Abstract
Type II DNA topoisomerases are divided into two families, IIA and IIB. Types IIA and IIB enzymes share homologous B subunits encompassing the ATP-binding site, but have non-homologous A subunits catalyzing DNA cleavage. Type IIA topoisomerases are ubiquitous in Bacteria and Eukarya, whereas members of the IIB family are mostly present in Archaea and plants. Here, we report the detection of genes encoding type IIB enzymes in which the A and B subunits are fused into a single polypeptide. These proteins are encoded in several bacterial genomes, two bacterial plasmids and one archaeal plasmid. They form a monophyletic group that is very divergent from archaeal and eukaryotic type IIB enzymes (DNA topoisomerase VI). We propose to classify them into a new subfamily, denoted DNA topoisomerase VIII. Bacterial genes encoding a topoisomerase VIII are present within integrated mobile elements, most likely derived from conjugative plasmids. Purified topoisomerase VIII encoded by the plasmid pPPM1a from Paenibacillus polymyxa M1 had ATP-dependent relaxation and decatenation activities. In contrast, the enzyme encoded by mobile elements integrated into the genome of Ammonifex degensii exhibited DNA cleavage activity producing a full-length linear plasmid and that from Microscilla marina exhibited ATP-independent relaxation activity. Topoisomerases VIII, the smallest known type IIB enzymes, could be new promising models for structural and mechanistic studies.
INTRODUCTION
DNA topoisomerases are essential for solving topological problems arising during DNA metabolic processes and are therefore critical for the preservation of genome stability (1–3). They can interconvert different topological forms of DNA and either catenate or decatenate DNA rings by generating transient single- (type I topoisomerases) or double-stranded (type II topoisomerases) DNA breaks in DNA backbones. Type II enzymes are especially important because of their unique capacity to catalyze the transfer of one DNA duplex through another. They are essential for resolving catenanes generated between daughter chromosomes at the end of DNA replication and also play a major role in relaxing positive supercoils generated by tracking processes occurring during transcription and replication. Accordingly, all cellular organisms, without exception, have at least one type II enzyme.
Type II topoisomerases are classified into two families, IIA and IIB, based on sequence and structural similarities (4–7). Archaeal and bacterial types IIA and IIB enzymes are heterotetramers composed of two different subunits (A and B), whereas type IIA enzymes from eukaryotes and their viruses are homodimers, with the B and A moieties fused into a single polypeptide. Several high-resolution structures of both types IIA and IIB enzymes have been solved, highlighting some structural similarities, but also large differences between these two families (8–11). The B subunits are homologous in the two families and contain a similar ATP-binding site located within a protein domain known as the Bergerat fold, which is characteristic of proteins of the GHKL (Gyrase, Hsp90, histidine Kinase, MutL) superfamily (4,12). Moreover, enzymes from the two families share two other functional domains: the Toprim domain displaying the Rossmann fold (involved in magnesium binding) and the Winged Helix Domain (WHD), which contains the active site tyrosine. However, the Toprim domain of type IIA topoisomerases is located within the B subunit, whereas in the IIB family, this domain is located within the A subunit. Furthermore, their WHDs are unrelated, despite the presence of the catalytic tyrosine in both cases (Figure 1). Overall, the A subunits of types IIA and IIB topoisomerases share neither sequence nor structural similarity. This suggests that these two enzyme families originated independently via the association of homologous B subunits with non homologous A subunits (5).
Figure 1.
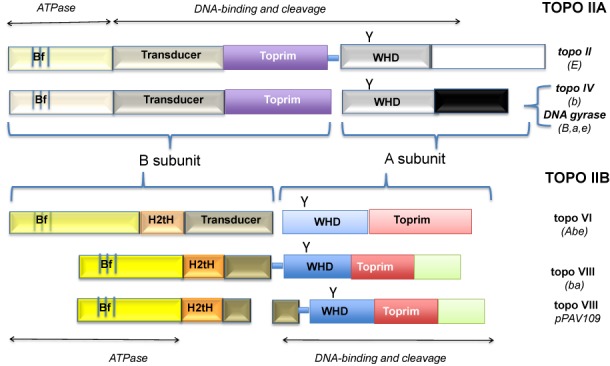
Domain organization of type II topoisomerases of the A and B families. Homologous domains are colored similarly, with color intensities differing according to the extent of sequence similarity. Bf: Bergerat fold (4) corresponding to the ATP-binding site with three conserved amino-acid signatures (small vertical bars), H2TH: Helix-2Turn-Helix domain, WHD: Winged Helix Domain; Y: catalytic tyrosine; triangle: catalytic site. The distributions of various type II topoisomerase subfamilies in the three domains of life, Archaea (A, a), Bacteria (B, b) and Eukarya (E, e) are indicated in brackets.
The origin and evolution of topoisomerases are rather puzzling. Ribosomal proteins exist in three distinct versions, corresponding to the three cellular domains, Archaea, Bacteria and Eukarya (13,14); however, each topoisomerase family exists in several versions (hereafter called subfamilies) that do not overlap with the three domains in phylogenomic analyses (5–7,15). In addition, several groups of viruses encode topoisomerases that are not related to those of their hosts but instead form monophyletic groups that branch in between cellular domains in phylogenetic analyses (5,7,15).
Several distinct subfamilies of type IIA enzymes are known, which differ in catalytic activity and/or quaternary structure. These include (i) DNA gyrases, present in all bacteria, in some members of the archaeal phylum Euryarchaeota and in eukaryotes with endosymbionts of cyanobacterial origin; (ii) topoisomerase IV enzymes, specific to bacteria; (iii) eukaryotic type II topoisomerases, present in all eukaryotic species; (iv) viral type II topoisomerases encoded by some Nucleocytoplasmic Large DNA Viruses (NCLDV) that infect eukaryotes; and (v) viral type II topoisomerases encoded by T4-like myoviruses that infect bacteria.
The distribution of type IIB topoisomerases is more limited than that of type IIA. This family includes archaeal type IIB enzymes, also called topoisomerase VI, with close homologs in Archaeplastida (plants, red and green algae), some protists and a few bacteria (7,15). Topoisomerases VI are relaxing enzymes devoid of gyrase activity (16), are the only type II topoisomerases present in most archaea, suggesting that they are critical for chromosome segregation. Topoisomerases VI are also the only enzymes capable of relaxing positive supercoils in two of the three major archaeal phyla (17). Thus, these enzymes are also essential for managing the waves of positive supercoiling generated during DNA replication and transcription.
In Eukarya, type IIA topoisomerases control topological stress generated by chromosome segregation, replication and transcription. However in plants, type IIB topoisomerases (close relatives of archaeal topoisomerase VI) control endoreduplication, a polyploidization process responsible for the enlargement of plant cells, which in turn determines plant size (18).
The inconsistent phylogenomic pattern of topoisomerases in the three domains of life is common to most proteins involved in DNA replication, recombination or repair (for the case of DNA polymerases, see (19)). Lateral gene transfers cannot account for the distribution of these enzymes within the classical tree of life based on the ribosome. Instead, this pattern requires more elaborate evolutionary scenarios. We proposed previously that proteins involved in DNA metabolic activities (including topoisomerases) originated first in the viral world and were later transferred ‘randomly’ to various cellular lineages (20). This scenario predicts that some contemporary viruses or related mobile elements (plasmids, transposons) still encode unique versions of DNA replication proteins, such as topoisomerases, that were never transferred to the ‘cellular world’ (except as integrated mobile elements) (7).
Here, we report the in silico discovery and preliminary biochemical characterization of a new DNA topoisomerase subfamily that supports the prediction outlined above. The genes encoding these proteins correspond to the fusion of the B and A subunits of type IIB topoisomerases. We identified these genes in 19 bacterial genomes and in three plasmids (two from bacteria and one from an archaeon). In bacterial genomes, these genes are located within integrated mobile elements related to conjugative plasmids. We purified three members of this new subfamily of type IIB enzymes (that we propose to call topoisomerase VIII): one from a mesophilic bacterium, one from a thermophilic bacterium and one from a bacterial plasmid. We show that the enzyme encoded by the bacterial plasmid exhibits ATP-dependent relaxation and decatenation activities, the hallmark of type II topoisomerases, whereas the other two enzymes, encoded by integrated elements, exhibit DNA cleavage activity, producing DNA double-stranded breaks, and/or ATP-independent relaxation activity.
MATERIALS AND METHODS
Plasmids and reagents
Negatively supercoiled plasmid pBR322 DNA was purchased from Fermentas life science and kDNA (kinetoplast DNA) from Topogen. The reverse-gyrase enzyme was kindly given by Prof. Marc Nadal. Primers for site-directed mutagenesis were synthesized by Sigma-Aldrich.
Sequence database search
The fusion of the topoisomerase VI protein subunits A and B was used as a query to carry out psi-BLAST and tBLASTn searches of different databases on the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). A multiple sequence alignment was obtained with PipeAlign (21) and manually corrected based on conserved motifs. Secondary structures predictions were performed with PSI-PRED (22).
Phylogenetic analysis
For phylogenetic analysis, 27 identified homologs of topoisomerases VIII were aligned with MUSCLE 3.8.31 (23) and manually inspected with Seaview 4.3.3 (24). The alignment was trimmed with BMGE software (25) and the BLOSUM30 matrix, resulting in 571 amino acid positions. ProtTest 3 (26) was used to determine the best substitution model for further analysis with the Akaike Information Criterion (AICc) criteria. Maximum likelihood analysis was preformed with RaxML 7.4.2 (27) and the combined tree-search/fast bootstrap method (‘-f a’) under the PROTGAMMALG model (four discrete rate categories) with 1000 fast bootstraps. Bayesian analysis was performed with MrBayes 3.0 (28) and the mixed amino acid model with four discrete rate categories, which supported the WAG model with 100% posterior probability. Four Markov chains starting with a random tree were run simultaneously for 1 million generations, sampling trees at every 100th generation. The first 2500 sampled trees (25%) were discarded as ‘burn in’.
Protein structure predictions
Structural predictions were performed with the Phyre2 webserver (29). The N- and C-terminal halves of the three topoisomerase VIII sequences (corresponding to the B and A topoisomerase VI subunits, respectively) were also modeled separately because conserved regions and motifs are scattered all along their alignment. The model was validated by SWISS-MODEL (30). Comparisons between different topoisomerases models were conducted with the PyMOL Molecular Graphics System (http://www.pymol.org).
Analysis of the genetic context and DotPlot analysis
The genomic context of identified topoisomerase VIII genes was analyzed with CLC Genomic Workbench as described previously (31). The topoisomerase VIII gene-containing region was considered to be potentially mobile when two criteria were satisfied: (i) the integrase-coding gene was present in proximity of the topoisomerase VIII gene; and (ii) the region containing both topoisomerase VIII- and integrase-coding genes was flanked by direct repeats (attachment [att] sites). Gepard (32) was used to generate dotplots for the analysis of large mobile genomic regions.
Recombinant protein expression and purification
The gene encoding the topoisomerase VIII from Ammonifex degensii (gene ID: 646359367), Microscilla marina ATCC 23134 (gene ID: 640218028) and Paenibacillus polymyxa M1 (gene ID: 2518132008) were synthesized by GenScript@. The synthetic genes of A. degensii and M. marina were cloned into a pUC57 vector, and amplified by PCR with primers containing Strep-Tag, NdeI and NotI restriction sites. The PCR products with the Strep-tags either at the 5′ or 3′ ends of the genes were cloned into a pET26b expression vector and the sequences of recombinant topoisomerase VIII clones were confirmed by DNA sequencing. One of each of the recombinant plasmids: MicNStrep.pET26b::topoisomerase, MicCStrep.pET26b::topoisomerase, AdegNStrep.pET26b::topoisomerase and AdegCStrep.pET26b::topoisomerase was used to transform Escherichia coli BL21 (DE3) strains (Novagen). The synthetic gene of P. polymyxa topoisomerase VIII was purchased directly with an N-terminal Strep-Tag. Bacteria were transformed with the various constructs and were grown subsequently in 1–4 l of Luria Broth (LB) medium containing kanamycin (50 μg/ml), at 16°C overnight, after a heat shock at 42°C at the beginning of growth phase (OD600 nm of 0.1). Induction was carried out with 0.5 mM isopropyl-d-1-thiogalactopyranoside (IPTG) when cell cultures reached an OD600 nm of 0.5. Cells were harvested by centrifugation, stored overnight at −80°C, and then suspended in a Tris–HCl 40 mM pH 8.0, NaCl 200–1000 mM, Dirhiotreitol (DTT) 1 mM, acide éthylène diamine tétraacétique (EDTA) 0.1 mM buffer containing protease inhibitors. Cell lysis was completed by sonication. The cell extracts were then centrifuged at 10 000 × g for 15 min at 4°C to remove cellular debris and aggregated proteins. Strep-tagged proteins from the soluble fraction were purified by gravity-flow chromatography on a Strep-Tactin column (IBA BioTAGnology) according to the manufacturer's recommendation and the eluted proteins were then run on a gel filtration column (Superdex™ 200 16/600, GE Healthcare) with an FPLC AKTA system. The column was equilibrated with buffer B (buffer A containing 10% (v/v) ethylene glycol or glycerol) and the protein was eluted in the same buffer. Topoisomerase VIII enzymes were detected by checking for the presence of a polypeptide with the expected size on an Sodium dodecyl sulfate (SDS)-polyacrylamide gel. Fractions containing topoisomerase VIII polypeptides were pooled and concentrated with Amicon 30 kDa cutoff concentrators (Millipore). Protein concentration was determined and the fractions were aliquoted and stored at −80°C.
In vitro mutagenesis
Plasmids bearing P. polymyxa and M. marina topoisomerase VIII mutant genes were generated with the QuikChange site-directed mutagenesis kit (Stratagene). After mutagenesis, plasmids were purified with Macherey Nagel minipreps kit and sequenced to ensure the absence of unwanted mutations.
Positively supercoiled pBR322 preparation
Positively supercoiled pBR322 was prepared by incubation of pBR322 plasmid for 30 min at 90°C in the presence of Sulfolobus solfataricus reverse-gyrase (RG1) in buffer containing 50 mM Tris–HCl [pH 8.0], 20 mM MgCl2, 1 mM adenosine triphosphate (ATP) and 1 mM DTT (33). The reaction was stopped with 10 mM EDTA and 200 mM NaCl and the positively supercoiled plasmid was purified with the Macherey-Magel NucleoSpin Gel and PCR Clean-up kit.
DNA relaxation and decatenation assays
Relaxation assays (per 20 μl) were performed with negatively or positively supercoiled pBR322 plasmid or kDNA (200 ng) and the indicated amount of enzyme in buffer containing 50 mM Tris [pH 8.0], 2.5 mM MgCl2, 0.1 mM EDTA and ATP (as indicated in the figure legends) for 40 min at 20–30°C or 10 min at 70°C depending on the topoisomerase host. The reaction also contained a final concentration of 0.30 M NaCl and ≈19% glycerol from the protein storage buffer. In some cases at the end of the reaction, an additional incubation of 30 min at 55°C, was done after addition of 2 μl sodium dodecyl sulfate (SDS) 10% and 2 μl proteinase K (1 mg/ml) to check for stabilized cleavable complexes.
Reactions were stopped with 2 μl 1(10% SDS):1(30% glycerol) stop buffer. The reactions were loaded on a 1% agarose gel in 0.5× Tris-Borate-EDTA (TBE) buffer (45 mM Tris–borate [pH 8.3], 1 mM EDTA). The gels were run at 50 V/cm for 4 h, stained with ethidium bromide (EtBr) and visualized with an imaging system.
RESULTS
Identification of DNA topoisomerase VIII, a new subfamily of type IIB topoisomerase
We routinely screen the NCBI non-redundant protein and environmental sequence databases by psi-BLAST (34) to search for new type IIB topoisomerase-encoding genes. During the course of this work, we used the B subunit of topoisomerase VI from particular archaea (e.g. Sulfolobus shibatae) as a query. This led to the detection of a divergent version of type IIB topoisomerases in which the Bergerat fold (ATP-binding site) and the Toprim domain were present in the same polypeptide. We used these unusual proteins as queries to carry out additional Psi-Blast searches. This retrieved a set of closely related proteins of similar sizes corresponding to the topoisomerase VI subunits B and A fused, in that order, into a single polypeptide (Figure 1). After two iterations, we found that genes encoding these atypical type IIB topoisomerases are present in 11 complete and 8 partial bacterial genomes (Supplementary Table S1). In addition, we found two genes encoding these proteins on plasmids (Supplementary Table S1): one in the halophilic archaeon Halalkalicoccus jeotgali B3 (plasmid 5) and the other in the firmicute P. polymyxa M1 (pPPM1a). We also identified a very similar protein encoded by two genes (separated by 41 bp) located on a plasmid in Paenibacillus alvei DSM 29 (pPAV109): one gene encodes a protein homologous to the N-terminal moiety of the topoisomerase VI B subunit, whereas the other encodes a protein homologous to the C-terminal moiety of the B subunit fused to a domain homologous to the topoisomerase VI subunit A (Figure 1). Moreover, we also detected small DNA fragments encoding these proteins in several whole genome shotgun and environmental sequence databases (not shown).
These atypical type IIB topoisomerases, with sizes between 695 and 882 amino acids, are annotated either as topoisomerase VI, hypothetical proteins, ATP-binding proteins, Toprim or topoisomerase (ATP hydrolyzing). They harbor the four conserved regions important for type II topoisomerase function, i.e. the Bergerat fold (ATP-binding site), the transducer domain, the WHD and the Toprim domains, as well as the small H2TH domain specific of the type IIB family (Figures 1 and 2). The putative active site tyrosine was adjacent to either another tyrosine (YY) or a phenylalanine (FY), with the exception of the enzymes from Roseobacter sp. AzwK-3b and Rhizobium gallicum in which the tyrosine is preceded by a methionine (MY). This can be considered as a hallmark of the type IIB topoisomerase family, because topoisomerase VI and Spo11 proteins always have YY or FY in their active sites. In contrast, this feature is usually not observed in the type IIA family. Moreover, the identified proteins displayed only a remote relationship with topoisomerase VI, and their similarity was restricted to specific amino-acid signatures within the conserved motifs (Figure 2 and Supplementary Figure S1). Overall, the identified group of enzymes is not more closely related to bona fide bacterial topoisomerase VI than it is to archaeal or eukaryal topoisomerase VI. In contrast, these atypical type IIB enzymes exhibit extensive sequence similarity with each other, throughout their entire sequence. Furthermore, these proteins share a specific amino-acid signature, ‘R V/I E L N A/S M’, in their C-terminal region (Supplementary Figure S1), which is not found in topoisomerases from the currently established families and subfamilies.
Figure 2.

Conserved amino-acid regions shared between topoisomerases VI and VIII enzymes. In red, amino acids shared between the two families; in blue, amino acids specific for the topoisomerase VIII sub-family; in green, amino acids specific for the topoisomerase VI sub-family. Underlined are alternative amino acids rarely found in otherwise strictly conserved motifs. Adeg: Ammonifex degensii topoisomerase VIII, Teth: Thermoanaerobacter ethanolicus topoisomerase VIII, Mmar: Microscilla marina topoisomerase VIII; pPol: Paenibacillus polymyxa M1 plasmid pPPM1a topoisomerase VIII; Sshi: Sulfolobus shibatae topoisomerase VI; Mmaz: Methanosarcina mazei topoisomerase VI; Anae: Anaeromyxobacter sp. topoisomerase VI; Atha: Arabidopsis thaliana topoisomerase VI. Adeg, Teth, Mmar, Anae and P. polymyxa are Bacteria, Sshi and Mmaz are Archaea, Atha is a eukaryote.
All these observations strongly suggest the ancient divergence of these enzymes from topoisomerase VI, as opposed to recent independent fusions of topoisomerase VI subunits A and B in various bacterial lineages. Based on these observations as well as the data presented below, we propose that these proteins should be considered as a distinct subfamily of type IIB enzymes named DNA topoisomerase VIII. This classification is consistent with the historical nomenclature of topoisomerases in which subfamilies of type I enzymes have been systematically given odd numbers (topoisomerases I, III and V) and type II enzymes even numbers (topoisomerases II, IV, VI and VIII) (3).
Topoisomerase VIII is present in three bacterial phyla: Firmicutes (Clostridia and Bacilli), Bacteroidetes (Sphingobacteria) and Proteobacteria (alpha and gamma). Notably, those present in evolutionarily close genera are grouped together in phylogenetic analysis, whereas topoisomerase VIII enzymes from various genera of Proteobacteria and Firmicutes sometimes display mixed phylogenetic patterns (Figure 3). The three topoisomerase VIII enzymes encoded by plasmids are grouped with cellular topoisomerase VIII enzymes from species that are closely related to the hosts of these plasmids. For example, topoisomerase VIII enzymes encoded by pPPM1a and pPAV109 from P. polymyxa and P. alvei, respectively, are grouped with the chromosomally-encoded topoisomerase VIII from Paenibacillus panacisoli. Similarly, the topoisomerase VIII encoded by the plasmid from a haloarchaeon clusters with a sequence present in a metagenome from a hypersaline lake (Figure 3). These observations are consistent with previous studies that have shown that mobile elements often co-evolve with their hosts (35,36).
Figure 3.
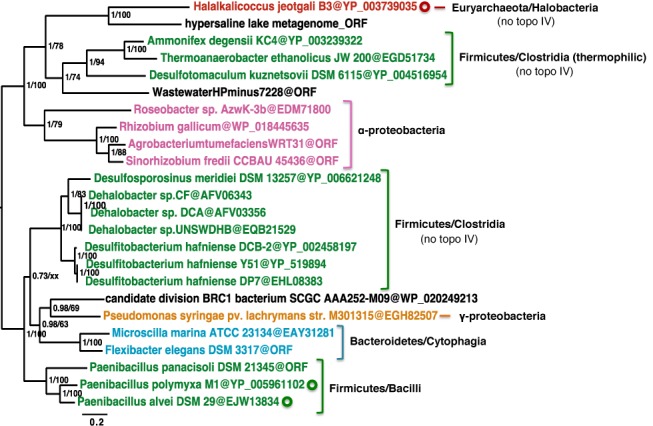
Phylogeny of topoisomerase VIII. Bayesian phylogeny of the 24 topoisomerase VIII homologs listed in Supplementary Table S1 and two topoisomerase VIII enzymes detected in metagenomes (571 amino acid positions). The tree was calculated with MrBayes (MIX model + gamma4). The scale bar shows the average number of substitutions per site. Values at nodes are posterior probabilities and bootstrap values calculated with the rapid bootstrap feature of RAxML (LG + gamma4) from heuristic searches of 1000 resampled datasets, when the same node was recovered. Topoisomerase VIII enzymes encoded by a plasmid are marked by circles.
Most bacteria carry two type IIA topoisomerases, DNA gyrase and DNA topoisomerase IV, which have specialized functions (3). However, there are cases (e.g. Thermotoga maritima and Mycobacterium tuberculosis) where one dual-function enzyme is sufficient (3). Therefore, we considered the possibility that topoisomerase VIII may compensate for the absence of topoisomerase IV in some species. However, we found no correlation between the presence of topoisomerase VIII and the presence/absence of topoisomerase IV in any of the species analyzed (Figure 3).
Topoisomerase VIII genes present in bacterial genomes are located within integrated conjugative plasmids
The presence of topoisomerase VIII-encoding genes on three different plasmids and the sporadic distribution of these genes in taxonomically distant bacteria suggested that the presumably ‘cellular’ (i.e. encoded on bacterial chromosomes) gene copies are, in fact, be carried by integrated mobile genetic elements (37). Indeed, our previous analysis showed that integrated mobile elements often carry genes encoding proteins involved in DNA metabolism (35,38). We performed a detailed analysis of the genetic context of the ‘cellular’ topoisomerase VIII-encoding genes to verify this hypothesis. Unfortunately, 12 of the 19 topoisomerase VIII-encoding bacterial genomes were only available as WGS (whole genome shotgun) libraries consisting of genomic contigs of variable lengths, complicating their analysis. Nonetheless, we were able to obtain evidence suggesting that at least 10 of the seemingly cellular topoisomerase VIII-encoding genes are encoded by mobile elements (Supplementary Table S1).
In several cases, the mobile elements could be detected via the identification of their integration sites. During recombination mediated by the element-encoded integrase, the target sequence (attachment site, attB) is duplicated, and flanks the element from both ends as direct repeats (attL and attR). One of the repeats is typically found next to a gene encoding the recombinase. We identified the precise targets of integration for seven of the elements. Two elements, AmmDeg-E1 (in A. degensii KC4) and RosAzw-E1 (in Roseobacter sp. AzwK-3b), are integrated in intergenic regions, whereas PaePan-E1 (in P. panacisoli DSM21345) and SinFre-E1 (in Sinorhizobium fredii CCBAU 45436) recombined with 3′-distal regions of protein-coding genes (Supplementary Table S1). In addition, elements AgrTum-E1 (in Agrobacterium tumefaciens WRT31), DesKuz-E1 (in Desulfotomaculum kuznetsovii DSM 6115) and RhiGal-E1 (in R. gallicum bv. gallicum R602sp) are integrated into tRNA-Thr, tRNA-Pro and tRNA-Cys genes, respectively (Figure 4).
Figure 4.
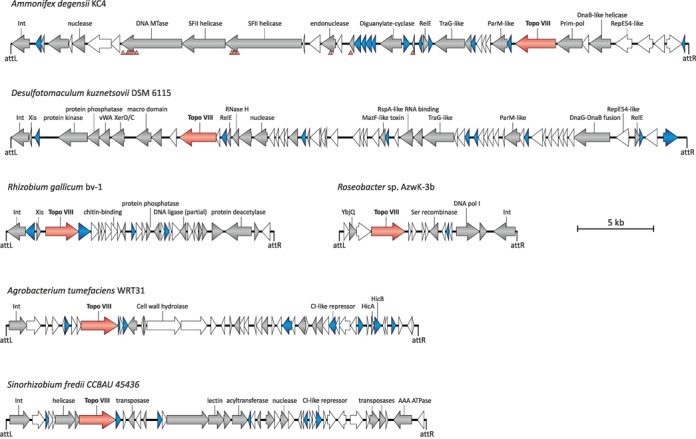
Topoisomerase VIII-coding genes in bacterial genomes are encoded within integrated mobile elements. Genomic organization of the six topoisomerase VIII-encoding integrated mobile elements. Open reading frames (ORFs) are depicted by arrows, and corresponding functions are indicated when possible. Topoisomerase VIII-coding genes are shown in red, genes encoding proteins with homologs in plasmids are depicted in gray, and blue arrows correspond to genes encoding DNA-binding proteins. Note that many of the DNA-binding proteins also have plasmid homologs. Positions of the left and right attachment sites (attL and attR, respectively) are also indicated. The positions of premature stop codons in several genes of the Ammonifex degensii KC4 element are depicted with red triangles under the genome map. Abbreviations: Int, integrase; MTase, methyltransferase; prim-pol, bifunctional primase-polymerase.
A search for putative attachment sites within topoisomerase VIII-encoding genomes did not result in the identification of additional integrated elements. This may be because some genomic contigs containing the integrated element were incomplete, the elements used another mechanism for integration, or the elements were in an advanced stage of decay, which would render the direct repeats unrecognizable due to mutations. Thus, we used an alternative approach involving the comparison of topoisomerase VIII-encoding and topoisomerase VIII-free closely related bacterial genomes. The integrated elements were expected to disrupt the collinearity of the corresponding genomic loci. DotPlot analysis showed that the genomic regions encoding topoisomerase VIII in Desulfosporosinus meridiei DSM 13257, Desulfitobacterium hafniense DCB-2 and D. hafniense Y51 are probably mobile. Notably, the elements in two D. hafniense strains are located in distinct genomic loci (Supplementary Figure S2). In addition, the topoisomerase VIII-encoding contig of D. hafniense DP7 is collinear throughout its length with the corresponding mobile region in D. hafniense DCB-2 and D. hafniense Y51. This suggests that this contig contains a mobile element which is related to the two other D. hafniense elements, DesHaf_DCB-2 and DesHaf_Y51.
The predicted integrated elements did not contain any viral signature genes (e.g. for virion components or genome packaging), suggesting that they are unlikely to be of viral origin. In contrast, many of the genes have homologs in bacterial plasmids (Figure 4). For example, 27 of the 42 open reading frames present in AmmDeg-E1 had plasmid homologs, indicating that the topoisomerase VIII-encoding elements are derived from integrative plasmids. We focused on elements for which exact boarders could be unequivocally defined to analyze genetic content in more detail (Figure 4). Elements PaePan-E1, AmmDeg-E1 and DesKuz-E1 and plasmids pPPM1a and pPAV109 encode both proteins typical of conjugative plasmids and integrative and conjugative elements (ICE) (31); e.g. all five elements encode TraG-like conjugal transfer coupling protein homologs. In addition, AmmDeg-E1 and DesKuz-E1 share homologs of ParM, a protein responsible for plasmid segregation upon cell division. Both elements also have several copies of toxin-antitoxin modules (RelE/B-like in AmmDeg-E1 and DesKuz-E1, HicA/HicB in AgrTum-E1 and MazE/F-like in AmmDeg-E1; Figure 4), which often ensure the stable maintenance of plasmids and ICEs (39). Several other proteins involved in DNA metabolism besides topoisomerase VIII are encoded within the predicted elements. AmmDeg-E1 and DesKuz-E1 encode homologs of the replication protein RepE of plasmid F, supporting further a link between the two integrated elements and conjugative plasmids. Interestingly, AmmDeg-E1 carries next to the RepE-like gene a homolog of the bifunctional primase-polymerase, which is typically found in various bacterial and archaeal mobile elements (40–42), whereas DesKuz-E1 contains at the same position a gene corresponding to a unique fusion of DnaG-like primase and DnaB-like helicase (Figure 4). The two smaller elements, RosAzw-E1 and RhiGal-E1, encode a homolog of family I DNA polymerase and a DNA ligase, respectively. Neither RosAzw-E1 nor RhiGal-E1 contains identifiable genes for components of the conjugal apparatus.
The elements harboring topoisomerase VIII-encoding genes appear to be at different stages of deterioration. At one end of the spectrum are the three plasmids, which probably rely on the encoded DNA metabolism proteins, including topoisomerase VIII, for their efficient replication. However, some of the integrated elements appear to be in the process of being inactivated. Analysis of the AmmDeg-E1 revealed that a number of genes, including those encoding superfamily II helicase, DNA methyltransferase, endonuclease, and two transposases, have accumulated mutations resulting in premature stop codons (Figure 4). However, we could identify the attachment sites (perfect direct repeats) flanking this element, suggesting that the insertion of this element into the bacterial chromosome was probably a recent event. At the opposite side of the spectrum are the elements for which the borders could not be defined, as in the case of M. marina ATCC 23134 (Supplementary Table S1). Notably, transposase may be involved in the process of element inactivation because topoisomerase VIII-encoding genes in Dehalobacter species CP, DCA and UNSWDHB are located next to truncated transposase genes.
Structural analysis of topoisomerases VIII
We performed structural modeling of the three topoisomerase VIII enzymes chosen for biochemical analyses (see below), including those from A. degensii KC4 (Adeg, 882 aa), M. marina ATCC 23134 (Mmar, 783 aa) and P. polymyxa M1 plasmid pPPM1a (pPpol, 751 aa). Structures of topoisomerase VI from S. shibatae (pdb code: 2ZBK; (10)) and Methanosarcina mazei (pdb code: 2Q2E; (9)) were used as templates. Despite their low sequence similarities, the three full-length topoisomerase VIII enzymes have the same modular organization as topoisomerase VI and contain five domains. Topoisomerase VIII contains the N-terminal Bergerat fold, followed by the S13-like H2TH and the ribosomal S5 domains 2-like fold that is present in the transducer domain of topoisomerase VI. This combination of domains is specific to the subunit B of type IIB topoisomerase. Topoisomerase VIII enzymes also possess a C-terminal WHD and Toprim domains; the organization of the Toprim domain is specific to type IIB topoisomerase subunit A (in type IIA enzymes, Toprim follows the transducer domain).
The ATP-lid in the ATP-binding site of topoisomerase VIII is highly conserved (Supplementary Figure S3). The switch lysine, which interacts with the γ-phosphate of the bound nucleotide is at position 446 in A. degensii, 374 in M. marinus and 369 in P. polymyxa topoisomerase VIII (Supplementary Figure S3). This lysine is conserved in all type IIA enzymes and topoisomerase VI transducer domains (K427 in S. shibatae topoisomerase VI). In the modeled structures, this switch lysine points away from the active site because the template used corresponds to the relaxed state conformation (9).
In regions corresponding to topoisomerase VI subunit B, the major difference between the two type IIB subfamilies is the length of the transducer domain, which is clearly smaller in topoisomerase VIII. The end of the last helix stops at nearly the middle of α11-helix of the S. shibatae topoisomerase VI structure (see alignment in Supplementary Figure S1 and Table S2). This corresponds to residue K450 which was identified as a hinge residue involved in helix bending that is crucial for conformational changes during the catalytic cycle (opening-closing of the N-gate) (10). The WHD domain of DNA topoisomerase VIII, a three-helix bundle, is smaller than the equivalent domain of topoisomerase VI (80 residues instead of 160: Supplementary Table S2). The first helix of this domain acts also as a junction between the transducer and the WHD domains (which corresponds to the junction between the B and A subunits). The structure of the Toprim domain is highly conserved and all the residues implicated in magnesium binding (E209, D261 and D263 in the S. shibatae structure) are spatially conserved.
Based on the structural similarities of the individual domains, we suggest that the architecture of the topoisomerase VIII homodimer is comparable to that of the topoisomerase VI heterotetramer. However, the C-terminal helix of the transducer and the N-terminal helix of the WHD domain are lacking in topoisomerase VIII; therefore, it is difficult to predict how the short junction characteristic of these enzymes organizes the orientation between these two domains (compare Figure 5 and Supplementary Figure S4). In addition, the β-turn-β in the topoisomerase VI Toprim domain responsible for the interactions between the two A subunits and the formation of a continuous β-sheet is not present in topoisomerase VIII. Therefore, it is difficult to propose a model for the full-length homodimer.
Figure 5.
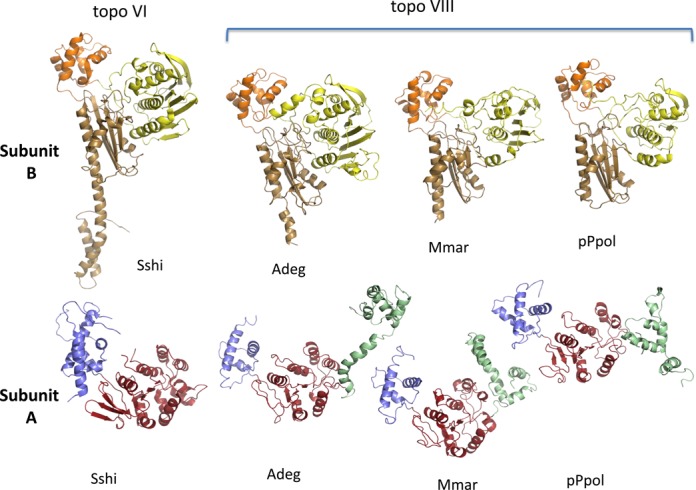
Ribbon representation of the crystal structure of Sulfolobus shibatae topoisomerases VI B (top) and A (bottom) subunits (Sshi; pdb code: 2ZBK) and molecular models of the N- (top) and C-terminal (bottom) domains of topoisomerase VIII from Ammonifex degensii KC4 (Adeg), Microscilla marina ATCC 23134 (Mmar) and Paenibacillus polymyxa M1 plasmid pPPM1a (pPpol). The Bergerat fold domain is shown in yellow, the H2TH in orange, the transducer in brown, the WHD in blue, the Toprim domain in red and the topoisomerases VIII specific C-terminal domain of the A subunit in pale green.
Topoisomerase VIII enzymatic activities
Impairment of the excision of integrated elements occasionally leads to permanent fixation of new functions within cellular lineages, which may even replace ancestral cellular counterparts. This route has been proposed for the origin of a novel nuclear protein in Dinoflagellates (43) and bacteriophage T3/T7-like RNA and DNA polymerases in mitochondria (44). With this in mind, we set out to verify experimentally whether plasmid-borne topoisomerase VIII possesses its predicted enzymatic activity and whether homologs encoded by (decaying) integrated elements retain these activities. For this purpose, we selected and purified three proteins encoded by elements in different stages of decay (see above): (i) we selected topoisomerase VIII from P. polymyxa M1 plasmid pPPM1a as a functional mobile element (45), (ii) the protein from AmmDeg-E1 of the thermophile A. degensii KC4 (46) as a case of recent integration, and (iii) topoisomerase VIII from the mesophile M. marina ATCC 23134 (47) as a representative of ancient integration.
We produced the proteins in E. coli strains carrying the synthetic topoisomerase VIII-encoding genes cloned into pET26-b vectors (see ‘Materials and Methods’ section). Most proteins were not soluble in classical expression conditions. However, soluble proteins were obtained in all cases by growing cells in early exponential phase at 42°C until OD 0.4–0.5 to induce endogenous chaperones, followed by overnight IPTG induction at 16°C. The topoisomerase VIII enzymes were purified by affinity chromatography and gel filtration (see ‘Materials and Methods’ section). The purity of the topoisomerase VIII preparations was similar to that of the purified S. shibatae DNA topoisomerase VI (Supplementary Figure S5). The three enzymes eluted as dimers during gel filtration (Supplementary Figure S6 for P. polymyxa topoisomerase VIII), indicating that these enzymes are homodimers, consistent with structural models.
We used a negatively supercoiled pBR322 plasmid DNA as a substrate and reaction mixtures containing magnesium with or without ATP to assay DNA relaxation activity (see ‘Materials and Methods’ section). We detected weak DNA relaxation activities with the enzymes of M. marina and P. polymyxa (Figure 6A and B). In contrast, the topoisomerase VIII of A. degensii only exhibited DNA cleavage activity in the presence of SDS and proteinase K, producing full-length linear plasmids, probably corresponding to the formation of a cleavable complex (Supplementary Figure S7, lanes 2–4). All these activities were magnesium dependent. Surprisingly, the relaxation activity of the M. marina enzyme was ATP independent (Supplementary Figure S7, compare lanes 2 and 3). However, the addition of 1 mM ATP inhibited slightly both the relaxation activity of the M. marina enzyme and the DNA cleavage activity of the A. degensii enzyme (Supplementary Figure S7, lanes 4). The relaxation activity of the M. marina protein was probably not due to a contamination, because it was abolished when the two tyrosines of the active site were replaced with phenylalanines in a recombinant protein (Figure 6A).
Figure 6.
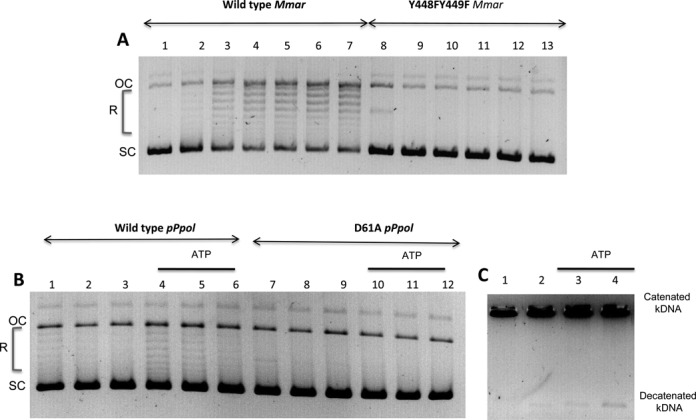
Relaxation and decatenation assays. (A) Relaxation assays of wild-type and double mutants of Microscilla marina (Mmar) topoisomerases VIII. Reactions were carried out without ATP and 200 ng of pBR322 was used as the substrate. Lane 1: control with no enzyme; lanes 2–7, wild-type (WT) with 1, 1.5, 2, 3, 6 and 12 pmol of enzyme, respectively; lanes 8–13, YY-FF double mutants with 0.5, 0.75, 1, 1.75, 3 and 6 pmol of enzyme, respectively. (B) Paenibacillus polymyxa (pPol) topoisomerase VIII relaxation assays with the wild-type enzyme; lanes 1–3, without ATP and with 10, 5, 2.5 pmol of enzyme, respectively, and lanes 4–6, with 1mM ATP and 10, 5, 2.5 pmol of enzymes, respectively; and with the D61A mutant lanes 7–9, without ATP, with 10, 5, and 2.5 pmol of enzyme, respectively, and lanes 10–12 with 1 mM ATP and 10, 5 and 2.5 pmol of enzyme, respectively. Supercoiled and relaxed (or open circular) topoisomers are noted as SC and R/OC, respectively. (C) Paenibacillus polymyxa topoisomerase VIII decatenation assays, with or without ATP; lane 1: no enzyme, lanes 2–4 with 0, 1 and 2 mM ATP, respectively and 20 pmol of enzyme. kDNA: kinetoplastid DNA.
Notably, the relaxation activity of the P. polymyxa topoisomerase VIII was lower than that of M. marina but ATP dependent, as expected of a type II DNA topoisomerase (Figure 6B and Supplementary Figures S8a, lanes 3, S8b, lanes 4 and 7). Substitution of the conserved aspartate in the P. polymyxa topoisomerase VIII Bergerat fold (D61), which is essential for ATP binding (D73 in E. coli DNA gyrase) abolished the ATP-dependent relaxation activity (Figure 6B). Treatment of the reaction products with proteinase K and SDS only produced linear DNA in the presence of ATP, suggesting the formation of a cleavable complex (Supplementary Figure S8, lane 6). Notably, we showed previously that the formation of a cleavable complex is ATP-dependent for S. shibatae DNA topoisomerase VI, whereas it is ATP independent for type IIA enzymes (48).
Some type II topoisomerases relax positively supercoiled DNA more efficiently than negatively supercoiled DNA (49). We thus tested the activity of the P. polymyxa DNA topoisomerase VIII on a positively supercoiled plasmid. This enzyme also exhibited low relaxation activity on positively supercoiled DNA (Supplementary Figure S8b). Finally, decatenation assays with kDNA as a substrate for P. polymyxa DNA topoisomerase VIII showed that this enzyme had a very weak decatenation activity, which was slightly stimulated by ATP (Figure 8C). The poor stability of the proteins precluded further enzymatic characterization.
DISCUSSION
We have identified a new group of type IIB topoisomerases that we call DNA topoisomerase VIII, which are encoded by either free or integrated plasmids. We partially characterized three proteins of this new subfamily. One of them exhibited only endonuclease activity, and the other two exhibited relaxation activity. In all cases, the relaxation activities detected with purified preparations of recombinant proteins were weak (complete relaxation was never observed). Furthermore, only the topoisomerase VIII encoded by the plasmid of P. polymyxa exhibited the expected ATP-dependent relaxation activity of type II topoisomerases. It is possible that the topoisomerase VIII of A. degensii and those of M. marina do not exhibit this activity because they are located on decaying elements and are no longer fully functional. The enzyme from P. polymyxa may exhibit the expected ATP-dependent relaxation activity because its activity is still required for the maintenance of this large plasmid (366 576 bp). The complete absence of relaxation activity in the case of the A. degensii enzyme is somewhat surprising because our in silico analysis suggests that the insertion of this gene into the bacterial chromosome is relatively recent. This indicates that the activity of enzymes encoded by mobile elements can be rapidly lost after integration; therefore, caution should be exercised when studying the biochemical properties of ‘cellular enzymes’ obtained by expressing genes present on cellular chromosome if the origin of these enzymes has not been carefully investigated.
We found that the structures of the Bergerat fold domains of the three topoisomerase VIII enzymes were rather variable (Figure 5). The rmsd values varied from 1.5 Å (A. degensii DNA topoisomerase VIII/S. shibatae topoisomerase VI) to 2.9 Å (A. degensii topoisomerase VIII/ M. marina topoisomerase VIII and M. marina topoisomerase VIII/S. shibatae topoisomerase VI). This variability may explain differences in the activities of the three proteins. However, the core fold is highly conserved (especially the ATP-binding pocket, see Supplementary Figure S3). The main difference involves an insertion in the A. degensii topoisomerase VIII and S. shibatae topoisomerase VI in the C-terminal end of the Bergerat fold domain that is not present in M. marina and P. polymyxa topoisomerase VIII. At present, we cannot make substantiated conclusions about the relationship between ATP-dependency and structural differences among topoisomerase VIII enzymes from the available information, especially given that two highly conserved enzymes, the DNA gyrases of M. tuberculosis and Mycobacterium leprae, have different activity spectra (50,51).
The ATP-independent relaxation activity of the M. marina topoisomerase VIII is reminiscent of that of bacterial DNA gyrase (52,53). Bates et al. suggested that the ancestral role of ATP in type II enzyme reactions was to prevent DNA double-strand breaks (DSBs) by controlling the separation of protein–protein interfaces (54). They suggested that the intersubunit interface is weaker in DNA gyrase than in other type II enzymes, explaining its ATP-independent relaxation activity (54). It is interesting that the three different forms of topoisomerase VIII studied here exhibited low ATP-stimulated relaxation, ATP-independent relaxation or produced DSBs. This suggests that the differences between these three enzymes results from subtle modifications of the protein-protein dimer interface.
The low activity of the P. polymyxa topoisomerase VIII in our in vitro assays may be due to the absence of potentially important accessory proteins required for optimal activity in vivo. For example, plant topoisomerase VI enzymes require two additional proteins, Midget and RLH1, for their activity, at least in vivo (55). Moreover, the cell division protein MukB stimulates the activity of E. coli topoisomerase IV in vitro (56). Other topoisomerases require post-translational modifications to achieve full catalytic strength (57). If additional proteins or processing enzymes are indeed required for the full activity of topoisomerase VIII, these accessory proteins are likely to be encoded by the plasmids bearing the topoisomerase VIII genes.
Origin and evolution of DNA topoisomerase VIII
Several families and subfamilies of DNA topoisomerases are widespread in the living world. Some are almost universal (such as topoisomerase IA) or present in all members of one or two cellular domains (such as DNA gyrases in Bacteria or type IIA topoisomerases in Bacteria and Eukarya) (7). In contrast, topoisomerase VIII enzymes appear to be rare, because they are encoded only in about 0.5% of currently available bacterial genomes and are not present in archaeal or eukaryotic genomes. Moreover, topoisomerase VIII is only present in a handful of representatives of three bacterial phyla. Furthermore, two of them, Proteobacteria and Firmicutes, correspond to the two phyla with by far the highest number of completely sequenced genomes. This very narrow distribution of topoisomerase VIII in the living world resembles the case of topoisomerase V, a unique member of type I topoisomerases (family C) that is only present in the archaeon Methanopyrus kandleri (58,59). It also resembles atypical topoisomerases found in the viral world, such as the heterotrimeric type IIA enzymes encoded by T4-like bacteriophages and the homodimeric type IIA enzymes encoded by some Megavirales (7). Strikingly, topoisomerase VIII enzymes are encoded by plasmids (two from bacteria and one from archaea), and in bacterial genomes, they are located within integrated mobile elements. This is therefore another example of a topoisomerase with an unusual phylogenomic distribution and complex evolutionary trajectory.
The fusion of two bacterial topoisomerase VI subunits in one bacterial phylum, followed by sporadic spread to other lineages by lateral gene transfer is one hypothesis that explains the rare and scattered distribution of topoisomerase VIII in Bacteria. However, this hypothesis seems unlikely considering the high divergence of primary sequence between topoisomerase VI and VIII. Notably, bona fide topoisomerase VI enzymes present in bacteria cannot be distinguished from their archaeal homologs and branch with archaeal DNA topoisomerase VI enzymes in phylogenetic analyses (38); in contrast, topoisomerase VI and VIII enzymes are so divergent that their amino-acid sequences cannot be reliably aligned for phylogenetic analyses. It is difficult to explain why the fusion protein of the two topoisomerase VI-like subunits (i.e. the ancestor of topoisomerase VIII) would have diverged so rapidly in one particular bacterial lineage but remained conserved during its dispersion in various bacterial lineages.
The divergence between topoisomerases VI and VIII enzymes suggests that the gene duplication at the origin of these two protein subfamilies occurred before the emergence of Archaea and Bacteria (Figure 7). In agreement with this view, topoisomerase VI was probably present in the last archaeal common ancestor (LACA) because this enzyme is now present in all archaea, with the exception of Thermoplasmatales (3). In contrast, the rarity of topoisomerase VIII in bacteria suggests that this protein was not present in the last bacterial common ancestor (LBCA), but was already encoded by plasmids at that time. Modern topoisomerase VIII thus probably originated before the diversification of Bacteria, by a fusion between the A and B subunits, following the duplication that initiated the divergence between topoisomerase VI and VIII enzymes (Figure 7). In this scenario, the sporadic distribution of topoisomerase VIII in the bacterial domain is easily explained by the co-evolution of bacterial plasmids bearing topoisomerase VIII-encoding genes with their hosts and the sporadic integration of these genes into bacterial genomes (37).
Figure 7.
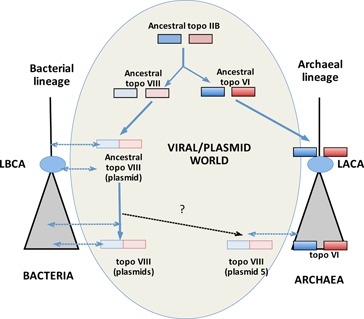
Proposed scenario for the evolution of the type IIB topoisomerase family. LBCA (last bacterial common ancestor), LACA (last archaeal common ancestor). Dotted double arrows indicate co-evolution of cellular and plasmidic lineages, with subsequent integration or loss of plasmidic topoisomerase VIII-encoding genes in cellular genomes. Black dotted arrow and question mark, possible transfer of a bacterial plasmid to the archaeal domain. For simplicity, both the presence of a few topoisomerase VI enzymes in Bacteria (transferred from Archaea) and the secondary split of the topoisomerase VIII gene in the plasmid PAV109 of Paenibacillus alvei are not indicated.
It has been suggested that DNA topoisomerases first originated and diverged into various families and subfamilies in the viral world (7). Viruses and plasmids are evolutionarily related and can be considered as the same sequence space within a greater viral world (60–62). Accordingly, it is possible that type IIB topoisomerases first originated and subsequently diversified into two subfamilies (VI and VIII) in this plasmid/viral world. Later on, topoisomerase VI were transferred to the cellular members of the branch of the tree of life leading to LACA, whereas plasmids encoding topoisomerase VIII co-evolved with ancestors of the bacterial domain (Figure 7). The presence of the topoisomerase VIII-encoding gene in the plasmid of a halophilic archaeon can also be explained by the presence of these plasmids in ancient archaeal organisms (LACA and close relatives). However, in this case, it is also possible that these plasmids were transferred from Bacteria to Archaea at the onset of the haloarchaeal lineage, which has experienced the introduction of around 1000 bacterial genes (63). Irrespective of the evolutionary scenario, the discovery of a subfamily of topoisomerases specifically encoded by plasmids confirms that mobile elements are potential reservoirs of novel proteins involved in DNA metabolism (7,36,42,64).
Possible physiological role of topoisomerase VIII
The role of topoisomerase VIII in plasmid physiology remains to be established. Several conjugative plasmids from Proteobacteria and Firmicutes encode enzymes related to bacterial topoisomerase III (a subfamily of type IA enzymes) (65). These plasmidic topoisomerase III enzymes are used either as decatenases to help the faithful segregation of these large plasmids with low copy number, or as swivels during conjugation to facilitate the unwinding of the donor strand that is transferred to the recipient host (65). Alternatively, they may also be involved in the resolution of hemicatenane structures during plasmid replication or recombination (66). Notably, the plasmids pPPM1a of P. polymyxa and pAV109 of P. alvei encode a DNA topoisomerase III, in addition to a topoisomerase VIII. Topoisomerase III may be involved in the resolution of hemicatenane structures, whereas topoisomerase VIII enzymes could be the decatenase and/or swivelase involved in the segregation of these large plasmids (>100 kb).
Interestingly, the genome of P. polymyxa contains nine loci dedicated to the synthesis of non-ribosomal peptides, one of which is located on the plasmid pPPM1a (67). Of note, some plasmids encode proteins containing pentapeptide repeats that interact with cellular topoisomerases, as demonstrated by fluoroquinolone resistance proteins (68). These peptides may serve as important antibiotics in the control of plant pathogens. We can speculate that some of the peptides encoded by the plasmid pPPM1a are also topoisomerase inhibitors. In that case, the plasmid-encoded topoisomerase III and topoisomerase VIII may also be resistant to these antibiotics, and serve as substitutes for antibiotic-sensitive cellular topoisomerases. Thus P. polymyxa may be an interesting model to study the role of topoisomerases in plasmid maintenance and transfer. It would also be relevant to screen Paenibacillus species for novel anti-topoisomerase inhibitors and/or new proteins interfering with topoisomerases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council (ERC) grant from the European Union's Seventh Framework Programme [FP/2007–2013 to P.F.]; Project EVOMOBIL [ERC Grant Agreement no. 340440 to P.F.]; Paul W. Zuccaire Foundation (to K.R.). Funding for open access charge: Institut Pasteur.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schoeffler A.J., Berger J.M. Topos: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 2.Wang J.C. Untangling the Double Helix. Harvard University: Cold Spring Harbor Laboratory Press; 2009. [Google Scholar]
- 3.Forterre P. Introduction and historical perspective. In: Pommier Y, editor. Topos and Cancer. Springer-Verlag: Humana press; 2011. pp. 1–52. [Google Scholar]
- 4.Bergerat A., de Massy B., Gadelle D., Varoutas P.C., Nicolas A., Forterre P. An atypical topoisomerases II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 5.Gadelle D., Filée J., Buhler C., Forterre P. Phylogenomics of type II Topos. Bioessays. 2003;25:232–242. doi: 10.1002/bies.10245. [DOI] [PubMed] [Google Scholar]
- 6.Corbett K.D., Berger J.M. Structure, molecular mechanisms, and evolutionary relationships in Topos. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 7.Forterre P., Gadelle D. Phylogenomics of Topos: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoeffler A.J., Berger J.M. Recent advances in understanding structure-function relationships in the type II topoisomerases mechanism. Biochem. Soc. Trans. 2005;33:1465–1470. doi: 10.1042/BST0331465. [DOI] [PubMed] [Google Scholar]
- 9.Corbett K.D., Benedetti P., Berger J.M. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerases VI. Nat. Struct. Mol. Biol. 2007;14:611–619. doi: 10.1038/nsmb1264. [DOI] [PubMed] [Google Scholar]
- 10.Graille M., Cladière L., Durand D., Lecointe F., Gadelle D., Quevillon-Cheruel S., Vachette P., Forterre P., van Tilbeurgh H. Crystal structure of an intact type II Topos: insights into DNA transfer mechanisms. Structure. 2008;16:360–370. doi: 10.1016/j.str.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 11.Laponogov I., Sohi M.K., Veselkov D.A., Pan X.-S., Sawhney R., Thompson A.W., McAuley K.E., Fisher L.M., Sanderson M.R. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 12.Dutta R., Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 13.Woese C.R., Kandler O., Wheelis M.L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U.S.A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lecompte O., Ripp R., Thierry J.-C., Moras D., Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 2002;30:5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forterre P., Gribaldo S., Gadelle D., Serre M.-C. Origin and evolution of Topos. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Bergerat A., Gadelle D., Forterre P. Purification of a Topos II from the hyperthermophilic archaeon Sulfolobus shibatae. A thermostable enzyme with both bacterial and eucaryal features. J. Biol. Chem. 1994;269:27 663–27 669. [PubMed] [Google Scholar]
- 17.Brochier-Armanet C., Gribaldo S., Forterre P. A Topos IB in Thaumarchaeota testifies for the presence of this enzyme in the last common ancestor of Archaea and Eucarya. Biol. Direct. 2008;3:54. doi: 10.1186/1745-6150-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugimoto-Shirasu K., Stacey N.J., Corsar J., Roberts K., McCann M.C. Topos VI is essential for endoreduplication in Arabidopsis. Curr. Biol. 2002;12:1782–1786. doi: 10.1016/s0960-9822(02)01198-3. [DOI] [PubMed] [Google Scholar]
- 19.Filée J., Forterre P., Sen-Lin T., Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- 20.Forterre P. The origin of DNA genomes and DNA replication proteins. Curr. Opin. Microbiol. 2002;5:525–532. doi: 10.1016/s1369-5274(02)00360-0. [DOI] [PubMed] [Google Scholar]
- 21.Plewniak F., Bianchetti L., Brelivet Y., Carles A., Chalmel F., Lecompte O., Mochel T., Moulinier L., Muller A., Muller J., et al. PipeAlign: A new toolkit for protein family analysis. Nucleic Acids Res. 2003;31:3829–3832. doi: 10.1093/nar/gkg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 23.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 25.Criscuolo A., Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abascal F., Zardoya R., Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 27.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 28.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley L.A., Sternberg M.J.E. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 30.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 31.Krupovic M., Forterre P., Bamford D.H. Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J. Mol. Biol. 2010;397:144–160. doi: 10.1016/j.jmb.2010.01.037. [DOI] [PubMed] [Google Scholar]
- 32.Krumsiek J., Arnold R., Rattei T. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 33.Bizard A., Garnier F., Nadal M. TopR2, the second reverse gyrase of Sulfolobus solfataricus, exhibits unusual properties. J. Mol. Biol. 2011;408:839–849. doi: 10.1016/j.jmb.2011.03.030. [DOI] [PubMed] [Google Scholar]
- 34.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krupovic M., Gribaldo S., Bamford D.H., Forterre P. The evolutionary history of archaeal MCM helicases: a case study of vertical evolution combined with hitchhiking of mobile genetic elements. Mol. Biol. Evol. 2010;27:2716–2732. doi: 10.1093/molbev/msq161. [DOI] [PubMed] [Google Scholar]
- 36.Soler N., Marguet E., Cortez D., Desnoues N., Keller J., van Tilbeurgh H., Sezonov G., Forterre P. Two novel families of plasmids from hyperthermophilic archaea encoding new families of replication proteins. Nucleic Acids Res. 2010;38:5088–5104. doi: 10.1093/nar/gkq236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forterre P. Darwin's goldmine is still open: variation and selection run the world. Front. Cell Infect. Microbiol. 2012;2:106. doi: 10.3389/fcimb.2012.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raymann K., Forterre P., Brochier-Armanet C., Gribaldo S. Global phylogenomic analysis disentangles the complex evolutionary history of DNA replication in Archaea. Genome Biol. Evol. 2014. doi:10.1093/gbe/evu004. [DOI] [PMC free article] [PubMed]
- 39.Wozniak R.A.F., Waldor M.K. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 40.Lipps G. The replication protein of the Sulfolobus islandicus plasmid pRN1. Biochem. Soc. Trans. 2004;32:240–244. doi: 10.1042/bst0320240. [DOI] [PubMed] [Google Scholar]
- 41.Krupovic M., Gonnet M., Hania W.B., Forterre P., Erauso G. Insights into dynamics of mobile genetic elements in hyperthermophilic environments from five new Thermococcus plasmids. PLoS One. 2013;8:e49044. doi: 10.1371/journal.pone.0049044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gill S., Krupovic M., Desnoues N., Béguin P., Sezonov G., Forterre P. A highly divergent archaeo-eukaryotic primase from the Thermococcus nautilus plasmid, pTN2. Nucleic Acids Res. 2014;42:3707–3719. doi: 10.1093/nar/gkt1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gornik S.G., Ford K.L., Mulhern T.D., Bacic A., McFadden G.I., Waller R.F. Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr. Biol. 2012;22:2303–2312. doi: 10.1016/j.cub.2012.10.036. [DOI] [PubMed] [Google Scholar]
- 44.Filée J., Forterre P. Viral proteins functioning in organelles: a cryptic origin. Trends Microbiol. 2005;13:510–513. doi: 10.1016/j.tim.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 45.Lal S., Romano S., Chiarini L., Signorini A., Tabacchioni S. The Paenibacillus polymyxa species is abundant among hydrogen-producing facultative anaerobic bacteria in Lake Averno sediment. Arch. Microbiol. 2012;194:345–351. doi: 10.1007/s00203-011-0763-0. [DOI] [PubMed] [Google Scholar]
- 46.Huber R., Rossnagel P., Woese C.R., Rachel R., Langworthy T.A., Stetter K.O. Formation of ammonium from nitrate during chemolithoautotrophic growth of the extremely thermophilic bacterium ammonifex degensii gen. nov. sp. nov. Syst. Appl. Microbiol. 1996;19:40–49. doi: 10.1016/S0723-2020(96)80007-5. [DOI] [PubMed] [Google Scholar]
- 47.Lewin R.A. A classification of flexibacteria. J. Gen. Microbiol. 1969;58:189–206. doi: 10.1099/00221287-58-2-189. [DOI] [PubMed] [Google Scholar]
- 48.Buhler C., Gadelle D., Forterre P., Wang J.C., Bergerat A. Reconstitution of DNA topoisomerase VI of the thermophilic archaeon Sulfolobus shibatae from subunits separately overexpressed in Escherichia coli. Nucleic Acids Res. 1998;26:5157–62. doi: 10.1093/nar/26.22.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McClendon A.K., Rodriguez A.C., Osheroff N. Human topoisomerase IIalpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 2005;280:39 337–45. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 50.Aubry A., Fisher L.M., Jarlier V., Cambau E. First functional characterization of a singly expressed bacterial type II topoisomerase: the enzyme from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2006;348:158–65. doi: 10.1016/j.bbrc.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 51.Matrat S., Petrella S., Cambau E., Sougakoff W., Jarlier V., Aubry A. Expression and purification of an active form of the Mycobacterium leprae DNA gyrase and its inhibition by quinolones. Antimicrob. Agents Chemother. 2007;51:643–8. doi: 10.1128/AAC.01282-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gellert M., Mizuuchi K., O'Dea M.H., Itoh T., Tomizawa J.I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. U.S.A. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugino A., Peebles C.L., Kreuzer K.N., Cozzarelli N.R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. U.S.A. 1977;74:4767–71. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bates A.D., Berger J.M., Maxwell A. The ancestral role of ATP hydrolysis in type II topoisomerases: prevention of DNA double-strand breaks. Nucleic Acids Res. 2011;39:6327–6339. doi: 10.1093/nar/gkr258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirik V., Schrader A., Uhrig J.F., Hulskamp M. MIDGET unravels functions of the Arabidopsis topoisomerase VI complex in DNA endoreduplication, chromatin condensation, and transcriptional silencing. Plant Cell. 2007;19:3100–10. doi: 10.1105/tpc.107.054361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayama R., Marians K.J. Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18826–18831. doi: 10.1073/pnas.1008140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chikamori K., Grozav A.G., Kozuki T., Grabowski D., Ganapathi R., Ganapathi M.K. DNA topoisomerase II enzymes as molecular targets for cancer chemotherapy. Curr. Cancer Drug Targets. 2010;10:758–71. doi: 10.2174/156800910793605785. [DOI] [PubMed] [Google Scholar]
- 58.Taneja B., Patel A., Slesarev A., Mondragón A. Structure of the N-terminal fragment of topoisomerases V reveals a new family of topoisomerases. EMBO J. 2006;25:398–408. doi: 10.1038/sj.emboj.7600922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forterre P. Topos V: a new fold of mysterious origin. Trends Biotechnol. 2006;24:245–247. doi: 10.1016/j.tibtech.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 60.Forterre P. The two ages of the RNA world, and the transition to the DNA world: a story of viruses and cells. Biochimie. 2005;87:793–803. doi: 10.1016/j.biochi.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 61.Koonin E.V., Dolja V.V. A virocentric perspective on the evolution of life. Curr. Opin. Virol. 2013;3:546–557. doi: 10.1016/j.coviro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krupovic M. Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses. Curr. Opin. Virol. 2013;3:578–586. doi: 10.1016/j.coviro.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 61.Nelson-Sathi S., Dagan T., Landan G., Janssen A., Steel M., McInerney J.O., Deppenmeier U., Martin W.F. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc. Natl. Acad. Sci. U.S.A. 2012;109:20 537–20 542. doi: 10.1073/pnas.1209119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolkenbrock S., Naumann B., Hippler M., Fetzner S. A novel replicative enzyme encoded by the linear Arthrobacter plasmid pAL1. J. Bacteriol. 2010;192:4935–4943. doi: 10.1128/JB.00614-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z., Hiasa H., Kumar U., DiGate R.J. The traE gene of plasmid RP4 encodes a homologue of Escherichia coli Topos III. J. Biol. Chem. 1997;272:19 582–19 587. doi: 10.1074/jbc.272.31.19582. [DOI] [PubMed] [Google Scholar]
- 66.Lee S.-H., Siaw G.E.-L., Willcox S., Griffith J.D., Hsieh T.-S. Synthesis and dissolution of hemicatenanes by type IA Topos. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3587–94. doi: 10.1073/pnas.1304103110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niu B., Rueckert C., Blom J., Wang Q., Borriss R. The genome of the plant growth-promoting rhizobacterium Paenibacillus polymyxa M-1 contains nine sites dedicated to nonribosomal synthesis of lipopeptides and polyketides. J. Bacteriol. 2011;193:5862–5863. doi: 10.1128/JB.05806-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vetting M.W., Hegde S.S., Wang M., Jacoby G.A., Hooper D.C., Blanchard J.S. Structure of QnrB1, a plasmid-mediated fluoroquinolone resistance factor. J. Biol. Chem. 2011;286:25265–25273. doi: 10.1074/jbc.M111.226936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


