618 | 0.01387 | 0.00137 | 0.84(0.75–0.93) | 0.30 |
| 9 | rs11622270 | 14 | 88051323 | PTPN21 | Intron | 0.81690 | 0.50960 | 0.49464 | 0.96(0.84–1.08) | 0.19 |
| 10 | rs12323840 | 14 | 88077179 | PTPN21 | Intron | 0.48740 | 0.66630 | 0.45789 | 0.91(0.70–1.18) | 0.04 |
| 11 | rs11159859 | 14 | 88097644 | ZC3H14 | Upstream | 0.18590 | 0.05759 | 0.02079 | 0.88(0.79–0.98) | 0.32 |
| 12 | rs2297124 | 14 | 88132920 | ZC3H14 | Intron | 0.45510 | 0.66880 | 0.44848 | 0.94(0.81–1.10) | 0.12 |
| 13 | rs11159862 | 14 | 88134030 | ZC3H14 | Intron | 0.98710 | 0.67820 | 0.71934 | 1.03(0.89–1.18) | 0.16 |
| 14 | rs12590101 | 14 | 88165603 | EML5 | Intron | 0.88060 | 0.67870 | 0.78283 | 1.02(0.89–1.17) | 0.16 |
| 15 | rs11159863 | 14 | 88178531 | EML5 | Intron | 0.88840 | 0.67970 | 0.67195 | 1.03(0.90–1.18) | 0.16 |
| 16 | rs1144921 | 14 | 88180048 | EML5 | Intron | 0.75630 | 0.55330 | 0.50406 | 0.95(0.82–1.11) | 0.12 |
| 17 | rs10132509 | 14 | 88273534 | EML5 | Intron | 0.02896 | 0.13820 | 0.01661 | 1.13(1.02–1.25) | 0.46 |
| 18 | rs11846241 | 14 | 88274107 | EML5 | Intron | 0.55970 | 0.78470 | 0.94721 | 1.00(0.89–1.11) | 0.26 |
| 19 | rs17260415 | 14 | 88281726 | EML5 | Intron | 0.04058 | 0.03751 | 0.00425 | 0.85(0.76–0.95) | 0.26 |
| 20 | rs10140896 | 14 | 88288291 | EML5 | Intron | 0.02532 | 0.06780 | 0.00647 | 1.15(1.04–1.27) | 0.48 |
| 21 | rs12880096 | 14 | 88288568 | EML5 | Intron | 0.02744 | 0.12310 | 0.01382 | 1.13(1.03–1.25) | 0.48 |
| 22 | rs7147796 | 14 | 88298322 | EML5 | Intron | 0.01437 | 0.21750 | 0.02019 | 1.12(1.02–1.24) | 0.48 |
| 23 | rs7157149 | 14 | 88301598 | EML5 | Intron | 0.01000 | 0.05079 | 0.00252 | 0.84(0.75–0.94) | 0.25 |
MAF: minor allele frequency; A1/2: alleles 1 and 2 (minor/major); 5 SNPs for replication and all significant P values are shown in bold.
The meta P values of all 23 shared SNPs in the two discovery datasets in the PTPN21/EML5 region were shown in Fig. 1A, and the LD structure in the combined datasets are shown in Fig. 1B. As seen in Fig. 1B, the 7 significant SNPs from PTPN21 gene shared very high LD (r2>0.8). They also had similar minor allele frequencies (MAF=0.30–0.33, as seen in Table 2) and odds ratios (OR=0.84, 95% CI: 0.75–0.94) (Table 2). It is therefore likely that they represented the same signal. The 6 significant SNPs from EML5 gene came from different LD blocks (r2=0.29–0.98), which also had very different LD with the 7 significant SNPs from PTPN21 gene (r2=0.27–0.74). These probably reflect independent signals within EML5 gene or between PTPN21 and EML5 genes. Based on this information, 5 SNPs (rs2274736, rs2401751, rs10150311, rs17260415 and rs7147796) were selected for replication.
Fig. 1.
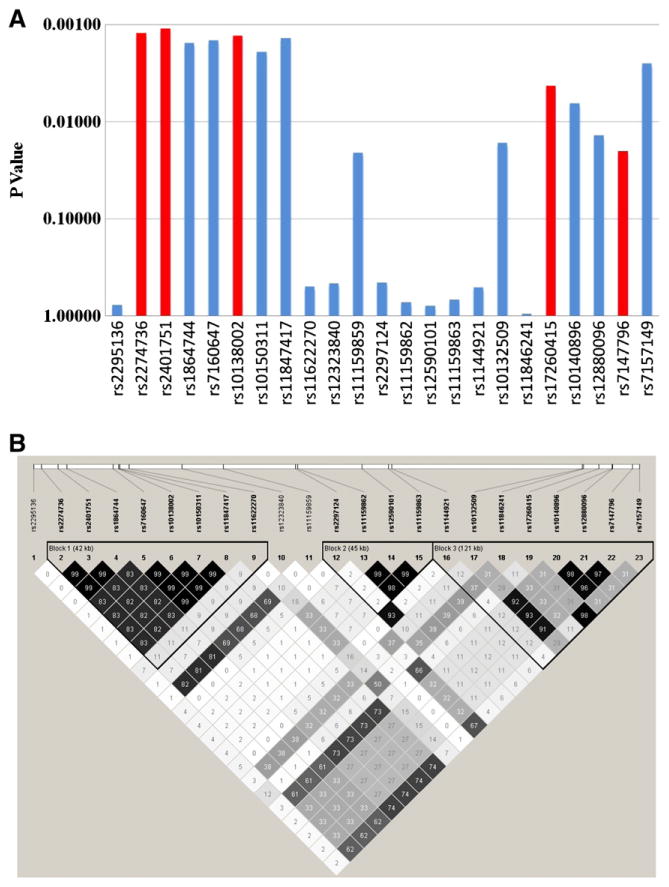
A. Meta-analysis of the CATIE and MGS-GAIN datasets. The markers selected for replication were highlighted. B. LD structure of the 23 markers typed in both CATIE and MGS-GAIN datasets. Pair-wise LD values (r2) were shown.
3.2. Meta-analyses of PTPN21/EML5 association in 13 replication datasets
Our data-mining and bioinformatic prioritization suggested that the PTPN21/EML5 region may be involved in schizophrenia. To verify the association, we requested and obtained data from 11 independent GWA datasets, in addition to our two IRISH samples. The information of the 13 replication datasets, plus the two data-mining datasets, CATIE and MGS-GAIN, were described in Table 1. To ensure the quality, we examined the intensity plots for these 5 markers. Initially, we genotyped the 5 SNPs in the IFAM and ICCSS samples. Although none of the SNPs reached significance in either individual sample (Fig. 2) or the combined samples, all the SNPs in our combined Irish samples showed the same direction of association for the same allele as those in the CATIE and MGS-GAIN datasets (data not shown). For further confirmation, we analyzed data from other 11 independent datasets (9 Caucasian and two African-American). The minor allele frequencies of all the datasets in this study were listed in supplementary Table S2. Although most of the individual datasets did not reach significance for these five SNPs, in the MGS-nonGAIN dataset, which was the largest amongst the replication datasets, the two non-synonymous SNPs (rs2274736 and rs2401751) were nominally significant (P=0.01046 and 0.01387, respectively), and one SNP (rs7147796) from the EML5 gene had the strongest signal (P=0.001912) (Fig. 2A, B, E). In ISC-Sweden2 dataset, one SNP (rs17260415) also showed nominal significant signal (P=0.03101) (Fig. 2D). Meta-analyses of the 13 replication datasets using GWAMA program showed that the two non-synonymous SNPs were most significantly associated with schizophrenia (rs2274736, OR=0.92, 95% CI: 0.86-0.97, P=5.45×10−3 and rs2401751, OR=0.92, 95% CI: 0.86-0.97, P=5.29×10−3). One SNP (rs7147796) from EML5 was also significant (OR=1.08, 95% CI: 1.02-1.14, P=6.43×10−3). These 3 markers remained significant after Bonferroni correction since we tested only 5 markers in the replication datasets. In the meta-analyses, we found no evidence for heterogeneity amongst the datasets, including the discovery datasets (Table 3). An analysis of combined replication datasets with PLINK program showed similar results (data not shown).
Fig. 2.
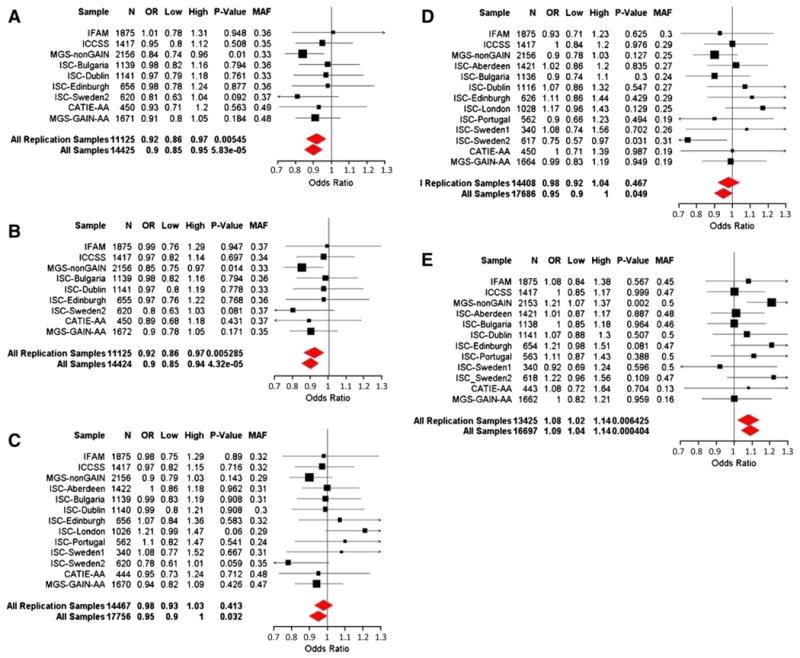
Meta-analyses of 5 SNPs from PTPN21/EML5 region for schizophrenia. A–E. rs2274736, rs2401751, rs10150311, rs17260415 and rs7147796. OR (Odds ratios) are reported for the minor alleles in the overall EA datasets.
3.3. rs2274736/rs2401751 and rs7147796 are independently associated with schizophrenia
As we observed that three SNPs in the PTPN21/EML5 region were significantly associated with schizophrenia, we tried to evaluate whether these signals are independent between these two genes. Since the 2 non-synonymous marker rs2274736 and rs22401751 were in high LD, we tested whether rs2274736/rs2401751 and rs7147796 were independently associated with schizophrenia. In the data-mining data sets, the LD between these two SNPs was relatively low (r2=0.33) and similar results were obtained in our combined European datasets (r2=0.33), including the MGS-GAIN and CATIE datasets. To perform this test, we first inferred the haplotypes from these two pairs of markers using the PLINK program for the combined European case–control datasets. We then evaluated whether those haplotypes sharing identical alleles at rs7147796, but different alleles at rs2274736/rs2401751, had different disease risks. If these haplotypes have significantly different risks, then the effects of these two markers would be at least partially independent. The results were shown in Table 4. We found that while the rs2274736–rs7147796 haplotypes sharing the same allele background at rs7147796 locus, i.e., haplotypes C–C and T–C, did not show a significantly different disease risks (P=0.266), haplotypes C–G and T–G showed significantly different risks for the disease (P=0.0188). In other words, rs2274736 had an effect independent of that of rs7147796. Similar results were also observed for haplotypes A–G and G–G for rs2401751–rs7147796 (P=0.0143). These results suggested that the association signals from the two genes may be at least partly independent.
Table 4.
Test of independent effects between rs2274736/rs22401756 and rs7147796a.
| Markers | Haplotype | Frequency | OR (Alternative) |
OR (Null) |
Sub-Null P | Null P |
|---|---|---|---|---|---|---|
| rs2274736–rs7147796 | CC | 0.029 | ref | ref | 0.2660 | 0.0244 |
| TC | 0.454 | 1.11 | ref | |||
| CG | 0.312 | 0.96 | 0.91 | 0.0188 | ||
| TG | 0.206 | 1.06 | 0.91 | |||
| rs2401756–rs7147796 | AC | 0.029 | ref | ref | 0.3290 | 0.0227 |
| GC | 0.454 | 1.10 | ref | |||
| AG | 0.311 | 0.95 | 0.91 | 0.0143 | ||
| GG | 0.206 | 1.05 | 0.91 |
In these conditional analyses, the null hypothesis is that the haplotypes having the same alleles at the conditioned locus, the second locus, rs7147796, have same effect on disease risk. The alternative hypothesis is that all haplotypes may have different disease risks. The sub-null hypothesis tests whether the two haplotypes sharing the conditioned allele have the same disease risks. Significant P values are shown in bold.
4. Discussion
GWA is a widely used study design for detecting genetic causes of complex diseases in recent years. Over 900 genes have been reported to be associated with a variety of complex diseases, including type 2 diabetes (Scott et al., 2007; Sladek et al., 2007; Zeggini et al., 2007), lung cancer (Amos et al., 2008; Spitz et al., 2008; Thorgeirsson et al., 2008), Parkinson's disease (Satake et al., 2009; Simon-Sanchez et al., 2009), rheumatoid arthritis (Raychaudhuri et al., 2008), and systemic lupus erythematosus (Graham et al., 2008). For schizophrenia, the results from GWA studies have been less successful. While several loci (ZNF804A, NRGN, TCF4 and the broad major histocompatibility region) were implicated (O'Donovan et al., 2008; Shi et al., 2009; Stefansson et al., 2009), many more remained to be identified. Of the many possible factors leading to these outcomes, insufficient power in these individual studies and the need to correct for a large number of markers tested may be important factors. Another possible reason is that schizophrenia may be etiologically heterogeneous and is caused by a combination of risk genes. Different combinations of risk genes may or may not present the same disease phenotypes, and the effects of these risk genes are too small to be detected in GWA studies. Since aggregated analyses indicated that there may be true findings among those markers passing nominal significance in GWA datasets (Purcell et al., 2009), how to identify those markers with true but small effects is a practical issue facing the field. There is therefore a pressing need for alternative methods for extracting information from GWA data sets. In a previous study, we adapted a two-stage approach, leading to the identification of the CMYA5 gene (Chen et al., 2010). This study is an extension of the same strategy. We report here the association of 2 non-synonymous markers in the PTPN21 gene with schizophrenia. This approach combines statistic and biological evaluations of markers, with an emphasis on potentially functional/causative variants. Non-synonymous mutations (altering amino acids) are relatively straight forward to interpret as potentially functional/causative variants. These emphases lead us to test the PTPN21 gene since it has 2 non-synonymous markers reaching nominal significance in our discovery datasets. It is worth noting that distinguishing the functional variants from nonfunctional ones is important in GWA studies since collapsing variants across different categories could increase noises and diminish statistical power. For example, schizophrenia cases may be enriched for functional variants, but inclusion of nonfunctional variants in analyses can dilute the signals (Li and Leal, 2008; Morris and Zeggini, 2010).
In this study, we tested a total of 5 SNPs in the replication datasets. Of these markers, 2 non-synonymous markers in PTPN21 gene are in high LD (r2=0.99). While the P values observed for these markers are modest, all of them remain significant after Bonferroni correction. The major reason for these modest P values may be due to small effects of these markers. On average, the association signals have an OR of 1.1, which requires a significantly large sample size to detect (Sham et al., 2000). In addition, we have observed 2 statistically independent signals. In the independence tests, we observed significantly different disease risks between haplotype pairs (CG–TG pair for rs2247436–rs7147796, AG–GG pair for rs2401751–rs7147796) with the same alleles at rs7147796 but different alleles at rs2274736/rs2401751. The insignificant results for haplotype pairs (CC–TC for rs2247436–rs7147796 and AC–GC for rs2401751–rs7147796) may be due to the low frequencies (0.029) of the CC and AC haplotypes (see Table 4). The observation of independent association signals makes the results more creditable since it less likely that 2 independent signals occur in a relatively small genomic region.
Our results are consistent with the findings of Shi et al. (Shi et al., 2009). In their study of the MGS sample, SNP rs1864744, which is in high LD with the two non-synonymous SNPs (r2=0.99 in our discovery datasets), was one of the top 12 candidates (P=2.18×10−6). The overall ORs of the two non-synonymous SNPs in our studies were 0.90 (95% CI 0.85–0.95), which was comparable to that observed for rs1864744 by Shi, et al. (OR=0.83 in European ancestry and 0.85 in combined European-ancestry and African American datasets). Surprisingly, the two non-synonymous SNPs from PTPN21 were not in the top list of their findings.
As for the EML5 gene, we found that rs7147796 showed significant association. This is also consistent with results reported by Shi et al. that rs10140896 from EML5 was the 3rd most significant marker (P=9.49×10−7) in MGS Caucasian subjects (Shi et al., 2009), since these 2 SNPs share a very high LD (r2=0.96) and have the same MAF (both MAF=0.48) from the combined CATIE and MGS-GAIN datasets (Fig. 1B and Table 2). It is very likely that these 2 SNPs represent the same signal. For both markers, the minor alleles were the risk alleles. The OR of rs7147796 in our combined datasets was 1.09, while for rs10140896 the OR was 1.22 in Shi et al.'s study. Due to the fact that the LD between the two non-synonymous SNPs and rs7147796 were relatively low (r2=0.33), the association signals are statistically independent.
PTPN21 (previously known as protein tyrosine phosphatase D1 (PTPD1)) functionally belongs to the protein tyrosine phosphatase nonreceptor (PTPN) family. Although the function of PTPN21 is not fully understood, it has been shown to interact with a Tec kinase family member, the epithelial and endothelial tyrosine kinase (Etk), and plays a role in the regulation of cell growth and differentiation (Jui et al., 2000). Other member of the PTPN family, such as PTPN22, has been consistently associated with multiple autoimmune disorders, including systemic lupus erythematosus (SLE) and rheumatoid arthritis (Kyogoku et al., 2004; Chung and Criswell, 2007). A few studies also showed that PTPN21 may be involved in the immune system (Mustelin, 2006), and is critical in signaling from many surface receptors, including the T cell antigen receptor (Han et al., 2000) and Fas/CD95 (Sato et al., 1995). The fact that the most consistent finding for schizophrenia is the 6p major histocompatibility region supports the involvement of the immune system in schizophrenia. Taken together, this information is supportive that PTPN21's function in the immune system may be relevant to schizophrenia.
Interestingly, Zietlin et al. reported that while there was no association of PTPN21 with an autoimmune syndrome, Graves' disease, the T allele of the non-synonymous SNP rs2274736 seemed to be a risk factor leading to younger age of onset for this disease (Zeitlin et al., 2006). In this study, our results also suggest that the T allele of rs2274736 is a risk allele for schizophrenia. In the literature, there were case reports that some Graves' disease patients had psychiatric syndromes like paranoid schizophrenia (Adams et al., 2004; Ogah et al., 2009), and this was partially due to the influence of high thyroid hormone on brain function.
The two non-synonymous SNPs (rs2401751 and rs2274736) reside in different exons of the PTPN21 gene. rs2401751 changes the 385th amino acid of the protein from leucine to phenylalanine, and rs2274736 changes 936th amino acid of the protein from valine to alanine. These changes alter the size and hydrophobicity of the residues that might affect the activities of the enzyme in ways we do not yet understand, contributing to the etiology of schizophrenia. These amino acid changes resulting from the two non-synonymous SNPs provide a great opportunity for further clarifying the pathophysiology of schizophrenia.
The EML5 gene is a homolog to the major microtubule-associated protein in dividing sea urchin embryos, and is expressed in post-mitotic neurons (in adult animals and during development). It potentially plays a significant role in microtubule assembly and cytoskeletal rearrangements during neuronal development and in the adult brain (O'Connor et al., 2004). It is plausible that abnormal cytoskeletal arrangements result in changes in neuronal development, thereby altering risk for schizophrenia.
Recently, we reported CMYA5 as a candidate gene for schizophrenia using a two-stage approach (Chen et al., 2010). With the same approach, here we provide evidence that two non-synonymous markers in the PTPN21 gene and one SNP in EML5 are significantly associated with schizophrenia. These studies demonstrate that there are many markers/genes with true but small effects for schizophrenia embedded in the GWA datasets. Given creative designs and appropriate sample sizes, these markers/genes can be eventually identified.
Supplementary Material
Acknowledgments
The authors thank the volunteers, patients and their family members for participating in this study. This study was supported in part by a research grant (07R-1770) from the Stanley Medical Research Institute and an Independent Investigator Award from NARSAD to XC, and by grants to investigators involved in the collection and analyses of the datasets from CATIE, GAIN, and Consortium (ISC). The principal investigators of the CATIE trial were Jeffrey A. Lieberman, T. Scott Stroup, and Joseph P. McEvoy. The CATIE trial was funded by a grant from the National Institute of Mental Health (N01 MH900001) along with MH074027 (PI PF Sullivan). Genotyping was funded by Eli Lilly and Company. The principle investigators for the MGS were Pablo Gejman and Douglas Levinson. MGS study was supported by funding from the National Institute of Mental Health and the National Alliance for Research on Schizophrenia and Depression. Genotyping of part of the sample was supported by GAIN and the Paul Michael Donovan Charitable Foundation. Genotyping was carried out by the Center for Genotyping and Analysis at the Broad Institute of Harvard and MIT with support from the National Center for Research Resources.
Role of the funding source: The funding agencies of this study have no roles in the design, execution and writing-up of this study.
Footnotes
Supplementary materials related to this article can be found online at doi:10.1016/j.schres.2011.06.023.
Contributors: Author contribution:
Jingchun Chen: Conducted the experiments, data-analyses, and wrote the paper;
Grace Lee: Conducted experiments;
Ayman H. Fanous: Involved in the collection and analyses of the ISC samples;
Zhongming Zhao: Involved in the data-mining of GWA datasets;
Peilin Jia: Involved in the analyses of ISC datesets;
Anthony O'Neill: Involved in the collection of Irish family and case control samples;
Dermot Walsh: Involved in the collection of Irish family and case control samples;
The International Schizophrenia Consortium: Contributed the ISC datasets;
Kenneth S Kendler: Involved in the collection of Irish samples and revision of the manuscript;
Xiangning Chen: designed the experiments, data-analyses, and wrote the paper.
Conflict of interest: The authors declare no conflict of interest.
The members of the International schizophrenia consortium:
Cardiff University: Michael C O'Donovan,6 George K Kirov,6 Nick J Craddock,6 Peter A Holmans,6 Nigel M Williams,6 Lyudmila Georgieva,6 Ivan Nikolov,6 N Norton,6 H Williams,6 Draga Toncheva,16 Vihra Milanova,17 Michael J Owen;6 Karolinska Institutet/University of North Carolina at Chapel Hill: Christina M Hultman,11,12 Paul Lichtenstein,11 Emma F Thelander,11 Patrick Sullivan;7 Trinity College Dublin: Derek W Morris,9 Colm T O'Dushlaine,9 Elaine Kenny,9 Emma M Quinn,9 Michael Gill,9 Aiden Corvin;9 University College London: Andrew McQuillin,8 Khalid Choudhury,8 Susmita Datta,8 Jonathan Pimm,8 Srinivasa Thirumalai,18 Vinay Puri,8 Robert Krasucki,8 Jacob Lawrence,8 Digby Quested,19 Nicholas Bass,8 Hugh Gurling;8 University of Aberdeen: Caroline Crombie,15 Gillian Fraser,15 Soh Leh Kuan,14 Nicholas Walker,20 David St Clair;14 University of Edinburgh: Douglas HR Blackwood,10 Walter J Muir,10 Kevin A McGhee,10 Ben Pickard,10 Pat Malloy,10 Alan W Maclean,10 Margaret Van Beck;10 Queensland Institute of Medical Research: Naomi R Wray,5 Stuart Macgregor,5 Peter M Visscher;5 University of Southern California: Michele T Pato,13 Helena Medeiros,13 Frank Middleton,21 Celia Carvalho,13 Christopher Morley,21 Ayman Fanous,13,22,23,24 David Conti,13 James A Knowles,13 Carlos Paz Ferreira,25 Antonio Macedo,26 M Helena Azevedo,26 Carlos N Pato;13 Massachusetts General Hospital: Jennifer L. Stone,1,2,3,4 Douglas M Ruderfer,1,2,3,4 Andrew N Kirby,2,3,4 Manuel AR Ferreira,1,2,3,4 Mark J Daly,2,3,4 Shaun M Purcell,1,2,3,4 Pamela Sklar;1,2,3,4 Stanley Center for Psychiatric Research and Broad Institute of MIT and Harvard: Shaun M Purcell,1,2,3,4 Jennifer L Stone,1,2,3,4 Kimberly Chambert,3,4 Douglas M Ruderfer,1,2,3,4 Finny Kuruvilla,4 Stacey B Gabriel,4 Kristin Ardlie,4 Jennifer L Moran,4 Mark J Daly,2,3,4 Edward M Scolnick,3,4 Pamela Sklar.1,2,3,4
References
- Adams DD, Knight JG, Manning P, Smith G. An informative case of Graves' disease with implications for schizophrenia. J Clin Lab Immunol. 2004;53:13–25. [PubMed] [Google Scholar]
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, Houlston RS. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Chen X, Wang X, Hossain S, O'Neill FA, Walsh D, Pless L, Chowdari KV, Nimgaonkar VL, Schwab SG, Wildenauer DB, Sullivan PF, van den OE, Kendler KS. Haplotypes spanning SPEC2, PDZ-GEF2 and ACSL6 genes are associated with schizophrenia. Hum Mol Genet. 2006;15:3329–3342. doi: 10.1093/hmg/ddl409. [DOI] [PubMed] [Google Scholar]
- Chen X, Lee G, Maher BS, Fanous AH, Chen J, Zhao Z, Guo A, van den OE, Sullivan PF, Shi J, Levinson DF, Gejman PV, Sanders A, Duan J, Owen MJ, Craddock NJ, O'Donovan MC, Blackman J, Lewis D, Kirov GK, Qin W, Schwab S, Wildenauer D, Chowdari K, Nimgaonkar V, Straub RE, Weinberger DR, O'Neill FA, Walsh D, Bronstein M, Darvasi A, Lencz T, Malhotra AK, Rujescu D, Giegling I, Werge T, Hansen T, Ingason A, Noethen MM, Rietschel M, Cichon S, Djurovic S, Andreassen OA, Cantor RM, Ophoff R, Corvin A, Morris DW, Gill M, Pato CN, Pato MT, Macedo A, Gurling HM, McQuillin A, Pimm J, Hultman C, Lichtenstein P, Sklar P, Purcell SM, Scolnick E, St CD, Blackwood DH, Kendler KS. GWA study data mining and independent replication identify cardiomyopathy-associated 5 (CMYA5) as a risk gene for schizophrenia. Mol Psychiatry. 2010 doi: 10.1038/mp.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SA, Criswell LA. PTPN22: its role in SLE and autoimmunity. Autoimmunity. 2007;40:582–590. doi: 10.1080/08916930701510848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, Burtt NP, Guiducci C, Parkin M, Gates C, Plenge RM, Behrens TW, Wither JE, Rioux JD, Fortin PR, Graham DC, Wong AK, Vyse TJ, Daly MJ, Altshuler D, Moser KL, Gaffney PM. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozeva D, Kirov G, Ivanov D, Jones IR, Jones L, Green EK, St Clair DM, Young AH, Ferrier N, Farmer AE, McGuffin P, Holmans PA, Owen MJ, O'Donovan MC, Craddock N. Rare copy number variants: a point of rarity in genetic risk for bipolar disorder and schizophrenia. Arch Gen Psychiatry. 2010;67:318–327. doi: 10.1001/archgenpsychiatry.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Williams S, Mustelin T. Cytoskeletal protein tyrosine phosphatase PTPH1 reduces T cell antigen receptor signaling. Eur J Immunol. 2000;30:1318–1325. doi: 10.1002/(SICI)1521-4141(200005)30:5<1318::AID-IMMU1318>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Jui HY, Tseng RJ, Wen X, Fang HI, Huang LM, Chen KY, Kung HJ, Ann DK, Shih HM. Protein-tyrosine phosphatase D1, a potential regulator and effector for Tec family kinases. J Biol Chem. 2000;275:41124–41132. doi: 10.1074/jbc.M007772200. [DOI] [PubMed] [Google Scholar]
- Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VE, Chang M, Ramos P, Baechler EC, Batliwalla FM, Novitzke J, Williams AH, Gillett C, Rodine P, Graham RR, Ardlie KG, Gaffney PM, Moser KL, Petri M, Begovich AB, Gregersen PK, Behrens TW. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet. 2004;75:504–507. doi: 10.1086/423790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinforma. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2010;34:188–193. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle JG, Dodd AF, McGrath JA, Wolyniec PS, Mitchell AA, Shetty AC, Sobreira NL, Valle D, Rudd MK, Satten G, Cutler DJ, Pulver AE, Warren ST. Microdeletions of 3q29 confer high risk for schizophrenia. Am J Hum Genet. 2010;87:229–236. doi: 10.1016/j.ajhg.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustelin T. Are other protein tyrosine phosphatases than PTPN22 associated with autoimmunity? Semin Immunol. 2006;18:254–260. doi: 10.1016/j.smim.2006.03.013. [DOI] [PubMed] [Google Scholar]
- O'Connor V, Houtman SH, De Zeeuw CI, Bliss TV, French PJ. Eml5, a novel WD40 domain protein expressed in rat brain. Gene. 2004;336:127–137. doi: 10.1016/j.gene.2004.04.012. [DOI] [PubMed] [Google Scholar]
- O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Cichon S, Nothen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Cloninger CR. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Ogah OS, Timeyin AO, Kayode OA, Otukoya AS, Akinyemi RO, Adeyemi FI. Graves' disease presenting as paranoid schizophrenia in a Nigerian woman: a case report. Cases J. 2009;2:6708. doi: 10.4076/1757-1626-2-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, Gianniny L, Korman BD, Padyukov L, Kurreeman FA, Chang M, Catanese JJ, Ding B, Wong S, van der Helm-van Mil AH, Neale BM, Coblyn J, Cui J, Tak PP, Wolbink GJ, Crusius JB, van der Horst-Bruinsma IE, Criswell LA, Amos CI, Seldin MF, Kastner DL, Ardlie KG, Alfredsson L, Costenbader KH, Altshuler D, Huizinga TW, Shadick NA, Weinblatt ME, de VN, Worthington J, Seielstad M, Toes RE, Karlson EW, Begovich AB, Klareskog L, Gregersen PK, Daly MJ, Plenge RM. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Korn JM, McCarroll SA, Altshuler D, Sklar P, Purcell S, Daly MJ. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, Kawaguchi T, Tsunoda T, Watanabe M, Takeda A, Tomiyama H, Nakashima K, Hasegawa K, Obata F, Yoshikawa T, Kawakami H, Sakoda S, Yamamoto M, Hattori N, Murata M, Nakamura Y, Toda T. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- Sato T, Irie S, Kitada S, Reed JC. FAP-1: a protein tyrosine phosphatase that associates with Fas. Science. 1995;268:411–415. doi: 10.1126/science.7536343. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Cherny SS, Purcell S, Hewitt JK. Power of linkage versus association analysis of quantitative traits, by use of variance-components models, for sibship data. Am J Hum Genet. 2000;66:1616–1630. doi: 10.1086/302891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe'er I, Dudbridge F, Holmans PA, Whittemore AS, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Crowe RR, Oksenberg JR, Mirel DB, Kendler KS, Freedman R, Gejman PV. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, Kruger R, Federoff M, Klein C, Goate A, Perlmutter J, Bonin M, Nalls MA, Illig T, Gieger C, Houlden H, Steffens M, Okun MS, Racette BA, Cookson MR, Foote KD, Fernandez HH, Traynor BJ, Schreiber S, Arepalli S, Zonozi R, Gwinn K, van der BM, Lopez G, Chanock SJ, Schatzkin A, Park Y, Hollenbeck A, Gao J, Huang X, Wood NW, Lorenz D, Deuschl G, Chen H, Riess O, Hardy JA, Singleton AB, Gasser T. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100:1552–1556. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di FM, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St CD, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Ophoff A, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietilainen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Borglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Bottcher Y, Olesen J, Breuer R, Moller HJ, Giegling I, Rasmussen HB, Timm S, Mattheisen M, Bitter I, Rethelyi JM, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, Gulcher JR, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA, Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, de FR, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Yoon JL, Shianna KV, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jonsson EG, Terenius L, Agartz I, Petursson H, Nothen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, St CD, Goldstein DB, Stefansson K, Collier DA. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF. The genetics of schizophrenia. PLoS Med. 2005;2:e212. doi: 10.1371/journal.pmed.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Lin D, Tzeng JY, van den OE, Perkins D, Stroup TS, Wagner M, Lee S, Wright FA, Zou F, Liu W, Downing AM, Lieberman J, Close SL. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Jia P, Fanous AH, Webb BT, van den Oord EJ, Chen X, Bukszar J, Kendler KS, Zhao Z. A multi-dimensional evidence-based candidate gene prioritization approach for complex diseases—schizophrenia as a case. Bioinformatics. 2009;25:2595–6602. doi: 10.1093/bioinformatics/btp428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Schizophrenia Consortium Stone. O'Donovan J, Gurling M, Kirov HM, Blackwood G, Corvin D, Craddock A, Gill N, Hultman M, Lichtenstein C, McQuillin P, Pato A, Ruderfer CN, Owen D, St MJ, Sullivan CD, Sklar PF, Purcell P, Stone S, Korn J, Macgregor J, Morris S, O'Dushlaine D, Daly C, Visscher PM, Holmans P, Scolnick E, Williams NM, Georgieva L. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsater A, Flex A, Aben KK, de VF, Mulders PF, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, McCarthy MI, Hattersley AT. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin AA, Heward JM, Brand OJ, Newby PR, Franklyn JA, Gough SC, Simmonds MJ. Use of Tag single nucleotide polymorphisms (SNPs) to screen PTPN21: no association with Graves' disease. Clin Endocrinol (Oxf) 2006;65:380–384. doi: 10.1111/j.1365-2265.2006.02608.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


