Abstract
The rapid growth of the number of protein sequences that can be inferred from sequenced genomes presents challenges for function assignment, as only a small fraction (currently <%) of have been experimentally characterized. Bioinformatics tools are commonly used to predict functions of uncharacterized proteins. Recently there has been significant progress in using protein structures as an additional source of information to infer aspects of enzyme function, which is the focus of this review. Successful application of these approaches has led to the identification of novel metabolites, enzyme activities, and biochemical pathways. We discuss opportunities to systematically elucidate protein domains of unknown function, orphan enzyme activities, dead-end metabolites, and pathways in secondary metabolism.
Keywords: enzyme function prediction, protein structures, homology modeling, docking, metabolic pathways
The challenge of protein function assignment
The rapid advances in genome sequencing technology have created enormous opportunities and challenges in defining the functional significance of encoded proteins. While the number of genome sequences continues to grow rapidly, experimentally verified functional annotations lag well behind and grow at a much slower pace. As of May 2014, the UniProtKB (TrEMBL and Swiss-Prot) database contained 56,010,222 sequences, but only 545,388 sequences (~%) are listed in Swiss-Prot, the manually annotated and reviewed portion of UniProtKB [1, 2], where experimental information about function is reported. High-throughput bioinformatics methods are clearly needed to bridge this gap, but many significant challenges remain for reliably predicting the functions of proteins using the most common approaches, which are based primarily on transferring the relatively small number of experimentally determined functions to large collections of proteins based on sequence similarity. The rates of misannotation in the major repositories of protein sequence information, such as GenBank and TrEMBL, are unknown but estimated to be large [3, 4].
One fundamental challenge is that there is no universal criterion sufficient to determine when a pair of proteins are likely to have the same or different functions; even if two proteins are highly homologous to one another and have similar structures, a change of only a few residues in the active site can change the functional specificity [5]. A second fundamental challenge is that annotation transfer, by definition, cannot identify new, uncharacterized protein functions. These challenges have motivated the development of diverse approaches to protein functional characterization and prediction. Such approaches use additional types of information beyond protein sequence, such as high-throughput metabolomics [6], RNA profiling [7-9], proteomics [10, 11], and phenotyping experiments [12], and orthogonal types of bioinformatics information such as genome organization (operons and gene clusters; domain fusions) and metabolic systems analysis [13].
In this review, we focus on the use of protein structure, in conjunction with other types of information, to aid function assignment, including the determination of novel functions and pathways. Structural information has been used to help elucidate many aspects of function including protein-protein interactions (e.g., scaffolding) and regulation, but our focus here is biochemical function; that is, the determination of enzymatic activities in vitro and in vivo.
Using structure to infer small molecule binding
From structure to function
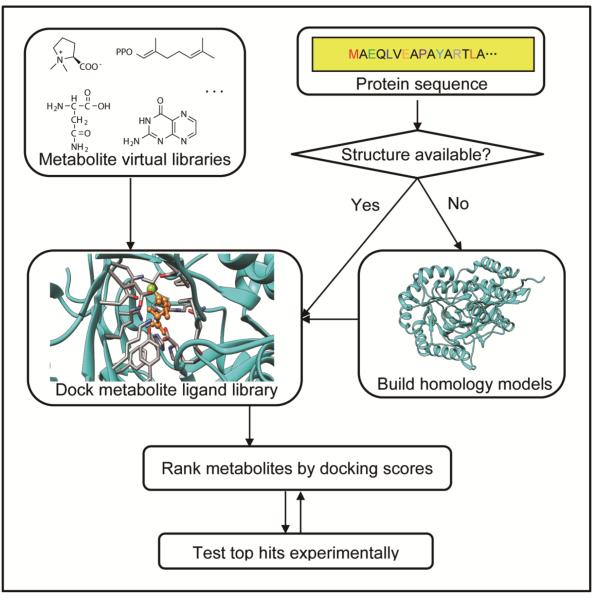
Structural genomics (see Glossary) efforts have generated a large number of structures for proteins with uncertain function. In the case of enzymes, these structures can be used to make inferences about function, either qualitatively, through inspection by an expert, or in more quantitative and automated ways. One class of methods generates functional hypotheses based on physicochemical similarity of the putative active site to the active sites of structurally and functionally characterized enzymes [14-18]. A second class of methods exploits computational tools developed primarily for computer-aided drug design to predict the substrates, products, or intermediates of an enzyme. Specifically, the strategy consists of docking (see Glossary) an in silico metabolite library against an enzyme active site and experimentally testing the top ranking metabolites to determine in vitro biochemical activity (Figure 1). A number of excellent reviews are available describing the algorithms used in docking programs and their limitations [19, 20], including their highly approximate treatment of key forces driving binding such as electrostatics, solvation, and entropy losses. Although such algorithms have been extensively benchmarked, and demonstrated their practical utility for computer-aided drug design, significant effort was required to test docking for enzyme-substrate recognition, resulting in various modifications to improve performance in this application [21-34]. Many metabolites are more highly charged than typical drug-like molecules; one successful approach for metabolite docking uses molecular mechanics-based scoring functions that treat electrostatics and solvation in a more realistic (and computationally expensive) [21, 35]. Shoichet and co-workers introduced the concept of docking "high energy intermediates" rather than substrates or products of enzymes, and demonstrated that this approach improved the ability to predict the binding mode of metabolites, and the ability to distinguish true substrates from false positives [30, 36].
Figure 1.
Structure based virtual metabolite docking protocol for enzyme activity prediction. When no structure has been experimentally determined for a protein sequence, a model can be built using a variety of comparative modeling methods, but only when the structure of a homologous protein is available that has ~30% of greater sequence identity to the protein of interest. Whether using a structure of a model, it is critical that active site metal ions and cofactors are present, and that catalytic residues are positioned appropriate for catalysis. Virtual metabolites libraries can be constructed and "docked" against the putative active sites of structures or models using computational tools more commonly employed in structure-based drug design (e.g., Glide, DOCK). The docking scoring functions can be used to rank the ligands according to their estimated relative binding affinities. Top scoring metabolites are typically inspected for plausibility (Is the predicted binding mode compatible with catalysis? Is the metabolite likely to be present in the relevant organism?), and then selected for experimental testing (in vitro enzymology). Protocols similar to that shown here have been used in retrospective and prospective studies [22-25, 27-33, 36, 39].
Even with these methodological improvements, there are numerous caveats to this approach, both fundamental and practical. A fundamental limitation is that docking methods can, at best, predict binding interactions, which is necessary but not sufficient for a ligand to be the substrate of an enzyme. In practice, experimental testing of top hits from metabolite docking frequently reveals many false positives, including weak substrates with very poor kcat (but reasonable KM), that is, metabolites that bind to the enzyme but are not efficiently turned over [27].
An important practical limitation of metabolite docking is that existing databases of metabolites are incomplete. A second practical limitation is that the structures used for docking must have ordered active sites, including any metal ions. However, it is possible to predict relatively small conformational changes associated with ligand binding, especially in side chains [37].
Another limitation for molecular mechanics-based scoring functions is that the electronic structures of transition states cannot be accurately described. In principle, combining quantum mechanics and molecular mechanics methods (QM/MM) can provide more accurate analysis of the mechanisms and specificities of enzymes. A proof-of-concept study has shown that such an approach may become practical for studying certain challenging aspects of enzyme specificity, compared to the more common use of quantum mechanical methods to investigate reaction mechanisms [38]. In the future, this type of approach may be particularly important when studying enzymes with intermediates that are radicals (e.g. P450 enzymes and radical SAM enzymes). However, such calculations are currently prohibitively expensive to be used in large scale.
Despite these limitations, metabolite docking has proven to be useful in practice for generating testable hypotheses about function, which have proven to be correct in many cases. Herman et al. [30, 36] and Fan et al. [28, 29, 39] docked the high-energy intermediates of metabolites and successfully predicted deaminase activity in several functionally uncharacterized enzymes of the amidohydrolase superfamily. Favia et al. [22] examined the ability of docking to identify cognate substrates of enzymes belonging to the short chain dehydrogenases/reductases superfamily. In several of these studies, subsequently determined co-crystal structures with metabolites confirmed the binding mode predicted by docking [23, 24, 27, 32, 33].
From sequence to function using homology models
Structural information can also be leveraged to help infer enzymatic function for proteins lacking structures. Although homology modeling (see Glossary) remains imperfect [40], models have been successfully used to infer aspects of function in a great many cases, including models of proteins based on the structures of proteins with which they have relatively low sequence identity (30% or lower); examples involving metabolite docking are discussed below [27, 28, 33]. The leverage of a single structure can be large; on average, each new structure determined by structural genomics efforts could be used to create models for hundreds or thousands of homologous sequences [41]. Pre-computed homology models can be obtained from databases such as SwissModel [42] and ModBase [43], which contain models for millions of protein sequences.
One of the simplest approaches to infer aspects of enzyme function, when no structure is available, is to identify putative active site residues in protein sequences by sequence alignment to proteins with solved structures. Changes in critical active site residues can suggest changes in the enzymatic reaction (e.g., changes in catalytic amino acids) or specificity. Constructing homology models can provide additional information about the predicted three-dimensional arrangement of active site residues. Catalytic and other critical active site residues are frequently very well conserved across homologs, facilitating accurate sequence alignment, and hence the accuracy of the models, for regions surrounding the active site; nonetheless, allowing some degree of receptor flexibility in the docking protocol can be helpful to address small errors in, for example, side chain positioning [24, 33, 37].
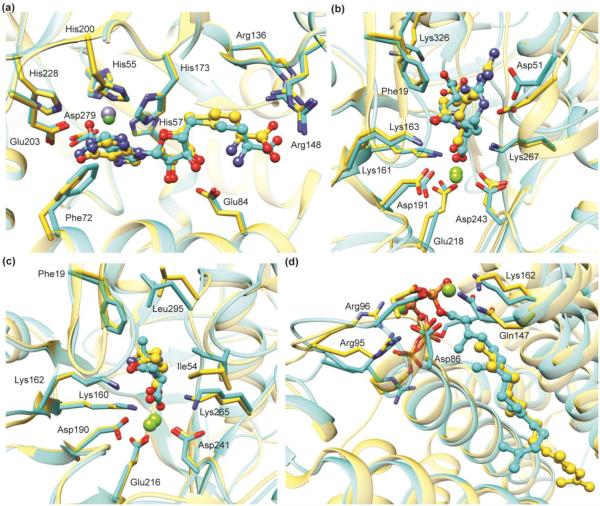
Homology models have been used to accurately predict the substrate specificity of enzymes in the enolase (Figures 2b and 2c) and isoprenoid synthase (Figure 2d) superfamilies [24, 25, 27, 33]. In each case, a structure of the enzyme was subsequently determined that confirmed the predicted binding mode, and in vitro enzymology confirmed that the ligands were proficient substrates. The examples in Figures 2c and 2d are taken from studies in which predictions were made for dozens of enzymes [27, 33], using homology models constructed based on template structures with sequence identities as low as 25%. That is, it is straightforward to automate the process of creating multiple homology models, all based on a particular template structure, for a series of homologous proteins in a multiple-sequence alignment and then dock against all of them [28].
Figure 2.
Predicted binding poses are in good agreement with subsequently determined experimental structures. Predicted ligand binding mode (cyan) superimposed with the X-ray crystal structure (gold) of: (a) S-adenosylhomocysteine deaminase (PDB: 2PLM); (b) N-succinyl-L-Arg racemase (PDB: 2P8C); (c) D-Ala-D-Ala epimerase (PDB: 3Q4D), and (d) a polyprenyl synthase (PDB: 4FP4). In (b), (c), and (d), the docking predictions were made using homology models based on crystal structures with 35%, 39%, and 29% sequence identity, respectively.
Finally, certain X-ray crystal structures can be used to help identify small molecule ligands in a complementary fashion. Almo and co-workers have estimated that 3-5% of all structures determined by the NYSGXRC structural genomics center contain organic ligands from the expression host that survived purification [44]. Unassigned electron density, at sufficient resolution, can be sufficient in some cases to infer the nature of the substrate, although determining the mass of the metabolite by mass spectroscopy provides a very useful constraint. This type of detective work led Almo and co-workers to discover a novel metabolite, carboxy-S-adenosyl-L-methionine, and a pathway that uses it to modify RNA [44]. In cases where the identity of the ligand remains ambiguous, metabolite docking may provide a useful way of identifying ligands that match the electron density and are predicted to have favorable binding interactions [45, 46].
Although the number of protein structures is much smaller than the number of protein sequences inferred from genome sequencing, and will undoubtedly remain so, a variety of complementary approaches have emerged to utilize these structures to make inferences concerning enzymatic function. At the present time, experimental testing remains essential, but the computational approaches can help guide the design of experiments, and focus attention on enzymes likely to have novel or unexpected activities. In favorable cases, homology modeling can be used to extend the use of structure-based methods to large numbers of proteins lacking experimental structures. A major challenge is automating the metabolite docking methods, which remain technically complex; the Metabolite Docker web resource (http://metabolite.docking.org/) [47, 48], and its application to metabolite docking, represents important progress in this direction.
Structural information in the context of pathways
As we have shown, a single structure (or model) of an enzyme can be used to make testable predictions concerning its potential substrate(s). However, in vitro activity does not, by itself, necessarily imply in vivo biochemical function. When enzymes can be placed into pathways or networks, additional information is available for predicting both in vitro and in vivo biochemical function.
In prokaryotes and certain eukaryotes, enzymes involved in pathways are frequently located in close proximity on the genome. In some cases, functionally related proteins also appear in certain organisms as gene fusions. A family of genome context analysis techniques takes advantage of these observations to infer functional relationships among genes, even when they do not share sequence similarity. These techniques have been exploited by databases such as Metacyc [49], MicrobesOnline [50], STRING [51], SEED [52] and IMG [53]. Although genome proximity is not a useful source of information for most eukaryotes, other types of experiments, such as interactome mapping by mass spectroscopy or other methods [54, 55], can be used in an analogous manner; that is, to develop hypotheses concerning proteins that have related functions.
Structural genomics efforts have added a structural perspective to biochemical pathways in certain organisms. The Joint Center for Structural Genomics has determined the structures of over 100 enzymes in the central metabolism of Thermotoga martima, and created homology models for hundreds of others [56]. In less well-studied organisms it would be rare to find entire pathways for which each enzyme has been structurally characterized, but as in the case of T. maritima it is frequently possible to create models for multiple enzymes in a putative pathway. In this context, metabolite docking can be expanded to pathway docking; that is, metabolite docking against multiple structures or models of proteins hypothesized to participate in a metabolic pathway or network [26, 32]. In addition to potentially increasing the in vivo relevance of the results, docking metabolites to multiple binding sites in the same pathway can also increase the reliability of in silico predictions of substrate specificity because the pathway intermediates are chemically similar even if the proteins involved are structurally unrelated. Put simply, the product of one enzyme is the substrate for another enzyme, and comparing the metabolite docking results can help to refine hypotheses concerning the individual protein functions as well as the overall pathway.
Pathway docking was first introduced by Kalyanaraman et al. to retrospectively ‘predict’ the intermediates in the glycolysis pathway in Escherichia coli [26]. In this proof-of-concept study, a large and diverse in silico metabolite library derived from Kyoto Encyclopedia of Genes and Genomes (KEGG) was docked against structures and homology models of ten enzymes in the glycolysis pathway. The ranks of the 'correct' substrates were all within the top 1% of the hit list, and in six out of ten cases, cognate substrates were ranked within the top 0.3%, i.e., among the top ~50 ligands.
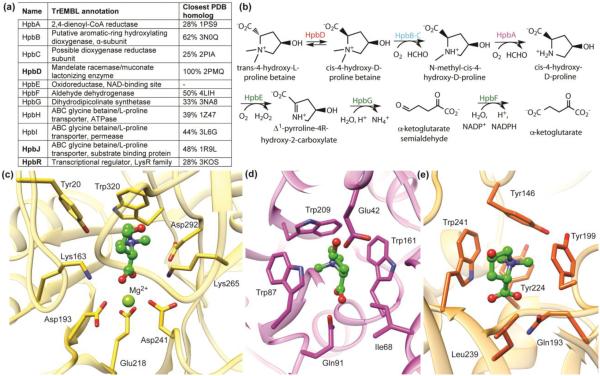
Zhao et al. performed a prospective application of the pathway docking method, which led to the discovery of new enzymes in the hydroxyproline betaine/proline betaine metabolism pathways (Figure 3) [31, 32]. The initial focus was an uncharacterized member of the enolase superfamily, HpbD, the apo structure of which was determined in a structural genomics effort. The genome contexts are similar for HpbD and its putative orthologs in ~20 organisms, suggesting a conserved pathway, and homology models could be created for many of these (Figure 3) Metabolite docking against the structure and several homology models suggested that the pathway involved catabolism of amino acid derivatives, especially N-modified proline derivatives. A model of a periplasmic binding protein encoded by a gene located close to HpbD was particularly informative and suggested that the binding site contained a cation-π cage composed of 3 Trp side chains (Figure 3); docking results strongly suggested that the cation would be a quaternary amine, specifically a betaine (N-trimethylated amino acid). The combined results led to the prediction of catabolic pathways for proline betaine and trans-4R-hydroxyproline betaine (both are important osmolytes in marine organisms), with HpbD performing inversion of stereochemistry at the Cα position [31, 32]. Subsequent in vitro enzyme assays and in vivo metabolomics experiments confirmed these predictions and elucidated aspects of the regulation of these pathways.
Figure 3.
Structure-guided discovery of new enzymes in a novel hydroxyproline betaine metabolism pathway. Panel (a) shows the name, TrEMBL annotation, and most similar homolog in the PDB for each protein in the pathway. The automated TrEMBL annotations are incorrect or imprecise for all proteins in the pathway. However, there is rich structural information that can be used for modeling and docking, as shown in the closest PDB homolog column. The pathway is shown in (b). Panels c-e show the binding site and/or active site of the three proteins (HpbD, HpbJ and HpbR, shown in bold in (a)) in the pathway, respectively, along with the docking-predicted binding mode for the ligand trans-4-hydroxy-L-proline betaine (ball-and-stick, green color). Both HpbJ and HpbR have a predicted cation-π cage, known for binding quaternary amines. In HpbD, two catalytic residues (Lys163 and Lys265) replace aromatic residues, leaving Trp320 as the key aromatic residue forming a cation-π interaction with the substrate.
Challenges and opportunities
No single computational or experimental approach alone is likely to "solve" the problem of predicting or determining the functions of the millions of currently uncharacterized enzymes, especially for the most challenging goal of identifying novel enzymatic activities and biochemical pathways. However, the combination of sequence-based (bioinformatics) and structure-based computational methods—together with high-throughput protein expression, enzyme assays, crystallography, metabolomics, phenotyping, and potentially many other approaches—can provide powerful approaches to generate and evaluate hypotheses. A major challenge and opportunity is the development of methods to optimally combine these disparate types of computational and experimental data to make functional inferences. Even in the context of pathway docking, functional inferences have thus far been made with the aid of human knowledge and intuition, but certain aspects of the data integration can certainly be automated and systematized. The scope of the potential applications of these integrated approaches is vast, and we highlight a few opportunities here.
Biosynthetic pathways for natural products
Natural products such as polyketides, non-ribosomal peptides, isoprenoids, alkaloids and ribosomally synthesized and posttranslationally modified peptides are structurally diverse secondary metabolites (see Glossary), many of which have biological activity and are used in modern medicine (erythromycin, vancomycin, taxol, morphine, duramycin, etc.). The biochemical pathways that create these natural products represent a challenge for function prediction because the chemical space is enormous; that is, the number of possible intermediates and end products of pathways in secondary metabolism is virtually limitless. Moreover, the experimental characterization of the structures of these secondary metabolites is often challenging due to frequently complex ring structures and stereochemistry. For these reasons, the elucidation of the biosynthetic pathways of these high value secondary metabolites remains challenging, even when the genome of the producing organism has been sequenced; for instance, only a small fraction of the tens of thousands of known alkaloids have their biosynthetic pathways fully elucidated [57, 58].
One area of rapid progress has been the prediction of templated biosynthetic pathways for polyketides and non-ribosomal peptides, due to the modular nature of the biosynthetic enzymes and their frequent occurrence in large gene clusters or operons. Sequence-structure-function relationships have been well characterized for certain classes of enzymes in these pathways, such as polyketide synthases and non-ribosomal peptide synthetases [59-61]. This knowledge has been harnessed in efforts to achieve combinatorial biosynthesis of novel polyketides and non-ribosomal peptides [62-64]. However, elucidating the biosynthetic pathway of non-templated natural products such as isoprenoids and alkaloids remains non-trivial.
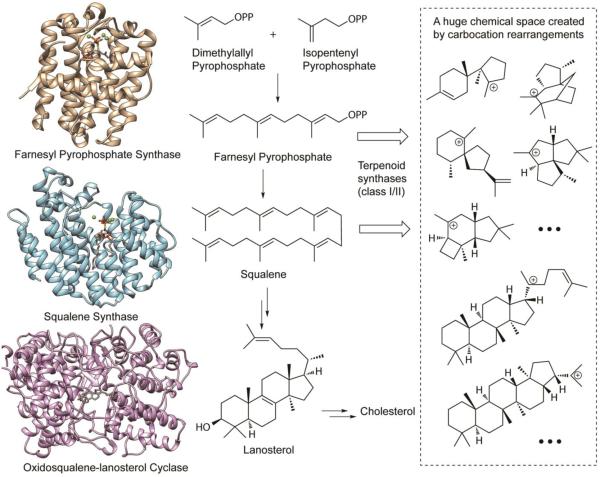
Isoprenoid biosynthesis pathways present both opportunities and challenges with respect to function prediction [65, 66]. In the biosynthesis of isoprenoids, isoprene units (C5) are assembled by polyprenyl transferases to give long chain terpenes such as geranyl pyrophosphate (C10), farnesyl pyrophosphate (C15), geranylgeranyl pyrophosphate (C20), and squalene (C30), which can then be converted into diverse carbon skeletons by terpenoid synthases (also called terpene cyclases), which are sometimes further modified by other enzymes such as S-adenosyl methionine (SAM) dependent methyl transferases. A paradigmatic isoprenoid pathway, the biosynthesis of cholesterol, is illustrated in Figure 4; the crystal structures of key enzymes in the pathway have been solved, including farnesyl pyrophosphate synthase (gold; PDB: 1RQI), squalene synthase (light blue; PDB: 3WEG) and oxidosqualene-lanosterol cyclase (magenta; PDB 1W6K).
Figure 4.
The biosynthesis of cholesterol: a paradigmatic isoprenoid pathway. Crystal structures of key enzymes in the pathway have been solved, including farnesyl pyrophosphate synthase (gold; PDB: 1RQI), squalene synthase (light blue; PDB: 3WEG), and oxidosqualene-lanosterol cyclase (magenta; PDB 1W6K). These crystal structures provide opportunities to predict functions of related enzymes of the isoprenoid synthase superfamily. However, function prediction for the terpenoid synthases (also called terpene cyclases) is extremely challenging due to the huge product chemical space created by carbocation rearrangements.
It is relatively straightforward to leverage structural information in order to predict the product specificities of the polyprenyl synthases. Product chain length has been shown to be determined primarily by the size of the cavity, and Wallrapp et al. [33] have shown that it is possible to predict chain length specificity for sequences lacking structures through a combination of homology modeling and docking. By contrast, predicting the product specificity of isoprenoid synthases is extremely challenging, because the number of possible products is enormous, and the enzymes must bind and stabilize several carbocations and transition states leading to a given product [67]. Despite these challenges, the potential impact of elucidating the sequence-structure-function relationships of isoprenoid synthases is very high, given the importance of these enzymes in the biosynthesis of complex, bioactive natural products and drugs.
Domains of unknown function
A high-value subset of functionally uncharacterized proteins is “Domains of Unknown Function” (DUFs). As the name suggests, no function is known for any member of a DUF protein family; thus, annotating even a single member of a DUF can have a large impact, by defining (in the case of enzymes) aspects of the biochemical capabilities. In Pfam 27.0, 26% (3885 out of 14831) Pfam families are DUFs, with "unknown function" or “uncharacterized protein” in their descriptions [68]. Structures are available in the PDB for proteins in 379 DUF families (as of 2013) [69].
The potential impact of the systematic, structure-guided study of DUFs is suggested by the recent work of Bastard et al. [70], who determined that the DUF849 Pfam family contains β-keto acid cleavage enzymes of diverse substrate specificity. In this work, 14 novel in vitro enzymatic activities of the DUF849 Pfam family have been revealed through an integrated strategy, combining bioinformatics analysis to cluster the protein sequences and structural analysis using both crystal structures and homology models. The structural analysis was primarily qualitative (e.g. whether the substrate is neutral, positively or negatively charged) but also supported by metabolite docking. High-throughput enzymatic screening confirmed many of the predictions and resulted in discovery of in vitro activities for 80 enzymes, including several novel functions; remarkably rapid progress for a protein family that was, until recently, entirely uncharacterized.
Missing links in metabolism orphan enzyme activities and dead-end metabolites
In addition to the many functionally uncharacterized enzymes, there are also many enzyme activities that have been identified but are not associated with any protein sequence. In fact, despite considerable efforts in the past few years [72-78], 20% (1042 out of 5294 [79], as of Feb 2014) of enzyme commission (EC) numbers are not associated with sequence data in any of the three major enzyme databases (Metacyc [49], Expasy [80] and Brenda [81]) and thus are described as orphan ECs. Other terms such as “orphan metabolic activities” and “orphan enzymes” have also been used to describe the phenomenon. The original publication dates for orphan ECs ranges from the 1950s to today, with a mean of 1977 [74, 75]. Many orphan ECs play biologically important roles, and could be an unexplored reservoir of new drug targets [74, 82].
Our incomplete understanding of metabolism is also reflected by "dead-end" metabolites. Metabolites in biochemical networks are generally linked to at least 2 enzymes; that is, each metabolite is both the product of one biochemical reaction and the substrate of another. Dead-end metabolites are those that currently can only be linked to 1 enzyme in an organism, and these can be readily identified by methods of automated metabolic network reconstruction [83]. For example, Mackie et al. recently identified, 127 potential dead-end metabolites in E. coli K-12 [84].
The number of orphan enzyme activities and dead-end metabolites will naturally decrease as new enzyme functions are discovered. However, the ability to identify holes in our understanding of metabolism in specific species suggests new structure-based approaches. Instead of the current approach where a candidate enzyme is studied for functional clues, one could dock substrates (or intermediates) corresponding to orphan enzyme reactions and dead-end metabolites to structures or models of many uncharacterized enzymes within the relevant organism(s).
Although enzyme function can be predicted from protein sequence or, as emphasized in this review, protein structure, the combination of these approaches with high-throughput experimental methods of studying metabolism and methods to computationally interrogate the metabolic networks of entire organisms is likely to be even more powerful. Integrated experimental and computational methods have great promise to systematically fill holes in our understanding of both primary and secondary metabolism.
Concluding remarks
In the sequence-structure-function paradigm, inferring function from structure has proven challenging, and many approaches to function prediction have not utilized structural information at all. In the case of enzymes, there has recently been rapid progress in experimental and computational approaches to inferring aspects of enzymatic activity from structure. Numerous challenges remain (Box 1), including the limitations of existing algorithms for metabolite docking and homology modeling, incomplete in silico databases of metabolites, and incomplete structural coverage of putative enzyme families, despite the advances made by high-throughput protein expression and structural biology (structural genomics). Nonetheless, structure-guided approaches have shown promise, particularly for the most challenging goal of identifying novel metabolites, enzyme activities, and biochemical pathways. As in drug discovery, where structural information is now routinely used to guide design, we believe that enzyme structures will prove to be an essential component of strategies for enzyme function prediction, not in isolation, but rather integrated with many other experimental and computational methods.
Box 1. Box captions.
Outstanding questions
When the binding site of an enzyme is unknown, and cannot be inferred from homologous proteins, can we predict the site using sequence- and/or structure-based methods? Can enzymes be readily identified from sequence or structure, compared to proteins that lack catalytic function?
How complete are existing in silico databases of metabolites, for specific organisms (e.g., E. coli, humans) and for life on Earth in general? Are there entirely new classes of secondary metabolites that have not yet been discovered?
How can we define the functions of an enzyme when it catalyzes multiple reactions? What is the best way to predict functions of such enzymes?
How can information from high-throughput metabolomics, protein interaction, and phenotyping experiments be optimally combined with sequence and structural information to infer enzyme activities and pathways or networks?
Among the ~50 million protein sequences identified from genome sequences thus far, how many enzyme activities exist? What fraction of enzymes have multiple activities in vitro and in vivo?
Highlights.
Of the >50 million protein sequences, <1% have experimentally determined functions.
Protein structures can provide clues to function such as the substrates of enzymes.
Homology modeling and ligand docking algorithms can help infer function from structure.
Recent successes include discovery of novel metabolites, enzymes, and pathways.
Acknowledgments
This work was part of the Enzyme Function Initiative supported by the National Institutes of Health Grant U54 GM093342. We thank John Gerlt (U. Illinois) for helpful discussions. We also thank Dr. Johannes Hermann and Dr. Frank Wallrapp for kindly sending us docked poses for Fig. 2a and Fig. 2d. MPJ is a consultant to Schrodinger LLC, which developed and distributes some of the software used in studies cited here.
Glossary Box
- Homology modeling
A computational technique that builds an atomic model of a target protein using its sequence and an experimental three-dimensional structure of a homologous protein (called the “template”). The quality of a homology model depends on the accuracy of the sequence alignment between target and template, which varies (loosely) with the sequence identity (roughly speaking, pairwise identity higher than 40% is ideal, and lower than 25% is poor).
- Ligand docking
A computational technique that predicts and ranks the binding poses of small molecule ligands to receptors (e.g. proteins). Docking usually consists of a sampling method that generates possible binding poses of a ligand in a binding site, and a scoring function that ranks these poses. Most scoring functions are empirical, and give only a crude estimate of the binding free energy of a ligand.
- Secondary metabolism
Biochemical pathways to produce organic molecules (i.e. secondary metabolites) that are not absolutely required for the survival of the organism. There are five particularly prevalent classes of secondary metabolites: isoprenoids, alkaloids, polyketides, non-ribosomal peptides and ribosomally synthesized and post-translationally modified peptides. Secondary metabolites are often restricted to a narrow set of species and have important ecological roles for the organisms that produce them. Many secondary metabolites are bioactive (antibacterial, anticancer, antifungal, antiviral, antioxidant, anti-inflammatory, anti-parasitic, anti-malaria, cytotoxic etc.) and have been used as drugs and drug leads.
- Structural genomics
An effort to determine the three-dimensional, atomic-level structure of every protein encoded by a genome through a combination of high-throughput experimental and modeling approaches. The determination of a protein structure though a structural genomics effort often precedes knowledge of its function, motivating the development of methods to infer function from structure.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.UniProtKB/Swiss-Prot protein knowledgebase release 2014_01 statistics. [Online]. Available: http://web.expasy.org/docs/relnotes/relstat.html.
- 2.UniProtKB/TrEMBL protein database release 2014_01 statistics. [Online]. Available: http://www.ebi.ac.uk/uniprot/TrEMBLstats.
- 3.Friedberg I. Automated protein function prediction - the genomic challenge. Briefings in Bioinformatics. 2006;7:225–242. doi: 10.1093/bib/bbl004. [DOI] [PubMed] [Google Scholar]
- 4.Schnoes AM, et al. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comp. Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seffernick JL, et al. Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different. J. Bacteriol. 2001;183:2405–2410. doi: 10.1128/JB.183.8.2405-2410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patti GJ, et al. Innovation: Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner EM. Monitoring gene expression: quantitative real-time rt-PCR. Methods Mol. Biol. 2013;1027:19–45. doi: 10.1007/978-1-60327-369-5_2. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, et al. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu AR, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 11.Meier M, et al. Proteome-wide protein interaction measurements of bacterial proteins of unknown function. Proc. Natl. Acad. Sci. U. S. A. 2013;110:477–482. doi: 10.1073/pnas.1210634110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs H, et al. Mouse phenotyping. Methods. 2011;53:120–135. doi: 10.1016/j.ymeth.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Bassel GW, et al. Systems analysis of plant functional, transcriptional, physical interaction, and metabolic networks. Plant Cell. 2012;24:3859–3875. doi: 10.1105/tpc.112.100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kufareva I, et al. Compound activity prediction using models of binding pockets or ligand properties in 3D. Curr. Top. Med. Chem. 2012;12:1869–1882. doi: 10.2174/156802612804547335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilmeier JP, et al. Rapid catalytic template searching as an enzyme function prediction procedure. PLoS One. 2013;8:e62535. doi: 10.1371/journal.pone.0062535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, et al. Understanding a substrate's product regioselectivity in a family of enzymes: a case study of acetaminophen binding in cytochrome P450s. PLoS One. 2014;9:e87058. doi: 10.1371/journal.pone.0087058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amin SR, et al. Prediction and experimental validation of enzyme substrate specificity in protein structures. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E4195–4202. doi: 10.1073/pnas.1305162110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carbonell P, Faulon JL. Molecular signatures-based prediction of enzyme promiscuity. Bioinformatics. 2010;26:2012–2019. doi: 10.1093/bioinformatics/btq317. [DOI] [PubMed] [Google Scholar]
- 19.Meng EC, et al. Automated docking with grid-based energy evaluation. J. Comput. Chem. 1992;13:505–524. [Google Scholar]
- 20.Wang RX, et al. Comparative evaluation of 11 scoring functions for molecular docking. J. Med. Chem. 2003;46:2287–2303. doi: 10.1021/jm0203783. [DOI] [PubMed] [Google Scholar]
- 21.Kalyanaraman C, et al. Virtual screening against highly charged active sites: Identifying substrates of alpha-beta barrel enzymes. Biochemistry. 2005;44:2059–2071. doi: 10.1021/bi0481186. [DOI] [PubMed] [Google Scholar]
- 22.Favia AD, et al. Molecular docking for substrate identification: the short-chain dehydrogenases/reductases. J. Mol. Biol. 2008;375:855–874. doi: 10.1016/j.jmb.2007.10.065. [DOI] [PubMed] [Google Scholar]
- 23.Xiang DF, et al. Functional annotation and three-dimensional structure of Dr0930 from Deinococcus radiodurans, a close relative of phosphotriesterase in the amidohydrolase superfamily. Biochemistry. 2009;48:2237–2247. doi: 10.1021/bi802274f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song L, et al. Prediction and assignment of function for a divergent N-succinyl amino acid racemase. Nat. Chem. Biol. 2007;3:486–491. doi: 10.1038/nchembio.2007.11. [DOI] [PubMed] [Google Scholar]
- 25.Kalyanaraman C, et al. Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening. Structure. 2008;16:1668–1677. doi: 10.1016/j.str.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalyanaraman C, Jacobson MP. Studying enzyme-substrate specificity in silico: a case study of the Escherichia coli glycolysis pathway. Biochemistry. 2010;49:4003–4005. doi: 10.1021/bi100445g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukk T, et al. Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4122–4127. doi: 10.1073/pnas.1112081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan H, et al. Assignment of pterin deaminase activity to an enzyme of unknown function guided by homology modeling and docking. J. Am. Chem. Soc. 2013;135:795–803. doi: 10.1021/ja309680b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hitchcock DS, et al. Structure-guided discovery of new deaminase enzymes. J. Am. Chem. Soc. 2013;135:13927–13933. doi: 10.1021/ja4066078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermann JC, et al. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448:775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar R, et al. Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine. MBio. 2014;5:e00933–00913. doi: 10.1128/mBio.00933-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao SW, et al. Discovery of new enzymes and metabolic pathways by using structure and genome context. Nature. 2013;502:698–702. doi: 10.1038/nature12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallrapp FH, et al. Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E1196–E1202. doi: 10.1073/pnas.1300632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rakus JF, et al. Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis. Biochemistry. 2009;48:11546–11558. doi: 10.1021/bi901731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacobson MP, et al. Force field validation using protein side chain prediction. J. Phys. Chem. B. 2002;106:11673–11680. [Google Scholar]
- 36.Hermann JC, et al. Predicting substrates by docking high-energy intermediates to enzyme structures. J. Am. Chem. Soc. 2006;128:15882–15891. doi: 10.1021/ja065860f. [DOI] [PubMed] [Google Scholar]
- 37.Sherman W, et al. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 38.Tian BX, et al. Predicting enzyme-substrate specificity with QM/MM methods: a case study of the stereospecificity of D-glucarate dehydratase. Biochemistry. 2013;52:5511–5513. doi: 10.1021/bi400546j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamat SS, et al. Enzymatic deamination of the epigenetic base N-6-methyladenine. J. Am. Chem. Soc. 2011;133:2080–2083. doi: 10.1021/ja110157u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moult J, et al. Critical assessment of methods of protein structure prediction (CASP) - round x. Proteins: Struct. Funct. Bioinform. 2014;82:1–6. doi: 10.1002/prot.24452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 42.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pieper U, et al. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2014;42:D336–346. doi: 10.1093/nar/gkt1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, et al. Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function. Nature. 2013;498:123–126. doi: 10.1038/nature12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Binkowski TA, et al. Assisted assignment of ligands corresponding to unknown electron density. J Struct Funct Genomics. 2010;11:21–30. doi: 10.1007/s10969-010-9078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lasker K, et al. Determining macromolecular assembly structures by molecular docking and fitting into an electron density map. Proteins: Struct. Funct. Bioinform. 2010;78:3205–3211. doi: 10.1002/prot.22845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irwin JJ, et al. Automated docking screens: a feasibility study. J. Med. Chem. 2009;52:5712–5720. doi: 10.1021/jm9006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The Metabolite Docker. [Online]. Available: http://metabolite.docking.org/
- 49.Caspi R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dehal PS, et al. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 2010;38:D396–D400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franceschini A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aziz RK, et al. SEED Servers: High-performance access to the SEED genomes, annotations, and metabolic models. PLoS One. 2012;7:e48053. doi: 10.1371/journal.pone.0048053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Markowitz VM, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babu M, et al. Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae. Nature. 2012;489:585–589. doi: 10.1038/nature11354. [DOI] [PubMed] [Google Scholar]
- 55.Havugimana PC, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, et al. Three-Dimensional structural view of the central metabolic network of Thermotoga maritima. Science. 2009;325:1544–1549. doi: 10.1126/science.1174671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zotchev SB. Alkaloids from marine bacteria. New Light on Alkaloid Biosynthesis and Future Prospects. 2013;68:301–333. [Google Scholar]
- 58.Ziegler J, Facchini PJ. Alkaloid biosynthesis: Metabolism and trafficking. Annu. Rev. Plant Biol. 2008;59:735–769. doi: 10.1146/annurev.arplant.59.032607.092730. [DOI] [PubMed] [Google Scholar]
- 59.Walsh CT, Fischbach MA. Natural products version 2.0: connecting genes to molecules. J. Am. Chem. Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keatinge-Clay AT. The structures of type I polyketide synthases. Nat. Prod. Rep. 2012;29:1050–1073. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- 61.Rottig M, et al. NRPSpredictor2--a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011;39:W362–W367. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Go MK, et al. Establishing a toolkit for precursor-directed polyketide biosynthesis: exploring substrate promiscuities of acid-CoA ligases. Biochemistry. 2012;51:4568–4579. doi: 10.1021/bi300425j. [DOI] [PubMed] [Google Scholar]
- 63.Williams GJ. Engineering polyketide synthases and nonribosomal peptide synthetases. Curr. Opin. Struct. Biol. 2013;23:603–612. doi: 10.1016/j.sbi.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wong FT, Khosla C. Combinatorial biosynthesis of polyketides--a perspective. Curr. Opin. Chem. Biol. 2012;16:117–123. doi: 10.1016/j.cbpa.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sacchettini JC, Poulter CD. Biochemistry - Creating isoprenoid diversity. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- 66.Christianson DW. Roots of biosynthetic diversity. Science. 2007;316:60–61. doi: 10.1126/science.1141630. [DOI] [PubMed] [Google Scholar]
- 67.Tantillo DJ. Biosynthesis via carbocations: theoretical studies on terpene formation. Nat. Prod. Rep. 2011;28:1035–1053. doi: 10.1039/c1np00006c. [DOI] [PubMed] [Google Scholar]
- 68.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goodacre NF, et al. Protein domains of unknown function are essential in bacteria. MBio. 2013;5:e00744–00713. doi: 10.1128/mBio.00744-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bastard K, et al. Revealing the hidden functional diversity of an enzyme family. Nat. Chem. Biol. 2014;10:42–49. doi: 10.1038/nchembio.1387. [DOI] [PubMed] [Google Scholar]
- 71.Gerlt JA, et al. The Enzyme Function Initiative. Biochemistry. 2011;50:9950–9962. doi: 10.1021/bi201312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamada T, et al. Prediction and identification of sequences coding for orphan enzymes using genomic and metagenomic neighbours. Mol. Syst. Biol. 2012;8:581. doi: 10.1038/msb.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith AAT, et al. The CanOE strategy: integrating genomic and metabolic contexts across multiple prokaryote genomes to find candidate genes for orphan enzymes. PLoS Comp. Biol. 2012;8:e1002540. doi: 10.1371/journal.pcbi.1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pouliot Y, Karp PD. A survey of orphan enzyme activities. BMC Bioinformatics. 2007;8:244. doi: 10.1186/1471-2105-8-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen LF, Vitkup D. Distribution of orphan metabolic activities. Trends Biotechnol. 2007;25:343–348. doi: 10.1016/j.tibtech.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 76.Ramkissoon KR, et al. Rapid identification of sequences for orphan enzymes to power accurate protein annotation. PLoS One. 2013;8:e84508. doi: 10.1371/journal.pone.0084508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watschinger K, Werner ER. Orphan enzymes in ether lipid metabolism. Biochimie. 2013;95:59–65. doi: 10.1016/j.biochi.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lespinet O, Labedan B. ORENZA: a web resource for studying ORphan ENZyme activities. BMC Bioinformatics. 2006;7:436. doi: 10.1186/1471-2105-7-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Enzyme database statistics. [Online]. Available: http://www.enzyme-database.org/stats.php.
- 80.Artimo P, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schomburg I, et al. BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA. Nucleic Acids Res. 2013;41:D764–772. doi: 10.1093/nar/gks1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lespinet O, Labedan B. Orphan enzymes could be an unexplored reservoir of new drug targets. Drug Discov. Today. 2006;11:300–305. doi: 10.1016/j.drudis.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 83.Karp PD, et al. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Briefings in Bioinformatics. 2010;11:40–79. doi: 10.1093/bib/bbp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mackie A, et al. Dead end metabolites - Defining the known unknowns of the E. coli metabolic network. PLoS One. 2013;8:e75210. doi: 10.1371/journal.pone.0075210. [DOI] [PMC free article] [PubMed] [Google Scholar]