Abstract
In this Review, we provide an overview of flatbed scanner based biomedical imaging and sensing techniques. The extremely large imaging field-of-view (e.g., ~600–700 cm2) of these devices coupled with their cost-effectiveness provide unique opportunities for digital imaging of samples that are too large for regular optical microscopes, and for collection of large amounts of statistical data in various automated imaging or sensing tasks. Here we give a short introduction to the basic features of flatbed scanners also highlighting the key parameters for designing scientific experiments using these devices, followed by a discussion of some of the significant examples, where scanner-based systems were constructed to conduct various biomedical imaging and/or sensing experiments. Along with mobile phones and other emerging consumer electronics devices, flatbed scanners and their use in advanced imaging and sensing experiments might help us transform current practices of medicine, engineering and sciences through democratization of measurement science and empowerment of citizen scientists, science educators and researchers in resource limited settings.
Introduction
Several consumer electronics based imaging and sensing solutions have been recently developed to address global health problems by creating low-cost and yet quite powerful point-of-care devices that exhibit unique advantages over their conventional counterparts. For example, mobile phone based systems have received special attention due to their ability to work even in remote locations and resource poor settings, and to acquire, process, evaluate, and transmit measurement data and results in real time1–21. In a similar way, conventional flatbed scanners, normally used for document or photo digital scanning, offer unique capabilities by providing, within a cost-effective design, an extremely large imaging field of view (e.g., ~600–700 cm2) while having a modest spatial resolution of <10 μm. Here, we review the use of conventional flatbed scanners for biomedical imaging and sensing applications. First, we give a short overview of the optical properties and imaging performance of these flatbed scanner based systems, with an emphasis on the important parameters for designing scientific experiments using these devices. Following this, we discuss some of the key examples of biomedical applications and imaging/sensing experiments that make use of digital scanners. Overall, flatbed scanners, along with other consumer electronics devices including e.g., mobile phones and emerging wearable computers create unique opportunities for democratization of measurement science1 empowering researchers and educators in developing world and resource limited institutions to conduct imaging, sensing and diagnostics related experiments with significantly reduced budgets, infrastructure and maintenance needs, potentially helping us transform how medicine, engineering and sciences are practiced globally. Furthermore, this broad research theme might also empower citizen science by converting everyday digital instruments into advanced measurement tools, helping us generate large quantities of high quality data through a global network of consumers, taking the lead as citizen scientists. Various successful implementations of citizen science have already emerged including for example to predict protein folding and structure22 or to diagnose malaria infected cells using crowd-sourced serious games, i.e., BioGames23,24. While many of these initial efforts have not involved physical measurements or experiments to be performed by the members of the citizen scientist crowd, with the emergence and spread of cost-effective and ubiquitous measurement tools that can be converted from consumer electronics devices, including flatbed scanners and mobile phones, a new level of citizen science would be feasible to distribute not only data analysis and simulations, but also experiments and data collection.
Imaging architectures and properties of flatbed scanners
The flatbed scanners were invented for and are extensively used for document scanning. This task required an imaging system that is capable of creating a digital image over a large field of view (FOV) of ~620 cm2, i.e., the size of an A4 or a US letter paper. This FOV criterion was satisfied by mechanically scanning a one dimensional opto-electronic sensor-array over the entire sample, thus reducing the sensor size, and circumventing the need for a several giga-pixel two dimensional sensor-array, as well as an optical system that is capable of imaging such a large field of view all at once. The spatial resolution of these scanning devices was initially not sufficient for micro-scale imaging tasks, however with the technology evolving to be able to scan photographs and films, relatively high resolution flatbed scanners entered the market.
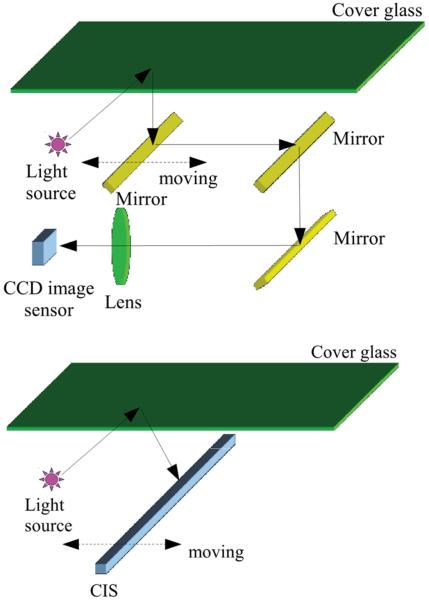
There are mainly two types of flatbed scanner technologies currently available: (1) The Charge-Coupled-Device (CCD) based systems, which utilize a single imaging lens in conjunction with the 1D sensor array; and (2) The Contact Image Sensor (CIS) based systems, where a Gradient Index (GRIN) lens array (also known as the self-focusing lens array) is used to create a unit magnification image (see e.g., Figure 1). In CCD based scanners the length of the CCD chip is usually on the order of a few centimeters (e.g., ~ 4 cm), thus demagnification is performed by the lens to be able to image the full FOV of the sample (e.g., A4 paper). Some of the recent scanners use trilinear CCDs for color imaging, where three rows of pixels are placed next to each other, each equipped with a color filter to be sensitive for the red, green, and blue parts of the spectrum, respectively. The light source used for illumination in these systems is generally a white fluorescent lamp. Recently, to eliminate warm up times, white light-emitting-diodes (LEDs) have also been used. The pixel size of linear CCDs used inside scanners is typically ~2–4 μm. The resolution of these systems are further enhanced by using a staggered pixel configuration, where an additional line of red, green, and blue sensitive pixels is placed next to the original ones with a half pixel shift. Since the CCD sensor pixels are usually more sensitive to light in the middle of the pixel26, this spatial sensitivity information together with the staggered configuration can be used to synthesize an image with a spatial resolution corresponding to half of the pixel pitch without sacrificing sensitivity27–29. Generally speaking, CCD based scanner systems have a longer depth of field (~1–2 mm), which makes them suitable to image samples placed outside the ideal focal plane of the lens, such as samples in Petri dishes. However, as the optical system is designed to image paper sheets, thicker objects can cause significant distortions, especially near the edges of the objects.30
Figure 1.
(Top) Sketch of the optical setup in a CCD based scanner. The image is relayed through several mirrors, and a single lens demagnifies the width of the field of view to fit it onto the CCD sensor active area. (Bottom) Sketch of the optical setup in a CIS based scanner. Imaging is done by a gradient index lens array under unit magnification, i.e., the sensor length equals the width of the field of view. The figure is taken from reference 25.
In CIS based scanner devices the sensor's length is the same as the length of the imaging area31. This is usually achieved by using several (e.g., 8 inside the CanoScan LIDE 200 scanner) shorter opto-electronic sensors placed next to each other. This leads to small gaps (~40 μm) at the sensor array connection points, which creates spatial discontinuities (i.e., dead spots). While in general negligible, this might need to be taken into account when designing a scientific experiment based on a CIS scanner. The pixel size of these sensors is on the order of 5–10 μm. Large format pixels have better noise characteristics and sensitivity, which in general reduce the scanning time. Due to the unit magnification of the imaging system, even with these large format pixels, the CIS scanner provides a resolution similar to CCD based scanners, which need demagnification. Furthermore, since the image of the sample plane is relayed through the GRIN lens array, the depth of field of a CIS based optical system is considerably smaller than CCD based scanners, (for example ~100 μm vs. 1–2 mm, respectively). This means that extremely thin sample holders and/or direct placement of the samples on the scanner glass are required to utilize this type of an imaging system in a biomedical imaging/sensing experiment. In CIS based systems, the scanner head is pushed to be in contact with the bottom surface of the scanner glass, thus keeping the focal plane slightly above the top surface. One possible way to raise the focal plane above the scanner glass, in order to accommodate e.g., thicker sample holders, would be to replace the scanner glass with a thinner one.
As with all imaging systems, the optical resolution and the depth of field depend on the numerical aperture (NA) of the optics. The smaller depth of field in CIS based systems is due to the higher NA of the lens, which also means that the optical resolution provided by the GRIN lens is higher compared to the CCD based systems, and the bottleneck of resolution is usually due to the large pixel size (creating pixelation); this also makes pixel super-resolution methods a possibility to digitally improve spatial resolution in scanner based imaging experiments, as recently demonstrated in Ref. 32.
Note also that the imaging characteristics of a GRIN lens array are fundamentally different than regular lenses. The array creates an image of the object with unit magnification, and each individual point of the object is imaged by multiple GRIN lenses in such a way that when the object and the sensor pixels are at the focal planes, the images created by the individual lenses overlap and create the final image.33 This also means that for out of focus samples the overlap of images relayed by neighboring lenses will not be perfect, hence several shifted copies of the object will appear in addition to the regular out of focus blur corresponding to each lens in the array. Therefore, for experimental systems using a CIS type scanner in conjunction with other optical elements, it is typical to remove the GRIN lens array, and use the bare sensor line to avoid such imaging artifacts25,34.
Illumination in CIS based systems is mostly done by red, green and blue LEDs, either by using several LEDs in a line along the lens array, or using a light rod to distribute the light emitted from a single multi-color LED at the end of the rod. Generally, there are no color filters above the pixels in CIS sensors, so the sensor is by itself monochromatic. The color image is created by blinking the red, green, and blue LEDs sequentially, and registering the responses of the detector for different illuminations.
In both CCD and CIS based scanner systems in order to gain the high resolution color image from the raw sensory information, the data need to be synchronized and post processed, which is usually automatically done by the scanner's chipset. Since scientific experiments usually deal with samples that the scanner was not designed to image, some of these post processing steps, that were optimized for text or photo scanning, can alter the readout data, thus reducing the linearity and the sensitivity of the measurement system. In order to address this problem one can either extract the raw sensory data from the scanner32, or simultaneously scan known objects and use them to digitally calibrate the system35,36. Getting access to raw sensory information is not straightforward, as regular scanner software applications do not provide this feature, and most device drivers are not open access. One possible solution is the use of the Linux scanner driver package: Scanner Access Now Easy (SANE). The scanner itself is controlled internally by the device's chipset, and the back-end of SANE package handles the communication between the chipset and the PC. By setting the values of the chipset's registers before sending the control data-stream to the scanner, it is possible to control the device according to the imaging needs, change the bit depth, gain and scanning speed, modify the shading table used to equalize the pixel's readout signal or turn on/off the built-in illumination.32,37
The large field of view of these scanner-based systems is a clear advantage, however in order to truly judge their capabilities for imaging, we also need to know their optical resolution. The spatial resolution of scanners is given in points per inch (ppi) by their vendors, however almost all of them claim the amount of pixels in the final interpolated image, and not the actual optical resolution achieved by the system. Even in the case of non-interpolated images, the “optical” dpi usually corresponds to the size of the image sensor's pixels. This means that due to the spatial sampling and under-sampling performed by the sensor array, the real optical resolution is generally lower. Also, due to the scanning nature of the system and the frequently used rectangular pixels, the optical resolution can be different in the sensor direction (where it is largely determined by the pixel size) compared to the movement/scanning direction (where the step size and accuracy of the motor play an important role for the final spatial resolution).32
Current flatbed scanners have a modest spatial resolution of <10 μm, however, together with their ultra-large field-of-view (~600 cm2), a pixel count of several giga-pixels can be achieved in each acquired image, resulting in a file size of hundreds of Megabytes. Fortunately, due to recent popularity of giga-pixel photography there are various digital solutions for handling and visualizing giga-pixel images using standard personal computers (see for example Deep Zoom38).
We should emphasize that for most scientific applications, selection of a scanner should be based on the ability of getting low level control of the device and the raw sensor data. CCD based scanners have larger depth of field, and are thus more suitable for samples within relatively thick sample holders (e.g., Petri dish), but are slightly more expensive. CIS based scanners, on the other hand, are cheaper, offer unique advantages for easier modification of their set-up due to the simpler optical system; however they have a shallower depth of field, and therefore are better suited for applications that require imaging of 2D samples in relatively thin sample holders.
Biomedical imaging and sensing applications utilizing flatbed scanners
Flatbed scanners are extensively used for biomedical imaging and sensing tasks, especially for ones that require extremely large fields of view. Among these are the digital imaging of colorimetric assays, absorption based assays, biological samples in large sample holders (e.g., Petri dishes, 96 well-plates) and large samples (brain slices, plant roots, etc.). Here we review some of these experiments that utilize the flatbed scanner as an imager to show the capabilities and the future potential of this consumer electronics device in measurement science.
Colorimetric assays
One of the most frequent uses of flatbed scanners in biomedical imaging is to digitally evaluate colorimetric assays. In this case, the resolution of the system is less important, as the size of the individual objects to be imaged is usually several millimeters in diameter. However the large FOV is necessary, as the total area of the assay can be several cm2. These assays provide inexpensive and yet powerful methods for chemical and biological sensing. They typically consist of several sensor elements, each of which contains a unique dye molecule. These sensor elements have a chemoselective color response upon binding of the target molecules. The absorbance spectra of the dye molecule changes after the exposure to the target chemical (analyte), and the resulting color change is then detected by the scanner. Although the process might be visible to the naked eye, digitization of the data is necessary in order to increase the assay sensitivity and to be able to quantify the analyte concentration, as well as to improve repeatability and remove human reading errors. We should also note that in addition to flatbed scanners, other consumer electronics devices (e.g., cellphones or wearable computers such as Google Glass) have been successfully utilized to fulfill this task (although over significantly smaller fields of view).15,39
In general, each sensor element can give a color response to several different chemicals. The arrays are designed to contain a combination of these sensor elements, where the color change can uniquely identify the analyte. The number of sensor elements per array has risen in the past decade from ~12 to >100.40–42 This creates an increasingly high dimensional measurement vector for detecting, differentiating and quantifying the concentration of the analyte, and makes the digitization and the computer assisted data processing essential. Due to the size and relative complexity of these tests, the flatbed scanner has emerged as a cost-effective solution for high-throughput digitization of the color information in these tests. In such colorimetric experiments, usually the images are acquired by using the factory provided scanner software, and the data are represented in various color spaces for post processing40,43–49. The most popular one is the direct use of the values at the red, green, and blue channels of the captured image; however alternatives, for example the CYMK (Cyan, Yellow, Magenta, Key), or the HSV (Hue, Saturation, Value) color spaces are also utilized50. The optimal choice of the color space representation51 depends on the expected color change of the sample of interest, and can be accordingly adjusted to achieve better signal readout characteristics52,53. In essence, all these solutions work with data that have already been postprocessed by the scanner, and converted into different representations to detect and quantify the color changes. Unfortunately, in implementations that use the factory software the raw bit depth of the sensor is usually lost and the output of the scan can vary with the target itself, creating sensitivity and quantification issues. Gaining low level access to the scanner allows the collection of the raw sensory data32, which can mitigate this problem. Calibration targets scanned together with the object/sample can also provide a possible solution to some of these issues35,36.
Various colorimetric assays were developed by the Suslick research group and were digitized using flatbed scanners. These assays were initially created to detect the presence of metal ligating vapors such as alcohols, amines, arenes, ethers, halocarbons, ketons, phosphines, etc. with better than 1 parts per million (ppm) sensitivity in air.40,46 These “Optoelectronic Noses” were proven to work with complex mixtures as well,46 were used to identify amines with ~0.1 ppm sensitivity45 and were further developed to differentiate closely related volatile organic compounds (VOC) forming a library of 100 VOCs43. In addition to these gas sensors, colorimetric sensor arrays have been created to recognize organic compounds in aqueous solutions with concentrations as low as 1 μM.48 Several applications of these colorimetric assays have been demonstrated, such as the differentiation and quality control of different types of beers and soft drinks47,49. Another advancement for these scanner-based colorimetric assays was the use of nanoporous pigments, which can improve the stability and durability of the array55. These pigments are made by immobilizing soluble dyes into organically modified porous silicates. Using these pigments, a new colorimetric assay was constructed to identify natural and artificial sweeteners at millimolar concentrations,44,56,57 and to detect toxic gases55. Carey et al. used a colorimetric essay in conjunction with a flatbed scanner to identify bacteria with 98.8% accuracy in less than 10 hours from the volatiles they produce (see Figure 2).54
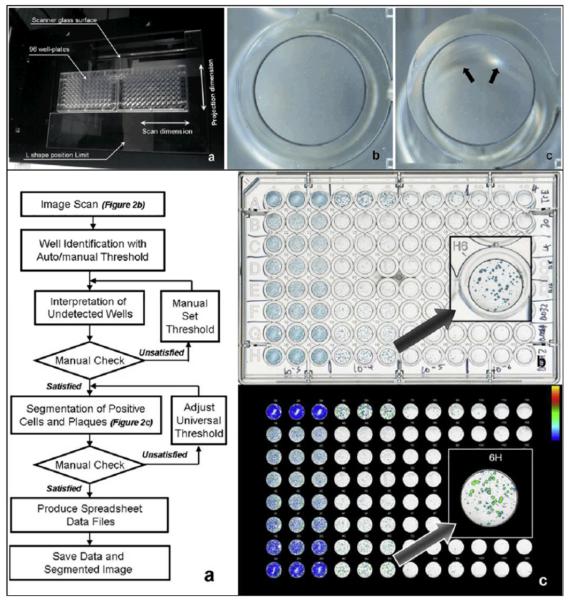
Figure 2.
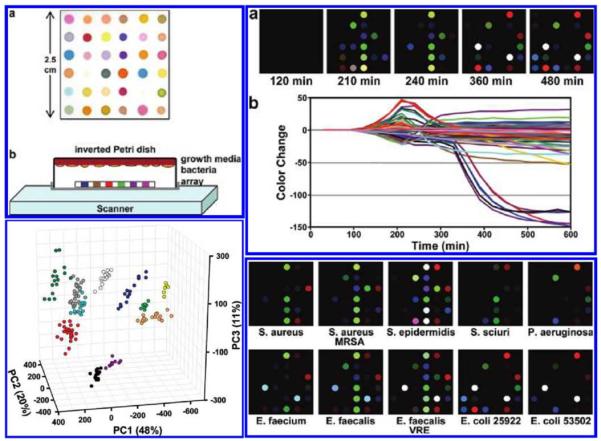
Flatbed scanner and a colorimetric sensor array used for the detection and identification of pathogenic bacteria grown on standard agar from the signatures created by the volatile organic compounds produced by the specimen. (Top Left) (a) The colorimetric sensor array and (b) the schematic of the apparatus containing an inverted Petri dish with the growth media, bacteria and the colorimetric sensor array placed on the scanner glass. (Top Right) (a) Color difference map of E. coli measurements captured over time from successive scans. (b) The color change plotted over time for each spot. (Bottom Right) The color difference maps for various bacterial strains. (Bottom Left) Principle component analysis (PCA) results for 10 strains of bacteria showing three principle components. Each color corresponds to a different strain. Reprinted (adapted) with permission from Reference 54. Copyright (2011) American Chemical Society.
Other research groups have also used flatbed scanners to digitize colorimetric assays. For example, they successfully quantified adsorbed elements and organic compounds on polyurethane foam with comparable results to traditional adsorption-spectrophotometric methods.58–60 Also, flatbed scanners have been used in conjunction with colorimetric assays for pH monitoring61,62, and to develop a dopamine biosensor63,64. Another emerging application is scanner based digitization of paper based, inkjet printed, microfluidic devices to perform chemical sensing65 and immunochemistry66.
Absorption based assays
Flatbed scanners have also been used to image absorption based assays. In these types of assays, generally a single dye is used to stain the sample, and the signal/transmission change corresponds to the concentration of the target chemical/analyte. For example, protein concentrations have been successfully measured using a flatbed scanner in conjunction with ponceau S.68 staining and the Biuret test69. As another example, Rahman et al. described a method named “ScanMIC” to perform minimum inhibitory concentration measurements on Gram-Negative bacteria, such as E. coli using a flatbed scanner. They demonstrated that this method is >92% accurate, and concluded that using a flatbed scanner can be labor saving and provide reliable results within a cost-effective platform.70 Also, an ionophore-based sensor was designed to measure potassium concentrations using a flatbed scanner.71
Metal nanoparticle based assays have also been used in conjunction with a flatbed scanner. For example, Taton et al. created a method for combinatorial DNA analysis, where they used oligonucleotide-modified gold nanoparticle probes, and a silver(I) reduction based signal amplification technique along with a flatbed scanner.72 This research from the Mirkin group has led to the Scanometric approach which was later used for the detection of proteins73, mercury ions74 and cancer markers75. The silver enhanced gold nanoparticle based sample preparation technique was also modified to perform direct and ELISA-like immunoassays on a flatbed scanner by Yeh et al.67 The system was able to achieve a detection limit of ~ 1 ng/mL for a target antibody (see Figure 3). The Scanometric approach offers some advantages over standard ELISA like immunoassays and fluorescence based microarrays due to its high sensitivity and selectivity.75 The procedure is based on increasing the scattered light from gold nanoparticle probes, which makes it ideal to be used in conjunction with a reflection (or transmission) based flatbed scanner to perform complex biomedical assays. Dual enlarged silver enhanced gold nanoparticles have also been used in conjunction with a scanner to perform the detection of Campylobacter jejuni.76
Figure 3.
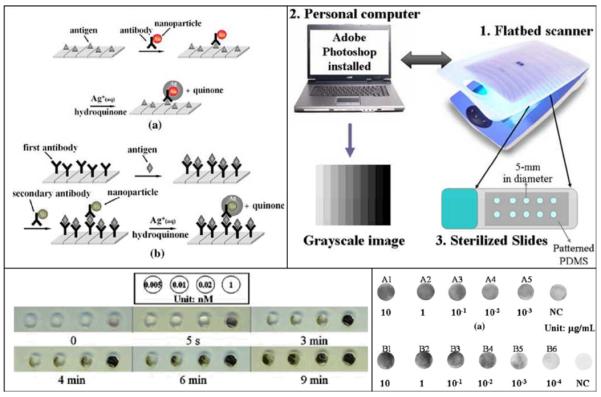
Immunoassay performed on a flatbed scanner. (Top left) Sketches of (a) two layered and (b) three layered (ELISA-like) immunoassay showing silver enhanced gold nanoparticle labeling technique. (Top right) Sketch of the instrumentation to detect the optical signal change caused by the silver precipitation. (Bottom left) Color change caused by the gold nanoparticle catalyzed silver precipitation during the two layered technique with various concentrations of IgG-AuNP. (Bottom Right) Gray level image taken by the scanner for different concentrations of two target proteins (A & B), and their comparison to negative control sample. Springer, Microfluids and Nanofluids, 6, 2008, 85–91 ”An immunoassay using antibody-gold nanoparticle conjugate, silver enhancement and flatbed scanner”; C.-H. Yeh, C.-Y. Hung, T. C. Chang, H.-P. Lin, and Y.-C. Lin; compiled from figures 1, 2, 3, 6, Copyright (2009) - (Reference 67). With permission from Springer Science and Business Media.
Imaging of large biomedical samples
Flatbed scanners have also been used for direct imaging of biological samples in large sample holders. By taking advantage of the large field of view, one can obtain a large dataset that can be used to infer results based on statistical features. Due to the low cost of flatbed scanners, these systems can be easily scaled up to conduct several experiments simultaneously, further increasing the throughput. This approach has been used to image e.g., virus plaques30, mammalian cell colonies77,78, bacterial colonies79–81 and C. elegans37,82 samples. In addition to scanner based systems, cellphone based and on-chip imaging technologies were also developed to achieve wide-field imaging of biological specimen (including e.g., sperms83,84, human blood cells6,9,85, CD4/CD8 cells86, viruses8, Papanicolu smears87–89, etc.) which in general achieve much better spatial resolution than what flatbed scanners are capable of providing.
The wide FOV that is provided by flatbed scanners can be rather important and useful for certain biomedical imaging tasks. For instance in the case of imaging a 96-well cell culture plate, using a 10X microscope objective-lens, to cover the FOV of a single well would take dozens of images, and imaging the whole plate area would take thousands. In practice some of the conventional high throughput automated imaging solutions only capture a limited subset of images from each well, and assume that the information on these images represents the entire area of the well plate.30. However, as Sullivan et al. pointed out, for samples that have spatially non-uniform patterns, (such as virus plaques), this method can yield unreliable measurements. By using a flatbed scanner, however, the whole surface area of each well can be imaged, and in conjunction with automated cell counting algorithms, the results can be rapidly obtained (see e.g., Figure 4).30
Figure 4.
A CCD based flatbed scanner used to evaluate a plaque assay measuring the infectivity and replication inhibition of various influenza virus strains in the presence of oseltamivir carboxylate. (Top) (a) Two 96-well plates are placed in the middle of the CCD based scanner to minimize the out-of-focus artifacts created by the wells placed far from the optical axis of the scanner lens. The artifact is depicted in (c), and can be compared to an image of a well taken from the middle of the plate shown in (b). (Bottom) (a) Image post processing algorithm for automatic evaluation of the scanned images. (b) Scanned image of the 96 well plate containing modified Madin-Darby canine kidney cells in triple columns infected with (from left to right) 103–106 of influenza virus A/Trieste/25/2007 and treated with 5 fold dilutions of oseltamivir carboxylate (20–0.0013nM,rows B to H) and row A as cell control with no oseltamivir carboxylate. (c) Positive population shown in a pseudo-colored image. Reprinted from Journal of Virological Methods, 179, Kate Sullivan, Johannes Kloess, Chen Qian, Donald Bell, Alan Hay, Yi Pu Lin, Yan Gu, “High throughput virus plaque quantitation using a flatbed scanner”, 81–89, Copyright (2012), with permission from Elsevier30.
Along the same lines, Levin-Reisman et al. also developed an automated system using several flatbed scanners to measure the growth rate and, in parallel, the delay in growth for thousands of organisms. By screening a library of E. coli deletion mutants, they were able to find a new bacteria growth phenotype with dramatically increased lag time upon starvation, and, as such, more resistant to antibiotics.81 This research shows the potential of large field of view automated imaging using scanners for tasks that would be extremely laborious to do with traditional imaging methods.
In a similar fashion, Stroustroup et al. combined several flatbed scanners within a temperature controlled incubator to measure C. elegans lifespan. This scanner-based automated imaging system not only creates large amounts of data, but also avoids the need to periodically remove the samples from the incubator to perform imaging (see Figure 5)37.
Figure 5.
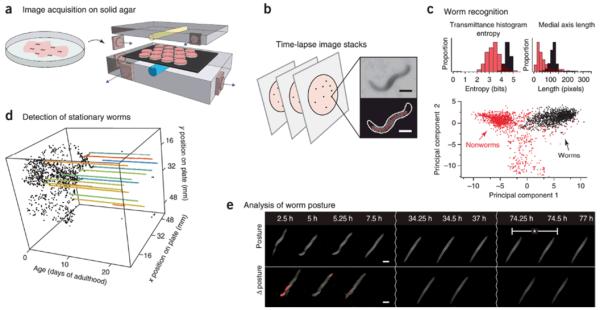
Images captured by several flatbed scanners inside an incubator for measurement of C. elegans lifespan. (a) Schematic showing Petri dishes containing age-synchronized C. elegans worms placed on the scanner. (b) Time-lapse images are captured by each scanner, and objects are segmented. Scale bars: 250μm. (c) Classification is done based on several morphological features; bar graphs show two examples. Bottom part shows a projection of the point cloud created by using 65 dimensional feature vectors to differentiate worms from non-worms. (d) The positions of the worms over time. Colored lines are stationary worms. (e) Graph of stationary animals over time, and their corresponding posture changes. The device can automatically gather death times from over 30,000 animals. Reprinted with permission from Macmillan Publishers Ltd: [Nature Methods] (Reference: 37), copyright (2013).
In certain experiments, the size of the biological sample itself (e.g., human tissue, bone or plant roots) might also require the use of an ultra-large field of view imaging system. For example, a high optical resolution (5600 dpi) flatbed scanner was used to create a complete map of a rat brain (~100 sections) in 10 hours. This device could resolve individual neurons labelled with gold intensified diaminobenzidine in thionin-stained rat brain sections.90 As another example, Dong et al. used a conventional flatbed scanner attached to a Rhizotron window to image plant roots and quantify their lengths91. Lobet and Draye also used a flatbed scanner in a similar approach, however they were able to vectorize the entire Rhizotron-grown root system, obtain position, morphology, and topology information about every root, and analyze spatial root water uptake patterns92. In another application, Fox et al. successfully used a flatbed scanner for forensic purposes by imaging human skeleton.93
Modified flatbed scanners used for biomedical imaging tasks
Recently flatbed scanners have also been modified to take further advantage of their large field of view, scanning mechanism, and their ability to capture giga-pixel images.32,25,34 As an example, Shimobaba et al. reported a modified scanner system to be used in an in-line digital holographic imaging setup25. They modified a CIS based scanner by removing the GRIN lens array and used an external laser as a light source. Their results demonstrated that the flat-bed scanner is capable of capturing fringe magnified digital holograms of objects that are spaced 30–50 centimeters away from the scanner's sensor plane. The overall spatial resolution of this system was ~8.8 μm, while the field of view was reduced to 2.2 cm × 2.9 cm due to fringe magnification.
One of the limitations of conventional flatbed scanners compared to traditional optical microscopes is their inability to directly perform fluorescent measurements. To provide a solution to this need, Göröcs et al. converted, for the first time a conventional flatbed scanner to a wide-field fluorescent imaging device to detect fluorescent objects inside optically dense media, such as whole blood. The key steps that were used to convert the scanner into a fluorescent imager involved taking complete, low-level control over the built-in chipset of the device. Maximizing the gain, optimizing the scanning speed, disabling all the on-chip data processing, and thus acquiring raw sensory information, were all essential to increase the sensitivity of the flatbed scanner to a level that it was able to detect the fluorescent emission from the specimen. The hardware of the scanner was also modified to convert the scanner into a fluorescent imaging device: fluorescent excitation was provided by an external, software controlled light source consisting of 600 LEDs in a 20 × 30 matrix positioned ~2 cm above the scanner glass (Figure 6).
Figure 6.
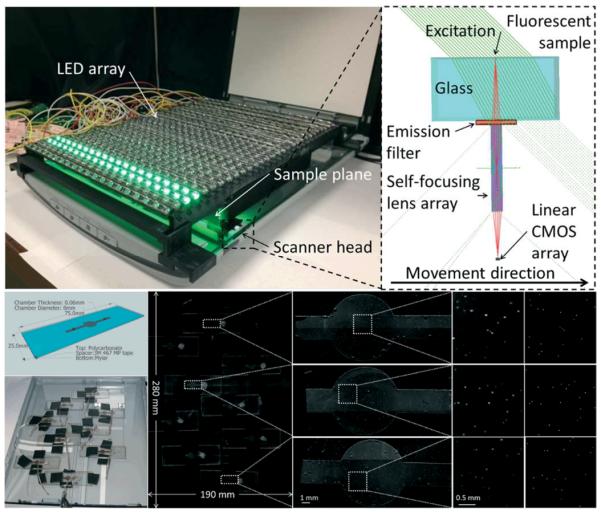
Flatbed scanner converted to a 2.2 giga-pixel fluorescent imaging device with a field of view of 532 cm2. (Top left) Photograph of the device consisting of a flatbed scanner fitted with an emission filter and an array of 600 LEDs providing uniform illumination over the entire field of view. (Top right) Diagram showing the light path inside the scanner head. The excitation arrives over a 45 degree angle to minimize the amount of light entering the scanner's gradient index lens. The light scattered by the sample is attenuated by the filter, while the emitted fluorescent light passes through it. (Bottom) Experiment designed to validate the performance of the system. Microfluidic chips filled with whole blood spiked with 10μm fluorescent particles are scattered over the field of view. Scanned image of the sample and (from left to right) increasing zooms to focus onto the chambers. Last column shows fluorescent microscope comparison images captured by a 4x objective-lens (NA=0.13). This scanner-based fluorescent imaging device achieved 98.8% accuracy with ~1% standard deviation for counting the particles inside the chambers. The whole field of view, 532 cm2, can be imaged in <5 minutes. Reproduced from Ref. 32 with permission from The Royal Society of Chemistry.
These LEDs were oriented in a 45° angle to minimize the excitation light entering into the image sensor, essentially creating a dark-field like excitation scheme as illustrated in Figure 6. A thin layer of custom made absorbing excitation filter was placed in front of the GRIN lens array to prevent the excitation light scattered by the sample to reach the image sensor. The extremely large field of view (532 cm2) of the scanner was utilized to image 2.2 mL of whole blood sample per scan, achieving more than 2 giga-pixels per image. The blood sample was spiked with 10 μm fluorescent particles, and placed into a relatively thin microfluidic chamber over the scanner glass. This scanner-based fluorescent imaging device was able to detect the fluorescent particles within the whole blood sample with 98.8% counting accuracy, and the entire scanning/imaging procedure took less than 5 min (see Figure 6). This flatbed scanner based fluorescent imaging platform was also capable of detecting white blood cells labeled with a fluorescent dye (EtBr).32
Conclusions
Using consumer electronics devices, such as cellphones or flatbed scanners, for biomedical imaging and sensing has several advantages over traditional microscopy approaches, including e.g., portability and cost effectiveness. In addition, these consumer electronics based devices can perform large field-of-view automated imaging tasks, which would be too laborious to achieve with conventional microscopes that traditionally require relatively expensive scanning systems. For this end, flatbed scanners offer unique capabilities due to their ultra large field of view (>500–600 cm2), which makes them excellent candidates for imaging large biomedical samples. As the consumer electronics market continues to grow, these emerging biomedical imaging systems will further improve, expanding their already diverse uses in biomedical imaging and sensing applications. In addition to mobile phones and other emerging consumer electronics devices including wearable computers, flatbed scanners and their use in advanced imaging and sensing experiments might help us transform current practices of medicine, engineering and sciences through democratization of measurement science and empowerment of citizen scientists, science educators and researchers in resource limited settings.
Acknowledgements
Ozcan Research Group at UCLA gratefully acknowledges the support of the Presidential Early Career Award for Scientists and Engineers (PECASE), Army Research Office (ARO) Life Sciences Division, ARO Young Investigator Award, National Science Foundation (NSF) CAREER Award, NSF CBET Division Biophotonics Program, NSF Emerging Frontiers in Research and Innovation (EFRI) Award, Office of Naval Research (ONR) and National Institutes of Health (NIH) Director's New Innovator Award DP2OD006427 from the Office of the Director, National Institutes of Health. This work is based upon research performed in a renovated laboratory by the National Science Foundation under Grant No. 0963183, which is an award funded under the American Recovery and Reinvestment Act of 2009 (ARRA).
Footnotes
Conflicts of Interest Statement A.O. is the co-founder of a start-up company (Holomic LLC) that aims to commercialize computational microscopy and diagnostics tool.
Notes and references
- 1.Ozcan A. Lab. Chip. 2014 DOI:10.1039/C4LC00010B. [Google Scholar]
- 2.Coskun AF, Ozcan A. Curr. Opin. Biotechnol. 2014;25:8–16. doi: 10.1016/j.copbio.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei Q, Nagi R, Sadeghi K, Feng S, Yan E, Ki SJ, Caire R, Tseng D, Ozcan A. ACS Nano. 2014 doi: 10.1021/nn406571t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erickson D, O'Dell D, Jiang L, Oncescu V, Gumus A, Lee S, Mancuso M, Mehta S. Lab. Chip. 2014 doi: 10.1039/c4lc00142g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang J, Wang X, Chao R, Ren Y, Hu C, Xu Z, Liu GL. Sens. Actuators B Chem. 2014;193:653–659. [Google Scholar]
- 6.Navruz I, Coskun AF, Wong J, Mohammad S, Tseng D, Nagi R, Phillips S, Ozcan A. Lab. Chip. 2013;13:4015–4023. doi: 10.1039/c3lc50589h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vashist SK, Mudanyali O, Schneider EM, Zengerle R, Ozcan A. Anal. Bioanal. Chem. 2013:1–15. doi: 10.1007/s00216-013-7473-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei Q, Qi H, Luo W, Tseng D, Ki SJ, Wan Z, Göröcs Z, Bentolila LA, Wu T-T, Sun R, Ozcan A. ACS Nano. 2013;7:9147–9155. doi: 10.1021/nn4037706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu H, Sencan I, Wong J, Dimitrov S, Tseng D, Nagashima K, Ozcan A. Lab. Chip. 2013;13:1282–1288. doi: 10.1039/c3lc41408f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coskun AF, Wong J, Khodadadi D, Nagi R, Tey A, Ozcan A. Lab. Chip. 2013;13:636–640. doi: 10.1039/c2lc41152k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.You DJ, Park TS, Yoon J-Y. Biosens. Bioelectron. 2013;40:180–185. doi: 10.1016/j.bios.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Tapley A, Switz N, Reber C, Davis JL, Miller C, Matovu JB, Worodria W, Huang L, Fletcher DA, Cattamanchi A. J. Clin. Microbiol. 2013;51:1774–1778. doi: 10.1128/JCM.03432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oncescu V, O'Dell D, Erickson D. Lab. Chip. 2013;13:3232–3238. doi: 10.1039/c3lc50431j. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H, Isikman SO, Mudanyali O, Greenbaum A, Ozcan A. Lab. Chip. 2012;13:51–67. doi: 10.1039/c2lc40864c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mudanyali O, Dimitrov S, Sikora U, Padmanabhan S, Navruz I, Ozcan A. Lab. Chip. 2012;12:2678–2686. doi: 10.1039/c2lc40235a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu H, Sikora U, Ozcan A. Analyst. 2012;137:2541–2544. doi: 10.1039/c2an35071h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen L, Hagen JA, Papautsky I. Lab. Chip. 2012;12:4240–4243. doi: 10.1039/c2lc40741h. [DOI] [PubMed] [Google Scholar]
- 18.Vella SJ, Beattie P, Cademartiri R, Laromaine A, Martinez AW, Phillips ST, Mirica KA, Whitesides GM. Anal. Chem. 2012;84:2883–2891. doi: 10.1021/ac203434x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balsam J, Ossandon M, Kostov Y, Bruck HA, Rasooly A. Lab. Chip. 2011;11:941–949. doi: 10.1039/c0lc00431f. [DOI] [PubMed] [Google Scholar]
- 20.Smith ZJ, Chu K, Espenson AR, Rahimzadeh M, Gryshuk A, Molinaro M, Dwyre DM, Lane S, Matthews D, Wachsmann-Hogiu S. PLoS ONE. 2011;6:e17150. doi: 10.1371/journal.pone.0017150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breslauer DN, Maamari RN, Switz NA, Lam WA, Fletcher DA. PLoS ONE. 2009;4:e6320. doi: 10.1371/journal.pone.0006320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper S, Khatib F, Treuille A, Barbero J, Lee J, Beenen M, Leaver-Fay A, Baker D, Popović Z, Players F. Nature. 2010;466:756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mavandadi S, Dimitrov S, Feng S, Yu F, Sikora U, Yaglidere O, Padmanabhan S, Nielsen K, Ozcan A. PLoS ONE. 2012;7:e37245. doi: 10.1371/journal.pone.0037245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mavandadi S, Dimitrov S, Feng S, Yu F, Yu R, Sikora U, Ozcan A. Lab. Chip. 2012;12:4102–4106. doi: 10.1039/c2lc40614d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimobaba T, Yamanashi H, Kakue T, Oikawa M, Okada N, Endo Y, Hirayama R, Masuda N, Ito T. Sci. Rep. 2013;3 doi: 10.1038/srep02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenbaum A, Luo W, Khademhosseinieh B, Su T-W, Coskun AF, Ozcan A. Sci. Rep. 2013;3 [Google Scholar]
- 27.Sheu LS, Truong T, Yuzuki L, Elhatem A, Kadekodi N. Proc. SPIE. 1984;0501:84–88. [Google Scholar]
- 28.Reulke R, Tempelmann U, Stallmann D, Cramer M, Haala N. Proc. ISPRS congress. Istanbul: 2004. [Google Scholar]
- 29.Holst GC. Sampling, Aliasing, and Data Fidelity for Electronic Imaging Systems, Communications, and Data Acquisition. PM55. JCD Publishing; 1998. [Google Scholar]
- 30.Sullivan K, Kloess J, Qian C, Bell D, Hay A, Lin YP, Gu Y. J. Virol. Methods. 2012;179:81–89. doi: 10.1016/j.jviromet.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Anderson EE, Wang W-L. Proc. SPIE. 1993;1901:173–181. [Google Scholar]
- 32.Göröcs Z, Ling Y, Yu MD, Karahalios D, Mogharabi K, Lu K, Wei Q, Ozcan A. Lab. Chip. 2013;13:4460–4466. doi: 10.1039/c3lc51005k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawazu M, Ogura Y. Appl. Opt. 1980;19:1105–1112. doi: 10.1364/AO.19.001105. [DOI] [PubMed] [Google Scholar]
- 34.Zheng G, Ou X, Yang C. Biomed. Opt. Express. 2014;5:1–8. doi: 10.1364/BOE.5.000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishizuka S, Washburn NR, Munson PJ. BioTechniques. 2006;40:442, 444, 446. doi: 10.2144/000112144. passim. [DOI] [PubMed] [Google Scholar]
- 36.Tan HY, Ng TW, Liew OW. BioTechniques. 2007;42:474–478. doi: 10.2144/000112402. [DOI] [PubMed] [Google Scholar]
- 37.Stroustrup N, Ulmschneider BE, Nash ZM, López-Moyado IF, Apfeld J, Fontana W. Nat. Methods. 2013;10:665–670. doi: 10.1038/nmeth.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. http://www.microsoft.com/silverlight/deep-zoom/
- 39.Feng S, Caire R, Cortazar B, Turan M, Wong A, Ozcan A. ACS Nano. 2014;8:3069–3079. doi: 10.1021/nn500614k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rakow NA, Suslick KS. Nature. 2000;406:710–713. doi: 10.1038/35021028. [DOI] [PubMed] [Google Scholar]
- 41.Lonsdale CL, Taba B, Queralto N, Lukaszewski RA, Martino RA, Rhodes PA, Lim SH. PLoS ONE. 2013;8:e62726. doi: 10.1371/journal.pone.0062726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bourzac K. MIT Technol. Rev. 2012 [Google Scholar]
- 43.Janzen MC, Ponder JB, Bailey DP, Ingison CK, Suslick KS. Anal. Chem. 2006;78:3591–3600. doi: 10.1021/ac052111s. [DOI] [PubMed] [Google Scholar]
- 44.Musto CJ, Suslick KS. Curr. Opin. Chem. Biol. 2010;14:758–766. doi: 10.1016/j.cbpa.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rakow NA, Sen A, Janzen MC, Ponder JB, Suslick KS. Angew. Chem. Int. Ed. 2005;44:4528–4532. doi: 10.1002/anie.200500939. [DOI] [PubMed] [Google Scholar]
- 46.Suslick KS, Rakow NA, Sen A. Tetrahedron. 2004;60:11133–11138. [Google Scholar]
- 47.Zhang C, Bailey DP, Suslick KS. J. Agric. Food Chem. 2006;54:4925–4931. doi: 10.1021/jf060110a. [DOI] [PubMed] [Google Scholar]
- 48.Zhang C, Suslick KS. J. Am. Chem. Soc. 2005;127:11548–11549. doi: 10.1021/ja052606z. [DOI] [PubMed] [Google Scholar]
- 49.Zhang C, Suslick KS. J. Agric. Food Chem. 2007;55:237–242. doi: 10.1021/jf0624695. [DOI] [PubMed] [Google Scholar]
- 50.Cantrell K, Erenas MM, de Orbe-Payá I, Capitán-Vallvey LF. Anal. Chem. 2010;82:531–542. doi: 10.1021/ac901753c. [DOI] [PubMed] [Google Scholar]
- 51.Hoffmann G. CIELab Color Space. [Google Scholar]
- 52.Martinez AW, Phillips ST, Carrilho E, Thomas SW, Sindi H, Whitesides GM. Anal. Chem. 2008;80:3699–3707. doi: 10.1021/ac800112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carrilho E, Phillips ST, Vella SJ, Martinez AW, Whitesides GM. Anal. Chem. 2009;81:5990–5998. doi: 10.1021/ac900847g. [DOI] [PubMed] [Google Scholar]
- 54.Carey JR, Suslick KS, Hulkower KI, Imlay JA, Imlay KRC, Ingison CK, Ponder JB, Sen A, Wittrig AE. J. Am. Chem. Soc. 2011;133:7571–7576. doi: 10.1021/ja201634d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim SH, Feng L, Kemling JW, Musto CJ, Suslick KS. Nat. Chem. 2009;1:562–567. doi: 10.1038/nchem.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lim SH, Musto CJ, Park E, Zhong W, Suslick KS. Org. Lett. 2008;10:4405–4408. doi: 10.1021/ol801459k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Musto CJ, Lim SH, Suslick KS. Anal. Chem. 2009;81:6526–6533. doi: 10.1021/ac901019g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Apyari VV, Dmitrienko SG, Ostrovskaya VM, Anaev EK, Zolotov YA. Anal. Bioanal. Chem. 2008;391:1977–1982. doi: 10.1007/s00216-008-2112-y. [DOI] [PubMed] [Google Scholar]
- 59.Apyari VV, Dmitrienko SG, Zolotov YA. Int. J. Environ. Anal. Chem. 2009;89:775–783. [Google Scholar]
- 60.Shishkin YL, Dmitrienko SG, Medvedeva OM, Badakova SA, Pyatkova LN. J. Anal. Chem. 2004;59:102–106. [Google Scholar]
- 61.Abbaspour A, Mehrgardi MA, Noori A, Kamyabi MA, Khalafi-Nezhad A, Soltani Rad MN. Sens. Actuators B Chem. 2006;113:857–865. [Google Scholar]
- 62.Capel-Cuevas S, Cuéllar MP, de Orbe-Payá I, Pegalajar MC, Capitán-Vallvey LF. Anal. Chim. Acta. 2010;681:71–81. doi: 10.1016/j.aca.2010.09.033. [DOI] [PubMed] [Google Scholar]
- 63.Abbaspour A, Khajehzadeh A, Ghaffarinejad A. The Analyst. 2009;134:1692–1698. doi: 10.1039/b901273g. [DOI] [PubMed] [Google Scholar]
- 64.Abbaspour A, Valizadeh H, Khajehzadeh A. Anal. Methods. 2011;3:1405–1409. [Google Scholar]
- 65.Abe K, Suzuki K, Citterio D. Anal. Chem. 2008;80:6928–6934. doi: 10.1021/ac800604v. [DOI] [PubMed] [Google Scholar]
- 66.Abe K, Kotera K, Suzuki K, Citterio D. Anal. Bioanal. Chem. 2010;398:885–893. doi: 10.1007/s00216-010-4011-2. [DOI] [PubMed] [Google Scholar]
- 67.Yeh C-H, Hung C-Y, Chang TC, Lin H-P, Lin Y-C. Microfluid. Nanofluidics. 2009;6:85–91. [Google Scholar]
- 68.Bannur SV, Kulgod SV, Metkar SS, Mahajan SK, Sainis JK. Anal. Biochem. 1999;267:382–389. doi: 10.1006/abio.1998.3020. [DOI] [PubMed] [Google Scholar]
- 69.Birch NC, Stickle DF. Clin. Chim. Acta. 2003;333:95–96. doi: 10.1016/s0009-8981(03)00168-2. [DOI] [PubMed] [Google Scholar]
- 70.Rahman M, Kühn I, Rahman M, Olsson-Liljequist B, Möllby R. Appl. Environ. Microbiol. 2004;70:2398–2403. doi: 10.1128/AEM.70.4.2398-2403.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lapresta-Fernández A, Capitán-Vallvey LF. Sens. Actuators B Chem. 2008;134:694–701. [Google Scholar]
- 72.Taton TA, Mirkin CA, Letsinger RL. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 73.Nam J-M, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 74.Lee J-S, Mirkin CA. Anal. Chem. 2008;80:6805–6808. doi: 10.1021/ac801046a. [DOI] [PubMed] [Google Scholar]
- 75.Kim D, Daniel WL, Mirkin CA. Anal. Chem. 2009;81:9183–9187. doi: 10.1021/ac9018389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao C, Gontard LC, Thuy Tram LL, Wolff A, Bang DD. Small. 2011;7:1701–1708. doi: 10.1002/smll.201100294. [DOI] [PubMed] [Google Scholar]
- 77.Bewes JM, Suchowerska N, McKenzie DR. Phys. Med. Biol. 2008;53:5991. doi: 10.1088/0031-9155/53/21/007. [DOI] [PubMed] [Google Scholar]
- 78.Dahle J, Kakar M, Steen HB, Kaalhus O. Cytometry A. 2004;60A:182–188. doi: 10.1002/cyto.a.20038. [DOI] [PubMed] [Google Scholar]
- 79.Clarke ML, Burton RL, Hill AN, Litorja M, Nahm MH, Hwang J. Cytometry A. 2010;77A:790–797. doi: 10.1002/cyto.a.20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gabrielson J, Hart M, Jarelöv A, Kühn I, McKenzie D, Möllby R. J. Microbiol. Methods. 2002;50:63–73. doi: 10.1016/s0167-7012(02)00011-8. [DOI] [PubMed] [Google Scholar]
- 81.Levin-Reisman I, Gefen O, Fridman O, Ronin I, Shwa D, Sheftel H, Balaban NQ. Nat. Methods. 2010;7:737–739. doi: 10.1038/nmeth.1485. [DOI] [PubMed] [Google Scholar]
- 82.Mathew MD, Mathew ND, Ebert PR. PLoS ONE. 2012;7:e33483. doi: 10.1371/journal.pone.0033483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Su T-W, Choi I, Feng J, Huang K, McLeod E, Ozcan A. Sci. Rep. 2013;3 doi: 10.1038/srep01664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Su T-W, Xue L, Ozcan A. Proc. Natl. Acad. Sci. 2012;109:16018–16022. doi: 10.1073/pnas.1212506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mudanyali O, Bishara W, Ozcan A. Opt. Express. 2011;19:17378–17389. doi: 10.1364/OE.19.017378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei Q, McLeod E, Qi H, Wan Z, Sun R, Ozcan A. Sci. Rep. 2013;3 doi: 10.1038/srep01699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Greenbaum A, Akbari N, Feizi A, Luo W, Ozcan A. PLoS ONE. 2013;8:e76475. doi: 10.1371/journal.pone.0076475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Greenbaum A, Feizi A, Akbari N, Ozcan A. Opt. Express. 2013;21:12469–12483. doi: 10.1364/OE.21.012469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Greenbaum A, Ozcan A. Opt. Express. 2012;20:3129–3143. doi: 10.1364/OE.20.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krout KE, Jenkins JM, Loewy AD. J. Neurosci. Methods. 2002;113:37–40. doi: 10.1016/s0165-0270(01)00471-x. [DOI] [PubMed] [Google Scholar]
- 91.Dong S, Neilsen D, Neilsen GH, Weis M. HortScience. 2003;38:1385–1388. [Google Scholar]
- 92.Lobet G, Draye X. Plant Methods. 2013;9:1. doi: 10.1186/1746-4811-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fox SC, Eliopoulos C, Moutafi I, Manolis SK. J. Forensic Sci. 2011;56:S154–S157. doi: 10.1111/j.1556-4029.2010.01596.x. [DOI] [PubMed] [Google Scholar]