Abstract
In vitro cell culture and animal models are the most heavily relied upon tools of the pharmaceutical industry. When these tools fail, the results are costly and have at times, proven deadly. One promising new tool to enhance preclinical development of drugs is Organs on Chips (OOCs), proposed as a clinically and physiologically relevant means of modeling health and disease. Bringing the patient from bedside to bench in this form requires that the design, build, and test of OOCs be founded in clinical observations and methods. By creating OOCs as models of the patient, the industry may be better positioned to evaluate medicinal therapeutics
Keywords: Organs on chips, microenvironment, patient, model, clinical, pharmacokinetics pharmacodynamics (PK/PD), induced pluripotent stem cells (iPSCs)
Introduction
Bringing a drug to market is estimated to cost over $1.3 billion, a significant contributor to pharmaceutical research and development costs in the United States that exceed $50 billion annually.1 However, despite this rising investment in drug discovery, the rate of FDA drug approval has remained constant over the last 60 years.2 This stagnation and the rising costs of drug discovery are in part due to the failure of drug development tools and techniques to evolve with our advances in basic science.
The pharmaceutical industry often identifies toxicities in drug candidates during clinical trials.3 The late discovery of potentially harmful effects or lack of efficacy is not due to a lack of funding for early drug screening. It is estimated that nearly 32% of the $1.3 billion required to bring a drug to market is spent on pre-clinical screening and testing.4 The high rate of late-stage drug failure may be due to ineffective screening methods that do not adequately to replicate the patient. Traditional cell culture methods do not accurately model organ microenvironments and the systemic effects of drugs. Similarly, animal models do not always fully recapitulate the effects of pharmacological agents in humans. It has been estimated that about a third of successful animal studies have translated to successful human clinical trials; this is in large part due to different methodologies used in animal testing vs clinical trials, biased animal study reporting, and the physiological differences among species.5, 6
There are a number of examples of preclinical successes that have been not only ineffective, but deadly in the clinic. Early animal models for Class I antiarrhythmic agents, including Encainide and Flecainide (Tambocor), suggested that these drugs were effective at suppressing irregular cardiac pacing.7 However, the Cardiac Arrhythmia Suppression Trial (CAST) of the late 1980’s later showed that patients taking Encainide and Flecainide had a 2.5 times greater risk of suffering a fatal cardiac event.8 Similarly, early successes in canine and rodent animal models were followed by the clinical failure of the Hepatitis B drug Fialuridine (FIAU), causing the death of a third of the patients involved in a 1993 clinical trial.9 More recently, despite evidence that the cancer drug Targretin reversed plaque build-up in mouse Alzheimer models, it proved ineffective in human Alzheimer patients.10 These costly, unsuccessful clinical trials have motivated the pharmaceutical industry to reevaluate the methodologies used to develop drugs; successes and failures alike need to be identified early in the preclinical stages of drug discovery.
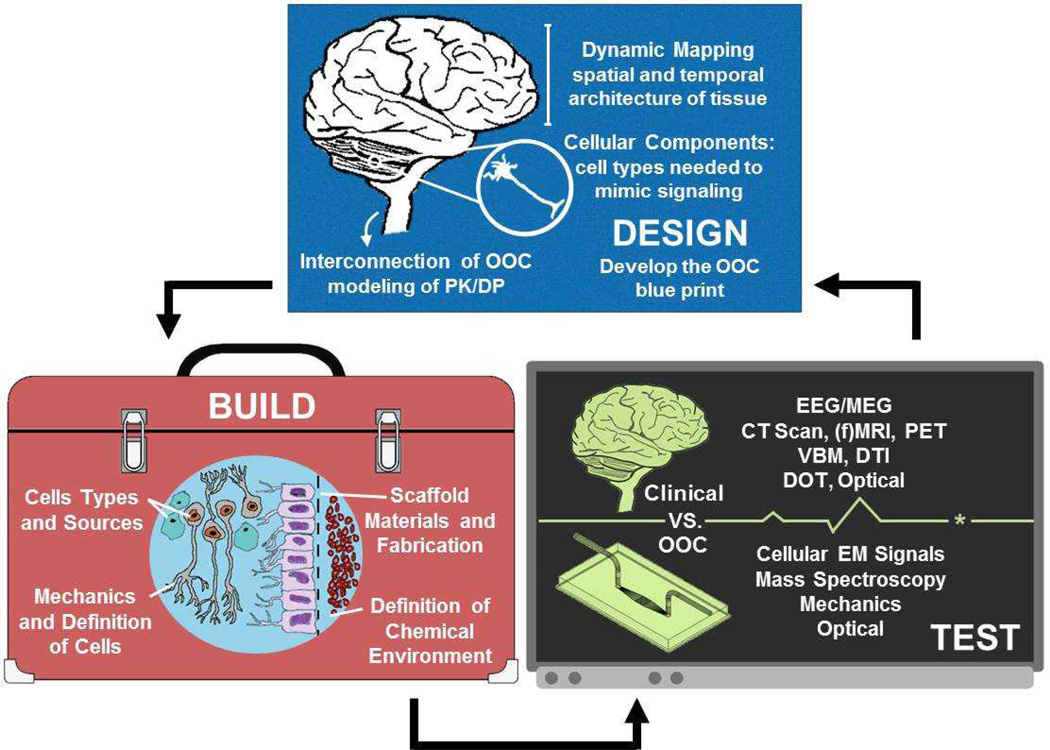
Preclinical evaluation methods might be more effectively designed if they model the patient. Patient-derived induced pluripotent stem cells (iPSCs) provide the patient-relevant foundation for in vitro disease modeling and drug testing; iPSCs are customizable cells that have the distinctive characteristics of the patient such as genetics, sex, age, and ethnicity.11 However, the platforms in which iPSCs are used must likewise mimic the dynamic, three dimensional structure of the tissues being modeled to achieve meaningful function. To accomplish this, Organs on Chips (OOCs) are the tools being designed to recapitulate the patient by mimicking the structure, function, and subsequent response to drugs or other foreign stimuli.12 For example, a recent study reported, a model of pulmonary edema using a “lung on a chip” which was able to screen for the functional effects of a known pathogen and mimic animal testing results of a new, potentially therapeutic agent to treat the disease.13 Clinical observations and data-driven design, and subsequent build, of OOCs whose readouts ultimately need to register with traditional clinical diagnostics (Figure 1). Organs on chips can provide the pharmaceutical industry with patient-relevant drug testing models if their design, build, and test reflect the successful replication of the treated patient.
Figure 1.
A depiction of the Design, Build, and Test algorithm as applied to the engineering of a Brain-on-Chip device. The OOC “blueprint” should illustrate the spatiotemporal arrangements of cells, then determine the dynamic architecture of the tissue, and finally understand the necessary organ systems to be included for physiologically relevant PK/PD. The build of an OOC must focus on defining the tools and materials from which the chips is build: ensuring the cell building blocks and 3D architecture in which they are assembled mimics that of the patient. The test of an OOC must likewise be inspired by the patient.
Design: Creating the Spatiotemporal Blueprints of OOCs
The goal of OOCs is to better model the patient by recapitulating the necessary structure-function relationships required to mimic the healthy and disease state of the afflicted organ. This is accomplished by using a multi-scale approach to recapitulate organ microenvironments in health and disease.14, 15 Recapitulating the microenvironments of organs requires design of the extracellular matrix, definition of the biochemical environment, engineering of biotic/abiotic interfaces in vitro, and control over the dynamic mechanical stimuli experienced in the organ.16 However, the fundamental building blocks of tissues and organs are specialized, collectively functioning populations of cells. Mono-culture models represent an isolated and limited functionality; without the signaling and metabolic interactions with other cell types. For example, much of the research on Diabetes Types I and II is exclusively focused on the insulin producing pancreatic beta-cells.17, 18 However, since the other pancreatic islet cell types (alpha-cells, delta-cells, etc.) and the pancreatic acinar cells are closely linked to beta-cells through their differentiation and structural proximity, it is inevitable that their interactions are functionally coupled.19 Similarly, all organs are vascularized; the endothelial cells that comprise the vasculature are known to communicate with and influence the development and function of tissues.20 Eventual development of OOCs with heterogeneous cell demographics is necessary to recapitulate the dynamic interactions that potentiate organ function.
Function, however, is derived from structure. For many tissues, the details of the microenvironment are poorly distinguished and therefore the mapping, or physiological blueprints, of organs is difficult. To create a better blueprint, traditional methods such as autopsy and histology can provide a first approximation of structure. However, these techniques are static representations of processed samples, lacking the ability to capture the dynamic nature of living tissue. The adaptive behavior of living tissues may be measured using non-destructive, real-time imaging techniques such as those normally used for spatiotemporal clinical diagnostics. Cardiac MRI, for example, is now capable of real-time imaging of heart-beats up to 50fps and Magnetic Resonance Spectroscopy (MRS) has been used in concert with MRI to monitor choline and lipid levels in brainstem glioma.21 Imaging techniques such as these offer greater insight into the spatiotemporal structure and composition of tissues, providing a more detailed, multidimensional map for the design of OOCs.
The detailed mapping of individual tissues or organs is, however, insufficient to model the patient. OOC design is enabled by considering the pharmacokinetics and pharmacodynamics (PK/PD) of a drug in a multi-organ system. The PK/PD of a drug describes both how a drug or toxin is transported, metabolized, impacts targeted and nontargeted tissues, and is eventually excreted. Drug metabolism by the liver, clearance via the kidneys, or absorption through the intestines, for example, play critical roles in the PK/PD of many drugs.22 Designing systems of interconnected OOCs to better model multi-organ systems in an effort to replicate patient PK/PD promises to be a challenge requiring a unique partnership between OOC designers and modelers.
Should connected OOCs be scaled by allometry, functional output, or simply by surface area or volume? Accurate scaling of connected OOCs is essential for the proper PK/PD of drugs in these systems. However, how to best scale OOCs still remains in question.23, 24 Mathematical models done in silico are the design tools that can help to address the scaling and design of in vitro OOC systems.25 By creating “biosimulations” that model drug PK/PD based on our knowledge of cellular molecular pathways, insight into targeting complexes, metabolism, transport, and the spatiotemporal functional effects of drugs can be gained in a “virtual patient” population.26 Only by mathematically understanding the systems and interactions being modeled in OOCs can we appropriately scale the lessons learned from trials with OOCs.
Build: Choosing the Materials and Tools for Building OOCs
Building materials and manufacturing techniques used to put the blueprints of OOCs into practice must also be founded in patient physiology. OOCs need to be built with materials and techniques that can mimic the microenvironment of the patient. While many current OOCs replicate some of the characteristics of the tissue they aim to model, the materials and tools used to build the next generation of chips must imitate the mechanical, chemical, and structural properties of a tissue they model.
The building materials of OOCs are the cells and biomaterial-based structures that collectively mimic the organ microenvironment. The development of iPSCs as a renewable source of customizable, patient specific cells has provided a promising cell source with other human cell lines that are commercially available to build OOCs around.27, 28 In contrast, although polydimethylsiloxane (PDMS) has served as the principal extracellular matrix (ECM) building material in current OOCs because of its low cost, optical properties, and widespread use in soft lithography for protein patterning,29, 30 it often inadequately models the tissues it is designed to mimic. PDMS, while a very useful material that has enabled the field of soft lithography since its genesis, suffers from a wide range of issues including its high stiffness and the transient nature of surface modifications which limit the length of time cells remain viable cultured on it.31 Most problematic, however, is the absorption of proteins, drugs, and hydrophobic molecules into PDMS which complicates the PK/PD and accurate dosing of drugs.32 The use of PDMS puts a significant limitation on the models we can achieve with OOCs.
Alternatives to PDMS may offer an opportunity to mimic organ microenvironments on OOCs while evading the difficulties described above. Hydrogels that are mechanically tunable and chemically functionalized for cell attachment, such as alginate, are an alternative building material to PDMS that better mimic soft tissue microenvironments and enable long term culture.33 Native ECM protein-based hydrogels assembled in extruded micro-tubes or fibers have been shown to guide aligned tissue formation of a variety of cell types in vitro.34 Similarly, protein or blended protein-synthetic polymer nanofibers are also chemically and mechanically tunable substrates with nanoscale features and alignment similar to native matrix.35,36 With these alternative materials, we can begin to model more complex physiological states on OOCs. Models of fibrosis in the heart, for example, can be achieved in vitro by mechanically altering the ECM stiffness37 or dynamic stretch38 of tissues. Similarly, by chemically functionalizing ECM with specific proteins and/or tuning the stiffness of the substrate, stem cell differentiation can be guided to study development.39, 40 Changes in extracellular matrix composition and mechanics influence and result from pathophysiological behavior of tissues; by controlling these properties of the ECM materials, we may better control the pathophysiology in OOCs.
OOC assembly requires a toolset of fabrication techniques suitable for mass manufacturing. The development of soft lithography has led to microcontact printing of substrates to control cell and tissue structure in vitro41, 42 as well as inexpensive and relatively simplistic manufacturing of microfluidics for drug discovery and cell culture.43, 44 These techniques, though suitable for the laboratory, are largely manual and require scalable automation to meet the requirements of mass manufacturing. However, recent advances in 3D printing techniques have enabled the direct fabrication of microvasculature and tissues using a number of synthetic and biological polymers printed with and without, embedded cells.45, 46 Manufacturing tools, such as 3D printing, that can simultaneously create the structure of an OOC while also organizing its cellular components, can bring OOCs from “boutique” laboratory tools to the pharmaceutical industry.
However, manufacturing at such a large scale requires the implementation of quality controls and standards not currently established in the field. For example, if reprogrammed iPSCs are to be the model cell source for an organ, whose organ are they modeling? The genotype, phenotype, and developmental maturity of iPSC derived cell lines must be consistent and defined for us to know what the clinical comparison of the OOC is.47 Similarly, the cell culture media for organs on chips needs to be standardized. The field still relies heavily on serum-based, specific growth factor-supplemented, and/or proprietary specialized media which makes the culture of heterogeneous cell populations or the connection of different OOCs into systems difficult. As individual OOCs are assembled into larger systems, their platforms and connections should be standardized and modularized to simplify assembly and allow for dynamic adjustments to the system. The manufacture of chips suitable for the pharmaceutical industry will require controls and standards to be established for all components of OOCs.
Test: Correlating OOCs and the Patient
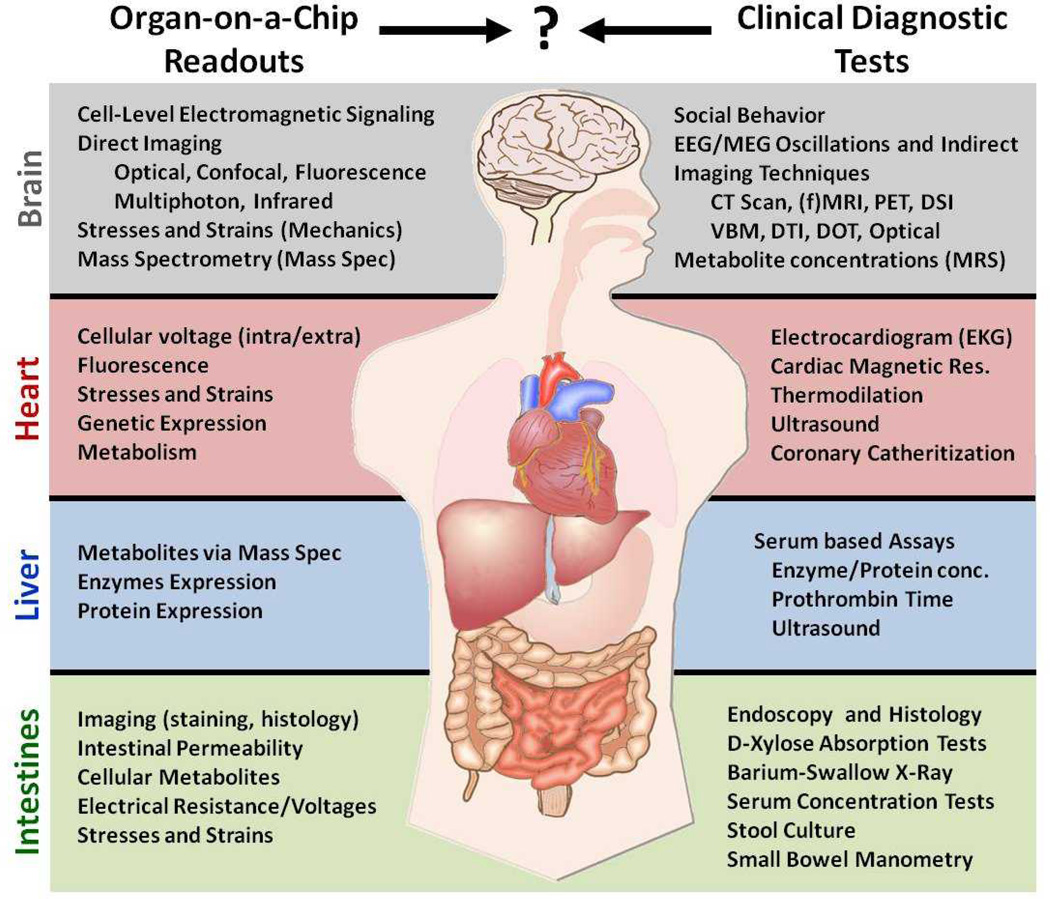
The goal of OOCs is to better mimic the human response of the patient in vitro; the readouts of OOCs must therefore map to the standard clinical diagnostics they are built to model. However, the correlation of OOC readouts to their clinical counterparts is difficult to make due to differences in measurement techniques (Figure 2). Because of these differences, there remains a data fusion problem between OOCs and the clinic. How can the data we obtain from OOCs be correlated to organ physiology and pathophysiology? By mapping the more simplified readouts of OOCs to their corresponding clinical diagnostics, we may better determine their significance. Subsequently, implementing control loop engineering techniques into OOCs will enable the manipulation of OOC readouts which can, when collected in real-time, more accurately model the living and dynamic patient.
Figure 2.
A comparison of various clinical function tests used for diagnostics against the typical readouts that are possible with Organ-on-Chip technologies for the Brain, Heart, Liver and Intestines. Although some OOC readouts can be related to functional tests (as with the Liver), the majority lack a firm relationship with a corresponding clinical diagnostic technique.
Because OOCs are simplified models of organs, their readouts are also simplified subsets of overall organ function measured in the clinic. The result is that OOCs only tell part of the story; interpreting their readouts in a clinical context is often difficult. For example, although electro/magneto encephalography (EEG/MEG) are principally measures of extracellular electric fields and currents, the exact mapping between organ-level recorded EEG/MEG and cellular-level electrode measurement commonly done in vitro is not clear.48 To interpret the more basic measurements of OOCs will require computational modeling to put the measurements in greater context. Data from organs on chips can inspire computational models in combination with clinical or other data and assumptions to build models of the patient. Calcium transients in a single cardiomyocyte, for example, can be used in a series of linked mathematical models to determine the resulting whole-heart function including clinical readouts such as imaging, stroke volume, ejection fraction, and cardiac output.49 By pairing the data obtained from OOCs with appropriately scaled, mathematical models of organ function, we can better understand their relevance to the clinic.
Although the computational models used to interpret OOCs may be complex, they will be represented by equations with inputs (OOC functional data) and outputs (organ level functional data). For the outputs of these models to mimic the dynamic states of health and disease, the inputs from OOCs must be controlled by opening and accessing the feedback loops that govern the physiology.50 Controlling OOCs in this way may offer systems capable of simulating a number of intra/extra-organ interactions.51 However, the field is currently ill-equipped to implement such control loops on chips due to the lack of real-time access to the inputs and outputs of OOC systems. Many common methods of analysis, such staining or antibody-based assays, can be destructive and suffer from significant time delays before readout. Therefore, OOC testing must move toward enabling real-time, non-destructive, and multi-modal biochemical read-write capabilities in order to actively control and monitor the biology on the chip.
There are many technologies that may help achieve real-time control over OOC inputs and outputs. Integrated electrode arrays,52 optogenetics,53 live fluorescence imaging, and various combinations of all these techniques can be used to stimulate and extract data from OOCs in real-time. More recently developed technologies such as functionalized nanoparticles and conductive polymers incorporated into bio-systems have provided targeted delivery of drugs, antigens, and other biomolecules to both perturb and screen/image systems.54, 55 Furthermore, connecting Mass Spectrometry (Mass Spec) and/or Ion Mobility Spectrometry (IMS) systems to OOCs can yield nearly instant insight into the molecular composition of circulating media or chip waste.56 By controlling the inputs of an OOC and being able to observe their effects in real-time, OOCs can be tested and treated as dynamically as the patient in the clinic.
Conclusion
The clinical failures of drugs have motivated a reassessment of how they are tested during the early stages of development. Organs on Chips are a developing toolset which may offer a more relevant means of drug testing to supplement standard cell culture and animal testing. The goal of OOCs is to better mimic the patient in vitro with the hope of providing the platform for in vitro clinical trials and customized, patient specific drug testing. Therefore, OOCs must reflect the complexity and uniqueness of the human physiology and pathology they are designed to model. Building standards and controls for the cells, ECM and synthetic materials, and fabrication tools need to be established to ensure that OOC manufacturing is sustainable and cost efficient. Finally, the readouts obtained from OOCs must translate to the clinic. This requires computational modeling to extrapolate data from OOCs to their clinical significance. By controlling the inputs and monitoring the outputs of OOCs in real-time, this clinical significance will be a better representation of the dynamic nature of the patient.
Acknowledgements
This work was funded by the Harvard School of Engineering and Applied Sciences, the Wyss Institute for Biologically Inspired Engineering, the National Institutes of Health (UH2 NS080728-01), and the Defense Advanced Research Projects Agency (W911NF-12-2-0036).
Citations
- 1.Kaitin KI. Clin Pharmacol Ther. 2010;87:356–361. doi: 10.1038/clpt.2009.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honig P, Lalonde R. Clinical pharmacology and therapeutics. 2010;87:247–251. doi: 10.1038/clpt.2009.298. [DOI] [PubMed] [Google Scholar]
- 3.Maxmen A. Nature. 2011;478:S16–S18. doi: 10.1038/478S16a. [DOI] [PubMed] [Google Scholar]
- 4.Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. Nature. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 5.Bart van dew Worp H, Howells DW, Sena ES, Porritt MJ, Rewell S, O’Collins V, Macleod MR. PLOS Med. 2010;7:e1000245. doi: 10.1371/journal.pmed.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hackman DG. JAMA. 2006;296:1731–1732. [Google Scholar]
- 7.Harrison DC, Winkle RW, Sami M, Mason J. Am Heart J. 1980;100:1046–1054. doi: 10.1016/0002-8703(80)90212-4. [DOI] [PubMed] [Google Scholar]
- 8.Echt DS, Liebson PR, Mitchell LB, Peters RW, Obias-Manno D, Barker AH, Arensberg D, Baker A, Friedman L, Greene HL, Huther ML, Richardson DW CAST Investigators. New Engl J Med. 1991;324:781–788. doi: 10.1056/NEJM199103213241201. [DOI] [PubMed] [Google Scholar]
- 9.Manning FJ, Swartz M, editors. Committee to Review the Fialuridine (FIAU/FIAC) Clinical Trials, Institute of Medicine. Review of the Fialuridine (FIAU) Clinical Trials. 1st edn. ch. 13. Washington: National Academies Press; 1995. pp. 98–99. [PubMed] [Google Scholar]
- 10.Osherovich L. SciBX. 2013;6 [Google Scholar]
- 11.Robinton DA, Daley GQ. Nature. 2013;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huh D, Hamilton GA, Ingber DE. Trends Cell Bio. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, Thorneloe KS, McAlexander MA, Ingber DE. Sci Transl Med. 2012;4:159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Science. 2010;328:1662. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grosberg A, Alford PW, McCain ML, Parker KK. Lab Chip. 2011;11:4165. doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huh D, Torisawa Y, Hamilton GA, Kim HJ, Ingber DE. Lab Chip. 2012;12:2156–2164. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- 17.Eizirk DL, Colli ML, Ortis F. Nat. Rev. Endocrinol. 2009;5:219–226. doi: 10.1038/nrendo.2009.21. [DOI] [PubMed] [Google Scholar]
- 18.Muoio DM, Newgard CB. Nat. Rev. Mol. Cell Biol. 2008;9:193–205. doi: 10.1038/nrm2327. [DOI] [PubMed] [Google Scholar]
- 19.Murtaugh LC. Development. 2007;134:427–438. doi: 10.1242/dev.02770. [DOI] [PubMed] [Google Scholar]
- 20.Ondine C, Melton DA. Nat Med. 2003;9:661–668. doi: 10.1038/nm0603-661. [DOI] [PubMed] [Google Scholar]
- 21.Pollack IF, Jakacki RI. Nature Rev. Neurology. 2011;7:495–506. doi: 10.1038/nrneurol.2011.110. [DOI] [PubMed] [Google Scholar]
- 22.Imura, Sato K, Yoshimura E. Anal. Chem. 2010;82:9983–9988. doi: 10.1021/ac100806x. [DOI] [PubMed] [Google Scholar]
- 23.Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, Matloff WJ. Lab on a Chip. 2013;13:3496–3511. doi: 10.1039/c3lc50243k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moraes C, Labuz JM, Leung BM, Inoue M, Chun T, Takayama S. Integrative Biology. 2013;5:1149–1162. doi: 10.1039/c3ib40040a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sung JH, Kam C, Shuler ML. Lab Chip. 2010;10:446–455. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- 26.Rullman JAC, Struemper H, Defranoux NA, Ramanujan S, Meeuwisse CML, van Elsas A. IEE Proc-Syst Biol. 2005;152:256–462. doi: 10.1049/ip-syb:20050053. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi K, Yamanaka S. Cell. 2006;4:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 28.Matins AM, Vunjak-Novakovic G, Reis RL. Stem Cells Rev and Rep. 2014:1550–8943. doi: 10.1007/s12015-013-9487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia Y, Whitesides GM. Angew Chem Int Ed. 1998;37:5650–5675. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 30.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 31.Mukhopadhyay R. Am Chem S. 2008;8:270–280. [Google Scholar]
- 32.Toepke MW, Beebe DJ. Lab Chip. 2006;6:1484–1486. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- 33.Agarwal A, Farouz Y, Nesmith AP, Deravi LF, McCain ML, Parker KK. Adv Funct Mater. 2013;23:3738–3746. doi: 10.1002/adfm.201203319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onoe H, Okitsu T, Itou A, Kato-Negishi M, Gojo R, Kiriya D, Sato K, Miura S, Iwanaga S, Kuribayashi-Shigetomi K, Matsunaga YT, Shimoyama Y, Takeuchi S. Nat Mater. 2013;12:584–590. doi: 10.1038/nmat3606. [DOI] [PubMed] [Google Scholar]
- 35.Badrossamay MR, McIlwee HA, Goss JA, Parker KK. Nano Lett. 2010;10:2257–2261. doi: 10.1021/nl101355x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badrossamay MR, Balachandran K, Capulli AK, Golecki HM, Agarwal A, Goss JA, Kim H, Shin K, Parker KK. Biomaterials. 2014;35:3188–3197. doi: 10.1016/j.biomaterials.2013.12.072. [DOI] [PubMed] [Google Scholar]
- 37.McCain ML, Lee H, Aratyn-Schaus Y, Kleber AG, Parker KK. PNAS. 2012;109:9881–9887. doi: 10.1073/pnas.1203007109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCain ML, Sheehy SP, Grosberg A, Parker KK. PNAS. 2013;110:9770–9775. doi: 10.1073/pnas.1304913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheehy SP, Grosberg A, Parker KK. Biomech Model Mechanobiol. 2012;11:1227–1239. doi: 10.1007/s10237-012-0419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 41.Wilbur JL, Kumar A, Kim E, Whitesides GM. Adv Mater. 1994;6:600–604. [Google Scholar]
- 42.Mrksich M, Dike LE, Tien J, Ingber DE, Whitesides GM. Exp Cell Res. 1997;235:305–313. doi: 10.1006/excr.1997.3668. [DOI] [PubMed] [Google Scholar]
- 43.Dittrich PS, Manz A. Nat. Rev Drug Discovery. 2006;5:210–218. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- 44.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Annu Rev Biomed Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 45.Wu W, DeConinck A, Lewis JA. Adv Funct Mater. 2011;23:178–183. [Google Scholar]
- 46.Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA. Adv Mater. 2014 doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 47.Sheehy SP, Pasqualini F, Grosberg A, Park SJ, Aratyn-Schaus Y, Parker KK. Stem Cell Reports. 2014;2:282–294. doi: 10.1016/j.stemcr.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buzaki G, Anasassiou CA, Koch C. Nature Rev. Neurosci. 2012;13:407–420. doi: 10.1038/nrn3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shim EB, Leem CH, Abe Y, Noma A. Phil Trans R Soc A. 2006;364:1483–1500. doi: 10.1098/rsta.2006.1782. [DOI] [PubMed] [Google Scholar]
- 50.Wikswo JP, Prokop A, Baudenbacher F, Cliffel D, Csukas M, Velkovsky B. IEEE Proc-Nanobiotechnol. 2006;153:81–101. doi: 10.1049/ip-nbt:20050045. [DOI] [PubMed] [Google Scholar]
- 51.LeDuc PR, Messner WC, Wikswo JP. Annu. Rev. Biomed. Eng. 2011;13:369–396. doi: 10.1146/annurev-bioeng-071910-124651. [DOI] [PubMed] [Google Scholar]
- 52.Blake AJ, Rodgers FC, Bassuener A, Hippensteel JA, Pearce TM, Pearce TR, Zarnowska ED, Pearce RA, Williams JC. J. Neurosci. Methods. 2010;189:5–13. doi: 10.1016/j.jneumeth.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yizhar, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 54.Nambiar S, Yeow JTW. Biosens Bioelectron. 2011;26:1825–1832. doi: 10.1016/j.bios.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 55.Sharma P, Brown S, Walter G, Santra S, Moudgil B. Adv Colloid Interface Sci. 2006;123:471–485. doi: 10.1016/j.cis.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 56.Enders JR, Goodwin CR, Marasco CC, Seale KT, Wikswo JP, Mclean JA. Spectrosc. Supp. Curr. Trends Mass Spectrom. 2011:18–23. [Google Scholar]