Abstract
Androgen receptor (AR) plays an important regulatory role in prostate cancer. AR's transcriptional activity is regulated by androgenic ligands, but also by post-translational modifications, such as SUMOylation. To study the role of AR SUMOylation in genuine chromatin environment, we compared androgen-regulated gene expression and AR chromatin occupancy in PC-3 prostate cancer cell lines stably expressing wild-type (wt) or doubly SUMOylation site-mutated AR (AR-K386R,K520R). Our genome-wide gene expression analyses reveal that the SUMOylation modulates the AR function in a target gene and pathway selective manner. The transcripts that are differentially regulated by androgen and SUMOylation are linked to cellular movement, cell death, cellular proliferation, cellular development and cell cycle. Fittingly, SUMOylation mutant AR cells proliferate faster and are more sensitive to apoptosis. Moreover, ChIP-seq analyses show that the SUMOylation can modulate the chromatin occupancy of AR on many loci in a fashion that parallels their differential androgen-regulated expression. De novo motif analyses reveal that FOXA1, C/EBP and AP-1 motifs are differentially enriched at the wtAR- and the AR-K386R,K520R-preferred genomic binding positions. Taken together, our data indicate that SUMOylation does not simply repress the AR activity, but it regulates AR's interaction with the chromatin and the receptor's target gene selection.
INTRODUCTION
Prostate cancer is a major health concern among men by being one of the most common cancers diagnosed and one of the most common causes of cancer death [(1), http://www.cancerresearchuk.org/]. The androgen receptor (AR) has an important role in the development and progression of prostate cancer and regardless of the progress in prostate cancer pathobiology, the AR remains the major druggable target for the advanced disease. AR is an androgen-regulated transcription factor which in prostate is activated by the binding of 5α-dihydrotestosterone. Subsequently, the AR moves to nucleus, binds to specific androgen response elements (AREs) on the regulatory regions of its target genes and in this way conveys the message of androgens directly to the level of gene programs (2–4). In addition to hormone binding, the AR activity is regulated by post-translational modifications, including SUMOylation (5). SUMOylation is a reversible modification in which small ubiquitin-related modifier protein (SUMO) 1, 2 or 3 is covalently attached to target proteins' specific lysine residues via an enzymatic E1→E2→E3 pathway analogous to ubiquitylation but with enzymes (E1, SAE1/2; E2, UBC9; E3, e.g. PIAS proteins) distinct from the ubiquitylation (6,7). The SUMOylation pathway does not generally target proteins for degradation, but regulates proteins' activity and changes their interactions with other protein and/or subcellular or nuclear localization (6,8,9).
Previous studies have shown that the N-terminal transactivation domain of AR is covalently modified by SUMOs at two conserved lysine (in human sequence K386 and K520) residues in an androgen-inducible and reversible fashion (5,10). Moreover, SUMOylation pathway components act as AR coregulators in transcription assays (10–12). Disruption of these sites increases the transcriptional activity of AR on compound ARE-driven promoters in reporter gene assays, suggesting that the modification is linked to transcriptional repression. However, very little is known about the importance and role of the AR SUMOylation in a genuine chromatin environment and regulation of endogenous AR target genes in prostate cancer cells. To study in a systematic genome-wide fashion the role of AR SUMOylation in prostate cancer chromatin environment, we used PC-3 cell lines that stably express wild-type (wt) or SUMOylation-deficient AR (AR-K386R,K520R; AR-2KR) and analyzed their androgen-regulated transcripts. We also compared the capabilities of these two AR forms to bind to chromatin by using chromatin immunoprecipitation coupled to deep sequencing (ChIP-seq). These genome-wide analyses that were additionally carried out in HEK293 cells stably expressing wtAR or AR-2KR revealed that the AR SUMOylation sites do not simply repress the AR activity on all target genes. The mutant also exhibited attenuated transcriptional activity on several genes and a group of target genes were insensitive to the SUMOylation. Interestingly, the genes differently expressed by androgen due to the AR SUMOylation sites are significantly enriched in cell proliferation and apoptosis pathways. Our cistrome analyses also show that the SUMOylation can regulate the receptor's chromatin occupancy in a locus-selective fashion.
MATERIALS AND METHODS
Cell culture
Stably AR-expressing PC-3 prostate cancer cells were maintained and generated as described in reference (13). Stably AR-expressing isogenic Flp-In-293 (HEK293) (Invitrogen) were maintained and generated as described in reference (14). In experiments steroid-depleted transfection medium was used (5% charcoal-stripped-FBS in F-12 for PC-3 cells and 2.5% charcoal-stripped-FBS in Dulbecco's modified Eagle's medium (DMEM) for HEK293). In both cell types, several clones were analyzed before continuing studies with one representative clone.
RT-qPCR analysis
PC-3 and HEK293 cells were seeded onto 6-well plates, grown 48 h in steroid-depleted transfection medium and then exposed to vehicle (EtOH) or 10 nM R1881 for 16 h (PC-3) or 24 h (HEK293). Total RNA of biological triplicates was extracted (TriPure isolation reagent, Roche) and converted to cDNA (Transcriptor First Strand cDNA synthesis Kit, Roche) according to manufacturer's instructions. Expression of AR target genes with specific primers (Supplementary Table S1) was measured by RT-qPCR as described in reference (15) using RPL13A (for PC-3) and GAPDH (for HEK293) mRNA levels to normalize the amounts of total RNA between the samples.
Microarray analysis
Total RNA of biological triplicates was collected, and hybridized to Illumina HumanHT-12 v3 (for HEK293 cells) or v4 (for PC-3 cells) Expression BeadChips (San Diego, CA, USA) at the Finnish Microarray and Sequencing Centre (Turku, Finland) using protocols recommended by the manufacturer. The Illumina BeadChIP data were analyzed using the Bioconductor associated packages (16). Data were preprocessed (bgAdjust), variance stabilizing transformed (vst) and robust spline normalized (rsn) with lumi package (17) and analyzed using the Linear Models for Microarray Data (limma) package (18) (empirical Bayes statistics with a Benjamini and Hochberg multiple test correction procedure). A gene was considered significantly changed, if it had the adjusted P-value <0.05 and fold change ≤0.7 or ≥1.4. Heat maps were generated by using heatmap.2 in the R package gplots. Ingenuity Pathway Analysis® (IPA) was used to identify biological processes enriched for differentially expressed genes. First, a core analysis was performed with two distinct lists (androgen-regulated genes in wtAR or AR-2KR expressing cells). The results were then compared to identify any distinct biological processes that were differentially regulated.
Cell proliferation and apoptosis assay
In proliferation assay, PC-3 cells were seeded in 96-wells and cells were grown 24 h before addition of 10 nM R1881 or vehicle (EtOH) and the amount of viable cells was measured using CellTiter-Glo® Luminescent Cell Viability Assay (Promega) according to the manufacturer's instructions. Similarly, 48 h and 72 h after exposure viable cells were measured. The results were calculated as percentage of cell growth using the first measurement as reference point. In apoptosis assay, cells were cultured in 96-wells and grown 24 h before treating with 1.5 μM camptothecin for 20 h. Apoptosis was measured using Cell Death Detection ELISAPlus kit (Roche) according to the manufacturer's instructions.
ChIP coupled to high-throughput sequencing (ChIP-seq)
The chromatin immunoprecipitation (ChIP) experiments and analyses were performed as previously described (15). Briefly, PC-3 (13) or HEK293 cells (14) were seeded onto 10-cm plates, grown 72 h in steroid-depleted transfection medium and exposed to vehicle (EtOH) or 10 nM R1881 for 1 h (PC-3) or 40 min (HEK293) before crosslinking with 1% (v/v) formaldehyde and harvesting for immunoprecipitation. ChIP-seq samples were immunoprecipitated with anti-AR antibody (19) and for quantitative PCR (qPCR)-based ChIP assays, samples were immunoprecipitated with anti-FOXA1 (Abcam, ab23738), anti-c-Jun (Santa Cruz Biotechnology, sc-1694), anti-C/EBPβ (Santa Cruz Biotechnology, sc-150) or rabbit-IgG (Santa Cruz Biotechnology, sc-2027). Specific primers used in qPCR analysis are listed in Supplementary Table S1. Single-end 50 bp ChIP-seq data were generated using Illumina Hiseq System (Illumina) in the EMBL Genomics Core Facility (Heidelberg, Germany). The quality of raw reads was analyzed by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and after that FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) was used for preprocessing (removal of artifacts, 50-bp trimming, quality filtering and collapsing identical reads). Remaining reads were aligned against the human reference genome version hg19 with Bowtie software 0.12.7 (20), allowing one mismatch and accepting only the best alignment (with essential command line arguments: -v 1 -k 1 –m 1 –f –S —best hg19). Resulting Sequence Alignment/Map (SAM) files were analyzed using HOMER software version 4.2 (21). Enriched binding sites were detected using findPeaks command with factor analysis strategy using 3-fold enrichment over input background read set (own input for each cell line) and finally selecting binding sites for which the normalized tag counts were ≥6. Matching the ENCODE Consortium guidelines, we sequenced two independent biological replicates per sample and gained ≥10 million uniquely mapped reads per replicate (22). Binding site overlaps between each replicate pair, identified using BEDTools (23), also fulfilled the ENCODE requirements for replicate similarity. Initial overlap analysis of wtAR- or AR-2KR- expressing cell binding sites was performed using BEDTools and non-shared binding sites were analyzed further using getDifferentialPeaks command of HOMER using a cutoff of 2-fold enrichment over the other sample to find final preferred binding sites for wtAR- and AR-2KR-expressing cells. HOMER software was also used to format the data for visualization in Integrative Genomics Viewer (IGV), genomic annotation of peak positions and for the de novo motif analysis. De novo discovery of the motifs (length 6,10,15,20) was performed on sequences covering ±100 bp of the AR peak center. Association of AR binding sites to nearby genes (±100 kb from transcription start site) and androgen-regulated genes was done with Anduril (Engine 1.2.18) (24) with NextBioentity (Ensembl Homo Sapiens 71.37) (Supplementary file 1).
Accession numbers
Bead array and ChIP-seq data are submitted to the NCBI Gene Expression Omnibus database (25) (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE54202.
RESULTS
Androgen target genes are differently modulated by AR SUMOylation
To study the role of the AR SUMOylation in genuine chromatin environment, we constructed PC-3 prostate cancer cell lines stably expressing wild-type AR (wtAR) or SUMOylation deficient AR (AR-2KR) in which SUMO acceptor sites K386 and K520 were converted to arginine residues (Supplementary Figure S1A), (13). Immunoblot analysis of the cells confirmed that the wtAR and the AR-2KR are equally expressed and that their expression levels do not exceed that of endogenous AR in LNCaP cells and their subcellular distributions are comparable (Supplementary Figure S1B and C). Based on transfected reporter gene activity in the PC-3 cells, mutation of the SUMOylation sites increases the transcriptional activity of AR on compound ARE-driven promoters as shown previously for cells of nonprostatic origin (Supplementary Figure S1D), (5). Moreover, our recent preliminary AR target gene analyses of isogenic HEK293 cell lines expressing wtAR or AR-2KR (at a level comparable to that of endogenous AR in VCaP prostate cancer cells) suggested that the effects of SUMOylation on the AR activity are target gene-dependent (14). We next compared gene expression profiles of the wtAR and AR-2KR PC-3 cells exposed to synthetic androgen (R1881) or vehicle by using Illumina HT-12 v4 Expression BeadChips. Comparison of our microarray data with those from VCaP cells showed a reasonable, ∼30% overlap of androgen-regulated genes (Supplementary Figure S2). The genome-wide analysis indicated that more genes were androgen-regulated in the wtAR cells (473) than in the AR-2KR cells (143), with 47% and 61% of these genes being up-regulated by androgen in the wtAR and AR2-KR cells, respectively, (Figure 1A, Supplementary file 2). The difference between wtAR and AR-2KR remains also if other P-value cut offs are applied (Supplementary Table S2). This was also seen in genome-wide gene expression analysis of the HEK293 wtAR and AR-2KR cells (368 versus 178 regulated genes; 49% versus 57% up-regulated by androgen) (Supplementary Figure S3A, Supplementary file 2). More interestingly, as shown by the Venn diagrams in Figure 1A, only a small portion of androgen-regulated genes were common to both cells. Unsupervised hierarchical clustering of the androgen-regulated genes in the PC-3 and the HEK293 cell lines further emphasized the differences in the androgen-regulated gene expression between the wtAR and the AR-2KR cells (Figure 1B, Supplementary Figure S3B). Expression of one gene from each cluster in PC-3 cells was validated by RT-qPCR analysis (Figure 1C). These analyses show that there are different classes of AR target genes regarding their sensitivity to the SUMOylation. Some genes, such as FKBP5, are insensitive to the AR SUMOylation sites, being similarly expressed in both cell lines. A subset of genes, such as CLDN8, require the SUMOylation sites, as they are robustly androgen-regulated only in the wtAR cells, whereas certain genes, such as ADRA1B, are contrastingly inhibited by the SUMOylation, as demonstrated by their enhanced expression in the AR-2KR cells. Similar AR target gene classes were seen and validated in the HEK293 cells (Supplementary Figure S3B and C). Thus, our unbiased genome-wide expression data indicate that the SUMOylation modulates the AR function in a target gene-selective fashion instead of simply repressing the AR activity, as previously suggested based on simple ARE-driven reporter gene assays (5,10).
Figure 1.

SUMOylation sites modulate the AR activity in a target gene-selective fashion. Stably wtAR- and AR-2KR-expressing PC-3 prostate cancer cells were treated with R1881 (10 nM) or vehicle (ethanol, EtOH) for 16 h and isolated RNAs were analyzed by Illumina Expression BeadChips as described in ‘Materials and Methods’ section. The genes that had adjusted P-value < 0.05 and fold-induction ≥1.4 or ≤0.7 were considered as androgen-regulated genes. (A) Venn diagrams showing androgen-up- and androgen-down-regulated genes in the wtAR- and the AR-2KR-expressing cells. (B) Heat map of androgen-regulated genes clustered by unsupervised hierarchical clustering. (C) RT-qPCR validation of one AR target gene of each cluster. The relative mRNA expressions represent the means (n = 3) ± SDs. Statistically significant differences (***P < 0.001, **P < 0.01 and *P < 0.05; Student's t-test) between the wtAR and the AR-2KR after androgen exposure are indicated.
AR SUMOylation regulates apoptosis sensitivity and cell proliferation of prostate cancer cells
The androgen-regulated genes of both wtAR and AR-2KR cells were next subjected to IPA to identify enriched molecular and cellular functions and pathways regulated by the AR SUMOylation. The androgen-regulated genes in the AR-2KR PC-3 cells were enriched for genes associated with such biological functions as cellular movement, cellular development, cell death and proliferation (Figure 2A). Generally, 13–37% of androgen-regulated genes in the AR-2KR cells were linked to these functions, whereas only 5–21% of the genes in the wtAR cells were linked to these functions. Many of the most differentially expressed genes due to AR mutation and androgen exposure were linked to several of these functions (Supplementary Figure S4). Examples of such genes include RASD1 (encoding a Ras-related protein that is stimulated by dexamethasone), EFEMP1 (encoding a member of the fibulin family of extracellular matrix glycoproteins), LRIG1 (encoding a transmembrane protein that interacts with epidermal growth factor receptor tyrosine kinases), and DUSP1 (encoding dual specificity protein phosphatase 1). In contrast, the androgen-regulated genes in the wtAR PC-3 cells showed enrichment in various biosynthesis-related canonical pathways, including androgen biosynthesis (Figure 2B), highlighting the differences in transcriptional regulation by wtAR and AR-2KR upon androgen stimulation.
Figure 2.

Androgen-regulated genes in the wtAR- and the AR-2KR-expressing PC-3 cells are partly enriched in different biological functions. Androgen-regulated genes from both cell types were subjected to Ingenuity Pathway Analysis® (IPA). Threshold for a gene list to be significantly involved in a particular biological function was P-value < 0.05 (or –log10 (B-H adjusted P-value) > 1.30). (A) Comparative analysis showing the five most significantly enriched molecular and cellular functions. Percentage and number of genes in each category overlapping the set of androgen-regulated genes are shown. (B) Heat map of hierarchically clustered biosynthesis-related canonical pathways.
Since the wtAR and the AR-2KR cells showed significant differences in their androgen-regulated expression of genes governing cellular proliferation and survival, we compared their growth rates and apoptosis sensitivities. Although androgen exposure increased the growth of both wtAR and AR-2KR cells, the AR-2KR-expressing PC-3 cells grew significantly faster than their wtAR counterpart cells (Figure 3A). Similarly, the SUMOylation-deficient AR expressing HEK293 cells grew faster than the wtAR-expressing cells (Supplementary Figure S5). These results are in line with our gene expression data showing that androgen-regulated genes, e.g. RASD1 and LRIG1, known to decrease cell proliferation were more robustly up-regulated in the wtAR expressing cells and that genes, such as EFEMP1, DUSP1, ADRA1B (encoding alpha-1B-adrenergic receptor that can function as a protooncogene), reported to increase proliferation were up-regulated in the AR-2KR expressing cells (Figure 3B, Supplementary file 2), (26–30). To assess the apoptosis sensitivity, we exposed our PC-3 cells to an apoptosis inducer camptothecin and monitored their cell death. The apoptosis assays showed that the AR-2KR cells are more sensitive to camptothecin-induced apoptosis than the wtAR cells (Figure 3C).
Figure 3.

Mutation of AR SUMOylation sites promotes cell proliferation and influences apoptosis sensitivity. (A) Amount of viable cells after vehicle or R1881 treatment was measured 48 h and 72 h after exposure using CellTiter-Glo® Luminescent Cell Viability Assay (Promega). Growth percentages represent the means (n = 5) ±SDs. (B) RT-qPCR analysis of examples of genes that were differentially expressed between the cell lines and previously shown to have a role in cell proliferation. (C) Cellular apoptosis, measured using Cell Death Detection ELISAPlus kit (Roche) after 20 h exposure to camptothecin (1.5 μM), and expressed as the mean relative level of apoptosis (±SD, n = 3). Statistically significant differences (***P < 0.001, **P < 0.01 and *P < 0.05; Student's t-test) between the wtAR and the AR-2KR cells after androgen exposure are indicated.
Collectively these results strongly suggest that the molecular and cellular functions identified with the IPA are biologically relevant and the SUMOylation is regulating the AR function in a gene pathway selective fashion.
Effect of SUMOylation on AR cistrome
To complement the AR target gene expression analyses, we next investigated whether SUMOylation influences the AR chromatin occupancy by identifying the genomic wtAR and AR-2KR binding sites using chromatin immunoprecipitation coupled to deep sequencing (ChIP-seq) analyses. Our biological replicates showed excellent concordance, as judged by tag density comparisons (Supplementary Figure S6). The ChIP-seq analyses revealed 19 400 high confidence AR binding sites (ARBs) in androgen exposed wtAR PC-3 cells and 23 059 ARBs in SUMOylation deficient AR cells. Based on initial overlap analysis shown in Figure 4A, most of these ARBs overlapped. Moreover, the tag density maps and average tag profiles of the wtAR and the AR-2KR occupancy showed that the two receptors were loaded to the same extent to them (Figure 4A and B). Further analysis of non-shared binding sites lead to identification of two other ARB groups, wtAR-preferred and AR-2KR-preferred sites, showing at least 2-fold difference between the occupancy of the wtAR and that of the AR-2KR. Based on these analyses, the mutation of the AR SUMOylation sites resulted in 2675 AR-2KR-preferred ARBs and 1561 wtAR-preferred ARBs, while 16 222 ARBs remained unchanged (Figure 4A and B).
Figure 4.
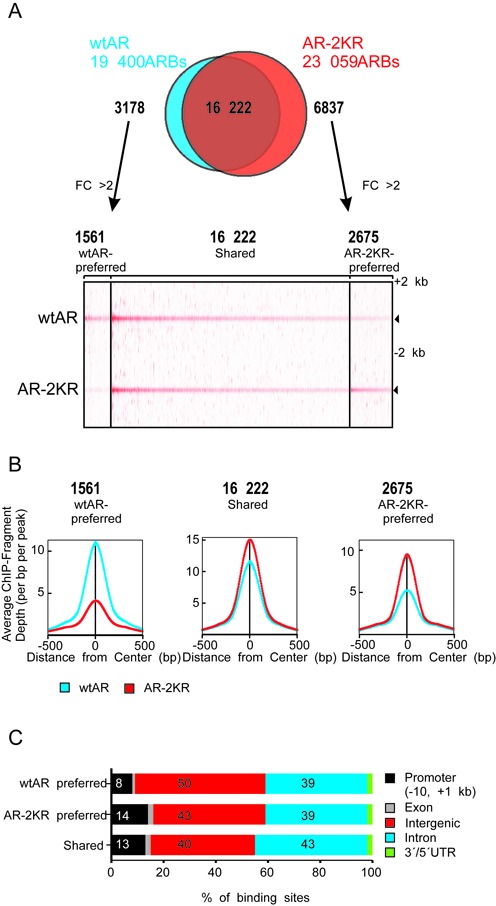
Genome-wide AR-binding events are influenced by AR SUMOylation sites. ChIP-seq analysis of AR chromatin interactions in the wtAR and the AR-2KR PC-3 cells after 1 h R1881 exposure. (A) Venn-diagram of AR binding events in wtAR- and AR-2KR-expressing cells (upper panel). Non-shared binding sites were analyzed further using getDifferentialPeak tool with >2-fold difference as described in ‘Materials and Methods’ section to achieve final preferred binding sites. Heat map showing wtAR and AR-2KR ChIP-seq tag densities for wtAR- and AR-2KR-shared and wtAR- or AR-2KR-preferred regions in a window ±2 kb (bottom panel). (B) Comparison of the wtAR and the AR-2KR average tag counts in ±500 bp from the centers of ARBs in three categories (shared/preferred). (C) Genomic distribution of the ARBs.
Similarly, ChIP-seq analysis of the wtAR and AR-2KR HEK293 cells revealed that 1473 AR-2KR-preferred ARBs and 806 wtAR-preferred ARBs, while 25 561 ARBs were insensitive to the mutation (Supplementary Figure S7A–C). Comparison of the wtAR PC-3 to the wtAR HEK293 ChIP-seq data showed 45% overlap between the ARBs (Supplementary Figure S7D). Similarly, ∼40% of wtAR PC-3 ARBs overlapped with the ARBs of VCaP prostate cancer cells expressing endogenous AR (Supplementary Figure S7D), (31). The overall distribution of the ARBs in relation to gene structures was very similar in our HEK293 and PC-3 cell models; most of the ARBs resided in introns or were intergenic, while only <14% resided within proximal promoter regions (−10 kb to +1 kb from transcription start sites, TSSs) (Figure 4C, Supplementary Figure S7E).
IPA of genes in the PC-3 cells that had a preferred ARB within ±100 kb of their TSS showed that the AR-2KR-preferred ARBs were significantly associated with cellular development, cellular growth and proliferation and cellular movement (Supplementary Figure S8A). Majority of the androgen-regulated genes both in the wtAR PC-3 cells (≥77%) and in the AR-2KR PC-3 cells (≥91%) harbored at least one ARB within ±100 kb of their TSS (Supplementary Figure S8B, Supplementary file 3). However, on the genome-wide level, there was no clear difference in the association of the wtAR- and AR-2KR-preferred ARBs and the receptor type-preferred androgen target genes (Supplementary Figure S8B), indicating that the differences in the cistromes between the wtAR and the AR-2KR cells cannot alone explain the observed differences in their transcript levels.
Although on the whole genome level the majority of the ARBs were insensitive to the AR SUMOylation sites, there were several androgen-regulated genes that harbored one or more wtAR- or AR-2KR-preferred ARBs (Figure 5). For example, CLDN8 that was androgen-regulated only in wtAR PC-3 cells did not show any clear ARB in the AR-2KR cells. Similarly, LRIG1 was significantly androgen-regulated only in wtAR cells and it contained a wtAR-preferred ARB. In contrast, ADRA1B and TENM4 that showed a significantly stronger response to androgen in the AR-2KR cells possessed one or more AR-2KR-preferred ARBs. FKBP5, on the other hand, is an example of an AR target of which androgen regulation is insensitive to the AR SUMOylation sites and of which 16 ARBs out of 18 were shared ones. Examples of the wtAR- and the AR-2KR-preferred loci in the HEK293 cells are given in Supplementary Figure S9.
Figure 5.

Examples of androgen-regulated loci in which binding of AR differs between the wtAR- and AR-2KR-expressing PC-3 cells. Peak tracks showing the occupancy of AR in vehicle- (green) and androgen-treated wtAR cells (blue) and that of AR-2KR in androgen-treated cells (red) in the regulatory regions of CLDN8 and LRIG1 (wtAR-preferred) and those of ADRA1B and TENM4 (AR-2KR-preffered). FKBP5 is an example showing highly similar occupancy by the two AR forms. Red bars show the positions of the identified ARBs. Boxed numbers refer to fold-induction by androgen as determined by the microarray analysis.
We next performed de novo motif analyses with all ARBs to identify over-represented transcription factor-binding motifs in the ARBs. As expected, the de novo ARE consensus (that closely resembles the ARE motif in the JASPAR-database) was the most enriched motif among all ARBs (in >60% of wtAR-, AR-2KR-preferred and shared ARBs) (Figure 6A and B). Further analysis revealed no clear differences in the number of AREs per ARE-containing ARBs between the preferred and shared ARBs, with the means (±SD) 1.28 ± 0.52, 1.20 ± 0.47 and 1.31 ± 0.56 for the wtAR-, the AR-2KR- and the shared ARBs, respectively. Other significantly enriched motifs were those for ERG, FOXA1, C/EBP and AP-1 (Figure 6A). While the de novo ARE and ERG motifs were fairly similarly enriched in all three ARB subsets, there were significant differences in the enrichment of FOXA1, C/EBP and AP-1 motifs to the three ARB subsets. Based on western blotting analysis, factors binding to these motifs (FOXA1, C/EBPβ and c-Jun, major AP-1 component) are expressed in our PC3 model cells (Supplementary Figure S10). Both FOXA1 and C/EBP motifs were found three times more often within the wtAR-preferred then AR-2KR-preferred ARBs, whereas the AP-1 motif was nearly three times more prevalent within the AR-2KR-preferred ARBs (Figure 6B and C, Supplementary Figure S11). As shown in Figure 6C, more pronounced loading of both the FOXA1 and the C/EBPβ, for example, onto SAFB and ANKRD17 loci in wtAR than AR-2KR cells is in line with the in silico predictions in panel B. Moreover, c-Jun appears to be more avidly loaded, for example, onto ADRA1B locus in AR-2KR- than in wtAR-expressing PC3 cells (Figure 6C). De novo analysis of the HEK293 data showed, in addition to the AREs, a similar over-enrichment of FOXA1 within the wtAR-preferred ARBs. GATA1 was also over-enriched in the wtAR-preferred ARBs, whereas EBF1 and PAX2 were more prevalent in the AR-2KR preferred ARBs (Supplementary Figure S12A and B). The differences in the enriched motifs between the PC-3 and the HEK293 cells likely reflect cell-specific differences in the expression of their cognate transcription factors. Compared to LNCaP and VCaP prostate cancer cells in which about half of the identified ARBs have a FOXA1 motif (31,32), the PC-3 ARBs and the HEK293 ARBs less frequently harbor a FOXA1 motif. This may be due to the lower amount of FOXA1 in the latter two cell lines (Supplementary Figure S10).
Figure 6.
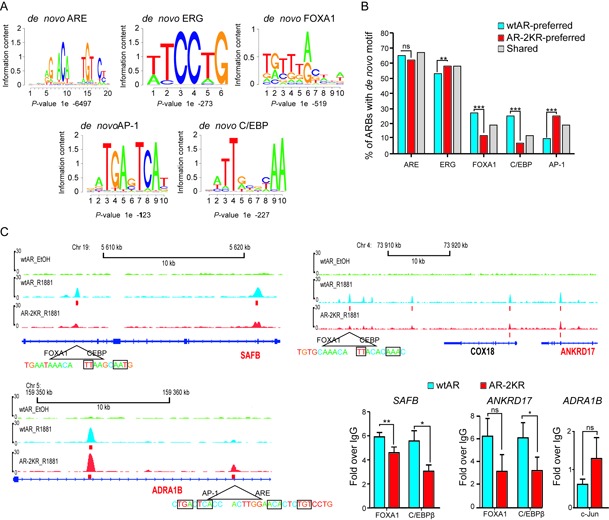
De novo motif analysis of the AR-binding sites (ARBs) identified in the PC-3 cells. De novo analysis was performed on ±100 bp of the AR peak center. (A) The top five motif matrices predicted by the de novo motif analysis by HOMER are shown. (B) Enrichment of de novo motifs in the wtAR-, the AR-2KR-preferred or the wtAR- and AR-2KR-shared ARBs. Statistically significant differences (***P < 0.001, **P < 0.01; Chi-square test) between wtAR and AR-2KR-preferred binding sites de novo motifs are indicated. (C) ChIP-seq track examples of the AR-binding events and the corresponding DNA sequences of the de novo motifs in SAFB, ANKRD17 and ADRA1B loci. Occupancies of FOXA1 and C/EBPβ in SAFB and ANKRD17 and c-Jun in ADRA1B locus after 1-h R1881 exposure were monitored using qChIP (bottom right hand panels). Results represent the means (n = 3) ± SDs and are shown as fold over to IgG-immunoprecipitated samples. Statistically significant differences (**P < 0.01 and *P < 0.05; Student's t-test) between the wtAR- and the AR-2KR-expressing cells are indicated.
Taken together, our ChIP-seq analyses suggest that SUMOylation regulates AR's interaction with the chromatin, leading to the observed differences in the AR cistromes in both PC3 and HEK293 cells. In addition, a number of transcription factor-binding motifs are differentially enriched within the wtAR- and AR-2KR-preferred binding locations.
DISCUSSION
A large number of transcription factors are known to be covalently modified by SUMOs, but the regulatory role of these modifications has only rarely been addressed in an unbiased genome-wide fashion (5, 15, 33, 34). Here, we have investigated the effect of AR SUMOylation sites on AR chromatin occupancy and endogenous target gene expression in a genome-wide fashion using stably wtAR- or SUMOylation site mutant receptor-expressing PC-3 and HEK293 cells. Already our recent expression analyses of a few AR target genes in the HEK293 cells suggested that the effect (enhancing, repressive or neutral) of SUMOylation on the AR activity is target gene-dependent (14). Our genome-wide analyses of androgen-regulated genes in the wtAR and the AR SUMOylation mutant cells indicate that the SUMOylation sites do not simply repress the AR activity on all target genes, as the mutant also exhibited attenuated transcriptional activity on a number of target genes. Moreover, not all AR target genes were influenced by the AR SUMOylation sites. The SUMOylation not merely affected the expression of androgen-induced genes, but it also influenced the ability of AR to repress its target genes. Our studies focused on the PC-3 prostate cancer cells, because the cell background better resembles the natural environment of AR function. A reasonable number (∼30%) of the androgen-regulated genes in our PC-3 wtAR model cells was the same as in VCaP prostate cancer cells expressing amplified levels of endogenous AR (a model of hormone-refractory prostate cancer) (GSE30316).
Pathway enrichment analysis of the genes differentially regulated by the wtAR and the AR SUMOylation mutant revealed that they are significantly associated with molecular and cellular functions of cellular movement, development, cell death and survival, cell morphology, and cellular growth and proliferation. Many genes with anti-proliferative effects, such as RASD1 and LRIG1 (26,27), showed significantly stronger androgen-up-regulation by the wtAR. On the other hand, genes, such as EFEMP1, DUSP1, ADRA1B (28–30), that were more robustly up-regulated by the SUMOylation-deficient AR are known to be associated with promotion of cell proliferation. Interestingly, the genes LRIG1, DUSP1 and RASD1 are all significantly up-regulated in prostate cancers compared to nonmalignant prostates (Supplementary Figure S13) (http://www.oncomine.org) (35).
SUMOylation also influenced the AR cistrome. Albeit the majority of ARBs (shared ARBs) were not affected by the AR SUMOylation mutation, our replicated genome-wide ChIP-seq analyses in PC-3 cells revealed that ∼10% of all identified ARBs were preferred either by wtAR (1561 ARBs) or the AR SUMOylation mutant (2675 ARBs). These preferred ARBs were similarly distributed to genomic elements as the shared ARBs; with the majority of the ARBs in all three groups residing in intergenic or intronic regions and only a very small proportion locating to the proximal promoter regions. These genomic distribution figures are similar to those of the ARBs recently reported from other prostate cancer cell lines (31,32,36,37). Moreover, ∼40% of PC-3 wtAR ARBs identified in this study overlap with those recently revealed in the VCaP cells.
General comparison of motif signatures within the gene repression- and activation-associated ARBs revealed remarkably similar features and the presence of AREs in both cases, suggesting that the AR-binding AREs also often mediate gene repression, not only activation, as recently seen by us and others for glucocorticoid target genes (15,38). Of the androgen-regulated genes in PC-3 cells, ∼80% and ∼90% of the wtAR and AR-2KR targets, respectively, are associated with at least one ARB. However, only 59/473 of the wtAR target genes showed wtAR-preferred ARBs and 52/143 of the AR-2KR target genes displayed AR-2KR-preferred ARBs, suggesting that the preferred ARBs do not alone explain the differences in the androgen-regulated transcriptomes between the wtAR and the AR-2KR cells. However, differences in the sampling points of RNA profiling (16 h) and ChIP-seq (1 h) and single time point data hamper direct comparison and correlation of the gene expression with the chromatin binding data. SUMOylation influences the AR's intranuclear mobility (based on FRAP assays) and this is also likely to be reflected on the AR's on-off kinetics in the chromatin (14). Moreover, different AR target genes display largely different mRNA induction kinetics. Nevertheless, already based on our single time point data, several AR target loci differentially regulated due to the AR SUMOylation sites, such as CLDN8, LRIG1, ADRA1B and TENM4, showed differences in their preferred ARBs and the level of ARB occupancy, which were in line with their androgen-regulated expression between the wtAR and the AR-2KR cells.
Interestingly, the PC-3 cells expressing SUMOylation mutant AR proliferated faster and were more sensitive to apoptosis than the wtAR-expressing cells. A similar difference was also detected with the HEK293 cells. These differences are perfectly in line with the altered androgen regulation of several genes influencing cell growth. Fittingly, SUMOylation modulates the target gene selection of progesterone receptor (PR) as well as glucocorticoid receptor (GR) such that their SUMO sensitive targets include genes associated with cell growth and proliferation (15,34). This is also the case with SUMOylation-defective microphthalmia-associated transcription factor (MITF) mutant associated with the predisposition to melanoma and renal carcinoma (39). As with both the AR and the GR, the MITF's chromatin occupancy is also influenced by the SUMOylation site mutation (39). Interestingly, however, our GR-expressing HEK293 model cells responded to the mutation of GR SUMOylation sites in a different fashion than our model AR cells, as the GR SUMOylation mutant cells displayed both more GR target genes and GR chromatin-binding sites (15). Also the attenuated ability of SUMOylation-deficient GATA-1 to induce genes controlling hematopoiesis has been linked to altered chromatin occupancy (40). Furthermore, the disruption of orphan nuclear receptor SF-1 (steroidogenic factor 1) SUMOylation in mice has intriguingly been linked to abnormal Hedgehog signaling and endocrine development (41). These data collectively suggest a wider role for SUMOylation in the regulation of target selection by sequence-specific transcription factors.
Our binding motif analyses indicate that the classic ARE is similarly overrepresented among the wtAR- and AR-2KR-shared, as well as the wtAR-preferred and the AR-2KR-preferred ARBs. However, there were significant differences in the enrichment of other transcription factor-binding motifs between the wtAR-preferred and the AR-SUMOylation mutant-preferred ARBs: motifs for pioneer transcription factor FOXA1 and C/EBP occurred more frequently among the wtAR-preferred ARBs, whereas AP-1 motif was more prevalent among the AR-2KR preferred ARBs. The motif analyses thus imply that SUMOylation may influence the AR chromatin occupancy and target gene selection via regulating interactions with other sequence-specific transcription factors, such as FOXA1 and AP-1, which may enhance or weaken the AR binding to the chromatin. In conclusion, our objective genome-wide analyses reveal that SUMOylation does not simply repress the AR activity, but the modification regulates the receptor's target gene selection and plays an important role in controlling the proliferative effects of androgens.
ACCESSION NUMBERS
Bead array and ChIP-seq data are accessible through GEO Series accession number GSE54202.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We wish to thank Merja Räsänen and Eija Korhonen for assistance with cell cultures and Merja Heinäniemi for her help with microarray data analysis and construction of deep sequencing pipeline. The EMBL GeneCore sequencing team, Beijing Genomics Institute and the Finnish Microarray and Sequencing Centre are also acknowledged for deep sequencing and microarray analyses.
FUNDING
Academy of Finland; Finnish Cancer Organisations; University of Eastern Finland Doctoral Programme in Molecular Medicine; University of Eastern Finland and the Sigrid Jusélius Foundation. Funding for open access charge: The Academy of Finland grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Makkonen H., Kauhanen M., Paakinaho V., Jääskeläinen T., Palvimo J.J. Long-range activation of FKBP51 transcription by the androgen receptor via distal intronic enhancers. Nucleic Acids Res. 2009;37:4135–4148. doi: 10.1093/nar/gkp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sampson N., Neuwirt H., Puhr M., Klocker H., Eder I.E. In vitro model systems to study androgen receptor signaling in prostate cancer. Endocr. Relat. Cancer. 2013;20:R49–R64. doi: 10.1530/ERC-12-0401. [DOI] [PubMed] [Google Scholar]
- 4.Green S.M., Mostaghel E.A., Nelson P.S. Androgen action and metabolism in prostate cancer. Mol. Cell. Endocrinol. 2012;360:3–13. doi: 10.1016/j.mce.2011.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poukka H., Karvonen U., Jänne O., Palvimo J. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc. Natl. Acad. Sci. U.S.A. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flotho A., Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 7.Hay R.T. Decoding the SUMO signal. Biochem. Soc. Trans. 2013;41:463–473. doi: 10.1042/BST20130015. [DOI] [PubMed] [Google Scholar]
- 8.Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K.M., Jackson S.P. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heun P. SUMOrganization of the nucleus. Curr. Opin. Cell Biol. 2007;19:350–355. doi: 10.1016/j.ceb.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 10.Kaikkonen S., Jääskeläinen T., Karvonen U., Rytinki M.M., Makkonen H., Gioeli D., Paschal B.M., Palvimo J.J. SUMO-specific protease 1 (SENP1) reverses the hormone-augmented SUMOylation of androgen receptor and modulates gene responses in prostate cancer cells. Mol. Endocrinol. 2009;23:292–307. doi: 10.1210/me.2008-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotaja N., Karvonen U., Jänne O.A., Palvimo J.J. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 2002;22:5222–5234. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poukka H., Aarnisalo P., Karvonen U., Palvimo J.J., Jänne O.A. Ubc9 interacts with the androgen receptor and activates receptor-dependent transcription. J. Biol. Chem. 1999;274:19441–19446. doi: 10.1074/jbc.274.27.19441. [DOI] [PubMed] [Google Scholar]
- 13.Kaikkonen S., Paakinaho V., Sutinen P., Levonen A., Palvimo J.J. Prostaglandin 15d-PGJ(2) inhibits androgen receptor signaling in prostate cancer cells. Mol. Endocrinol. 2013;27:212–223. doi: 10.1210/me.2012-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rytinki M., Kaikkonen S., Sutinen P., Paakinaho V., Rahkama V., Palvimo J.J. Dynamic SUMOylation is linked to the activity cycles of androgen receptor in the cell nucleus. Mol. Cell. Biol. 2012;32:4195–4205. doi: 10.1128/MCB.00753-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paakinaho V., Kaikkonen S., Makkonen H., Benes V., Palvimo J.J. SUMOylation regulates the chromatin occupancy and anti-proliferative gene programs of glucocorticoid receptor. Nucleic Acids Res. 2014;42:1575–1591. doi: 10.1093/nar/gkt1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentleman R., Carey V., Bates D., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du P., Kibbe W.A., Lin S.M. Lumi: a pipeline for processing illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 18.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 19.Karvonen U., Kallio P.J., Jänne O.A., Palvimo J.J. Interaction of androgen receptors with androgen response element in intact cells. roles of amino- and carboxyl-terminal regions and the ligand. J. Biol. Chem. 1997;272:15973–15979. doi: 10.1074/jbc.272.25.15973. [DOI] [PubMed] [Google Scholar]
- 20.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10 doi: 10.1186/gb-2009-10-3-r25. R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furey T.S. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ovaska K., Laakso M., Haapa-Paananen S., Louhimo R., Chen P., Aittomaki V., Valo E., Nunez-Fontarnau J., Rantanen V., Karinen S., et al. Large-scale data integration framework provides a comprehensive view on glioblastoma multiforme. Genome Med. 2010;2:65. doi: 10.1186/gm186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaidyanathan G., Cismowski M.J., Wang G., Vincent T.S., Brown K.D., Lanier S.M. The ras-related protein AGS1/RASD1 suppresses cell growth. Oncogene. 2004;23:5858–5863. doi: 10.1038/sj.onc.1207774. [DOI] [PubMed] [Google Scholar]
- 27.Thomasson M., Wang B., Hammarsten P., Dahlman A., Persson J.L., Josefsson A., Stattin P., Granfors T., Egevad L., Henriksson R., et al. LRIG1 and the liar paradox in prostate cancer: a study of the expression and clinical significance of LRIG1 in prostate cancer. Int. J. Cancer. 2011;128:2843–2852. doi: 10.1002/ijc.25820. [DOI] [PubMed] [Google Scholar]
- 28.Wu J., Liu S.L., Zhu J.L., Norton P.A., Nojiri S., Hoek J.B., Zern M.A. Roles of tissue transglutaminase in ethanol-induced inhibition of hepatocyte proliferation and alpha 1-adrenergic signal transduction. J. Biol. Chem. 2000;275:22213–22219. doi: 10.1074/jbc.M000091200. [DOI] [PubMed] [Google Scholar]
- 29.Pigazzi M., Manara E., Beghin A., Baron E., Tregnago C., Basso G. ICER evokes Dusp1-p38 pathway enhancing chemotherapy sensitivity in myeloid leukemia. Clin. Cancer Res. 2011;17:742–752. doi: 10.1158/1078-0432.CCR-10-0886. [DOI] [PubMed] [Google Scholar]
- 30.Song E.L., Hou Y.P., Yu S.P., Chen S.G., Huang J.T., Luo T., Kong L.P., Xu J., Wang H.Q. EFEMP1 expression promotes angiogenesis and accelerates the growth of cervical cancer in vivo. Gynecol. Oncol. 2011;121:174–180. doi: 10.1016/j.ygyno.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Sahu B., Laakso M., Ovaska K., Mirtti T., Lundin J., Rannikko A., Sankila A., Turunen J.P., Lundin M., Konsti J., et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30:3962–3976. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massie C.E., Lynch A., Ramos-Montoya A., Boren J., Stark R., Fazli L., Warren A., Scott H., Madhu B., Sharma N., et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30:2719–2733. doi: 10.1038/emboj.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cubenas-Potts C., Matunis M.J. SUMO: a multifaceted modifier of chromatin structure and function. Dev. Cell. 2013;24:1–12. doi: 10.1016/j.devcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knutson T.P., Daniel A.R., Fan D., Silverstein K.A., Covington K.R., Fuqua S.A., Lange C.A. Phosphorylated and sumoylation-deficient progesterone receptors drive proliferative gene signatures during breast cancer progression. Breast Cancer Res. 2012;14 doi: 10.1186/bcr3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma N.L., Massie C.E., Ramos-Montoya A., Zecchini V., Scott H.E., Lamb A.D., MacArthur S., Stark R., Warren A.Y., Mills I.G., et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell. 2013;23:35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Robinson J.L., Macarthur S., Ross-Innes C.S., Tilley W.D., Neal D.E., Mills I.G., Carroll J.S. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 2011;30:3019–3027. doi: 10.1038/emboj.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chng K.R., Chang C.W., Tan S.K., Yang C., Hong S.Z., Sng N.Y., Cheung E. A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. EMBO J. 2012;31:2810–2823. doi: 10.1038/emboj.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhlenhaut N.H., Barish G.D., Yu R.T., Downes M., Karunasiri M., Liddle C., Schwalie P., Hubner N., Evans R.M. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol. Cell. 2013;49:158–171. doi: 10.1016/j.molcel.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertolotto C., Lesueur F., Giuliano S., Strub T., de Lichy M., Bille K., Dessen P., d'Hayer B., Mohamdi H., Remenieras A., et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480:94–98. doi: 10.1038/nature10539. [DOI] [PubMed] [Google Scholar]
- 40.Lee H.Y., Johnson K.D., Fujiwara T., Boyer M.E., Kim S.I., Bresnick E.H. Controlling hematopoiesis through sumoylation-dependent regulation of a GATA factor. Mol. Cell. 2009;36:984–995. doi: 10.1016/j.molcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee F.Y., Faivre E.J., Suzawa M., Lontok E., Ebert D., Cai F., Belsham D.D., Ingraham H.A. Eliminating SF-1 (NR5A1) sumoylation in vivo results in ectopic hedgehog signaling and disruption of endocrine development. Dev. Cell. 2011;21:315–327. doi: 10.1016/j.devcel.2011.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


