Abstract
Mammalian mRNAs are generated by complex and coordinated biogenesis pathways and acquire 5′-end m7G caps that play fundamental roles in processing and translation. Here we show that several selenoprotein mRNAs are not recognized efficiently by translation initiation factor eIF4E because they bear a hypermethylated cap. This cap modification is acquired via a 5′-end maturation pathway similar to that of the small nucle(ol)ar RNAs (sn- and snoRNAs). Our findings also establish that the trimethylguanosine synthase 1 (Tgs1) interacts with selenoprotein mRNAs for cap hypermethylation and that assembly chaperones and core proteins devoted to sn- and snoRNP maturation contribute to recruiting Tgs1 to selenoprotein mRNPs. We further demonstrate that the hypermethylated-capped selenoprotein mRNAs localize to the cytoplasm, are associated with polysomes and thus translated. Moreover, we found that the activity of Tgs1, but not of eIF4E, is required for the synthesis of the GPx1 selenoprotein in vivo.
INTRODUCTION
Eukaryotic mRNAs, synthetized by RNA polymerase II (polII), are characterized by the presence of a 7-methylguanosine (m7G) cap structure at their 5′-end (1). Nuclear RNA capping serves as an important quality control checkpoint and is an essential determinant of mRNA expression, stability and biogenesis (2). The m7G cap promotes splicing of cap-proximal introns, 3′-end formation, nucleocytoplasmic export, control of RNA stability and translation (3–7). Two major cap-binding proteins mediate these processes: the cap-binding complex (CBC) in the nucleus, composed of the CBP20 and CBP80 subunits (3), and the translation initiation factor eIF4E in the cytoplasm (8–10). The binding of both CBC and eIF4E is highly specific for the m7G cap, and other modified caps are poorly recognized by these proteins (11–14). PolII small nuclear (snRNAs) and a few small nucleolar RNAs (snoRNAs) also acquire m7G caps in the nucleus but become further methylated at guanosine position 2 to generate the 2,2,7-trimethylguanosine cap (m3G or TMG cap) (15). TMG modification of snRNAs is performed in the cytoplasm by the trimethylguanosine synthase 1 (Tgs1) which catalyzes the m7G to m3G hypermethylation. After export to the cytoplasm, precursor snRNAs are assembled with Sm core proteins by the Survival of Motor Neuron (SMN) complex (16). Binding of SMN and Sm proteins is a prerequisite for further recruitment of Tgs1. The m3G cap together with the Sm ring represent bipartite signals that promote nuclear import of snRNPs for final maturation and assembly into the active spliceosome (17,18). The transfer of TMG-capped snRNPs to the nucleus is governed by snurportin which interacts specifically with m3G and Sm proteins, but not with m7G cap structures (19,20). Cap hypermethylation, therefore, represents a critical step in determining the fate of sn- and snoRNAs.
Unlike snRNAs, hypermethylation of m7G-capped snoRNAs takes place in the nucleus, and snoRNAs transit to the Cajal bodies (CB) in a PHAX (phosphorylated adaptor for RNA export)-dependent manner where they are hypermethylated by nuclear Tgs1 (21,22). In this case, Tgs1 is recruited to the snoRNA by the core proteins Nop56 and Nop58 for the box C/D snoRNAs, and by dyskerin for the H/ACA snoRNAs (23,24). Two Tsg1 isoforms have been identified; the long form (LF) is mainly involved in snRNA hypermethylation and is present in both the nucleus and cytoplasm, whereas a short nuclear isoform (SF) is dedicated to snoRNA maturation (23). In the nucleoplasm, Tgs1 isoforms are present in a large multiprotein complex that contains ribonucleoprotein (RNP) maturation, transport and assembly factors (15,22,25).
Selenoprotein mRNAs constitute an interesting class of mRNAs. Indeed, because of the presence of an in-frame UGA codon, recoded as selenocysteine (Sec) and otherwise read as a stop codon, they are submitted to distinctive biogenesis and translation pathways (26,27). Several cis- and trans-acting factors participate in the co-translational recoding of UGA Sec codons. Pivotal in this process are the SElenoCysteine Insertion Sequence (SECIS), a stem-loop in the 3′ untranslated region (3′UTR) of selenoprotein mRNAs, and the SECIS-binding protein 2 (SBP2) (28–31). We have previously shown that the assembly mechanism of selenoprotein mRNPs, in particular in the 3′UTR region, is partly common with that of snRNP, snoRNP particles and the telomerase (27). SBP2 which undergoes nucleocytoplasmic shuttling (32,33), binds to components of this conserved RNP assembly machinery linked to the protein chaperone Hsp90 (27). Besides, SBP2 shares functional RNA binding motifs with primary core proteins of sn- and snoRNPs (27,34), suggesting that selenoprotein mRNPs, sn- and snoRNPs could also share a common 5′ maturation pathway.
In this work, we demonstrate that several mammalian selenoprotein mRNAs, in contrast to other cellular mRNAs, undergo a 5′-end maturation pathway similar to that of sn- and snoRNPs and bear hypermethylated caps.
MATERIALS AND METHODS
Cell culture
HEK293FT, HeLa and U2OS cells were cultured at 37°C in 5% CO2 in Dulbecco's modified Eagle's media (DMEM) containing 10% Fetal Calf Serum (FCS), 1% penicillin–streptomycin and 10 nM sodium selenite. One percent geneticine was added to HEK293FT cells. Cells were transfected using Turbofect (Fermentas) and siRNA inactivation was done for 48 h using 30 or 100 nM of siRNA and Lipofectamine 2000 (Invitrogen) following the manufacturer's conditions. siTGS1, siSMN, siSBP2 and sieIF4E RNAs and sicontrol were ON-TARGET plus smart pools of four different siRNAs (Thermo Scientific Dharmacon), siNop58 RNAs were from Santa Cruz. For rescue experiments, cells were transfected for 24 h after siRNA inactivation with peGFP-Tgs1 (21,24), non-tagged plenti6/V5-SBP2 or pcDNA5-eIF4E expression plasmids and grown for an additional 24 h treatment. Cells were extracted with HNTG buffer (20 mM HEPES–NaOH pH 7.9, 150 mM NaCl, 1% Triton, 10% glycerol, 1 mM MgCl2, 1 mM EGTA (Ethylene glycol-bis(2-aminoethylether)-N,N,N',N'-tetraacetic acid), 1 mM PMSF (phenylmethylsulfonyl fluoride), anti-protease cocktail from Roche). Subcellular fractionation was performed according to (35).
Generation of stable cell lines and induced protein expression
Flp-InTM T-RexTM 293 system (Invitrogen) was used according to the manufacturer's instructions to generate stable cell lines with regulated expression of 3xHA-GPx1 (HA-GPx1) and 3xHA-GPx1mutCys (HA-Gpx1Cys). Stable clones containing the required open reading frame under the control of the cytomegalovirus/tetO2 hybrid promoter were selected by culturing in selective medium containing 250 μg/ml of hygromycin (InvivoGen) and 15 μg/ml of blasticidin (InvivoGen). Resistant colonies were expanded and tested for doxycycline-regulated protein expression. Protein expression was induced by addition of 0.5–1 μg/ml of doxycycline in culture medium and in the presence of 10 nM of sodium selenite.
Stable isogenic HeLa cells expressing the Flag-tagged Tgs1 proteins were obtained with HeLa Flp-In cells, by co-transfection of the parental cells with pcDNA5-3X Flag-Tgs1 (SF or LF) and a Flippase expression vector with Lipofectamine and Plus reagent (Invitrogen). Stable clones were then selected with 50 mM hygromycin B (Calbiochem), picked individually, expanded and characterized by western blots. Individual clones usually expressed similar levels of the tagged protein. DNA cloning was performed with the Gateway system (Invitrogen).
Immunopurification, co-immunoprecipitation and western blotting
Immunopurification of endogenous SBP2 complexes was performed as described in (27). For co-immunoprecipitation experiments, cell extracts (100–300 μl) were incubated with 50 μl of protein A-sepharose (Sigma) coupled to the indicated antibody for 2 h at 4°C in NT2 buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.05% NP40, 1 mM DTT (Dithiothreitol), 400 μM VRC (Vanadyl ribonucleoside complexes), 100 U RNasin/ml, anti-protease cocktail) or with anti-GFP magnetic beads (Miltenyi) in IPP150 (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% NP-40, 0.5 mM PMSF, 2 mM VRC). Beads alone were used as a control. Beads were washed with NT2 or IPP150, respectively, resuspended in Laemmli buffer, analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by western blot. Antibodies used were: anti-His (H3) mouse monoclonal (Santa Cruz), anti-SBP2 rabbit polyclonal (27), anti-Tgs1 mouse monoclonal (24), anti-Nop58 (C-20) polyclonal goat antibodies (Santa Cruz), anti-SMN (2B1) mouse monoclonal (Santa Cruz), anti-rpS21 goat polyclonal (Santa Cruz) and anti-AspRS rabbit polyclonal (36). For RNA IP, extracts were first clarified on protein A-sepharose to eliminate unspecific RNA binders. For microarray experiment, co-immunoprecipitation was performed according to (37).
TMG RIP and qRT-PCR analysis
Trimethyl-capped RNAs were immunoprecipitated using a strictly specific rabbit polyclonal anti-m3G (TMG) serum (Synaptic Systems) also referred to as R1131 (38,39). Ten microliters of serum was coupled to 50 μl of protein A-Sepharose beads saturated with 20 μg of both purified bovine serum albumin (BSA) and total yeast tRNA in NT2 buffer for 18 h at 4°C. The immobilized antibody was incubated with 200 μg of pre-cleared total RNAs prepared from HEK293FT cells (Tri-reagent, Euromedex) in a total volume of 1 ml for 2 h at 4°C. Beads were washed six times in NT2 buffer, the bound RNA was extracted by phenol/chloroform and precipitated. After DNase treatment, RNAs were reversed transcribed using AMV-RT (Q-Biogen) and cDNAs were amplified by classical or quantitative RT-PCR (qRT-PCR). Level of mRNAs after siRNA treatment was measured directly after RNA extraction using the same qRT-PCR method. qRT-PCR was performed on MX3005P (Stratagene) using the Maxima SYBR Green PCR kit (Fermentas). Oligonucleotides used for qRT-PCR are listed in Supplementary Table S1.
Anti-FLAG immunoprecipitation and microarray analysis
Anti-Flag Tgs1 LF and Tgs1 SF immunoprecipitations were done with M2 antibody-coated beads (Sigma-Aldrich). HeLa cells stably expressing Tgs1 LF, Tgs1 SF or parental cells as control, were extracted with lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton, antiprotease cocktail) for 30 min at 4°C and centrifuged for 10 min at 10 000 rpm at 4°C. Extracts were incubated with M2 beads for 2 h at 4°C. Beads were washed twice in HNTG and three times in phosphate buffered saline (PBS), RNAs from beads were homogenized in Trizol and purified with RNeasy mini kit (Qiagen). cDNA were prepared and labeled as recommended by the manufacturer (Affymetrix). RNAs were analyzed using human gene 1.0ST microarray covering all annotated transcripts (Affymetrix). Files were analyzed and normalized with expression consol software (Affymetrix). The 3XFlag-Tgs1 IPs were performed in duplicates, and the control IP (non-tagged parental cell line) was done in triplicate. Microarray data are accessible at GEO GSE57625.
Recombinant proteins and GST pull-down assays
Radiolabeled proteins were synthesized in the presence of 35S-Met in rabbit reticulocyte lysate (TNT; Promega). Binding was performed with 5 μg GST-tagged protein in 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA and 0.1% NP-40. Washing was performed with the same buffer. GST- and His-tagged proteins were produced by standard procedure and purified using glutathione sepharose (GE Healthcare) and Ni-NTA agarose (Qiagen), respectively. GST-4EK119A protein was expressed and purified as described by (40). For GST pull-down experiments purified GST-4EK119A or GST (200 μg) was bound to 200 μl of MagneGST beads (Promega) and incubated with 100 μg of HEK293 total RNA in RNP buffer (20 mM Tris–HCl pH 7.5, 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, 10% glycerol, 100 U RNasin/ml, anti-protease cocktail) for 2 h at 4°C. Beads were washed five times with RNP buffer. The RNAs present in the flow-through or on the beads were extracted by phenol/chloroform and precipitated.
Yeast-two hybrid interaction tests
For yeast-two hybrid interaction (Y2H) assays, appropriate pGBKT7 (DB) or pGADT7 (AD) plasmids were co-transformed into AH109 (Clontech Laboratories, Inc.) and plated on triple selective media (-Leu -Trp -Ade).
Polysome analysis
Polysome profiles were analyzed on sucrose gradients. HEK293 cells were cultivated up to 80% confluence. 2 × 106 cells were collected, washed in 3 volumes PBS and incubated in the presence of 150 μg/ml cycloheximide for 20 min on ice. Lysis of the cells was performed in polysome buffer (PB) containing 10 mM HEPES–NaOH pH 7.5, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 100 units RNasin/ml, 400 μM VRC, protease inhibitors and 150 μg/ml cycloheximide or in polysome denaturing buffer (PDB) containing 10 mM HEPES–NaOH pH 7.5, 10 mM potassium acetate, 0.5 mM magnesium acetate, 5 mM DTT. Cytoplasmic extracts were loaded onto a linear 7–47% sucrose gradient prepared in PB or PDB. Polysomes were separated by 2.5 h centrifugation at 37 000 rpm using a Beckmann SW41 rotor. Gradients were monitored by following absorbance at 254 nm. RNAs were extracted and analyzed by RT-qPCR as described previously.
RESULTS
A subclass of selenoprotein mRNAs harbor hypermethylated caps and show reduced affinity for eIF4E
Having previously established that selenoprotein mRNPs and sn-, snoRNPs share a common 3′ assembly pathway (27), we asked whether selenoprotein mRNAs could consequently undergo similar 5′ maturation events. Indeed, unlike mRNAs, PolII sn- and snoRNAs that are modified co-transcriptionally with a 5′ m7G cap, subsequently acquire a trimethylguanosine cap m2,2,7 (TMG) structure (15). We asked whether selenoprotein mRNAs could also bear similar cap modifications and could be recognized by the canonical translational machinery. A prerequisite to cap-dependent translation is the recognition of the eukaryotic mRNA cap structure by the translation initiation complex eIF4F (composed of the three subunits eIF4E, eIF4A and eIF4G). eIF4E specifically recognizes the m7G moiety of the 5′ m7GpppN cap of eukaryotic mRNAs and was shown to poorly recognize TMG caps (41). Therefore, we first examined the ability of selenoprotein mRNAs to be recognized by eIF4E.
We performed glutathione-S-transferase (GST) pull-down experiments using total RNAs from HEK293FT cells and a high-affinity mutant of eIF4E, GST-eIF4EK119A (40). This mutant was developed for specific isolation of 5′ m7G-capped mRNAs and showed strict specificity but 10-fold higher affinity for the m7G cap. The RNA content of eIF4E bound and unbound fractions was determined by qRT-PCR analysis. For the detection of selenoprotein mRNAs, we used primers complementary to 12 out of the 25 selenoprotein mRNAs characterized in mammals (Figure 1A). β-actin, HPRT (hypoxanthine guanine phosphoribosyltransferase) and LDHA (lactate dehydrogenase A) mRNAs were used as the m7G-capped controls, U3 snoRNA and U2 snRNA for TMG-capped controls. Coherently, an average of 75% of the canonical β-actin, HPRT and LDHA mRNAs were recovered in the eIF4E bound fraction, whereas 74% of sn-, snoRNAs were found in the unbound fraction (Figure 1A). The results revealed that selenoprotein mRNAs showed differential binding patterns to eIF4E. The selenoprotein mRNAs of SelR, glutathione peroxidases 1 and 4 (GPx1, GPx4), SelM and SelW showed a distribution pattern similar to that of sn-, snoRNAs with only 20–35% of the mRNA recovered in the eIF4E bound fraction (Figure 1A). SelT, SelO, thioredoxin reductase 1 (TrxR1), Sel15, SelK selenoprotein mRNAs showed an intermediate pattern with over 50% of the mRNAs in the bound fraction (Figure 1A), whereas selenoprotein mRNAs comprising glutathione peroxidase 3 (GPx3) and selenoprotein N (SelN) were enriched up to 70% in the eIF4E bound fraction with patterns similar to non-selenoprotein mRNAs (Figure 1A).
Figure 1.
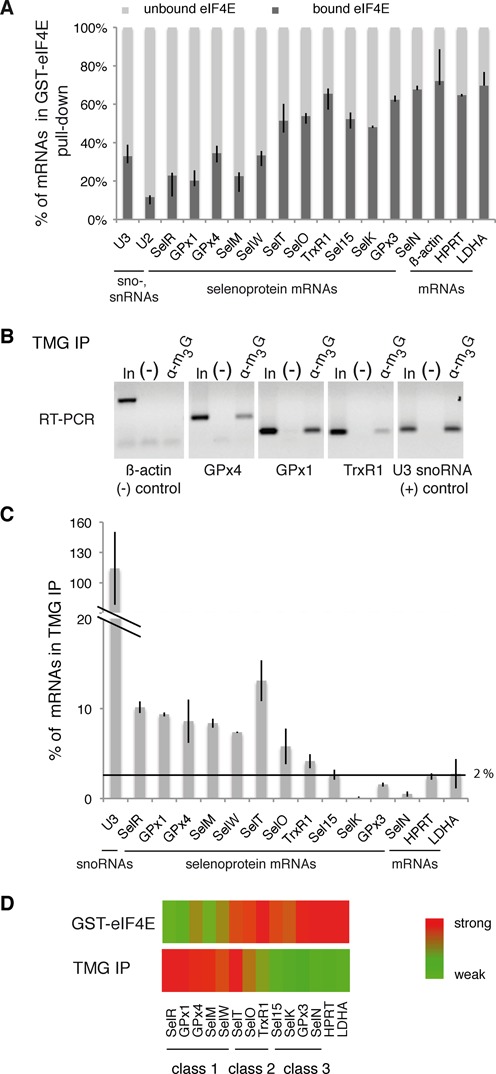
The cap of selenoprotein mRNAs is hypermethylated and poorly recognized by eIF4E. (A) Binding of recombinant GST-eIF4EK119A to HEK293 total RNA. GST-eIF4EK119A (or GST alone) was bound to glutathione beads and incubated with the extract. The percentage of mRNAs present in the bound and unbound fractions were determined separately by qRT-PCR and normalized to 100%. Asymmetric error bars represent the minimum and maximum observations for three independent biological replicates, reflecting intrinsic variability. SelR, GPx1, GPx4, SelM, SelW, SelT, SelO, TrxR1, Sel15, SelK, GPx3 and SelN are selenoprotein mRNAs. U3 snoRNA was used as a positive control, β-actin, HPRT and LDHA are housekeeping mRNAs used as negative controls. (B and C) Total RNA extracted from HEK293FT cells was immunoprecipitated with anti-TMG serum (α-m3G). Bound RNA was analyzed by (B) RT-PCR or (C) qRT-PCR. (−) Control without antibodies. In: input 10%. The graph represents the percentage of mRNAs in IP compared with the input RNA. Error bars represent standard deviation of an average of three independent experiments. The horizontal line represents the level of housekeeping mRNA binding (1–2% in average). See also Supplementary Figure S1 for specificity controls. (D) Heatmap representation of mRNA binding in GST-eIF4E pull-down and TMG IP experiments. The binding scale is represented to the right, maximum binding values in each set of experiments are represented in red and minimal binding values in green. Heat maps were generated with the MeV software. The three classes of selenoprotein mRNAs are indicated.
To explain the differential binding to eIF4E, we asked whether some of the selenoprotein mRNAs could bear 5′ modifications similar to sn- and snoRNAs as a trimethylguanosine cap m2,2,7 (TMG) structure. To answer this question, RNAs extracted from HEK293FT cells were immunoprecipitated with the highly specific anti-TMG cap R1131 serum that was demonstrated not to recognize monomethylated caps (38,39) (see also Supplementary Figure S1 for specificity validation of the antibody). The immunoprecipitated RNAs were extracted and analyzed by RT-PCR. Results showed that anti-TMG antibodies recognized GPx1, GPx4 and TrxR1 selenoprotein mRNAs as well as U3 snoRNA used as the control but not the m7G-capped β-actin mRNA, indicating that the cap of these endogenous selenoprotein mRNAs is hypermethylated (Figure 1B). Quantitative RT-PCR analysis revealed that seven selenoprotein mRNAs, namely SelR, GPx1, GPx4, SelM, SelW, SelT and SelO, were specifically immunoprecipitated with anti-TMG antibody (Figure 1C) while TrxR1 was only weakly recognized. These results correlate with the GST-eIF4E pull-down experiments, as these TMG-capped selenoprotein mRNAs correspond to those poorly recognized by eIF4E. TrxR1, SelT and SelO, that show an intermediate binding pattern of eIF4E, are consistently recognized in the TMG-IP (Figure 1A, C). Four selenoprotein mRNAs were not pulled-down over the background (Sel15, GPx3, SelN and SelK) in TMG immunoprecipitations (Figure 1C); they were consistently bound by eIF4E (Figure 1A). As expected, the U3 snoRNA positive control was fully retained but not the control HPRT or LDHA housekeeping mRNAs (Figure 1C). A significant fraction (5–15%) of the TMG-capped selenoprotein mRNAs was recovered in the anti-TMG immunoprecipitation. This precipitation is weaker compared with U3 snoRNA (100%) but is nevertheless consistent with previously reported levels for snRNAs (39).
Determinants such as the larger size, under-representation of selenoprotein mRNAs versus sno-, snRNAs as well as the shorter half-life of selenoprotein mRNAs compared to sn(o)RNAs may contribute to the lower recovery of selenoprotein mRNAs in the immunoprecipitation. It is also envisageable that different mRNA folded structures in the 5′UTR could lead to differential recognition of individual mRNAs by the antibody.
TMG-IP experiments, in combination to the GST-eIF4E pull-down results, suggest that a substantial fraction of selenoprotein mRNAs possesses a trimethylated guanosine cap. Altogether, our results reveal an inverse relationship between eIF4E binding and TMG capping (Figure 1D) and suggest the existence of three classes of selenoprotein mRNAs. In the first class, <50% of the selenoprotein mRNAs are bound to eIF4E and the TMG-IP efficiency is >5%. These mRNAs include SelR, GPx1, GPx4, SelM and SelW (Figure 1D); they harbor a hypermethylated cap and are not recognized efficiently by the translation factor eIF4E. A second class of selenoprotein mRNAs showed an intermediate pattern with >50% of the mRNAs bound by eIF4E and >5% TMG-IP efficiency; this class includes selenoprotein mRNAs such as SelT, SelO and TrxR1 that can possibly bear both types of caps (Figure 1D). Finally, the third class represents selenoprotein mRNAs—Sel15, SelK, GPx3 and SelN—(Figure 1D) bound with <5% in the TMG-IP, that are m7G-capped and recognized by eIF4E. Selenoprotein mRNAs thus appear to be subjected to differential 5′ processing events.
Tgs1 hypermethylates the cap of selenoprotein mRNAs
The trimethylguanosine synthase 1 (Tgs1) converts the m7G cap of sn- and snoRNA precursors into the functional TMG cap (21,24). This modification step plays a critical role in targeting sn- and snoRNA precursors to their final maturation and functional site (15). Tgs1 is present in a large U3 snoRNA processing complex that also contains components of the conserved RNP assembly complex linked to Hsp90 (27). Because selenoprotein mRNAs are assembled into mRNPs by the same molecular machinery (27), we asked whether Tgs1 was linked to selenoprotein mRNP biogenesis. Tgs1 exists as two molecular species: a full-length, mostly cytoplasmic (Tgs1 LF) and a shorter nuclear isoform (Tgs1 SF) (23). First, we examined if Tgs1 could be associated to selenoprotein mRNPs in vivo and performed immunoprecipitation of RNA–protein complexes from HEK293FT cells with an anti-Tgs1 antibody that recognizes both isoforms (Figure 2A). RNAs associated with endogenous Tgs1 were detected by RT-PCR. Several selenoprotein mRNAs, such as GPx1 and 4 as well as TrxR1 were specifically associated with Tgs1 in vivo, whereas this was not the case for the β-actin mRNA; U3 snoRNA was the positive interaction control that binds both Tgs1 isoforms (Figure 2A). Whether the interaction of Tgs1 could be generalized to all selenoprotein mRNAs was investigated by analyzing the RNA content of the Tgs1 immunoprecipitation reaction (IP) by microarrays. Because we did not have antibodies specific for either form of Tgs1, we used two HeLa cell lines that stably express either Tgs1 LF or Tgs1 SF with a 3XFlag tag, and performed the IPs using anti-Flag beads, using the parental cell line to perform control IPs (37). On average, selenoprotein mRNAs were significantly enriched in the IPs versus non-selenoprotein mRNAs that were not retained (Table 1). We found that both Tgs1 LF and Tgs1 SF associated to selenoprotein mRNAs (Table 1 and Figure 2B); likewise and as expected from previous studies (23), Tgs1 LF preferentially recognized snRNAs whereas Tgs1 SF bound more strongly to snoRNAs in our experiment (Table 1 and Figure 2B, (37)). To date, 25 selenoprotein genes have been identified in humans (42). Among the 21 mRNAs expressed in our experiment, 14 were bound by Tgs1 LF and/or SF (Figure 2B), and 7 were not detected: TrxR1, SelH, SelI, SPS2, DI2, TrxR3 and GPx1-2. Binding efficiencies were variable, the strongest signals being observed for SelT, Sel15, GPx1-1 and SelM selenoprotein mRNAs for which the enrichment was similar to that seen for snRNAs and snoRNAs (Figure 2B). TrxR1 results show differences with those obtained in the endogenous Tgs1 RNA IP experiments (Figure 2A) most likely because qRT-PCR is more sensitive and specific than microarrays for the detection of the endogenous population of TrxR1 mRNAs. Binding of Tgs1 does not appear to strictly correlate with TMG-capping, suggesting that recruitment of the enzyme does not necessarily lead to cap modification. This is similar to the case of intronic snoRNAs which bind Tgs1 without subsequent cap modification. Altogether our results show that selenoprotein mRNAs interact with the two isoforms of Tgs1. The affinity of Tgs1 LF for selenoprotein mRNAs appears only slightly higher than for Tgs1 SF and resembles that of snRNAs.
Figure 2.
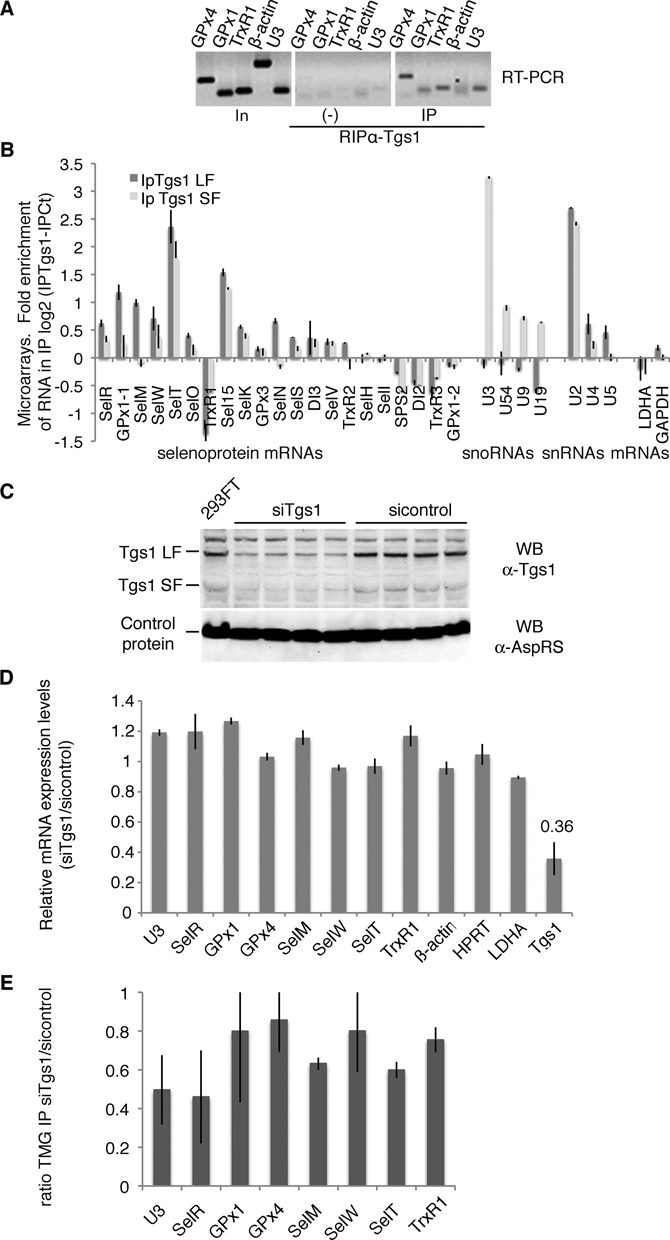
Tgs1 is involved in selenoprotein mRNA cap hypermethylation. (A) Tgs1 interacts with selenoprotein mRNAs in vivo. HEK293FT cells transfected with SBP2 were immunoprecipitated using anti-Tgs1 antibodies. Bound RNAs were detected by RT-PCR. In: input 15%; (−) no antibodies; U3: positive control; β-actin: negative control. (B) Tgs1 LF and SF associate with selenoprotein mRNAs. Isogenic HeLa cells stably expressing 3XFlag tagged Tgs1 LF and SF were used for anti-FLAG IPs, the RNA content was analyzed on microarrays. The control was the parental cell line that did not express any tagged protein. The graph represents the fold RNA enrichment in the IP in a log2 scale. Dark bars: Tgs1 LF IP; gray bars: Tgs1 SF IP. Data of all the expressed selenoprotein mRNAs are represented followed by examples of snoRNAs (U3 to U19), snRNAs (U2, U4, U5) and housekeeping mRNAs (LDHA and GAPDH). GPx1-1 and GPx1-2 are splice variants of GPx1. (C) Tgs1 inactivation by siRNA. Tgs1 mRNA was decreased to 36% which resulted in 35% of residual Tgs1 protein. Western blot detection of Tgs1 was performed using anti-Tgs1 antibodies; anti-AspRS antibodies were used as a control. siRNAs directed against firefly luciferase were used as controls (sicontrol). (D) siTgs1 did not have any effect on the steady state level of selenoprotein mRNAs. qRT-PCR was used to determine relative expression levels by the ΔΔCt method. (E) siTgs1 reduces hypermethylation efficiency. RNA-IP using anti-TMG serum was performed as described in Figure 1 under siTgs1 and sicontrol conditions. IP ratios between siTgs1 and sicontrol are represented by the histogram and deduced from Supplementary Figure S2. Error bars represent standard deviation of an average of three independent experiments.
Table 1. Microarray analysis of RNA immunoprecipitations using Tgs1 SF and LF isoforms.
| log2 (IP Tgs1-IPCt) | Tgs1-LF-Cont | Tgs1-SF-Cont |
|---|---|---|
| Seleno mRNA (20 genes) | 0.32 (0.0038) | 0.16 (0.0001) |
| snRNA (9 genes) | 0.82 (0.0031) | 0.25 (0.0031) |
| C/D snoRNA (112 genes) | −0.34 (0.0033) | 1.2 (0.0019) |
| H/ACA snoRNA (108 genes) | −0.11 (0.0037) | 0.61 (0.0001) |
| mRNA (18685 genes) | −0.022 (0.2612) | −0.027 (0.1910) |
Average of fold enrichment for each RNA family in IP (Tgs1 SF or LF) versus control measured in duplicate microarray analysis for Tgs1SF and Tgs1 LF RNA IPs. P-values were obtained from Mann–Whitney tests and are indicated in brackets. The average expression level of the entire set of selenoprotein mRNAs expressed in the HeLa cell line is indicated (20 expressed mRNAs among the 25 selenoprotein mRNA genes). The average expression level of the entire set of non-selenoprotein mRNAs is also indicated.
Having shown the interaction of Tgs1 with selenoprotein mRNAs, we further investigated whether this enzyme was indeed responsible for hypermethylation of the selenoprotein mRNA cap. To this end, Tgs1 was knocked-down by RNA interference (RNAi) in HEK293FT cells (Figure 2C). Tgs1 mRNA was decreased to 36% (Figure 2D), which resulted in 35% of residual Tgs1 protein (Figure 2C). The drop of Tgs1 did not affect selenoprotein mRNA steady state levels (Figure 2D) but led to a global reduction of TMG-IP efficiency in correlation with the level of Tgs1 knock-down (Figure 2E and Supplementary Figure S2). Individual selenoprotein mRNAs responded differentially to Tgs1 inactivation. TMG-IP efficiency dropped down to 45% in the case of SelR (Figure 2E), comparably to the U3 snoRNA positive control. Milder effects were observed in the case of GPx1, GPx4, SelW and TrxR1 as the TMG-IP efficiency was only reduced to 80%. This can be attributed to residual Tgs1 and suggests that its depletion does not affect all selenoprotein mRNAs to the same extent. Hierarchy of selenoprotein expression and variations in selenoprotein mRNA stability have been reported in numerous studies, particularly with regard to the glutathione peroxidase family (43–46). Individual selenoprotein mRNAs respond in a unique fashion to various imbalances, including Tgs1 depletion. Altogether, these results indicate that Tgs1 is involved in hypermethylation of the cap of the majority of selenoprotein mRNAs. Our data therefore conclude that the cap hypermethylation activity of Tgs1 is thus not only restricted to that of sn- and snoRNA substrates.
Tgs1 is recruited to selenoprotein mRNAs predominantly via the assembly chaperone SMN but also by the core protein Nop58
The recruitment mode of Tgs1 is dependent on the nature of its RNA targets. In the case of sn- and snoRNAs, the best characterized Tgs1 targets, two different strategies allow Tgs1 isoforms to be recruited to the m7G cap. Recruitment of Tgs1 LF to snRNAs is dependent on both a major assembly factor, the Survival of Motor Neuron (SMN) complex, and the core protein SmB, whereas the core proteins Nop56 and Nop58 recruit Tgs1 SF to box C/D snoRNAs (23,47,48). To determine if Tgs1 could be recruited to selenoprotein mRNAs via one of these two pathways, we separately inactivated expression of the Nop58 and SMN proteins by RNAi. This resulted in residual levels of 2 and 8% of Nop58 and SMN proteins, respectively, but did not affect expression of Tgs1 LF, SF and of the selenoprotein biosynthesis factor SBP2 (Figure 3 and Supplementary Figures S3A and S4A). Hypermethylation of the selenoprotein mRNA cap was affected in each case (Figure 3A and B). Cap hypermethylation was decreased in Nop58 depleted-cells by 60–80% (Figure 3A), but was totally abolished after siSMN treatment (Figure 3B). The steady state levels of selenoprotein mRNAs, Tgs1 and SBP2 mRNAs, as measured by qRT-PCR were not significantly affected in both cases (Figure 3C and D). As expected, Nop58 depletion strongly affected the stability of U3 snoRNA (Figure 3C) while SMN knock-down deregulated expression of U2 (Figure 3D). This is consistent with earlier reports showing that SMN deficiency or knock-down altered the stoichiometry of snRNAs in mammalian cells and could even increase their level, particularly in the case of U2 snRNA (49). We conclude that both Nop58 and SMN participate in hypermethylation of the selenoprotein mRNA cap. The strong effect of SMN inactivation suggests that recruitment of Tgs1 LF likely constitutes the major pathway.
Figure 3.
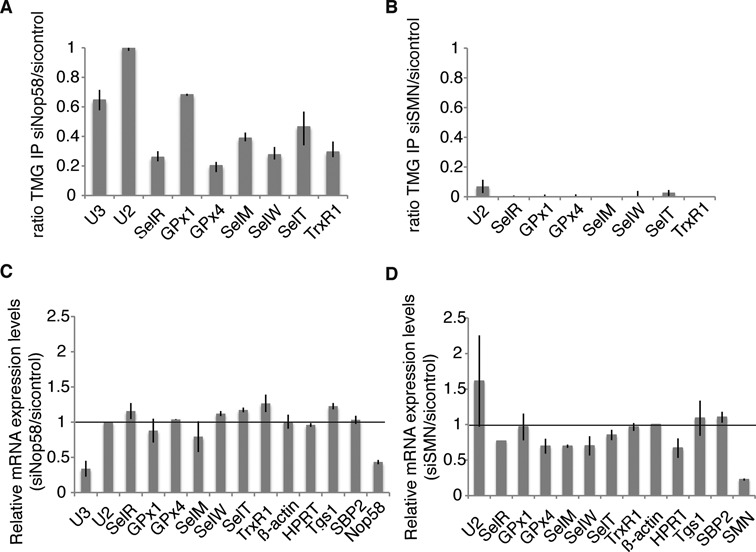
Nop58 and SMN contribute to selenoprotein mRNA hypermethylation. (A and B) Nop58 and SMN inactivation. RNA-IP using anti-TMG serum was performed as described in Figure 1 under siNop58 or siSMN and sicontrol conditions. IP ratios between siNop58, siSMN and sicontrol conditions are represented by the histogram bars and deduced from Supplementary Figures S3B and S4B, respectively. (C and D) siNop58 and siSMN have no effect on the steady state level of selenoprotein mRNAs nor on Tgs1 or SBP2 mRNAs. Relative expression levels were determined by qRT-PCR. U2 snRNA and β-actin were used as normalizers, respectively.
SBP2 interacts with Nop58 and SMN in vivo and recruits Tgs1
Next, we asked how Tgs1 isoforms, SMN and Nop58 could be recruited to selenoprotein mRNAs. As SBP2 plays central roles in selenoprotein biosynthesis by binding to the selenoprotein mRNA SECIS element (26), we first tested if SBP2 could interact with Tgs1. Endogenous protein complexes associated with SBP2 were immunoprecipitated from HeLa cell extracts using antibodies against the N-terminal region of SBP2. Western blotting using anti-Tgs1 antibody revealed the association of SBP2 with endogenous Tgs1 LF, and little, if any, with Tgs1 SF (Figure 4A). No association was seen with the control protein Hsp70 (Figure 4A). To confirm this finding, we co-transfected SBP2 with either GFP-Tgs1 LF or GFP-Tgs1 SF and immunoprecipitated the total cell lysates with anti-GFP antibodies. As shown in Figure 4B, SBP2 associated with Tgs1 LF in vivo, while binding to Tgs1 SF was not detected. Y2H tests confirmed protein–protein interactions between SBP2 and both Tgs1 LF and Tgs1 SF (Figure 4C). Finally, in vitro binding assays between 35S-labeled SBP2 proteins expressed in micrococcal nuclease treated rabbit reticulocyte lysate (RRL) and the recombinant His-Tgs1 LF protein produced in Escherichia coli, showed quantitative binding of SBP2 to Tgs1 LF. However, little interaction was observed between His-SBP2 and 35S-Tgs1 SF (Figure 4D). Similar results were obtained using 35S-labeled proteins translated in RNase-treated bacterial S30 lysates (Figure 4D). Recombinant SBP2 protein preparations, produced from bacterial heterologous systems, do not contain snRNAs, snoRNAs and corresponding RNP proteins, nor components dedicated to synthesis of mammalian selenoprotein. This excludes the possibility that the protein–protein interactions shown are RNA dependent or mediated via protein components of the sn-, sno- and selenoprotein mRNP assembly machinery. Thus, Tgs1 LF and SBP2 associate in vivo and in vitro and this association is RNA independent. Because SMN and Nop58 interact with Tgs1 and appear to be required for selenoprotein mRNA cap-hypermethylation in vivo (Figure 3A and B), we next analyzed whether Nop58 and SMN also interacted with SBP2. SBP2 was co-transfected with Nop58-YFP in HEK293FT cells and we immunoprecipitated the total cell lysates with anti-GFP beads. As shown in Figure 4E, SBP2 interacted with Nop58 in vivo. We also verified the interaction in vitro by GST pull-down experiments (Figure 4F) and found that (35S-Met)-SBP2 produced in RRL or bacterial S30 extracts bound strongly to GST-Nop58 independently of RNA. In addition, RNA-IPs in Nop58-YFP transfected cells revealed that GPx1 and GPx4 mRNAs were specifically associated with Nop58 in vivo (Supplementary Figure S5). To verify the link between SMN and selenoprotein mRNPs, we co-transfected GFP-SBP2 or GFP-SMN and SBP2 in HEK293FT cells for co-IP analysis. We found that GFP-SBP2 was able to interact with endogenous SMN and conversely that GFP-SMN interacted with transfected SBP2 in vivo (Figure 4G); these interactions were resistant to RNase A treatment and are therefore RNA independent (Figure 4G). GST pull-down experiments confirmed the interaction between (35S-Met)-SBP2 and GST-SMN in vitro (Figure 4H). We conclude that SBP2 plays a central role by interacting with both SMN, Nop58 and Tgs1. The recruitment of Tgs1 is likely to be dependent on the formation of the ternary complexes between SBP2/SMN/Tgs1 LF on one side, and SBP2/Nop58/Tgs1 SF on the other.
Figure 4.
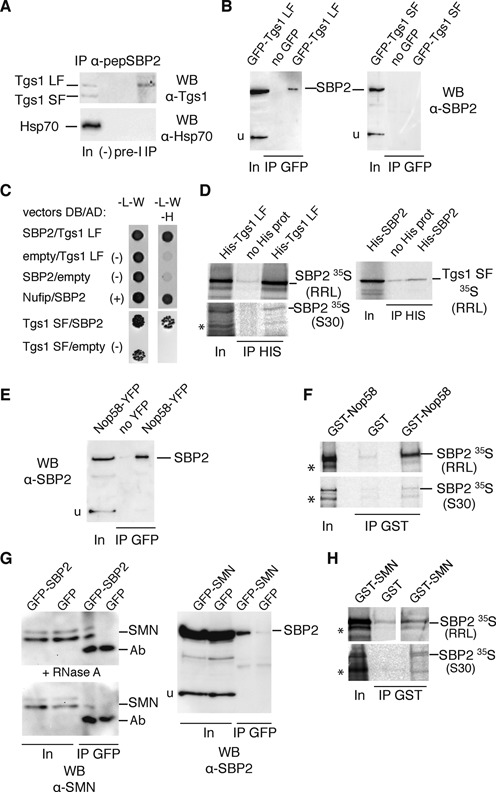
Tgs1 is recruited to selenoprotein mRNAs via interactions with SBP2. (A) Immunopurification of endogenous SBP2 from HeLa cytoplasmic extracts using antipeptide antibodies (α-pepSBP2) against amino acids 380–852. In: input 4%; (−) beads without antibodies; PreI: beads with preimmune serum; IP: beads with antibodies. (B) Co-immunoprecipitations using anti-GFP beads and HEK293FT cells transfected with SBP2 and GFP-Tgs1 or SBP2 alone (no GFP). (C) Y2H interaction tests performed in S. cerevisiae between SBP2 and Tgs1 LF or SF. (−) Controls with empty DNA binding (DB) or activation domain (AD) fusion vectors; (+) positive interaction control between SBP2 and Nufip. (D) Binding of recombinant His-Tgs1 to in vitro translated (35S-Met)-SBP2 using rabbit reticulocyte lysate (RRL) and bacterial S30 extracts (S30). His-Tgs1 was bound to protein A-Sepharose using anti-His antibodies. (E) Co-immunoprecipitations using anti-GFP beads and HEK293FT cells transfected by SBP2 and Nop58-YFP. (F) Binding of recombinant GST-Nop58 to in vitro translated (35S-Met)-SBP2 using RRL and S30 extracts. GST-Nop58 (or GST alone) was bound to glutathione beads and incubated with the extract. (G) Co-immunoprecipitations using anti-GFP beads and HEK293FT cells transfected by GFP-SBP2 or GFP-SMN and SBP2 without tag. (+ RNase A) co-immunoprecipitation performed in the presence of RNase A. (H) Binding of recombinant GST-SMN (or GST alone) to in vitro translated (35S-Met)-SBP2 using RRL and S30 extracts. Asterisks denote frequently encountered hSBP2 proteolysis fragments. u represents non-specific protein signal revealed by the anti-SBP2 antibodies and that serves as an internal control. Ab: cross-reactivity signal from antibodies bound to the beads.
Hypermethylated-capped selenoprotein mRNAs localize to the cytoplasm and are polysome-associated
The discovery of hypermethylated-capped selenoprotein mRNAs raises the fundamental question of their ability to be present and translated in the cytoplasm. Indeed, since the TMG cap is a part of the nuclear localization signal for snRNAs, it could also be envisaged that hypermethylation leads to sequestration of selenoprotein mRNAs in the nucleus. We thus performed subcellular fractionation of HEK293 cells (Figure 5A) followed by TMG-IP experiments and determined the percentage of each TMG-capped mRNA in the cytoplasm compared with the nucleus (Figure 5B). To assess the quality of the nuclear-cytoplasmic fractions, we have performed western blot analysis using antibodies directed against the transcription factor ZNF143 (a strictly nuclear protein (50)) and the cytoplasmic ribosomal protein rpS21 (Figure 5A). Results showed that globally selenoprotein mRNAs are more abundant in the cytoplasmic than the nuclear compartment; indeed 70–84% of TMG-capped selenoprotein mRNAs are found in the cytoplasm (Figure 5B). As expected, both U3 snoRNA and U6 snRNA were predominantly immunoprecipitated from the nuclear fraction. U6 was used as a control because it does not exit the nucleus during biogenesis; although it is not TMG-capped, it is always recovered in TMG-IPs because of its interaction with U4 snRNA (51). Non-selenoprotein mRNA controls were not recognized by the anti-TMG antibody. These results strongly suggest that hypermethylated-capped selenoprotein mRNAs are present in the cytoplasm, a prerequisite to their translation. To evaluate the ability of selenoprotein mRNAs to associate with actively translating ribosomes, we analyzed the polysome distribution of endogenous selenoprotein mRNAs. Cytoplasmic extracts of cycloheximide-treated HEK293 cells (blocking translation elongation) were fractionated on linear 7–47% sucrose gradients and the abundance of individual mRNAs in each fraction was measured by qRT-PCR (Figure 6A–C). The profiles revealed that all the endogenous selenoprotein mRNAs tested, except TrxR1, sedimented to fractions of lower molecular weight than non-selenoprotein mRNAs. The peak of selenoprotein mRNA population was found in fractions 10–26 in close proximity to the 80S monosomes (Figure 6B), whereas β-actin, HPRT and LDHA mRNAs sedimented in the heavier fractions 5–16 containing the polysomes (Figure 6C). As expected, U3 snoRNA was not found in polysomes (Figure 6C). These results indicate that fewer ribosomes are loaded on selenoprotein mRNAs, consistent with elongation pausing at the UGA Sec codon as previously reported for GPx4 (52). TrxR1 selenoprotein mRNA is the only mRNA found in heavier polysome fractions (see Figure 6B). This correlates with the fact that TrxR1 is the sole mRNA tested for which the UGA Sec codon is at the antepenultimate position before the stop codon; in all the other selenoprotein mRNAs of our test panel, the UGA Sec codon resides between 39 (SelW) and 285 nucleotides (SelR) downstream of the start codon. Worth of note, the proportion of free versus ribosome bound mRNAs is higher for selenoprotein than non-selenoprotein mRNAs. We next tested whether selenoprotein mRNAs present in polysomes bear hypermethylated caps. TMG-IP experiments were performed on pooled fractions 6-26 (polysomes) or 28-41 (RNP) that contain free or non-polysome associated RNAs. We reasonably considered that the two pools contained 100% of the RNAs. Immunoprecipitation yields dropped importantly after fractionation of the RNAs on sucrose gradients; nevertheless, the results clearly showed the presence of the hypermethylated selenoprotein mRNAs in the polysome pool (Figure 6D). Indeed, between 50% (SelT) and 80% (SelR) of the recovered TMG-capped mRNAs was present in the polysome fraction, the rest of the TMG-capped selenoprotein mRNAs being in the RNP fractions (Figure 6D). As expected, U3 snoRNA and U2 snRNA were predominantly recovered from the RNP pool (Figure 6D). The non-selenoprotein mRNA controls were not recognized by anti-TMG antibodies (see also Figure 1C). When RNA fractionation was performed under low magnesium concentration leading to ribosome dissociation into subunits (Figure 6A), the signal of selenoprotein mRNA TMG-IP was shifted to the RNP fractions (Figure 6E). Altogether these results show that hypermethylated selenoprotein mRNAs are found in polysomes and are therefore translated.
Figure 5.
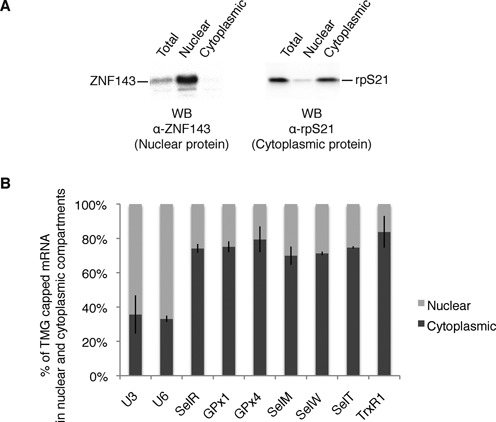
Hypermethylated selenoprotein mRNAs are more abundant in the cytoplasm. (A) Western blot analysis of the nuclear and cytoplasmic extracts using antibodies directed against the nuclear restricted ZNF143 transcription factor and the cytoplasmic rpS21. (B) RNA was extracted from total, nuclear and cytoplasmic extracts of HEK293FT cells; RNA-IPs using anti-TMG antibodies were performed and analyzed as described in Figure 1. The relative percentage of TMG-capped mRNA in both compartments is represented. Error bars represent standard deviation of an average of two independent experiments. U3 snoRNA and U6 snRNA were used as controls for the quality of the fractionation.
Figure 6.
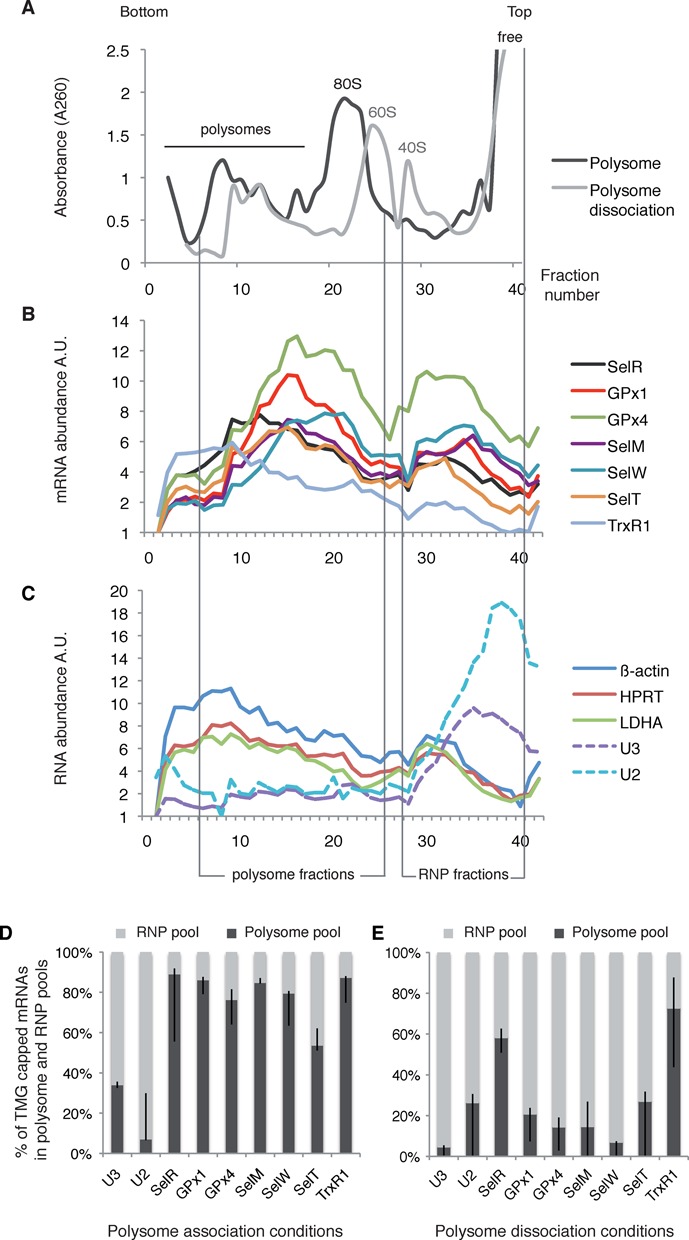
Polysome distribution of endogenous HEK293 selenoprotein mRNAs. (A) Cytoplasmic extracts from HEK293 cell were fractionated onto 7–47% (w/v) linear sucrose gradient and collected in 40 fractions. Typical absorbance profiles are shown and the positions of the polysomes, 80S, 60S, 40S ribosomal subunits as well as free RNA are indicated. Fractionation was performed in polysome association (black profile) or low magnesium dissociation conditions (gray profile). (B and C) The RNA content of each fraction was analyzed by qRT-PCR and the relative mRNA abundance was represented in arbitrary units A.U. Vertical bars indicate the position of the polysome and RNP fractions that were pooled and analyzed in (D). (B) Sedimentation profiles of selenoprotein mRNAs; (C) non-selenoprotein mRNAs, U3 snoRNA and U2 snRNA are represented. (D and E) Hypermethylated selenoprotein mRNAs co-fractionate with polysomes. RNA-IP using anti-TMG antibodies and qRT-PCR analysis was performed as described in Figure 1 in polysome association (D) and dissociation conditions (E). The amount of RNA immunoprecipitated from the polysome and RNP pool were determined separately by qRT-PCR and normalized to 100%. Error bars represent standard deviation of an average of two independent experiments.
Tgs1 activity, but not eIF4E, is required in vivo for translation of GPx1
In order to determine the functional importance of the TMG-cap modification by Tgs1 on selenoprotein synthesis in vivo, we constructed two stable cell lines capable of expressing HA tagged wild-type (wt) and mutant GPx1 selenoproteins. GPx1 was chosen as a model of class 1 selenoprotein mRNAs which bear a TMG cap and are poorly recognized by eIF4E. The mRNAs contained the complete natural 5′ and 3′UTRs and were under the control of a tetracycline inducible promoter (HA-GPx1 and HA-GPx1Cys, respectively). In HA-GPx1 cell lines, synthesis of GPx1 was selenium dependent (Supplementary Figure S6) and relied on translational recoding. In the HA-GPx1Cys mutant, the Sec codon was mutated to Cys. Expression of this construct is thus independent of the recoding mechanism. Translation of GPx1Cys was indeed unaffected by the selenium level (Supplementary Figure S6); it was used as a non-selenoprotein control. This system allowed us to analyze the effects of the depletion of biogenesis or translation factors on de novo synthesis of the GPx1 selenoprotein. We first inhibited expression of Tgs1 by siRNA using pools of four different, non-overlapping, siRNAs and two different siRNA conditions. After 48 h of siRNA treatment, the level of Tgs1 mRNA was reduced down to 52% (siRNA2) and the corresponding Tgs1 protein levels to 62% (Figure 7A). At this stage, expression of HA-GPx1 was selectively induced for an additional 12 h and we monitored the impact of Tgs1 depletion on de novo selenoprotein synthesis (Figure 7B). Partial Tgs1 depletion resulted in a statistically significant 47% drop of HA-GPx1 selenoprotein synthesis compared to the endogenous control protein AspRS that remained unaffected (Figure 7B). With a similar efficacy of siRNA treatments, the drop in protein synthesis was 66% in the case of SBP2 knock-down, a protein factor essential to the Sec incorporation process in vivo and no effect was observed in samples treated with control siRNAs (Figure 7B). Both the siTgs1 and siSBP2 effects could be rescued by transfection of SBP2 and Tgs1 expression plasmids 24 h after knockdown. Restoration of Tgs1 and SBP2 wt levels (Figure 7A) re-established 89 and 100% of HA-GPx1 expression, respectively (Figure 7B). Interestingly, depletion of neither Tgs1 nor SBP2 affected translation of the HA-GPx1Cys mutant (Figure 7B). In this case, synthesis of the HA-GPx1Cys protein does not rely on translational recoding events and becomes independent of the presence of SBP2, in agreement with depletion of SBP2 having no effect on protein production arising from this construct (Figure 7B). Altogether these results suggest that Tgs1 does play a functional role in selenoprotein synthesis in vivo, most likely by its direct action on selenoprotein mRNA cap hypermethylation.
Figure 7.
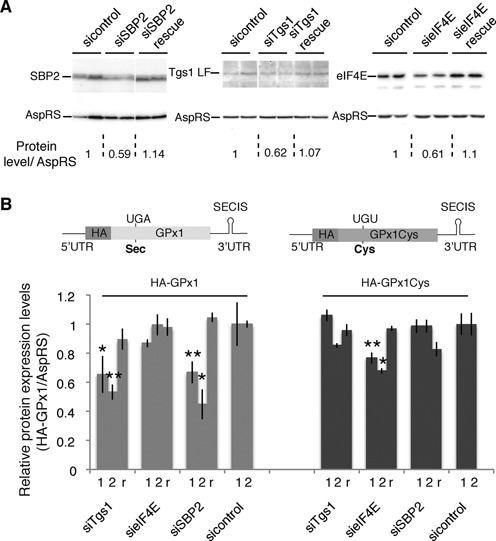
Tgs1, but not eIF4E, is required for de novo synthesis of GPx1. Stable cell lines expressing HA-GPx1 and HA-GPx1Cys mutant proteins under the control of an inducible promoter were used to monitor de novo selenoprotein synthesis following 48 h of Tgs1, eIF4E and SBP2 knock-down (siTgs1, sieIF4E and siSBP2). Two different siRNA conditions using 30 and 100 nM siRNA (marked 1 and 2 below the histogram) were analyzed for each gene. r indicates rescue experiments of siRNA2 conditions by transfection of a plasmid 24 h after siRNA. (A) Western blot analysis of siTgs1, sieIF4E and siSBP2 efficiency using antibodies directed against the targeted proteins. Levels of protein compared with the control are indicated below the panels and normalized against AspRS. (B) Quantification results of de novo HA-Gpx1 protein expression levels by western blot analysis using anti-HA antibodies. Normalization was performed using AspRS antibodies. Gray bars: HA-GPx1; dark bars: HA-Gpx1Cys. Error bars represent standard deviation of an average of four independent experiments. Asterisks indicate statistically significant differences between siTgs1, sieIF4E, siSBP2, rescues and corresponding sicontrol conditions. *P < 0.05 and **P < 0.005 based on Student‘s t-test. Expression constructs HA-GPx1 and HA-GPx1Cys, integrated in the stable cell lines, flanked by their natural GPx1 5′ and 3′UTRs as well as SECIS RNA, are represented above the histograms.
Because we showed that hypermethylation affected the recognition by eIF4E, we also examined the effects of eIF4E knock-down, using pools of four different siRNAs, on the in vivo expression of GPx1 using the same HA-GPx1 and HA-GPx1Cys inducible stable cell lines (Figure 7). eIF4E mRNAs were reduced to 58%. In this case, a 40% depletion of the eIF4E protein (Figure 7A) hardly affected HA-GPx1 selenoprotein synthesis, in agreement with our data showing that eIF4E only poorly recognized the GPx1 mRNA. In contrast, translation of the HA-GPx1Cys mutant was more affected and reduced between 23% (siRNA1) and 30% (siRNA2) (statistically significant, P = 0.0002 and P = 0.02 for siRNA1 and siRNA2, respectively) (Figure 7B). In this case, protein synthesis is independent of UGA Sec recoding and relies on canonical translation mechanisms. The results of eIF4E inactivation show opposite effects compared to the siTgs1 and siSBP2 control conditions that solely impacted UGA recoding (Figure 7B). The mild effect observed is consistent with the existence of strong homeostatic control mechanisms of eIF4E activity; indeed knockdowns have been reported to cause only minor reduction in translation (53). Rescue experiments of sieIF4E (Figure 7A) restored 98% of HA-Gpx1Cys expression (Figure 7B). These results suggest that eIF4E is not essential for the translation of hypermethylated-capped selenoprotein mRNAs.
DISCUSSION
Cellular mRNAs are known to be m7G-capped (1). Here we showed that mammalian mRNAs encoding selenoproteins bear a hypermethylated cap. Some mRNAs in Caenorhabditis elegans, Ascaris lumbricoides, Ciona intestinalis and Euglena gracilis acquire TMG caps but by spliced-leader (SL) trans-splicing, thus a radically different pathway than described here (54–57). Some Rev/RRE-dependent HIV-1 RNAs can also be TMG-capped by Tgs1, TMG capping representing in this case a regulation mechanism for selective expression (58). To assess the impact of TMG capping on selenoprotein mRNA export, stability and translation is challenging. Hypermethylation of selenoprotein mRNA cap may serve different purposes: dedicated assembly pathway for selenoprotein mRNPs and/or control of selenoprotein expression at the level of translation initiation or UGA Sec recoding. Our experiments reveal that selenoprotein mRNAs are subjected to differential 5′ processing and translational initiation events. Although some are substrates for Tgs1, harbor hypermethylated caps and are not recognized efficiently by the translation factor eIF4E, others are m7G-capped and recognized by eIF4E. These differences between selenoprotein mRNAs are not surprising as differential expression and stability regulation mechanisms have been reported for various subsets of selenoprotein mRNAs under conditions as different as selenium levels, type of SBP2–SECIS interactions, and sensitivity to NMD (nonsense mediated decay) (44,59–63). Control of gene-specific selenoprotein expression and differential regulation of UGA Sec recoding of a subset of selenoprotein mRNAs were recently shown to be selenium-dependent and mediated by the degree of tRNA[Ser]Sec Um34 methylation (43). Likewise, it is conceivable that cap hypermethylation could also be part of a regulation process of selenoprotein expression. As suggested by our results, hypermethylation does not necessarily occur equally on all selenoprotein mRNAs, possibly regulating translation initiation in a way that could contribute to the mechanisms of hierarchy of selenoprotein expression. Mechanisms and factors influencing the fate of selenoprotein mRNAs are far from being understood and so far no unified mechanism could be proposed for all selenoprotein mRNAs, at any stage of their mRNA life cycle. Our findings point to the existence of an additional level of complexity in the mechanism of selenoprotein synthesis residing at the stage of mRNP assembly, processing and translation initiation (Figure 8). This process is governed by the interactions between SBP2, Tgs1 and selenoprotein mRNAs. We established that the hypermethylase Tgs1 is recruited to selenoprotein mRNAs predominantly via SMN but also Nop58, and therefore probably belongs to two possible ternary complexes: SBP2/Nop58/Tgs1 or SBP2/SMN/Tgs1 (Figure 8). Interestingly, oxidative stress was identified as a novel regulation mechanism for selective selenoprotein synthesis (64). It was shown to induce nuclear shuttling of SBP2 and selenocysteine incorporation factors, and to promote assembly of selenoprotein mRNPs (32,33). It is conceivable that oxidative stress could also regulate hypermethylation of selenoprotein mRNAs by stimulating recruitment of the nuclear isoform of Tgs1.
Figure 8.
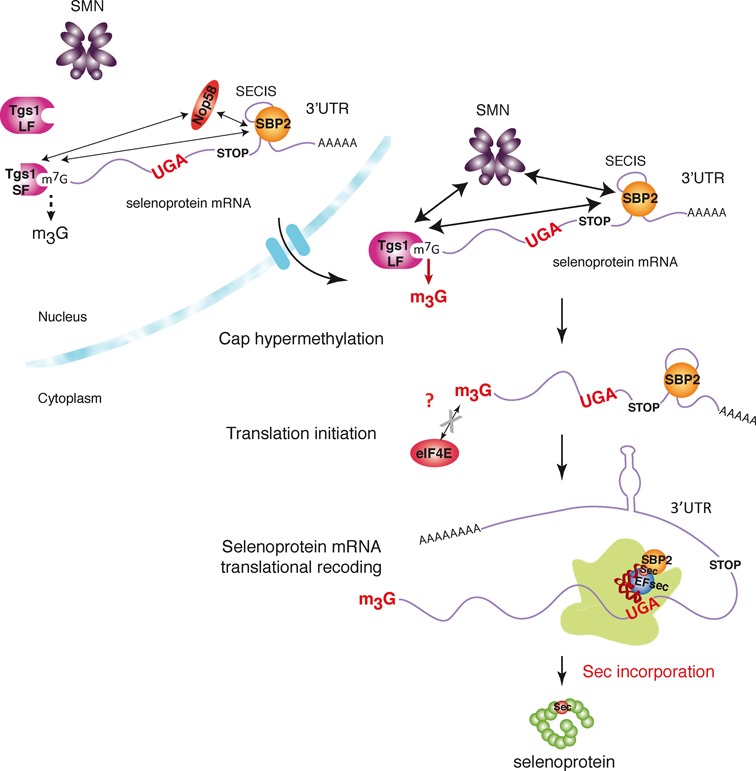
Model for cap hypermethylation and translation of selenoprotein mRNPs. The caps of selenoprotein mRNAs are hypermethylated by Tgs1. Tgs1 isoforms (Tgs1 SF and Tgs1 LF) interact with SBP2 and selenoprotein mRNAs and are recruited predominantly via the ternary complex SBP2/SMN/Tgs1 LF (thick arrows) but also via SBP2/Nop58/Tgs1 SF (thin arrows). Hypermethylated-capped selenoprotein mRNAs are localized to the cytoplasm and are polysome-associated but show reduced affinity for eIF4E. Tgs1 activity is required for Sec incorporation or translation initiation.
SBP2 is a member of the L7Ae family of RNA-binding proteins (65) that include the archaeal L7Ae and eukaryotic 15.5 kDa core proteins of the box C/D sRNPs and snoRNPs, respectively. We have previously shown that the structural motifs of the SECIS and box C/D RNAs as well as binding strategies governing the interaction of SBP2, L7Ae and 15.5 kDa to their cognate RNAs are similar and are controlled by a common conserved RNA chaperone complex (27,34,66). Interestingly, it is the L7Ae and 15.5 kDa proteins that play a key role in box C/D RNP assembly in Archaea and Eukarya, respectively, by specifically recognizing their cognate RNA motif. Moreover, in the case of U3 snoRNA, assembly and maturation was shown to be dependent on a large multiprotein complex that contains, in addition to the core proteins, components of the RNA chaperone complex but also RNA processing factors including Tgs1 (22,25). Likewise, our present data suggest that SBP2 could trigger the assembly of the cap modification complex by recruiting Nop58 or SMN in a similar manner, and this would ultimately lead to the binding of Tgs1 methylase for modification. Tgs1 SF is strictly nuclear but Tgs1 LF is present both in the nucleus and the cytoplasm. This suggests that m7G hypermethylation could occur in either of the two cellular compartments via two alternative pathways and could obey a fast and potentially reversible regulation process (Figure 8). Altogether, our results demonstrate that selenoprotein mRNAs indeed undergo a sophisticated assembly and 5’end maturation pathway similar to that of sn- and snoRNAs, and which involve common maturation factors and core proteins.
However, unlike TMG-capped sn- or snoRNAs, our results also show that hypermethylated-capped selenoprotein mRNAs can be found in the cytoplasm and in actively translating ribosomes, excluding nuclear retention as a consequence of cap hypermethylation. Hypermethylation by Tgs1 rather seems to be required for translational recoding events, as its knock-down reduced expression of GPx1. Our current data do not allow us to differentiate whether Tgs1 activity is required for Sec incorporation or translation initiation (Figure 8). It is conceivable that both mechanisms could be linked for optimal selenoprotein translation. Specialized translation initiation factors or alternative mechanisms are therefore likely to be involved in hypermethylated mRNA recognition. One possibility would be an IRES-dependent translation. Consistently, earlier reports showed that the cricket paralysis virus internal ribosome entry site is able to support Sec incorporation in a luciferase reporter construct, although the incorporation efficiency was decreased (67). Our results also showed a reduced affinity of the translation initiation factor eIF4E for selenoprotein mRNA cap structures. However, eIF4E variants that can accommodate the TMG cap were identified in the genome of C. elegans (68). Whether such a situation prevails for some selenoprotein mRNAs will be the aim of future investigations.
ACCESSION NUMBER
NCBI Gene Expression Omnibus accession number: GSE57625.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank R. Lührmann for the gift of HeLa cell extracts and antibodies, C.H. Hagedorn for the pGST-eIF4E plasmid, A. Schweigert for technical assistance, E. Westhof for invaluable input and constant support, P. Carbon and G. Eriani for helpful discussions and critical reading of the manuscript and J. Steitz for useful comments and critical reading of the manuscript.
Authors contributions: L.W, A.S.G.B., C.V., A.K., E.B. and C.A. designed research. A.S.G.B., L.W., C.V., M.L., A.T., S.B., F.M. and C.A. performed research. L.W., A.S.G.B., C.V., F.M., A.K., E.B. and C.A. analyzed data. C.A. wrote the paper.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
Present address:
Laurence Wurth, Gene Regulation, Stem Cells and Cancer Programme, Centre for Genomic Regulation and UPF, 08003 Barcelona, Spain.
FUNDING
Centre national de la recherche scientifique (CNRS); Agence Nationale de la Recherche [BLAN06–2_134682, ANR-2011-svse8 02501 to F.M.]. French Ministry of Research and the National Research Fund, Luxembourg [to L.W.]; Contract Doctoral of the University of Strasbourg [A.S.G.]. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shatkin A.J. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 2.Topisirovic I., Svitkin Y.V., Sonenberg N., Shatkin A.J. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA. 2011;2:277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 3.Izaurralde E., Lewis J., Gamberi C., Jarmolowski A., McGuigan C., Mattaj I.W. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 4.Kohler A., Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell. Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell P., Tollervey D. mRNA turnover. Curr. Opin. Cell Biol. 2001;13:320–325. doi: 10.1016/s0955-0674(00)00214-3. [DOI] [PubMed] [Google Scholar]
- 6.Wilusz C.J., Wormington M., Peltz S.W. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 7.Houseley J., Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 8.Fortes P., Inada T., Preiss T., Hentze M.W., Mattaj I.W., Sachs A.B. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell. 2000;6:191–196. [PubMed] [Google Scholar]
- 9.McKendrick L., Thompson E., Ferreira J., Morley S.J., Lewis J.D. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol. Cell. Biol. 2001;21:3632–3641. doi: 10.1128/MCB.21.11.3632-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonenberg N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem. Cell Biol. 2008;86:178–183. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- 11.Calero G., Wilson K.F., Ly T., Rios-Steiner J.L., Clardy J.C., Cerione R.A. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol. 2002;9:912–917. doi: 10.1038/nsb874. [DOI] [PubMed] [Google Scholar]
- 12.Marcotrigiano J., Gingras A.C., Sonenberg N., Burley S.K. Cocrystal structure of the messenger RNA 5’ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 13.Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., Mattaj I.W. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 14.Darzynkiewicz E., Stepinski J., Ekiel I., Jin Y., Haber D., Sijuwade T., Tahara S.M. Beta-globin mRNAs capped with m7G, m2.7(2)G or m2.2.7(3)G differ in intrinsic translation efficiency. Nucleic Acids Res. 1988;16:8953–8962. doi: 10.1093/nar/16.18.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matera A.G., Terns R.M., Terns M.P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell. Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 16.Chari A., Paknia E., Fischer U. The role of RNP biogenesis in spinal muscular atrophy. Curr. Opin. Cell Biol. 2009;21:387–393. doi: 10.1016/j.ceb.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Fischer U., Luhrmann R. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science. 1990;249:786–790. doi: 10.1126/science.2143847. [DOI] [PubMed] [Google Scholar]
- 18.Fischer U., Sumpter V., Sekine M., Satoh T., Luhrmann R. Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J. 1993;12:573–583. doi: 10.1002/j.1460-2075.1993.tb05689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber J., Cronshagen U., Kadokura M., Marshallsay C., Wada T., Sekine M., Luhrmann R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 1998;17:4114–4126. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamm J., Darzynkiewicz E., Tahara S.M., Mattaj I.W. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990;62:569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- 21.Verheggen C., Lafontaine D.L., Samarsky D., Mouaikel J., Blanchard J.M., Bordonne R., Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–2745. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boulon S., Verheggen C., Jady B.E., Girard C., Pescia C., Paul C., Ospina J.K., Kiss T., Matera A.G., Bordonne R., et al. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol. Cell. 2004;16:777–787. doi: 10.1016/j.molcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 23.Girard C., Verheggen C., Neel H., Cammas A., Vagner S., Soret J., Bertrand E., Bordonne R. Characterization of a short isoform of human Tgs1 hypermethylase associating with small nucleolar ribonucleoprotein core proteins and produced by limited proteolytic processing. J. Biol. Chem. 2008;283:2060–2069. doi: 10.1074/jbc.M704209200. [DOI] [PubMed] [Google Scholar]
- 24.Mouaikel J., Verheggen C., Bertrand E., Tazi J., Bordonne R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell. 2002;9:891–901. doi: 10.1016/s1097-2765(02)00484-7. [DOI] [PubMed] [Google Scholar]
- 25.Watkins N.J., Lemm I., Ingelfinger D., Schneider C., Hobach M., Urlaub H., Luhrmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Allmang C., Wurth L., Krol A. The selenium to selenoprotein pathway in eukaryotes: more molecular partners than anticipated. Biochim. Biophys. Acta. 2009;1790:1415–1423. doi: 10.1016/j.bbagen.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Boulon S., Marmier-Gourrier N., Pradet-Balade B., Wurth L., Verheggen C., Jady B.E., Rothe B., Pescia C., Robert M.C., Kiss T., et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Copeland P.R., Fletcher J.E., Carlson B.A., Hatfield D.L., Driscoll D.M. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lescure A., Allmang C., Yamada K., Carbon P., Krol A. cDNA cloning, expression pattern and RNA binding analysis of human selenocysteine insertion sequence (SECIS) binding protein 2. Gene. 2002;291:279–285. doi: 10.1016/s0378-1119(02)00629-7. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi A., Schmitt D., Chapple C., Babaylova E., Karpova G., Guigo R., Krol A., Allmang C. A short motif in Drosophila SECIS Binding Protein 2 provides differential binding affinity to SECIS RNA hairpins. Nucleic Acids Res. 2009;37:2126–2141. doi: 10.1093/nar/gkp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinzy S.A., Caban K., Copeland P.R. Characterization of the SECIS binding protein 2 complex required for the co-translational insertion of selenocysteine in mammals. Nucleic Acids Res. 2005;33:5172–5180. doi: 10.1093/nar/gki826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papp L.V., Lu J., Striebel F., Kennedy D., Holmgren A., Khanna K.K. The redox state of SECIS binding protein 2 controls its localization and selenocysteine incorporation function. Mol. Cell. Biol. 2006;26:4895–4910. doi: 10.1128/MCB.02284-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Jesus L.A., Hoffmann P.R., Michaud T., Forry E.P., Small-Howard A., Stillwell R.J., Morozova N., Harney J.W., Berry M.J. Nuclear assembly of UGA decoding complexes on selenoprotein mRNAs: a mechanism for eluding nonsense-mediated decay? Mol. Cell. Biol. 2006;26:1795–1805. doi: 10.1128/MCB.26.5.1795-1805.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clery A., Bourguignon-Igel V., Allmang C., Krol A., Branlant C. An improved definition of the RNA-binding specificity of SECIS-binding protein 2, an essential component of the selenocysteine incorporation machinery. Nucleic Acids Res. 2007;35:1868–1884. doi: 10.1093/nar/gkm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frugier M., Ryckelynck M., Giege R. tRNA-balanced expression of a eukaryal aminoacyl-tRNA synthetase by an mRNA-mediated pathway. EMBO Rep. 2005;6:860–865. doi: 10.1038/sj.embor.7400481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pradet-Balade B., Girard C., Boulon S., Paul C., Azzag K., Bordonne R., Bertrand E., Verheggen C. CRM1 controls the composition of nucleoplasmic pre-snoRNA complexes to licence them for nucleolar transport. EMBO J. 2011;30:2205–2218. doi: 10.1038/emboj.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luhrmann R., Appel B., Bringmann P., Rinke J., Reuter R., Rothe S., Bald R. Isolation and characterization of rabbit anti-m3 2,2,7G antibodies. Nucleic Acids Res. 1982;10:7103–7113. doi: 10.1093/nar/10.22.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tycowski K.T., Aab A., Steitz J.A. Guide RNAs with 5’ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr. Biol. 2004;14:1985–1995. doi: 10.1016/j.cub.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Choi Y.H., Hagedorn C.H. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3’ poly(A) end phenotype. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7033–7038. doi: 10.1073/pnas.1232347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcotrigiano J., Gingras A.C., Sonenberg N., Burley S.K. X-ray studies of the messenger RNA 5’ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Nucleic Acids Symp. Ser. 1997:8–11. [PubMed] [Google Scholar]
- 42.Kryukov G.V., Castellano S., Novoselov S.V., Lobanov A.V., Zehtab O., Guigo R., Gladyshev V.N. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 43.Howard M.T., Carlson B.A., Anderson C.B., Hatfield D.L. Translational redefinition of UGA codons is regulated by selenium availability. J. Biol. Chem. 2013;288:19401–19413. doi: 10.1074/jbc.M113.481051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun X., Li X., Moriarty P.M., Henics T., LaDuca J.P., Maquat L.E. Nonsense-mediated decay of mRNA for the selenoprotein phospholipid hydroperoxide glutathione peroxidase is detectable in cultured cells but masked or inhibited in rat tissues. Mol. Biol. Cell. 2001;12:1009–1017. doi: 10.1091/mbc.12.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss Sachdev S., Sunde R.A. Selenium regulation of transcript abundance and translational efficiency of glutathione peroxidase-1 and -4 in rat liver. Biochem. J. 2001;357:851–858. doi: 10.1042/0264-6021:3570851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moriarty P.M., Reddy C.C., Maquat L.E. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol. Cell. Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paushkin S., Gubitz A.K., Massenet S., Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:305–312. doi: 10.1016/s0955-0674(02)00332-0. [DOI] [PubMed] [Google Scholar]
- 48.Mouaikel J., Narayanan U., Verheggen C., Matera A.G., Bertrand E., Tazi J., Bordonne R. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep. 2003;4:616–622. doi: 10.1038/sj.embor.embor863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Myslinski E., Krol A., Carbon P. ZNF76 and ZNF143 are two human homologs of the transcriptional activator Staf. J. Biol. Chem. 1998;273:21998–22006. doi: 10.1074/jbc.273.34.21998. [DOI] [PubMed] [Google Scholar]
- 51.Mottram J., Perry K.L., Lizardi P.M., Luhrmann R., Agabian N., Nelson R.G. Isolation and sequence of four small nuclear U RNA genes of Trypanosoma brucei subsp. brucei: identification of the U2, U4, and U6 RNA analogs. Mol. Cell. Biol. 1989;9:1212–1223. doi: 10.1128/mcb.9.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fletcher J.E., Copeland P.R., Driscoll D.M. Polysome distribution of phospholipid hydroperoxide glutathione peroxidase mRNA: evidence for a block in elongation at the UGA/selenocysteine codon. RNA. 2000;6:1573–1584. doi: 10.1017/s1355838200000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanagiya A., Suyama E., Adachi H., Svitkin Y.V., Aza-Blanc P., Imataka H., Mikami S., Martineau Y., Ronai Z.A., Sonenberg N. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol. Cell. 2012;46:847–858. doi: 10.1016/j.molcel.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liou R.F., Blumenthal T. trans-spliced Caenorhabditis elegans mRNAs retain trimethylguanosine caps. Mol. Cell. Biol. 1990;10:1764–1768. doi: 10.1128/mcb.10.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Doren K., Hirsh D. mRNAs that mature through trans-splicing in Caenorhabditis elegans have a trimethylguanosine cap at their 5’ termini. Mol. Cell. Biol. 1990;10:1769–1772. doi: 10.1128/mcb.10.4.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maroney P.A., Denker J.A., Darzynkiewicz E., Laneve R., Nilsen T.W. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: a role for spliced leader addition in translational efficiency. RNA. 1995;1:714–723. [PMC free article] [PubMed] [Google Scholar]
- 57.Hastings K.E. SL trans-splicing: easy come or easy go. Trends Genet.: TIG. 2005;21:240–247. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 58.Yedavalli V.S., Jeang K.T. Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14787–14792. doi: 10.1073/pnas.1009490107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lei X.G., Evenson J.K., Thompson K.M., Sunde R.A. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J. Nutr. 1995;125:1438–1446. doi: 10.1093/jn/125.6.1438. [DOI] [PubMed] [Google Scholar]
- 60.Low S.C., Grundner-Culemann E., Harney J.W., Berry M.J. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J. 2000;19:6882–6890. doi: 10.1093/emboj/19.24.6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Squires J.E., Stoytchev I., Forry E.P., Berry M.J. SBP2 binding affinity is a major determinant in differential selenoprotein mRNA translation and sensitivity to nonsense-mediated decay. Mol. Cell. Biol. 2007;27:7848–7855. doi: 10.1128/MCB.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Budiman M.E., Bubenik J.L., Miniard A.C., Middleton L.M., Gerber C.A., Cash A., Driscoll D.M. Eukaryotic initiation factor 4a3 is a selenium-regulated RNA-binding protein that selectively inhibits selenocysteine incorporation. Mol. Cell. 2009;35:479–489. doi: 10.1016/j.molcel.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Latreche L., Duhieu S., Touat-Hamici Z., Jean-Jean O., Chavatte L. The differential expression of glutathione peroxidase 1 and 4 depends on the nature of the SECIS element. RNA Biol. 2012;9:681–690. doi: 10.4161/rna.20147. [DOI] [PubMed] [Google Scholar]
- 64.Touat-Hamici Z., Legrain Y., Bulteau A.L., Chavatte L. Selective up-regulation of human selenoproteins in response to oxidative stress. J. Biol. Chem. 2014;289 doi: 10.1074/jbc.M114.551994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koonin E.V., Bork P., Sander C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme. Nucleic Acids Res. 1994;22:2166–2167. doi: 10.1093/nar/22.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allmang C., Carbon P., Krol A. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA. 2002;8:1308–1318. doi: 10.1017/s1355838202020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Donovan J., Copeland P.R. The efficiency of selenocysteine incorporation is regulated by translation initiation factors. J. Mol. Biol. 2010;400:659–664. doi: 10.1016/j.jmb.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keiper B.D., Lamphear B.J., Deshpande A.M., Jankowska-Anyszka M., Aamodt E.J., Blumenthal T., Rhoads R.E. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J. Biol. Chem. 2000;275:10590–10596. doi: 10.1074/jbc.275.14.10590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


