Abstract
Imaging of protein–protein and RNA–protein interactions in vivo, especially in live animals, is still challenging. Here we developed far-red mNeptune-based bimolecular fluorescence complementation (BiFC) and trimolecular fluorescence complementation (TriFC) systems with excitation and emission above 600 nm in the ‘tissue optical window’ for imaging of protein–protein and RNA–protein interactions in live cells and mice. The far-red mNeptune BiFC was first built by selecting appropriate split mNeptune fragments, and then the mNeptune-TriFC system was built based on the mNeptune-BiFC system. The newly constructed mNeptune BiFC and TriFC systems were verified as useful tools for imaging protein–protein and mRNA–protein interactions, respectively, in live cells and mice. We then used the new mNeptune-TriFC system to investigate the interactions between human polypyrimidine-tract-binding protein (PTB) and HIV-1 mRNA elements as PTB may participate in HIV mRNA processing in HIV activation from latency. An interaction between PTB and the 3′long terminal repeat region of HIV-1 mRNAs was found and imaged in live cells and mice, implying a role for PTB in regulating HIV-1 mRNA processing. The study provides new tools for in vivo imaging of RNA–protein and protein–protein interactions, and adds new insight into the mechanism of HIV-1 mRNA processing.
INTRODUCTION
RNA–protein and protein–protein interactions are fundamental biological processes. Molecular imaging in live subjects is critical for understanding these interactions (1). However, in vivo imaging of protein–protein interactions remains a difficult task (2), and a method to image RNA–protein interactions in live animals has not been developed until now.
Fluorescence complementation (FC) is a valuable tool for investigating molecular interactions in physiological environments (3,4). The FC assay relies on the reconstruction of a reporter fluorescent protein from its two divided non-fluorescent fragments. Two types of FC systems have been developed: bimolecular fluorescence complementation (BiFC), which is a reliable tool for studying protein–protein interactions (5,6), and trimolecular fluorescence complementation (TriFC), which is used to investigate RNA–protein interactions (7). BiFC and TriFC are simple, powerful and promising tools that have been incorporated into several systems to visualize protein–protein and protein–RNA interactions in live cells (8,9). However, long-wavelength-spectrum FC systems for in vivo imaging remain to be developed.
One of the main obstacles to molecular imaging in live animals is the opacity of tissues to excitation light below 600 nm (10). A tissue ‘optical window’ between 600 and 1200 nm is feasible for in vivo imaging (11). Current red BiFC systems use mRFP1-Q66T (excitation and emission wavelengths of 549/570 nm), mCherry (587/610 nm) and mlumin (587/621 nm) (12–14) for live cell imaging, none of which can be excited above 600 nm. Most recently, we have reported a red mCherry TriFC system for imaging of RNA–protein interactions, but the system still cannot be excited above 600 nm for in vivo imaging (15). BiFC and TriFC systems with spectra within a tissue ‘optical window’ for live-body imaging are highly desirable.
Here, we aimed to construct long-wavelength-spectrum FC systems for in vivo imaging of protein–protein and RNA–protein interactions. mNeptune, a far-red monomerized Neptune variant, was used to develop the new FC systems because of its good optical properties (16). mNeptune has an excitation peak at 600 nm and an emission peak at 650 nm, both within the tissue ‘optical window’. mNeptune is also brighter than other red fluorescent proteins (RFPs) (i.e. mKate and mCherry) when excited at 633 nm for in vivo imaging.
In this study, we firstly built mNeptune BiFC systems by selecting appropriate split mNeptune fragments. The new mNeptune BiFC system was verified by imaging protein–protein interactions in live cells and mice. We then built a mNeptune-based TriFC system for monitoring mRNA–protein interactions in live subjects. Several known protein–RNA interactions, such as the interactions between influenza viral NS1 protein and the 5′ untranslated region (UTR) of nucleocapsid protein (NP) messenger ribonucleic acid (mRNA) and matrix protein (M) mRNA (17), were used to validate the new mNeptune-based TriFC system in live cells and live mice. Because it has been suggested that human polypyrimidine-tract-binding protein (PTB) plays roles in human immunodeficiency virus (HIV) activation from latency, which may be related to HIV mRNA processing (18,19), the interactions between HIV-1 mRNA elements [i.e. 5′long terminal repeat (LTR), 3′LTR and cis-acting repressive sequences (CRS)], and PTB was investigated using the new mNeptune-based TriFC system. A direct interaction between PTB and the 3′LTR region of HIV-1 mRNAs was found and imaged in live cells and mice.
MATERIALS AND METHODS
Construction of plasmid expression vectors
The coding genes of split mNeptune fragments were inserted into the pMN159 and pMC160 plasmids of the mCherry BiFC system (13) by substituting the mCherry fragments with the mNeptune fragments to obtain the pMN116, pMN155 and pMN169 and pMC117, pMC156 and pMC170 plasmids (see Supplementary Figure S1). To construct the mNeptune-BiFC system, the N-terminal sequences encoding mNeptune, MN116 (amino acid residues (aa) 1–116), MN155 (aa 1–155) and MN169 (aa 1–169), were fused to the C-terminus of either enhanced green fluorescent protein (EGFP) or bJun. The C-terminal sequences encoding mNeptune, MC117 (aa 117–244), MC156 (aa 156–244) and MC170 (aa 170–244), were fused to the N-terminus of EGFP, bFos or bFos179-193 (mbFos). The coding regions were connected by the linker sequence GGGGSGGGGS.
To construct the mNeptune-TriFC system, the MN155 fragment was fused to the C-terminus of the influenza A viral protein NS1 or the cellular protein PTB. The MC156 fragment was fused to the N-terminus of the MS2 coat protein (MCP). The MCP protein recognition site (ms2) was inserted into pECFP-C1. The 5′UTRs of the influenza NP and M mRNAs, the 5′ and 3′LTRs of HIV-1 mRNA and the CRSs of HIV-1 envelope (env) mRNA were inserted into pECFP-C1 following the ms2 sequence.
Cell culture and transfection
HeLa and 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100-U/ml penicillin and 100-μg/ml streptomycin and cultured at 37°C in 5% CO2. The day before transfection, the cells were plated on 35-mm diameter dishes and grown to 70–80% confluence. The cells were co-transfected with the expression vectors indicated in each experiment using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. The transfected cells were incubated at 37°C (5% CO2) for 5–6 h followed by 18–24 h at 30°C (5% CO2) until imaging.
Fluorescence microscopy and image processing and data analysis
Cells were imaged with an inverted Delta Vision Restoration Microscopy System (Applied Precision, WA, USA) equipped with a cooled CCD camera. GFP fluorescence was measured by excitation at 490/20 nm and emission at 538/19 nm. Red mNeptune BiFC and TriFC signals were measured with excitation at 640/20 nm and emission at 685/40 nm. Cyan fluorescent protein (CFP) fluorescence was used as an internal control to measure the BiFC and TriFC efficiency, with excitation at 430/20 nm and emission at 470/20 nm. The fluorescence ratios of mNeptune/GFP and mNeptune/CFP were quantified for every cell expressing the corresponding proteins after subtraction of the background fluorescence. Background fluorescence was calculated as the mean intensity of areas of the image with no cells. About 60 cells from 10 microscopic fields for each sample were evaluated to quantify the fluorescence intensity. Statistical analyses were performed using the SPSS14.0 software (SPSS Company, IL, USA). All P-values <0.05 were considered as statistically significant.
Fluorescence imaging of live mice
The in vivo fluorescent images were obtained using the Maestro 2 in vivo imaging system (CRi, Woburn, MA, USA). 293T cells transiently expressing the BiFC or TriFC systems were first imaged under microscope and analyzed ex vivo (in tubes and on a 96-well black plate) with the Maestro 2 in vivo imaging system. The transfection efficiency was determined as 60–70% for the two plasmids co-transfection (BiFC) and 50–60% for the three plasmids co-transfection (TriFC) by the number of cells with red fluorescence versus the total cell number. Then, different amounts (104, 105, 106, 107) of cells were injected subcutaneously into 5–7-week-old BALB/c-nu mice. The mice were imaged within 5 min of implanting the cells because the cells will diffuse in the subcutaneous layer in live mice. For fluorescence imaging of mNeptune, a 576–621-nm band-pass filter was used for excitation, and a longpass filter over 635 nm was used for emission. For enhanced CFP (ECFP) imaging, a 435–480-nm band-pass filter was used for excitation, and a longpass filter over 490 nm was used for emission. The emission light spectra of FC from the split mNeptune fragments were recorded to identify the correct emission spectra for mNeptune (Supplementary Figure S2). The spectral fluorescence images consisting of the spectra from BiFC and TriFC signals and autofluorescence spectra were then eliminated based on their spectral patterns using Maestro 3.0 software (CRi, Woburn, MA, USA). The fluorescence intensities (counts) of the regions of interest were measured to quantify the BiFC and TriFC signals with the measured panel of the Maestro 3.0 software. Statistical analyses for about six mice were performed using the SPSS14.0 software and P-values <0.05 were considered statistically significant.
mRNA quantification by quantitative reverse transcriptase-polymerase chain reactions and mRNA stability assays
Total RNA was extracted using the EZNA Total RNA Kit I (Omega Bio-Tek, GA, USA) according to the manufacturer's instructions. Then mRNAs were reverse transcribed with PrimeScript™ RT reagent Kit with genomic deoxyribonucleic acid (gDNA) Eraser (Takara, Dalian). mRNA levels were quantified by real-time polymerase chain reaction (PCR) using iTaq universal SYBR green supermix PCR reagent (BIO-RAD, CA, USA). Real-time PCR was carried out with a BIO-RAD CFX Connect Real-Time System. Analysis was performed using Bio-Rad CFX manager 2.1 software (BIO-RAD, CA, USA). The data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The primer sequences used for quantitative reverse transcriptase (RT) PCR are as follows: NP 5′UTR forward, 5′-CTGCTGCCCGACAACCA-3′; NP 5′UTR reverse, 5′-CGATTTTGATGTCACTCAG-3′; GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′; GAPDH reverse, 5′-GAAGATGGTGATGGGATTTC-3′.
For mRNA stability assays, cells were treated with 0.5 μg/ml of actinomycin D to stop mRNA transcription 24 h after transfection. Cells were harvested at different time points upon actinomycin D treatment, and total RNA was isolated with EZNA Total RNA Kit I (Omega Bio-Tek, Norcross, GA, USA). Then, mRNAs were reverse transcribed with PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Dalian) whereas U6 snRNA was reverse transcribed with U6 snRNA qPCR Primer Set (RIBOBIO, Guangzhou). mRNA levels were quantified by quantitative real-time RT-PCRs. Values were normalized to the stable U6 snRNA (20). The NP 5′UTR mRNA was amplified using the same primers in quantitative RT-PCR assays above and the U6 snRNA was amplified with the primers provided by U6 snRNA qPCR Primer Set (RIBOBIO, Guangzhou).
Immunoprecipitation experiments
For the immunoprecipitation assay, cells co-expressing the PTB protein and HIV RNA sequences were lysed for co-immunoprecipitation with a PTB antibody (Abcam) or with IgG1 as a control. The co-immunoprecipitated pellet was extracted, and a portion of it was used to detect HIV RNAs, while the other portion was used for protein analysis. The RNA present in the immunoprecipitates was extracted as described previously (21). The isolated RNA pellet was resuspended in nuclease-free water and analyzed with a One Step RT-PCR kit (Takara). For the PCR, we used primers specific for the HIV-1 3′LTR, 5′LTR or for the cellular housekeeping GAPDH gene. For western blot analysis, the samples were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis and then transferred onto polyvinylidene fluoride (PVDF) membranes. The PVDF membranes were incubated with PTB antibody, followed by horseradish peroxidase-conjugated rabbit anti-goat IgG. The signal was detected with a (3,3’-diaminobenzidine) DAB horseradish peroxidase color development kit (Beyotime).
RESULTS
Construction and application of the mNeptune-BiFC system
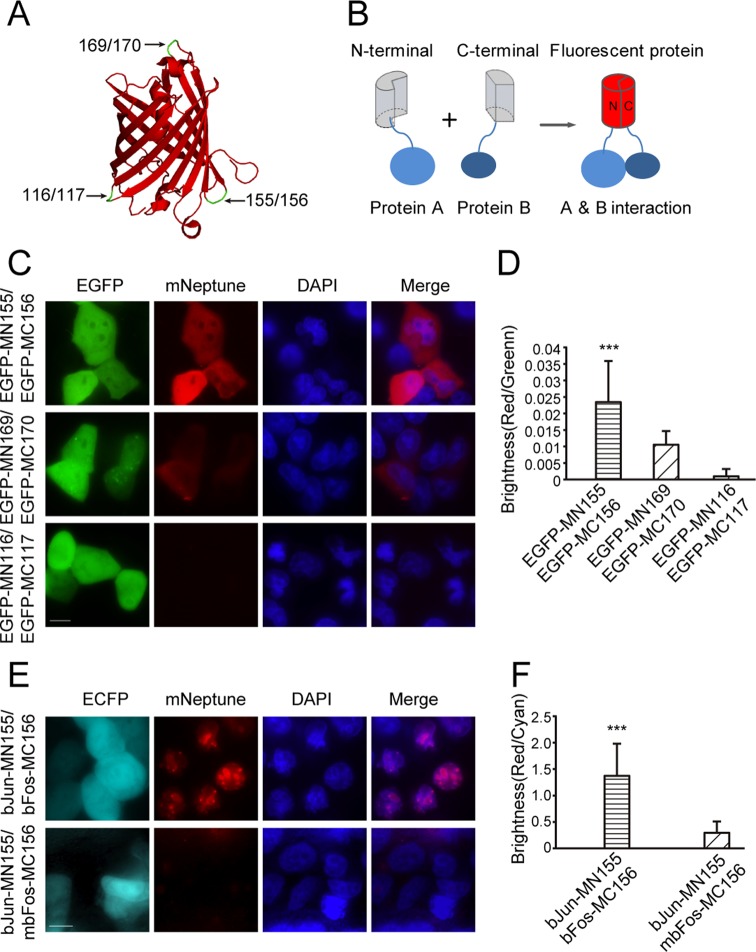
Based on the crystal structure of mNeptune (PDB Accession No. 3IP2) (16) and sequence alignments with other RFPs (mKate and mLumin), three sites between amino acids 116/117, 155/156 and 169/170 within the loops of the barrel-like structure were selected to split mNeptune to develop the BiFC system (Figure 1A). The schematic principle of the BiFC system is shown in Figure 1B. Two interacting proteins, A and B, are fused to N- and C-terminal fragments of a fluorescent protein, respectively. When A and B interact, the N- and C-terminal fragments are brought close together to facilitate reconstruction of the fluorescent protein. The coding sequences of the split mNeptune fragments were used to construct the plasmids pMN116, pMC117, pMN155, pMC156, pMN169 and pMC179 for the mNeptune-BiFC system (Supplementary Figure S1). The coding sequence of the EGFP was inserted into these plasmids to express N-terminal (MN)-EGFP or C-terminal (MC)-EGFP fusion proteins. The weak dimerization of EGFP (22) was used to test the BiFC system. EGFP also served as a reporter to indicate the expression of the mNeptune fragments as an internal control to calculate the BiFC efficiency. When the MN-EGFP and MC-EGFP pairs were co-expressed in HeLa cells, the combination of EGFP-MN155 and EGFP-MC156 produced bright red fluorescence, indicating a functional BiFC signal (Figure 1C). Wider cell populations with low magnifications are also shown in Supplementary Figure S3A. The combination of MN169-EGFP and MC170-EGFP also yielded red fluorescence, although the signal was weaker (Figure 1C). The combination of EGFP-MN116 and EGFP-MC117 produced only the EGFP signal but no red fluorescence. The transient expression of either pMN or pMC alone or their co-expression in HeLa cells also did not result in a red BiFC signal (Supplementary Figure S4). The relative BiFC efficiencies of the MN and MC fragments of mNeptune were calculated by dividing the intensity of the red fluorescence by the intensity of the green fluorescence upon co-expression of MN-EGFP and MC-EGFP after subtraction of the background fluorescence. As shown in Figure 1D, the BiFC efficiency of the combination of the MN155 and MC156 fragments was significantly higher than the combinations of the other fragments. These results demonstrate that mNeptune split at the 155/156 site could be used as a new red BiFC system.
Figure 1.
Construction of the mNeptune-BiFC systems. (A) The split sites (arrows) in the loop regions of the β-barrel structure of mNeptune are indicated. (B) The schematic principle of the BiFC system. (C) BiFC signals (mNeptune channel) were detected in HeLa cells by weak dimerization of EGFP for the different split mNeptune fragments. Scale bar: 10 μm. (D) Quantitative analysis of the BiFC efficiency in (C) was calculated by dividing the red fluorescence intensity by the green fluorescence intensity after subtracting the background fluorescence. (E) Images of the interactions between bJun and bFos or mbFos based on the MN155 and MC156 fragments in the BiFC system. (F) Quantitative analysis of the BiFC signals in (E) based on the fluorescence intensity ratio of BiFC/ECFP (red/cyan). All the quantitative analysis data are given as the mean + SD (n = 60). Statistical significance was evaluated using a two-tailed Student's t-test. *** indicates P < 0.01.
To verify the validity of the mNeptune-BiFC system we imaged the interactions between bFos and bJun, which are the basic region-leucine zipper domains of the transcription regulatory proteins Fos and Jun. The bJun and bFos encoding sequences were inserted into pMN155 and pMC156, respectively. When bJun-MN155 and bFos-MC156 were co-expressed in the same cells, a strong red BiFC signal was observed (Figure 1E) that exhibited a specific nucleoli localization pattern. The images in Supplementary Figure S3B also showed the BiFC assay of wider cell populations with low magnification. When mbFos (a leucine zipper deleted to eliminate the interaction between bFos and bJun) was fused to MC156 and co-expressed with bJun-MN155, only a very weak fluorescence signal was detected (Figure 1E). The ECFP plasmid, pECFP-C1, was also co-transfected into cells as an internal control during these experiments. Statistical analysis based on the fluorescence intensity ratio of BiFC/ECFP (red/cyan) demonstrated that the fluorescence of bJun-MN155 and bFos-MC156 was significantly higher than that of bJun-MN155 and mbFos-MC156 (Figure 1F). Therefore, the newly constructed mNeptune-BiFC system could be used to image and investigate protein–protein interactions.
In vivo imaging of protein–protein interactions in the mNeptune-BiFC system
The mNeptune-BiFC system was then tested by imaging the protein–protein interactions of bFos and bJun in live mice. 293T cells co-expressing either MN155-bJun/MC156-bFos/ECFP or MN155-bJun/MC156-mbFos/ECFP were analyzed in vitro using the Maestro 2 in vivo imaging system. To quantify the fluorescence accurately, the cell samples were placed onto 96-well black plates for detection. As shown in Figure 2A, the red BiFC fluorescence of the cells expressing MN155-bJun/MC156-bFos was greater than that of the cells expressing MN155-bJun/MC156-mbFos (Figure 2A, left). The two cell lines displayed similar CFP signals (Figure 2A, right). Statistical analysis based on the fluorescence intensity ratio of BiFC/ECFP (red/cyan) demonstrated that the fluorescence signal of MN155-bJun and MC156-bFos was significantly higher than that of MN155-bJun and MC156-mbFos (Figure 2B), in agreement with the microscopy analysis.
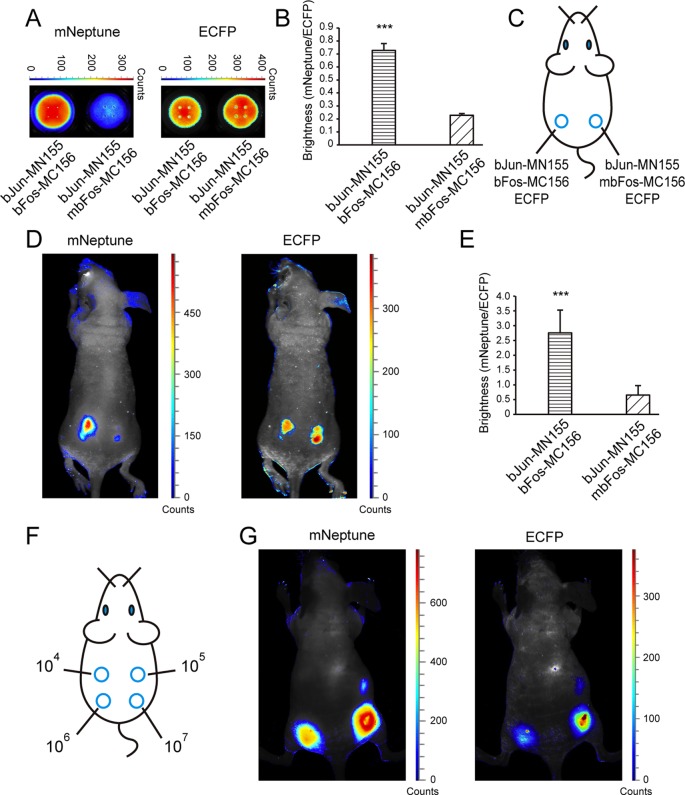
Figure 2.
In vivo imaging of protein–protein interactions in the mNeptune-BiFC system. (A) The BiFC signal (mNeptune channel) and CFP signal (ECFP channel) of 293T cell samples deposited in 96-well black plates co-expressing MN155-bJun, MC156-mbFos and ECFP, or co-expressing MN155-bJun, MC156-mbFos and ECFP. (B) Quantitative analysis of the BiFC signals in (A) based on the fluorescence intensity ratio of mNeptune/ECFP. (C) Implantation locations of the 293T cells transiently co-expressing combinations of fusion proteins as indicated. (D) Imaging of the interaction of bJun and bFos in the mNeptune-BiFC system in live mice injected subcutaneously with cells. mNeptune BiFC signals for the interactions bJun–MN155 and bFos–MC156 or mbFos–MC156 (mouse on the left) and ECFP signals corresponding to the interactions at the same locations (mouse on the right). (E) Quantitative analysis of the BiFC efficiencies for the interaction of bJun and bFos or mbFos in live mice using the fluorescence intensity ratio of BiFC/ECFP (mNeptune/ECFP). Quantitative analysis data are given as the mean + SD (n = 5). The statistical significance was evaluated using a two-tailed Student's t-test. *** indicates P< 0.01. (F) Implantation locations of different amounts (104, 105, 106, 107) of 293T cells co-expressing MN155-bJun and MC156-bFos. (G) mNeptune BiFC signals for different amounts of 293T cells co-expressing MN155-bJun and MC156-bFos (mouse on the left) and ECFP signals corresponding to the cells at the same locations (mouse on the right).
Cells co-expressing the split proteins were injected subcutaneously into mice (Figure 2C) for imaging using the Maestro in vivo imaging system. In live mice, the BiFC signal of the interaction between MN155-bJun and MC156-bFos was detected and imaged easily (Figure 2D, left in the mNeptune channel). There was limited red fluorescence in mice injected with bJun-MN155 and mbFos-MC156. The CFP fluorescence signals were similar, confirming that equal numbers of cells were injected at the different locations in the mouse (Figure 2D, right). Quantitative analysis of the fluorescence intensity ratio of BiFC/ECFP (red/cyan) in mice also revealed that the fluorescence signal of MN155-bJun and MC156-bFos was statistically significantly greater than that of MN155-bJun and MC156-mbFos (Figure 2E). Different amounts (104, 105, 106, 107) of 293T cells co-expressing MN155-bJun and MC156-bFos fusion proteins were injected subcutaneously into live mice (Figure 2F) for fluorescence imaging to test the in vivo sensitivity of the new mNeptune-BiFC system. As shown in Figure 2G, the red fluorescence signal from 105 cells co-transfected with the BiFC constructions could be discriminated from the background with the Maestro 2 in vivo imaging system. Considering that the co-transfection efficiency was 60–70% for the two BiFC plasmids, there should be 6–7 × 104 cells with BiFC signals being imaged in the live mouse.
Construction and application of the mNeptune-TriFC system
The split mNeptune fragments were used as the reporter to construct a long-spectrum TriFC system in which fluorescence was recovered by the formation of two pairs of protein–RNA interactions (7). As shown in Figure 3A, the MC156 fragment was fused to the bacteriophage MS2 coat protein (MCP) and tethered to its stem–loop RNA operator (ms2) by an MCP–ms2 interaction. The MN155 fragment was fused to an RNA-binding protein candidate. The ms2 sequence and the RNA sequence of interest were linked in the backbone plasmid pECFP-C1. The mNeptune-TriFC system (including plasmids pMN155, pMC156-MCP and pECFP-C1-ms2) was constructed based on this design. If the candidate RNA-binding protein interacted with the RNA sequence of interest, the re-association of the two mNeptune fragments would produce a red TriFC signal (Figure 3A).
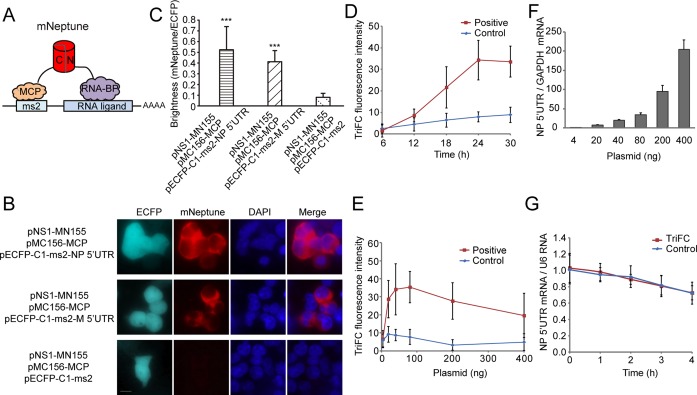
Figure 3.
Construction of the mNeptune-TriFC system. (A) Schematic illustration of the TriFC constructs. N and C (red cylinder) indicate the N-terminal and C-terminal ends, respectively, of the mNeptune fragments; MCP: bacteriophage MS2 coat protein; RNA-BP: RNA binding protein; ms2: stem–loop RNA operator. (B) Detection of the influenza virus NS1–NP 5′UTR and NS1–M 5′UTR interaction in live 293T cells. Cells were transfected with plasmids expressing MCP fused to the C-terminal complementing portion of mNeptune (pMC156-MCP) and the NS1 protein fused to the N-terminal complementing portion of mNeptune (pNS1-MN155). The reporter mRNAs contained the ms2 operator and the 5′UTR sequences of the nucleocapsid protein (NP) or matrix protein (M) mRNAs. Scale bar: 10 μm. (C) Quantitative analysis of TriFC assays for the NP/M 5′UTR and NS1 interaction. The statistical analysis was based on the fluorescence intensity ratio of TriFC/ECFP (mNeptune/ECFP). The data are given as the mean + SD (n = 60). The statistical significance was evaluated using a two-tailed Student's t-test. *** indicates significantly different from the control groups without NP/M 5′UTR, P< 0.01. (D) TriFC assay of NS1–NP 5′UTR interaction in live 293T cells transfected with pNS1-MN155, pMC156-MCP and pECFP-C1-ms2-NP 5′UTR (positive) or pECFP-C1-ms2 (control) at different post-transfection time points. (E) TriFC assay of NS1–NP 5′UTR interaction in live 293T cells when co-transfected with pNS1-MN155 (40 ng), pMC156-MCP (40 ng) and different amounts of pECFP-C1-ms2-NP 5′UTR. (F) The levels of NP 5′UTR mRNA were determined by quantitative real-time RT-PCR in the TriFC assays with different amounts of the pECFP-C1-ms2-NP 5′UTR plasmids corresponding to (E). The levels of NP 5′UTR mRNA are normalized to the GAPDH mRNA. (G) mRNA stability was tested in the TriFC assay. NP 5′UTR mRNA remaining following 0, 1, 2, 3 and 4 h of 0.5 μg/ml of actinomycin D treatment were determined by quantitative real-time RT-PCR. The values are normalized to the U6 snRNA.
The interactions between the influenza A virus NS1 protein and the 5′UTR of the NP and M mRNAs were used to test the newly constructed mNeptune-based TriFC system. The NS1 coding gene and the 5′UTR of the influenza viral NP and M mRNAs were cloned into TriFC plasmid vectors (Supplementary Figure S1) and co-transfected into 293T cells. As expected, red TriFC fluorescence signals were observed in the interaction assays of both NS1-NP 5′UTR and NS1-M 5′UTR (Figure 3B). ECFP fluorescence confirmed that the inserted RNA sequences were expressed correctly. Images from the TriFC assay of wider cell populations with low magnification are shown in Supplementary Figure S5. When the NS1 protein or the 5′UTR of the NP or M mRNAs was absent, no red TriFC signal was detected (Figure 3B). Quantitative analysis based on the fluorescence intensity ratio of mNeptune/ECFP demonstrated that the TriFC signal of cells co-expressing NS1 and the 5′UTR of the NP or M mRNAs was significantly higher than that of cells expressing NS1 but no 5′UTR of the NP or M mRNAs (Figure 3C).
The NS1–NP 5′UTR interaction was also used to optimize the experimental conditions for the new mNeptune-TriFC system. TriFC signals of the NS1–NP 5′UTR interaction in live 293T cells transfected with pNS1-MN155, pMC156-MCP and pECFP-C1-ms2-NP 5′UTR were detected at different post-transfection time points. The result showed that the TriFC signal could be discriminated from background at about 12 h after transfection and the signal increased from 12–24 h after transfection (Figure 3D). The TriFC signal plateaued at about 24 h, which should be suitable for signal detection. At transfection time points greater than 24 h, the TriFC signal did not increase obviously, but the background signal increased.
Plasmid usage and ratio was also tested for the new mNeptune-TriFC system. For a given quantity of the MN and MC plasmids (e.g. 40 ng or 80 ng in a 35-mm dish), different amounts of the plasmids that produce the RNA adapter were used to test the mNeptune-TriFC system. As shown in Figure 3E, when 40 ng of either of the pNS1-MN155 and pMC156-MCP plasmids was used, different amounts of the pECFP-C1-ms2-NP 5′UTR plasmids (pNS1-MN155: pMC156-MCP: pECFP-C1-ms2-NP 5′UTR of 1:1:0.1, 1:1:0.5, 1:1:1, 1:1:2, 1:1:5 and 1:1:10) were co-transfected into about 105 293T cells for TriFC assays. The levels of NP 5′UTR mRNA were also determined by quantitative real-time RT-PCR in the TriFC assays with different amounts of the pECFP-C1-ms2-NP 5′UTR plasmids (Figure 3F). The levels of NP 5′UTR mRNA were normalized to the GAPDH mRNA. The results of TriFC assays showed that when the amount of pECFP-C1-ms2-NP 5′UTR (e.g. 20 ng) was half that of pNS1-MN155 and pMC156-MCP, a distinct TriFC signal was acquired. The TriFC signal increased with increases in the quantity of plasmid pECFP-C1-ms2-NP 5′UTR, which produces more NP 5′UTR mRNA, and peaked when pNS1-MN155: pMC156-MCP: pECFP-C1-ms2-NP 5′UTR was 1:1:2. When plasmid pECFP-C1-ms2-NP 5′UTR was used in higher amounts, the TriFC signal became lower and decreased with further increases in the redundant NP 5′UTR mRNA.
We also tested the mRNA stability when using the TriFC system to image mRNA–protein interactions. 293T cells transfected with pECFP-C1-ms2-NP 5′UTR or co-transfected with pNS1-MN155, pMC156-MCP and pECFP-C1-ms2-NP 5′UTR were treated with actinomycin D to stop mRNA transcription 24 h after transfection. Cells were harvested at different time points upon actinomycin D treatment, and mRNA levels of NP 5′UTR mRNA were quantified by quantitative real-time RT-PCRs. Values were normalized to the stable U6 snRNA. The results showed that the presence of fluorescent protein fragments in the TriFC system did not significantly affect the stability of NP 5′UTR mRNA (Figure 3G).
In vivo imaging of protein–mRNA interactions in the mNeptune-TriFC system
The mNeptune-TriFC system was then tested for imaging protein–mRNA interactions in live mice. The NS1–NP 5′UTR interaction was visualized in live mice. 293T cells co-expressing NS1 and the NP 5′UTR mRNA in the TriFC system were analyzed in vitro using the Maestro 2 in vivo imaging system. As shown in Figure 4A, the red TriFC fluorescence of cells co-expressing NS1 and the NP 5′UTR mRNA was greater than that of cells expressing NS1 but no NP 5′UTR mRNA (Figure 4A, left). The CFP signals of the cell samples were similar (Figure 4A, right). Quantitative analysis based on the fluorescence intensity ratio of TriFC/ECFP (red/cyan) demonstrated that the TriFC signal of cells co-expressing NS1 and the NP 5′UTR mRNA was significantly higher than that of cells expressing NS1 but no NP 5′UTR mRNA (Figure 4B).
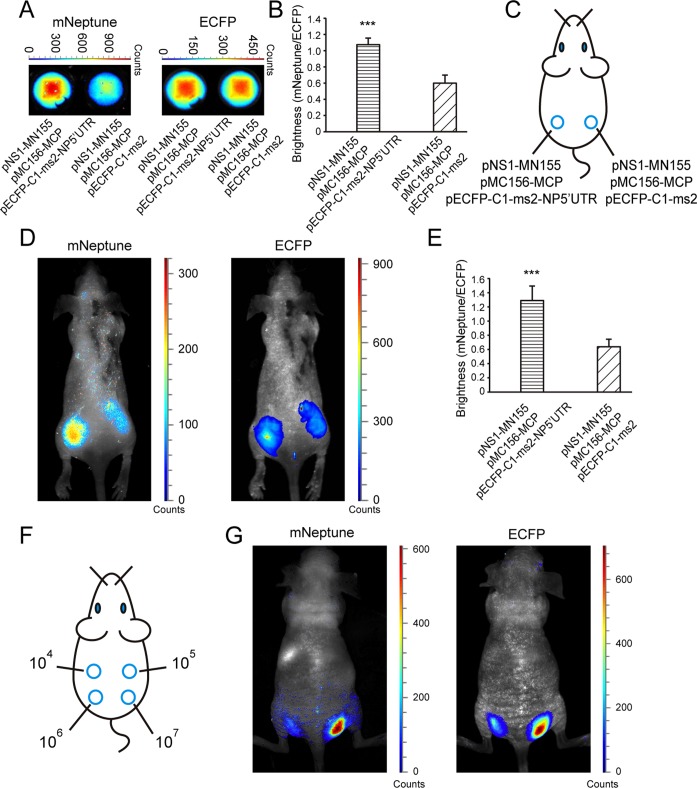
Figure 4.
In vivo imaging of protein–mRNA interactions in the mNeptune-TriFC system. (A) The TriFC signal (mNeptune channel) and CFP signal (ECFP channel) of cell samples deposited on 96-well black plates co-expressing NS1 protein and NP 5′UTR mRNA or without NP 5′UTR mRNA. (B) Quantitative analysis of the TriFC signals in (A) based on the fluorescence intensity ratio of mNeptune/ECFP. (C) Implantation locations of the 293T cells transiently co-transfected with combinations of plasmids as indicated. (D) Imaging of the interaction of NS1 and the 5′UTR of NP mRNAs in the mNeptune-TriFC system in live mice injected subcutaneously with the 293T cells. mNeptune TriFC signals for the interaction of NS1 and NP 5′UTR or without NP 5′UTR mRNA (mouse on the left) and ECFP signals corresponding to the interactions at the same locations (mouse on the right). (E) Quantitative analysis of TriFC efficiencies for the interaction of NS1 and NP 5′UTR mRNAs in live mice based on the fluorescence intensity ratio of TriFC/ECFP (mNeptune/ECFP). Quantitative analysis data are given as the mean + SD (n = 6). The statistical significance was evaluated using a two-tailed Student's t-test. *** indicates P< 0.01. (F) Implantation locations of different amounts (104, 105, 106, 107) of 293T cells co-expressing NS1 and the NP 5′UTR mRNA in the TriFC system. (G) mNeptune TriFC signals of different amounts of 293T cells with the interaction of NS1 and NP 5′UTR in the TriFC system (mouse on the left) and ECFP signals corresponding to the cells at the same locations (mouse on the right).
The cells co-transfected with TriFC plasmids were injected subcutaneously into mice (Figure 4C) and examined with the Maestro 2 in vivo imaging system. In live mice, the TriFC signal of the interaction between NS1 and the NP 5′UTR mRNAs was detected and imaged as shown in Figure 4D (left). When the NP mRNAs were absent, only a very low red fluorescence was detected in the mice. The CFP signals of the cells were similar, indicating that equal numbers of cells were injected at the different locations in the mouse (Figure 4D, right). Quantitative analysis of the fluorescence intensity ratio of TriFC/ECFP (red/cyan) in the mice demonstrated that the red TriFC fluorescence signal was statistically higher than in the control in which the NP 5′UTR mRNA was absent (Figure 4E). Different amounts (104, 105, 106, 107) of 293T cells co-expressing NS1 and the NP 5′UTR mRNA in the TriFC system were also injected subcutaneously into live mice (Figure 4F) for fluorescence imaging to test the in vivo sensitivity of the new TriFC system. As shown in Figure 4G, the red fluorescence signal from 106 cells co-transfected with the TriFC constructions could be visualized with the Maestro 2 in vivo imaging system. Considering that the co-transfection efficiency was 50–60% for the three TriFC plasmids, there should be 5–6 × 105 cells with TriFC signals being imaged in the live mouse.
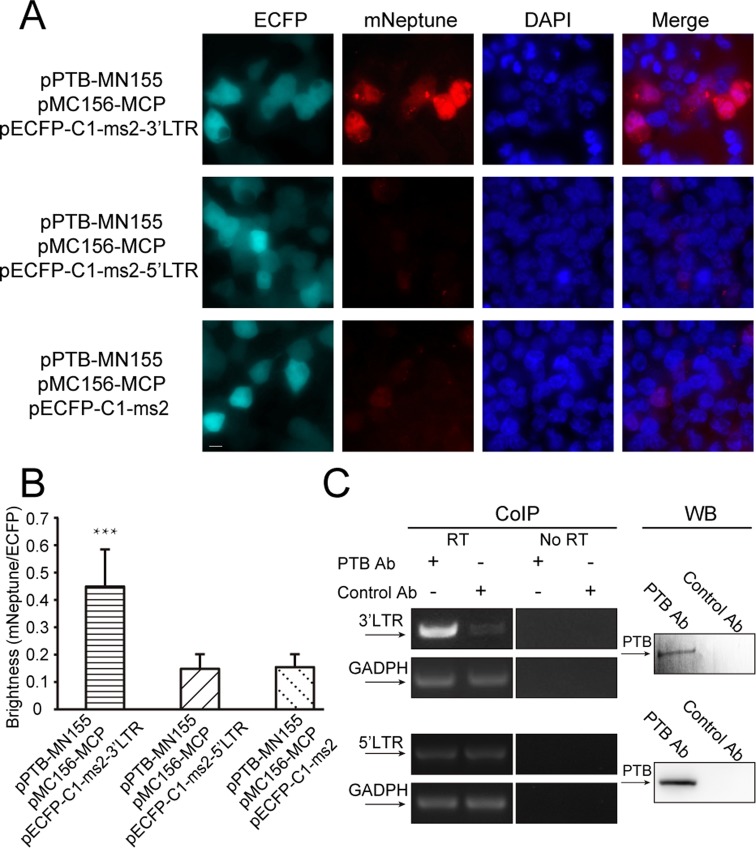
Imaging of the interactions between PTB and the 3′LTR of HIV mRNAs using the mNeptune-TriFC system
The mNeptune-TriFC system was next used to study unknown protein–mRNA interactions in live cells. Interactions between PTB and the HIV-1 mRNAs were tested using the mNeptune-TriFC system. The 3′LTR (untranslated region) and 5′LTR of HIV-1 mRNAs exist in all synthesized viral mRNAs and play important roles in viral mRNA processing. We assessed the interactions of PTB with the 3′3′LTR and 5′LTR of HIV-1 mRNAs. In the TriFC system, cells co-expressing PTB and the 3′LTR of HIV-1 mRNAs displayed a strong red TriFC signal, while cells co-expressing PTB and the 5′LTR of HIV-1 mRNAs did not produce a red TriFC signal (Figure 5A). The imaging and quantification (Figure 5B) demonstrated that PTB associates with the 3′LTR but not the 5′LTR of HIV-1 mRNA. We used co-immunoprecipitation experiments to verify the interaction between PTB and the 3′LTR of HIV-1 mRNA (Figure 5C). Co-immunoprecipitation experiments also showed there is no interaction between PTB and 5′LTR of HIV-1 mRNA. The known interaction between PTB and the CRS region of HIV-1 env mRNA (16) was also imaged in live cells using the mNeptune-TriFC system (Supplementary Figure S6).
Figure 5.
Imaging the interactions between PTB and the 3′LTR and 5′LTR of HIV-1 mRNAs in live cells. (A) TriFC (mNeptune channel) and ECFP signals of 293T cells co-expressing PTB and the 5′LTR or 3′LTR sequences of HIV mRNAs in the TriFC system. Cells were transfected with plasmids expressing MC156-MCP and PTB-MN155 fusion proteins. The reporter mRNAs contained the ms2 operator and the 5′LTR or 3′LTR sequences of HIV mRNAs. Scale bar: 10 μm. (B) Quantitative analysis of TriFC efficiency for the interactions between PTB and the 5′LTR or 3′LTR sequences of HIV mRNAs. Statistical analysis was based on the fluorescence intensity ratio of TriFC/ECFP. The data are given as the mean + SD (n = 60). Statistical significance was evaluated using a two-tailed Student's t-test. *** indicates significantly different from the control groups without 3′LTR or 5′LTR RNA sequences, P< 0.001. (C) Verification of the interaction between PTB and the 3′LTR of HIV-1 mRNAs. Co-immunoprecipitation (CoIP) assays were performed with cell lysate and anti-PTB antibody or control antibody IgG1. The binding of PTB to the 3′LTR of HIV-1 mRNAs was detected by reverse transcription (RT) followed by PCR amplification using primers specific to the 3′LTR of HIV-1 mRNAs and the housekeeping GAPDH mRNA. No amplification was detected in the ‘No RT’ control samples. CoIP experiments also showed there is no interaction between PTB and 5′LTR of HIV-1 mRNA. Western blotting (WB) was performed to monitor the presence of PTB in the samples used in the CoIP assays.
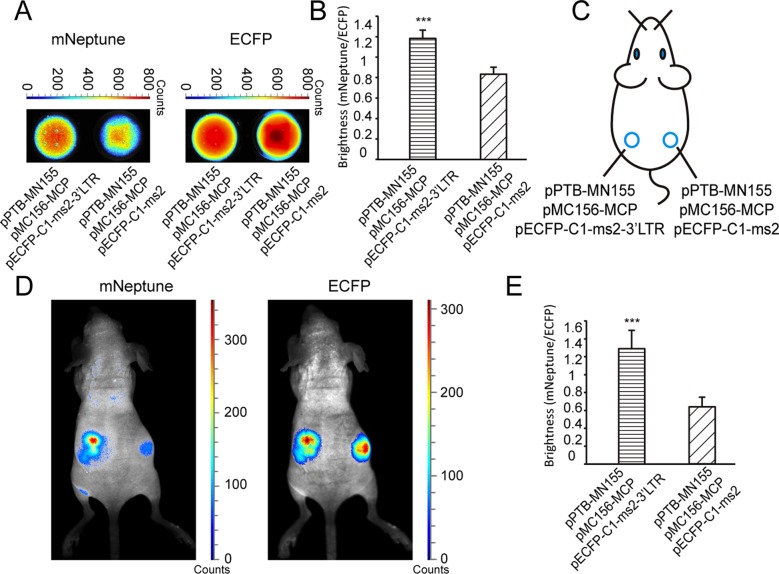
In vivo imaging of interactions between PTB and the 3′LTR of HIV mRNAs
The interaction between PTB and the 3′LTR of HIV-1 mRNA was imaged in live mice using the mNeptune-TriFC system. 293T cells co-expressing PTB and the 3′LTR of HIV-1 mRNA in the TriFC system were analyzed in vitro using the Maestro in vivo imaging system. As shown in Figure 6A, the red TriFC fluorescence of cells co-expressing PTB and the 3′LTR of HIV-1 mRNA was greater than that of cells expressing PTB but not the 3′LTR of HIV-1 mRNA (Figure 6A, left). The CFP signals of the cells were similar (Figure 6A, right). Quantitative analysis based on the fluorescence intensity ratio of TriFC/ECFP (red/cyan) demonstrated that the TriFC signal of cells co-expressing PTB and the 3′LTR of HIV-1 mRNA was significantly higher than that of cells expressing PTB but no 3′LTR of HIV-1 mRNA (Figure 6B).
Figure 6.
In vivo imaging of the interaction between PTB and the 3′LTR of HIV-1 mRNAs in the TriFC system. (A) TriFC signal (mNeptune channel) and CFP signal (ECFP channel) of cell samples on 96-well black plates co-expressing PTB and the 3′LTR of HIV-1 mRNAs or without 3′LTR of HIV-1 mRNAs. (B) Quantitative analysis of the TriFC signals in (A) based on the fluorescence intensity ratio of mNeptune/ECFP. (C) Implantation locations of the 293T cells transiently co-transfected with combinations of plasmids as indicated. (D) Imaging of the interaction between PTB and the 3′LTR of HIV-1 mRNAs in the mNeptune-TriFC system in live mice injected subcutaneously with the 293T cells. mNeptune TriFC signals for the interaction of PTB and the 3′LTR of HIV-1 mRNAs or without 3′LTR (mouse on the left) and ECFP signals corresponding to the interactions at the same locations (mouse on the right). (E) Quantitative analysis of TriFC efficiencies for the interaction of PTB and the 3′LTR of HIV-1 mRNAs in live mice based on the fluorescence intensity ratio of TriFC/ECFP (mNeptune/ECFP). Quantitative analysis data are given as the mean + SD (n = 4). Statistical significance was evaluated using a two-tailed Student's t-test. *** indicates P< 0.01.
Cells that were co-transfected with the TriFC plasmids were injected subcutaneously into mice (Figure 6C) for imaging with the Maestro 2 in vivo imaging system. As shown in Figure 6D, the interaction between PTB and the 3′LTR of HIV-1 mRNA yielded a red TriFC signal in live mice and when the 3′LTR of HIV-1 mRNA was absent, little red fluorescence was detected. The CFP signals of the cells were similar, confirming that equal numbers of cells were injected at different locations in the mouse (Figure 6D, right). Statistical analysis demonstrated that the red TriFC fluorescence of PTB and the 3′LTR of HIV-1 mRNA was significantly greater than the fluorescence in the absence of the 3′LTR of HIV-1 mRNA (Figure 6E).
DISCUSSION
In vivo imaging could help elucidate physiological and pathological mechanisms and contribute to drug evaluation. Several methods based on luciferase have been developed for imaging protein–protein interactions (23–26) in live subjects. However, no methods based on fluorescent proteins or, more importantly, methods to image RNA–protein interactions in live animals are currently available. In this study, the far-red mNeptune-TriFC system (600 nm/650 nm) was developed as a tool for in vivo imaging of RNA–protein interactions. To our knowledge, this is the first report of RNA–protein interactions being visualized successfully in live mice. The mNeptune-BiFC system also provided a new fluorescent protein-based system for in vivo imaging of protein–protein interactions. These FC systems with excitation and emission in the ‘tissue optical window’ are suitable for in vivo imaging. Because of the good optical properties of mNeptune and the superiority of protein-fragment complementation techniques, the mNeptune-BiFC and TriFC systems could become very useful and powerful tools to study protein–protein and RNA–protein interactions in live subjects. These new FC systems are also an addition to the tool set for evaluating novel drugs to inhibit RNA–protein and protein–protein interactions. In our experiments, the mNeptune-BiFC and TriFC signals could be observed in live mice without eliminating the autofluorescence signals of the mice (Supplementary Figure S7). The ECFP signals, on the other hand, could be detected by the Maestro 2 in vivo imaging system only after eliminating the autofluorescence spectra.
The new mNeptune TriFC system was used to investigate interactions between the cellular protein PTB and HIV-1 mRNA elements including 5′LTR, 3′LTR and CRS. The interaction between PTB and the 3′LTR of HIV-1 mRNAs was found in the mNeptune TriFC system. PTB is a multifunctional RNA chaperone that mediates mRNA transportation, splicing and translation. PTB contributes to the activation of latent HIV-1 viruses in resting CD4+ T cells from patients on (highly active antiretroviral therapy) HAART (19), although further studies of the mechanism are needed. PTB was reported to bind to the CRS region of HIV-1 env mRNA (also see Supplementary Figure S6), suggesting that PTB may play a role in regulating the processing of CRS-containing RNAs (18). However, whether and how PTB regulates the processing of HIV-1 mRNAs without the CRS sequence (i.e. multi-spliced short HIV-1 mRNAs) remains unclear. Multi-spliced short HIV-1 mRNAs have been found in the nucleus of resting CD4+ T cells, including the mRNAs of the important viral proteins Tat and Rev. Overexpression of PTB in resting CD4+ T cells induces cytoplasmic accumulation of HIV-1 mRNAs and virus production (19). The 3′LTR exists in all HIV-1 mRNAs and contains a polyadenylation signal (AATAAA) and polyadenylation addition sites critical for viral RNA synthesis (27). The interaction of PTB and the 3′LTR of HIV-1 mRNAs could function as a bridge for PTB and viral mRNA processing (including multi-spliced HIV-1 mRNAs), which is related to the HIV activation from latency.
In conclusion, we have constructed new far-red BiFC and TriFC systems with excitation and emission within the ‘tissue optical window’ and shown that they are good tools for in vivo imaging of protein–protein and mRNA–protein interactions. The new TriFC system was used to demonstrate the interaction of PTB protein and the 3′LTR of HIV-1 mRNA, which implies a role for PTB in regulating HIV-1 mRNA processing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
FUNDING
National Nano Project [2011CB933600 to Z.Q.C. and Z.P.Z.]; National Natural Science Foundation of China [31070774 to Z.Q.C.]; Chinese Academy of Sciences (CAS) [KSCX2-EW-Q-15 to Y.H., S.W., Z.Z., X.M., and Z.Q.C.]; CAS [KJZD-EW-TZ-LO4 to X.E.Z.]; Funding for open access charge: National Natural Science Foundation of China [31070774].
Conflict of interest statement. None declared.
REFERENCES
- 1.Massoud T.F., Gambhir S.S. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 2.Luker G.D., Sharma V., Pica C.M., Dahlheimer J.L., Li W., Ochesky J., Ryan C.E., Piwnica-Worms H., Piwnica-Worms D. Noninvasive imaging of protein-protein interactions in living animals. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6961–6966. doi: 10.1073/pnas.092022399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shyu Y.J., Hu C.D. Fluorescence complementation: an emerging tool for biological research. Trends Biotechnol. 2008;26:622–630. doi: 10.1016/j.tibtech.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Kerppola T.K. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu C.D., Chinenov Y., Kerppola T.K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 6.Hu C.D., Kerppola T.K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rackham O., Brown C.M. Visualization of RNA-protein interactions in living cells: FMRP and IMP1 interact on mRNAs. EMBO J. 2004;23:3346–3355. doi: 10.1038/sj.emboj.7600341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stohr N., Lederer M., Reinke C., Meyer S., Hatzfeld M., Singer R.H., Huttelmaier S. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozawa T., Natori Y., Sato M., Umezawa Y. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat. Methods. 2007;4:413–419. doi: 10.1038/nmeth1030. [DOI] [PubMed] [Google Scholar]
- 10.Stamatas G.N., Southall M., Kollias N. In vivo monitoring of cutaneous edema using spectral imaging in the visible and near infrared. J. Invest. Dermatol. 2006;126:1753–1760. doi: 10.1038/sj.jid.5700329. [DOI] [PubMed] [Google Scholar]
- 11.Tromberg B.J., Shah N., Lanning R., Cerussi A., Espinoza J., Pham T., Svaasand L., Butler J. Non-invasive in vivo characterization of breast tumors using photon migration spectroscopy. Neoplasia. 2000;2:26–40. doi: 10.1038/sj.neo.7900082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jach G., Pesch M., Richter K., Frings S., Uhrig J.F. An improved mRFP1 adds red to bimolecular fluorescence complementation. Nat. Methods. 2006;3:597–600. doi: 10.1038/nmeth901. [DOI] [PubMed] [Google Scholar]
- 13.Fan J., Cui Z., Wei H., Zhang Z., Zhou Y., Wang Y., Zhang X.-E. Split mCherry as a new red bimolecular fluorescence complementation system. Biochem. Biophys. Res. Commun. 2008;367:47–53. doi: 10.1016/j.bbrc.2007.12.101. [DOI] [PubMed] [Google Scholar]
- 14.Chu J., Zhang Z., Zheng Y., Yang J., Qin L., Lu J., Huang Z.L., Zeng S., Luo Q. A novel far-red bimolecular fluorescence complementation system that allows for efficient visualization of protein interactions under physiological conditions. Biosens. Bioelectron. 2009;25:234–239. doi: 10.1016/j.bios.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Yin J., Zhu D., Zhang Z., Wang W., Fan J., Men D., Deng J., Wei H., Zhang X.-E., Cui Z. Imaging of mRNA-protein interactions in live cells using novel mCherry trimolecular fluorescence complementation systems. PLoS ONE. 2013;8:e80851. doi: 10.1371/journal.pone.0080851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin M.Z., McKeown M.R., Ng H.L., Aguilera T.A., Shaner N.C., Campbell R.E., Adams S.R., Gross L.A., Ma W., Alber T., Tsien R.Y. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem. Biol. 2009;16:1169–1179. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park Y.W., Katze M.G. Translational control by influenza virus: Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J. Biol. Chem. 1995;270:28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 18.Black A.C., Luo J., Chun S., Bakker A., Fraser J.K., Rosenblatt J.D. Specific binding of polypyrimidine tract binding protein and hnRNP A1 to HIV-1 CRS elements. Virus Genes. 1996;12:275–285. doi: 10.1007/BF00284648. [DOI] [PubMed] [Google Scholar]
- 19.Lassen K.G., Ramyar K.X., Bailey J.R., Zhou Y., Siliciano R.F. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006;2:e68. doi: 10.1371/journal.ppat.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Song M.G., Kiledjian M. Transcript-specific decapping and regulated stability by the human Dcp2 decapping protein. Mol. Cell. Biol. 2008;28:939–948. doi: 10.1128/MCB.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peritz T., Zeng F., Kannanayakal T.J., Kilk K., Eiríksdóttir E., Langel U., Eberwine J. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- 22.Shaner N.C., Steinbach P.A., Tsien R.Y. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 23.De A., Gambhir S.S. Noninvasive imaging of protein-protein interactions from live cells and living subjects using bioluminescence resonance energy transfer. FASEB J. 2005;19:2017–2019. doi: 10.1096/fj.05-4628fje. [DOI] [PubMed] [Google Scholar]
- 24.De A., Loening A.M., Gambhir S.S. An improved bioluminescence resonance energy transfer strategy for imaging intracellular events in single cells and living subjects. Cancer Res. 2007;67:7175–7183. doi: 10.1158/0008-5472.CAN-06-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De A., Ray P., Loening A.M., Gambhir S.S. BRET3: a red-shifted bioluminescence resonance energy transfer (BRET)-based integrated platform for imaging protein-protein interactions from single live cells and living animals. FASEB J. 2009;23:2702–2709. doi: 10.1096/fj.08-118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dragulescu-Andrasi A., Chan C.T., De A., Massoud T.F., Gambhir S.S. Bioluminescence resonance energy transfer (BRET) imaging of protein-protein interactions within deep tissues of living subjects. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12060–12065. doi: 10.1073/pnas.1100923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freed E.O., Martin M.A. Fields Virology. 2007. HIVs and their replication; pp. 2107–2136. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.