Abstract
As critical DNA structures capping the human chromosome ends, the stability and structural polymorphism of human telomeric G-quadruplex (G4) have drawn increasing attention in recent years. This work characterizes the equilibrium transitions of single-molecule telomeric G4 at physiological K+ concentration. We report three folded states of telomeric G4 with markedly different lifetime and mechanical stability. Our results show that the kinetically favored folding pathway is through a short-lived intermediate state to a longer-lived state. By examining the force dependence of transition rates, the force-dependent transition free energy landscape for this pathway is determined. In addition, an ultra-long-lived form of telomeric G4 structure with a much stronger mechanical stability is identified.
INTRODUCTION
In addition to the canonical double helix B-form structure, DNA can adopt other structures, such as the duplex structures Z-DNA, S-DNA, as well as triplex and tetraplex, etc. (1,2). The tetraplex structures formed on G-rich sequences, referred to as G-quadruplex (G4), have drawn extensive attention recently. Increasing evidences have suggested that such structures are ubiquitous in the genome and may play various crucial regulatory functions such as regulation of chromosome stability and gene transcriptions (3,4). More than 300 000 putative quadruplex sequences have been found in the human genome (5,6) and many of them are located within the biologically functional regions such as telomeres, the ribosomal DNA, the immunoglobulin switch region (7) and promoters (8).
The human telomeric G4 forms in a specialized region of eukaryotic chromosomes, called telomeres, at the ends of the chromosomes. Human telomeric DNA consists of several kilobases of double-stranded DNA (dsDNA) tandem repeats of 5′-TTAGGG sequence, ended with a terminal single-stranded 3′ overhang of 100–200 nucleotides (9). The terminal single-stranded DNA (ssDNA) has the potential to fold into G4 structures which caps the chromosomal ends (1). Besides protection of the 3′ overhang end of chromosome, telomeric G4 plays a critical role in inhibiting the activity of telomerase (10). Telomerase is a telomere binding enzyme, which elongates and regulates the length of telomeres (11). The telomerase activity has been reported to be much higher in cancer cells than in normal cells (12). Small molecules stabilizing telomeric G4 have been shown to effectively inhibit telomerase activity and has an anti-cancer activity (13). Thus, the stability of telomeric G4 is also crucial for anti-cancer drug developments.
Telomeric G4 is made of multiple stacked planes each consisting of four coplanar guanines linked through Hoogsteen base pairing with a centrally located monovalent cation, K+ or Na+ (14,15). The structures of telomeric G4 are highly polymorphic: different structures have been reported, including the parallel G4 with four parallel strands (14), the anti-parallel G4 with two diagonal anti-parallel strands (15) and hybrid G4 with mixed parallel and anti-parallel strands (16–18). The formation of G4 structures is strongly dependent on the type of monovalent cations. Because of the significantly higher intracellular concentration of K+ (typically ∼100 mM) than Na+ (typically ∼10 mM), K+ induced G4 structures are considered as the predominant structures under physiological conditions.
Many studies have been directed to measure the stability of K+ induced telomeric G4 using a variety of experimental approaches including circular dichroism (CD), UV absorption spectrum and fluorescence resonance energy transfer (FRET) (19–22). In such studies, the melting curve of telomeric G4 has been measured by changing temperature or K+ concentration. The free energy cost of unfolding of K+ induced telomeric G4, ΔG0, has been reported in a wide range of 3.4–14.8 kcal mol−1 (20,23,24). Consistent with telomeric G4 as a stable structure, a fast folding rate of 16–50 s−1 and a slow unfolding rate of 1.3 × 10−3 s−1 have been reported by quick changing K+ concentration (25,26).
As multiple structures may co-exist in the K+ solutions, more recently efforts have been directed to investigate dynamics of single telomeric G4 molecules in order to detect different species including short lifetime transitions. Multiple folded conformations have been reported by tracking the dynamics of the single-molecule FRET (smFRET) efficiency of a single tethered DNA containing the telomeric repeats (22). However, in that study destabilizing low K+ concentrations (<10 mM) were used to increase the transition rates in order to obtain sufficient number of transitions for statistical analysis. In physiological level of ∼100 mM K+, due to slow unfolding rates, smFRET has been applied to study the populations of folded G4 structures instead of transition dynamics, which have revealed existence of multiple folded conformations (22,27). The studies of the dynamics for telomeric G4 have not been carried out in physiological concentration range of K+ (∼100 mM).
The slow unfolding rate of telomeric G4 structures in physiological K+ concentration can be overcome by applying forces to the G4 molecules. The so-called Bell's model (28) describes the force-dependent unfolding rate as:
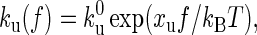 |
(1) |
where 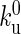 is the unfolding rate at zero force, xu is the distance from the native state to the transition state, kB is Boltzmann constant and T is the absolute temperature. The Bell's model shows that unfolding rate exponentially increases with force.
is the unfolding rate at zero force, xu is the distance from the native state to the transition state, kB is Boltzmann constant and T is the absolute temperature. The Bell's model shows that unfolding rate exponentially increases with force.
When the molecule is stretched with force linearly increasing with time at a loading rate of r, an unfolding force distribution, p(f), can be derived from the Bell's model:
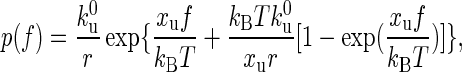 |
(2) |
which predicts a single force peak located at 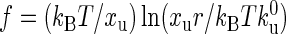 (29).
(29).
In previous single-molecule stretching experiments, telomeric G4 unfolding in 100 mM K+ has been achieved by stretching the molecule at certain loading rates, resulting in quick telomeric G4 unfolding typically at forces ∼21 pN at a loading rate of 5.5 pN/s (30). Folding of unfolded telomeric G4 was achieved by dropping force to a much lower value. Because unfolding and folding transitions do not occur at the same force, such experimental procedure does not determine ΔG0 directly. Instead, a non-equilibrium approach based on the Jarzynski's theorem (31) has been used to estimate ΔG0 to be around 9.8 ± 0.4 kcal mol−1 (30), which is close to the higher boundary reported from previous melting curve measurements. As applying Jarzynski's theorem requires a large number of stretch-relax procedures on the same G4 tether and an assumption that work involved in each stretch-relax procedure is close to kBT, ΔG0 obtained by this method still needs to be confirmed by more direct equilibrium measurements.
In this work, we directly observed the equilibrium transition of single-molecule telomeric G4 under constant forces at physiological relevant K+ concentration of 100 mM, using ultra-stable magnetic tweezers (32). From the long time trace that contains numerous folding and unfolding transitions at constant forces, multiple folded structures formed on telomeric G4 sequence have been identified and characterized by their lifetimes and mechanical stability.
MATERIALS AND METHODS
DNA construct and NMR
All DNA oligonucleotides were purchased form Integrated DNA Technologies (IDT). The 5′-thiol labeled 1449 bp and 5′-biotin labeled 601 bp dsDNA handles were prepared by PCR using DreamTaq DNA polymerase (Thermo Scientific) on lambda phage DNA template (New England Biolabs, NEB) using 5′-thiol and 5′-biotin primers. Both dsDNA handles have high GC content (>60%) to prevent DNA melting when DNA is held at high forces or during DNA overstretching transition (33–36). PCR products were purified using PureLink PCR purification kit (Invitrogen) and digested with BstXI restriction enzyme (NEB). Telomeric G-rich oligo, dsDNA handles were ligated using T4 DNA ligase (NEB). The ligated product (2042 bp with 26 nt) was purified by gel extraction with PureLink kit (Invitrogen). More details of the DNA constructs are included in SI: ‘DNA oligonucleotides’ and SI: Figure S1. 1D nuclear magnetic resonance (NMR) experiment was conducted on a Bruker AVANCE 600 MHz spectrometer at 25°C. JR-type pulse sequence was used for water signal suppression.
Single DNA stretching experiments
A flow chamber of 10–20 μl in volume was constructed on a (3-Aminopropyl)triethoxy silane (APTES) (Sigma-Aldrich) functionalized coverslip (32 × 24 mm). Thiol-end of DNA was covalently attached to the amine group of APTES via sulfo-SMCC crosslinker (Thermo Scientific). The APTES coverslip was first reacted with 1 mg/ml sulfo-SMCC (dissolved in 1× phosphate buffered saline (PBS) pH 7.4 buffer) for 30 min. After washing out unbound sulfo-SMCC, thiol-labeled G4 DNA construct was introduced into the chamber and incubated for 30 min. The chamber was then blocked with BSA solution (10 mg/ml BSA, 1mM 2-mercaptoethanol, 1 × PBS pH 7.4 buffer) for more than 2 h before experiments. After DNA constructs were bound to the surface, 2.8 μm-diameter streptavidin-coated paramagnetic beads (Dynal M-280, Life technologies) were introduced to attach to the biotin end of DNA. Finally, the buffer was changed to assay buffer (10 mM Tris-HCl, pH 8.0, 100 mM KCl) for single-molecule stretching experiments.
Ultra stable vertical magnetic tweezers built in lab were used to stretch the DNA constructs using a pair of magnets on the top of the chamber (Figure 1A). The magnetic tweezers were controlled by in-house-written LabVIEW program (National Instruments). The extension change of the construct was recorded with a sampling rate of ∼200 frames per second. Force was controlled by changing the distance between the permanent magnets and flow chamber. Loading rate control was achieved by moving the magnets through a programmed trajectory. The magnetic tweezers have a spatial resolution for bead stuck on surface of ∼2 nm, and the force calibration has a relative error of <10% (32). More details of the magnetic tweezers design, the force and loading rate control, as well as the force calibration were detailed in our previous publications (32,37). All experiments were carried out at our lab room temperature of 21−23 °C.
Figure 1.
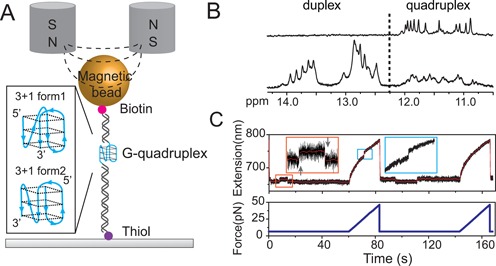
Experimental setup and procedure. (A) Schematic diagram of G4 DNA and magnetic tweezers. The 26 mer ssDNA of four-repeat human telomeric sequence d(TTGGG(TTAGGG)3TTT) is sandwiched bewteen a lower 1449 bp and an upper 601 bp dsDNA handles. The DNA construct is tethered between a 2.8 μm-diameter paramagnetic bead via biotin-streptavidin linkage and an amine functionalized coverslip surface through covalent cross-linker sulfo-SMCC. Inset shows two possible hybrid-type G4 structures that may form on the sequence (16,17). (B) Imino proton NMR spectrum of telomeric G4 sequence d(TTGGG(TTAGGG)3TTT) (top) and the 1:1:1 mixture of three sequences, d(CGAGTCTGTGCACAAGGTGC), d(CTACTGACCTGGCTGC) and d(CTTGTGCACAGACTCGTTGGG(TTAGGG)3TTTGCAGCCAGGTCAGTAGCGAC) (bottom). The mixture is expected to form a G-quadruplex in the centre flanked by two 16-bp Watson–Crick duplexes at the 5′- and 3′-ends respectively (underlined sequences). (C) Typical force responses of G4 DNA in two repeating stretching cycles (original and smoothed extension data are shown in black and red, respectively). In each cycle, a constant force was maintained at 6.5 pN for 60 s then increased to 50 pN at a constant loading rate of 2 pN/s. In the first cycle, at the constant force of 6.5 pN, two extension states with an extension difference of ∼6 nm were observed, indicated by an unfolding transition (up-arrow) followed by a refolding transition (down-arrow) in the zoom-in inset in the orange rectangle. During the subsequent force-increase scan at 2 pN/s, a typical G4 unfolding indicated by a sudden extension jump with a step size of ∼8 nm occurred at ∼25 pN (marked in cyan rectangle). In the second cycle after force was jumped back to 6.5 pN, a refolding transition and a following unfolding transition were observed. In the subsequent force-increase scan at 2 pN/s, G4 unfolding was not observed.
RESULTS
Folding and unfolding telomeric G4 at constant forces
In our experiments, a single-stranded telomeric G4 DNA formed on 5′TTGGG(TTAGGG)3TTT sandwiched between two dsDNA handles was stretched using ultra-stable magnetic tweezers (Figure 1A) (37). In each experiment, a single-tether attachment was ensured by the highly specific B-to-S transition of the dsDNA handles during DNA overstretching characterized by ∼1.68-fold extension increase in a narrow force range of ∼65 pN (33–36).
The sequence used in our experiment was confirmed to form G4 by NMR experiment (Figure 1B, top panel). To confirm the G4 formation in the context of single-molecule mechanical experiments, i.e. when this sequence is embedded between duplex handles, we performed NMR experiments on a construct containing short Watson–Crick duplexes at the 5′- and 3′-ends of the same G-rich fragment. Indeed, the NMR spectrum of this construct (Figure 1B, bottom panel) showed imino proton signals from 10.5 to 14 ppm, corresponding to the formation of both G-quadruplex (10.5–12 ppm) and Watson–Crick duplexes (12–14 ppm).
Figure 1C shows the force responses of the telomeric G4 formed in our DNA construct in two representative repeating stretching cycles. In each cycle, the tether was held at a low constant force of 6.5 ± 0.6 pN for 1 min followed by stretching at a constant loading rate of 2 pN/s up to ∼50 pN. During 1 min of holding the DNA tether at ∼6.5 pN, stepwise extension changes with a step size of ∼6 nm were observed (Figure 1B, inset). In the subsequent constant loading rate stretching, two types of force responses were observed: (i) If the DNA was in the lower extension state prior to the constant loading rate stretching, an abrupt unfolding transition with a step size of ∼8 nm was observed at a force of ∼20 pN (Figure 1C, first stretching cycle). (ii) If the DNA was in the longer extension state prior to the constant loading rate stretching, such unfolding step was not observed (Figure 1C, second stretching cycle). These results indicate that the stepwise extension changes at ∼6.5 pN are spontaneous telomeric G4 folding and unfolding transitions, and suggest that it is possible to observe equilibrium transitions of structures formed in the telomeric G4 sequence at low forces.
The unfolding force distribution p(f) obtained from constant loading rate stretching can be fitted with p(f) predicted by Equation (2) with xu = 0.8 ± 0.2 nm and 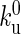 = 0.009 ± 0.008 s−1 (average ± standard deviation) (SI: Figure S2). A peak unfolding force of 17 ± 5 pN (average ± standard deviation) agrees with the value of 21 ± 1 pN reported in previous works conducted at a similar loading rate (5.5 pN/s) (30,38). The histogram of the unfolding step sizes can be fitted with Gaussian distribution with a peak value of 8 ± 2 nm (average ± standard deviation) at the force range (Figure S2), which is consistent with unfolding of 21 nucleotides based on the known force response of ssDNA and the size of G4 (39) (SI: ‘Force-extension curve for ssDNA’).
= 0.009 ± 0.008 s−1 (average ± standard deviation) (SI: Figure S2). A peak unfolding force of 17 ± 5 pN (average ± standard deviation) agrees with the value of 21 ± 1 pN reported in previous works conducted at a similar loading rate (5.5 pN/s) (30,38). The histogram of the unfolding step sizes can be fitted with Gaussian distribution with a peak value of 8 ± 2 nm (average ± standard deviation) at the force range (Figure S2), which is consistent with unfolding of 21 nucleotides based on the known force response of ssDNA and the size of G4 (39) (SI: ‘Force-extension curve for ssDNA’).
Dynamics of folding and unfolding transitions reveal three folded telomeric G4 conformations
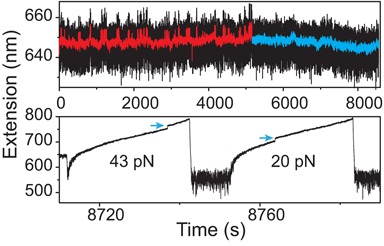
The dynamics of telomeric G4 folding and unfolding at constant forces was investigated. Two representative time traces of the extension fluctuation of a telomeric G4 DNA construct at two different forces (5 and 7 pN) are shown in Figure 2A, where G4 folding and unfolding transitions are indicated by sudden stepwise extension changes detected by a noise beating algorithm (40). At 5 pN, the folded state (the shorter extension state) is predominant; while at 7 pN, the unfolded state (the longer extension state) becomes predominant.
Figure 2.
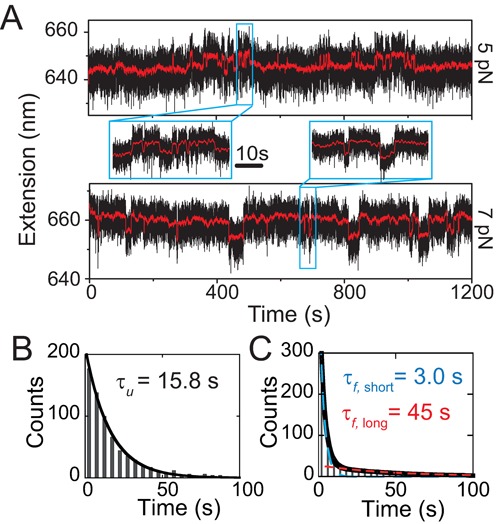
Equilibrium folding and unfolding dynamics under constant forces. (A) Representative time traces (black dots) of the extension fluctuation of a single telomeric G4 tether recorded at 5.0 ± 0.5 pN (upper panel) and 7.0 ± 0.7 pN (lower panel). The red lines show smoothed time traces by adjacent average of data using a 0.5 s sliding time window. Insets show zoom-in time traces containing rapid folding and unfolding transitions. (B) Histogram of the dwell time of the longer extension state (i.e. unfolded G4) recorded at 5.0 ± 0.5 pN, which is fitted by a single-exponential decay function with time constant of τu = 15.8 ± 0.4 s (black curve, average ± standard error). (C) Histogram of the dwell time of the shorter extension state (i.e. folded G4) recorded at the same force, which cannot be fitted by a single-exponential decay function. Therefore, a double-exponential decay function with two time constants of τf, short = 3.0 ± 0.1 s and τf, short = 45 ± 4 s was used for the fitting (black curve), which is the sum of two individual single-exponential fitting using the respective time constants. Data in panels (B) and (C) include 683 folding events and 684 unfolding events collected from 13 independent telomeric G4 DNA tethers.
The dwell time histograms of the unfolded state obtained at each force can be fitted with a single exponential decay function 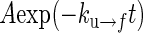 , where ku → f indicates the folding rate constant and the reciprocal τu = 1/ku → f is the lifetime of the unfolded state. τu at different forces are fitted to be 15.8 ± 0.4 s (5 pN), 53 ± 5 s (6 pN) and 101 ± 11 s (7 pN), respectively (Figure 2B and SI: Figures S3 and S4). Here the errors are fitting standard errors. At each force, data recorded from at least nine independent tethers are combined together to obtain the final fitting.
, where ku → f indicates the folding rate constant and the reciprocal τu = 1/ku → f is the lifetime of the unfolded state. τu at different forces are fitted to be 15.8 ± 0.4 s (5 pN), 53 ± 5 s (6 pN) and 101 ± 11 s (7 pN), respectively (Figure 2B and SI: Figures S3 and S4). Here the errors are fitting standard errors. At each force, data recorded from at least nine independent tethers are combined together to obtain the final fitting.
In contrast to the folding transition, the dwell time histograms of the folded states cannot be fitted with single exponential function while a double-exponential function 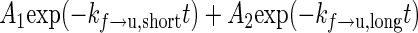 with two rate constants kf → u, short and kf → u, long can. Two distinct lifetimes of the folded states are determined to be (τf, short, τf, long) = (3.0 ± 0.1, 45 ± 4 s) at 5 pN (Figure 2C). At 6 and 7 pN, they are fitted to be (2.1 ± 0.2, 25.0 ± 6.3 s) and (3.2 ± 0.5, 16.6 ± 4.9 s), respectively (SI: Figure S4). Hereafter the folded state characterized by the lifetime of several seconds is referred to as the short-lived state, and the one characterized by a lifetime of tens of seconds is referred to as the long-lived state. The zoom-in time traces in Figure 2A show rapid unfolding and folding transitions of the short-lived state.
with two rate constants kf → u, short and kf → u, long can. Two distinct lifetimes of the folded states are determined to be (τf, short, τf, long) = (3.0 ± 0.1, 45 ± 4 s) at 5 pN (Figure 2C). At 6 and 7 pN, they are fitted to be (2.1 ± 0.2, 25.0 ± 6.3 s) and (3.2 ± 0.5, 16.6 ± 4.9 s), respectively (SI: Figure S4). Hereafter the folded state characterized by the lifetime of several seconds is referred to as the short-lived state, and the one characterized by a lifetime of tens of seconds is referred to as the long-lived state. The zoom-in time traces in Figure 2A show rapid unfolding and folding transitions of the short-lived state.
Besides the short- and long-lived states, a third folded state characterized by an ultra-long lifetime was identified. In the long time trace recorded at ∼5 pN (Figure 3, upper panel), stepwise extension fluctuations similar to those in Figure 2A were observed in the first 5100 s (red smoothed data for clarity). However, after 5100 s, the stepwise fluctuations disappeared and the G4 DNA remained folded for 1 h (blue smoothed data for clarity). The ultra long lifetime of this folded state indicates that it is different from the long-lived state discussed in the previous paragraph. Hereafter this folded state is referred to as the ultra-long-lived state.
Figure 3.
Ultra long-lived G4 state. Upper panel shows a long time trace of a telomeric G4 DNA extension fluctuation at ∼5 pN (original and smoothed data are shown in black and red, respectively). In the first 5100 s, stepwise fluctuation with similar kinetics to Figure 2A was observed (red smoothed data for clarity). After 5100 s, the tether extension remained at the folded extension state for 1 h (cyan smoothed data for clarity). Lower panel shows the extension change of the ultra-long lived telomeric G4 being stretched at 2 pN/s, where unfolding occurred at ∼43 pN (the first stretch). After subsequent refolding at 1 pN for 10 s, unfolding occurred at a much lower force of ∼20 pN.
This state has a remarkable mechanical stability, unfolding at 43 pN (Figure 3, lower panel, the first stretch) during a subsequent constant loading rate stretching of 2 pN/s, which is significantly higher than the peak unfolding force of 17 ± 4 pN revealed from the histogram in Figure 1C. After this ultra long-lived structure was unfolded, the tether was refolded at 1 pN for 20 s. In the next constant loading rate stretch it was unfolded at ∼20 pN. Similar long time measurements (1–4 h) were repeated for 15 times and such ultra-long-lived state was observed for 6 times, which remained folded in 5–7 pN for at least 1000 s. Unfolding these six ultra long-lived structures at 2 pN/s revealed unfolding forces in the range of 42 ± 5 pN (average ± standard deviation). Together with results revealed in Figures 2 and 3, we conclude that at least three folded states characterized by different lifetimes and mechanical stability exist.
G4 folding to the long-lived state is through the short-lived state as an intermediate
The folding kinetics can also be obtained by examining the time evolution of folding probability pfold(t), which was obtained by the following procedure. Firstly, a folded telomeric G4 was unfolded by constant loading rate stretching at 2 pN/s, followed by jumping to a lower force f for refolding by holding the tether for certain time t at the force. Successful refolding was indicated by an unfolding event in a subsequent constant loading rate stretching. Repeating such procedure N times, the pfold(t) was obtained by M/N, where M is the number of successful folding (SI: Figure S5). At each force f and holding time t, such stretching-folding procedure was repeated for more than 100 times from more than four independent tethers to obtain pfold(t). Within our holding time scale of <120 s, the formation of the ultra long-lived state is negligible (SI: Figure S6).
Figure 4A shows that pfold(t) obtained at 5 pN increases with time and nearly reaches a steady state at >60 s with a probability of ∼0.8. A sequential model where folding is through the short-lived state as an intermediate state to the long-lived state was considered to explain the result, plus a branched folding to the ultra long-lived state although it does not play a role in pfold(t) obtained within a time scale of 120 s. In this model, the transition rates can be directly determined by dwell time analysis at the same force as shown in Figure 2B-C: k12 = ku → f,k21 = kf → u, short, k32 = kf → u, long, k23 = k21Nlong/Nshort (SI: Table S1). Here, Nlong and Nshort are the numbers of the long-lived and short-lived states counted from the dwell time histogram, which ratio equals to the ratio of transition rates of k23 and k21. With these model parameters, pfold(t) was solved from the Master equations using an in-house written Matlab code based on the Runge–Kutta method (SI: ‘Kinetic models’), which was then compared with the experimental data. Using the rates determined at 5 pN, the predicted pfold(t) from the sequential model (Figure 2A, solid line) reasonably agrees with the experimental data. Similar analysis at 6 and 7 pN also show good agreement between the experimental data and the predictions by the sequential model. An alternative parallel model with competitive parallel folding into different folded states was also considered; however, it does not fit the experimental data as good as the sequential model (SI: Figure S7).
Figure 4.
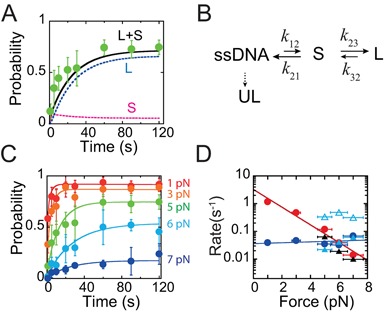
Telomeric G4 folding and unfolding kinetics and stability. (A) Green solid circles indicate experimental measured pfold(t) at 5 pN. Black curve is the predicted pfold(t) based on the sequential model in (B), which is the sum of the probabilities of the long-lived state (L, blue curve) and the short-lived state (S, purple curve). (B) Sketch of the sequential transition model, in which the long-lived state, short-lived state and ultra-long-lived state are indicated by ‘L’, ‘S’ and ‘UL’, respectively. (C) Filled circles of different colors indicate experimental pfold(t) measured at different forces. Each data point was calculated based on more than 100 refolding/unfolding cycles from at least four molecules. The solid lines with corresponding colors are best fitting based on the Master equation of the sequential model. (D) The fitted k12 (red solid circles) and k32 (blue solid circles) at different forces. The black triangles are ku → f determined in dwell time analysis at constant forces of 5, 6 and 7 pN, respectively. The open and solid cyan triangles are kf → u, short and kf → u, long of the short-lived (open triangle) and long-lived (solid triangle) states, respectively, which are directly determined by dwell time analysis. X-axis error bars reflect a ∼10% error in force determination. Y-axis error bars are the standard deviation.
Force-dependent telomeric G4 folding and unfolding kinetics and stability
In order to obtain the force dependence of folding and unfolding rates in a broader force range, the theoretical predicted pfold(t) based on the sequential model and experimental data obtained at different forces were compared, treating the transition rates as fitting parameters. The five parameters in the sequential model can be reduced based on the knowledge of the rates that were directly measured in 5–7 pN. Due to the transient nature of the short-lived state, the outward transition rates from the short-lived state (k21 and k23) are much faster that the inward transition rates (k12 and k32). In 5–7 pN, k21 ∼k23 have a weak dependence on force and are about 10-folds faster than (k12 and k32). This makes the short-lived state quickly relaxed to a steady state in a few seconds (Figure 4A, pink curve).
Simply set k23 = k21 = αk32 (α ≥ 5), the parameter pair (k12, k32) were varied to search for the best fitting values. We found that the best fitting values of (k12, k32) are insensitive to change of α > 5 (SI: ‘Kinetic models’), and the values obtained at α = 10 are plotted in Figure 4D. The results show that the folding rate k12 (solid red circles) has a strong force dependence: increasing force from 1 to 7 pN, it exponentially decreases by more than 100-folds. In contrast, unfolding rate k32 is much less sensitive to force change.
The force-dependence of the transition rates contains information of the transition energy barrier, which can be understood by the Bell's model (Equation 1). The sensitivity of k12 and the insensitivity of k32 to force imply a large transition distance for folding to the short-lived state and a small transition distance for the unfolding from the long-lived stated to the short-lived state. Fitting to the Bell's model the zero force transition rates (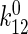 ,
, 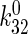 ) and transition distances (
) and transition distances (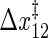 ,
, 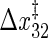 ) are respectively determined to be (1.6−6.7, 0.02−0.07 s−1 with 95% confidence) and (3.0 ± 0.7, 0.2 ± 0.6 nm) (average ± standard deviation), respectively.
) are respectively determined to be (1.6−6.7, 0.02−0.07 s−1 with 95% confidence) and (3.0 ± 0.7, 0.2 ± 0.6 nm) (average ± standard deviation), respectively.
In the force range 5–7 pN, the fitted k12(f) overlaps with the folding rate directly measured by dwell time analysis (solid black triangles), as shown in Figure 4D. As expected, the fitted k32(f) also overlaps with the unfolding rate of the long-lived state (solid cyan triangles) determined by the dwell time analysis. The unfolding rate of the short-lived state determined by dwell time analysis is also plotted for comparison (open cyan triangles).
The force dependent free energy cost for unfolding the short- and long-lived states to ssDNA, ΔGssDNA, short(f) = GssDNA(f) − Gshort(f) and ΔGssDNA, long(f) = GssDNA(f) − Glong(f), are calculated from the transition rates directly obtained at three constant forces (5, 6 and 7 pN) from dwell time analysis by 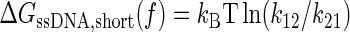 and
and 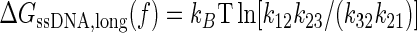 , respectively.
, respectively.
ΔG(f) between ssDNA and a folded state contains a force-independent term that is determined by molecular interactions, ΔG0, and a force-dependent term that comes from the different conformational force responses between a folded G4 and the unfolded ssDNA, which can be analytically expressed as (41):
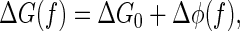 |
(3) |
where 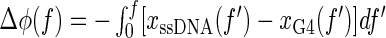 is the conformational energy change between unfolded ssDNA and the folded telomeric G4.
is the conformational energy change between unfolded ssDNA and the folded telomeric G4.
Due to complex stacking and hydrogen bond formation between the bases, the force response of ssDNA cannot be ideally described by the commonly used worm-like-chain or the freely-joint-chain polymer models. Up to date, the ssDNA force-extension curve xssDNA(f) is much better described by a phenomenological force–extension relation proposed by Cocco et al. (39) (SI: ‘Force-extension curve for ssDNA’). Folded G4 can be approximated as a rigid body with a size of l0 ∼1.7 nm estimated from NMR structure. Its projected length along the force direction is given by: 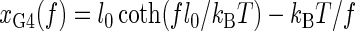 . Based on these force-extension curves and ΔG(f) data, ΔG0 of unfolding short- and long-lived state are calculated to be: 3.6 ± 0.6 kBT and 5.9 ± 0.4 kBT (2.1 ± 0.4 kcal mol−1 and 3.5 ± 0.2 kcal mol−1, average ± standard error), respectively.
. Based on these force-extension curves and ΔG(f) data, ΔG0 of unfolding short- and long-lived state are calculated to be: 3.6 ± 0.6 kBT and 5.9 ± 0.4 kBT (2.1 ± 0.4 kcal mol−1 and 3.5 ± 0.2 kcal mol−1, average ± standard error), respectively.
DISCUSSION
In summary, this work has studied the equilibrium folding and unfolding transitions of a single telomeric G4 in physiological potassium concentration using magnetic tweezers. Several important results have been obtained, including identifying three folded structures with distinct life times and mechanical stability. Our observation of a short-lived and a long-lived state for G4 formed in a time scale of several minutes is consistent with previous smFRET studies carried out at similar K+ concentrations that reported two folded G4 populations in the absence of force, which were indicted by different levels of FRET efficiency (22,27). In particular, the ultra long-lived state with a high mechanical stability has not been reported in previous experiments. Besides, the force dependent free energy cost of unfolding the telomeric G4 for both the short- and long-lived forms, ΔG(f), is obtained based on direct equilibrium measurements.
Previous studies have reported a wide range of ΔG0 in 3.4−14.8 kcal mol−1 (23). The value determined from this study is close to the lower boundary of this range. While the causes of such big variation of ΔG0 from different experiments have remained unclear, we reason that it may be related to the polymorphism of the telomeric G4. Different forms of telomeric G4 may be kinetically separated, which may lead to different G4 population distributions in different experiments depending on the exact folding protocol. This is consistent with a recent study reporting that the population distribution of multiple telomeric G4 structures folded by a slow thermal annealing procedure is distinct from that obtained by quick quench folding (42). It is possible that at a longer time scale, other structures such as ultra long-lived state observed in our study may accumulate, leading to a higher free energy cost for unfolding.
Our data have suggested that folding to the long-lived state is through the short-lived state as an intermediate, which agrees with the conclusion made in a previous smFRET study at a low KCl concentration (2 mM) (22). The presence of an intermediate state between ssDNA and G4 has also been suggested in several previous biochemical, single-molecule and simulation studies, which has been proposed to be a partially folded structure involving 3-folded strands and one peeled strand (38,43,44). The existence of a short-lived intermediate state is in general consistent with these earlier works.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online including [1–5].
Acknowledgments
We thank Daniela Rhodes and Michael Stone for stimulating discussions.
FUNDING
National Research Foundation through the Mechanobiology Institute Singapore [to J.Y.]; Ministry of Education of Singapore [MOE2012-T3-1-001 to J.Y. and A.T.P.]. Funding for open access charge: National Research Foundation through the Ministry of Education of Singapore [MOE2012-T3-1-001 to J.Y.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wells R.D. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Lipps H.J., Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Lam E.Y.N., Beraldi D., Tannahill D., Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013;4:1796. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 8.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moyzis R.K., Buckingham J.M., Cram L.S., Dani M., Deaven L.L., Jones M.D., Meyne J., Ratliff R.L., Wu J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. U.S.A. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zahler A.M., Williamson J.R., Cech T.R., Prescott D.M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 11.Greider C.W., Blackburn E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 12.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 13.De Cian A., Lacroix L., Douarre C., Temime-Smaali N., Trentesaux C., Riou J.-F., Mergny J.-L. Targeting telomeres and telomerase. Biochimie. 2008;90:131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 16.Ambrus A., Chen D., Dai J., Bialis T., Jones R.A., Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luu K.N., Phan A.T., Kuryavyi V., Lacroix L., Patel D.J. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim K.W., Amrane S., Bouaziz S., Xu W., Mu Y., Patel D.J., Luu K.N., Phan A.T. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane A.N., Chaires J.B., Gray R.D., Trent J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mergny J.L., Phan A.T., Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 21.Ying L., Green J.J., Li H., Klenerman D., Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J.Y., Okumus B., Kim D.S., Ha T. Extreme conformational diversity in human telomeric DNA. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaires J.B. Human telomeric G-quadruplex: thermodynamic and kinetic studies of telomeric quadruplex stability. FEBS J. 2010;277:1098–1106. doi: 10.1111/j.1742-4658.2009.07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Risitano A., Fox K.R. Stability of intramolecular DNA quadruplexes: comparison with DNA duplexes. Biochemistry. 2003;42:6507–6513. doi: 10.1021/bi026997v. [DOI] [PubMed] [Google Scholar]
- 25.Gray R.D., Chaires J.B. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008;36:4191–4203. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y., Kan Z., Zeng Z., Hao Y., Chen H., Tan Z. Determining the folding and unfolding rate constants of nucleic acids by biosensor. Application to telomere G-quadruplex. J. Am. Chem. Soc. 2004;126:13255–13264. doi: 10.1021/ja048398c. [DOI] [PubMed] [Google Scholar]
- 27.Long X., Parks J.W., Bagshaw C.R., Stone M.D. Mechanical unfolding of human telomere G-quadruplex DNA probed by integrated fluorescence and magnetic tweezers spectroscopy. Nucleic Acids Res. 2013;41:2746–2755. doi: 10.1093/nar/gks1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell G.I. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 29.Evans E., Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koirala D., Dhakal S., Ashbridge B., Sannohe Y., Rodriguez R., Sugiyama H., Balasubramanian S., Mao H. A single-molecule platform for investigation of interactions between G-quadruplexes and small-molecule ligands. Nat. Chem. 2011;3:782–787. doi: 10.1038/nchem.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarzynski C. A nonequilibrium equality for free energy differences. Phys. Rev. Lett. 1997;78:2690–2693. [Google Scholar]
- 32.Chen H., Fu H., Zhu X., Cong P., Nakamura F., Yan J. Improved high-force magnetic tweezers for stretching and refolding of proteins and short DNA. Biophys. J. 2011;100:517–523. doi: 10.1016/j.bpj.2010.12.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cluzel P., Lebrun A., Heller C., Lavery R., Viovy J.L., Chatenay D., Caron F. DNA: an extensible molecule. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 34.Smith S.B., Cui Y., Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 35.Fu H., Chen H., Marko J.F., Yan J. Two distinct overstretched DNA states. Nucleic Acids Res. 2010;38:5594–5600. doi: 10.1093/nar/gkq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X., Chen H., Le S., Rouzina I., Doyle P.S., Yan J. Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching by single-molecule calorimetry. Proc. Natl. Acad. Sci. U.S.A. 2013;110:3865–3870. doi: 10.1073/pnas.1213740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H., Zhu X., Cong P., Sheetz M.P., Nakamura F., Yan J. Differential mechanical stability of filamin A rod segments. Biophys. J. 2011;101:1231–1237. doi: 10.1016/j.bpj.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koirala D., Ghimire C., Bohrer C., Sannohe Y., Sugiyama H., Mao H. Long-loop G-quadruplexes are misfolded population minorities with fast transition kinetics in human telomeric sequences. J. Am. Chem. Soc. 2013;135:2235–2241. doi: 10.1021/ja309668t. [DOI] [PubMed] [Google Scholar]
- 39.Cocco S., Yan J., Léger J.-F., Chatenay D., Marko J.F. Overstretching and force-driven strand separation of double-helix DNA. Phys. Rev. E. Stat. Nonlin. Soft Matter Phys. 2004;70:011910. doi: 10.1103/PhysRevE.70.011910. [DOI] [PubMed] [Google Scholar]
- 40.Aggarwal T., Materassi D., Davison R., Hays T., Salapaka M. Detection of steps in single molecule data. Cell. Mol. Bioeng. 2012;5:14–31. doi: 10.1007/s12195-011-0188-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rouzina I., Bloomfield V.A. Force-induced melting of the DNA double helix 1. Thermodynamic analysis. Biophys. J. 2001;80:882–893. doi: 10.1016/S0006-3495(01)76067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long X., Stone M.D. Kinetic partitioning modulates human telomere DNA G-quadruplex structural polymorphism. PLoS One. 2013;8:e83420. doi: 10.1371/journal.pone.0083420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bončina M., Lah J., Prislan I., Vesnaver G. Energetic basis of human telomeric DNA folding into G-quadruplex structures. J. Am. Chem. Soc. 2012;134:9657–9663. doi: 10.1021/ja300605n. [DOI] [PubMed] [Google Scholar]
- 44.Li W., Hou X., Wang P., Xi X., Li M. Direct measurement of sequential folding pathway and energy landscape of human telomeric G-quadruplex structures. J. Am. Chem. Soc. 2013;135:6423–6426. doi: 10.1021/ja4019176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


