Abstract
Sampling – the process of collecting, preparing, and introducing an appropriate volume element (voxel) into a system – is often under appreciated and pushed behind the scenes in lab-on-a-chip research. What often stands in the way between proof-of-principle demonstrations of potentially exciting technology and its broader dissemination and actual use, however, is the effectiveness of sample collection and preparation. The power of micro- and nanofluidics to improve reactions, sensing, separation, and cell culture cannot be accessed if sampling is not equally efficient and reliable. This perspective will highlight recent successes as well as assess current challenges and opportunities in this area.
Introduction
As Richard Feynman famously declared in 1959, “there’s plenty of room at the bottom.1” Now that we are well on our way toward manipulating systems on the micron and sub-micro scales, how do we use these remarkable technologies to learn about the world around us? Micro- and nanofluidics provide exquisite control over small volumes of fluids. Their capabilities to react, separate, culture cells, and otherwise interrogate materials in unprecedented ways can open new views of the world around us if only we can effectively collect, prepare, and introduce appropriate bits of our world into them.
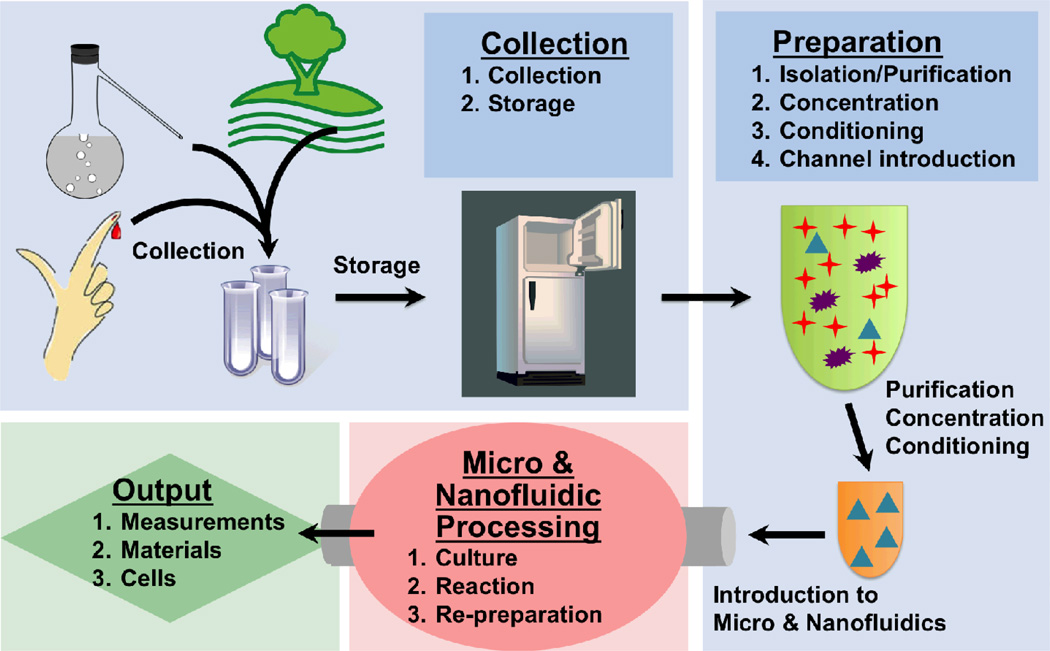
For real-world applications a voxel of interest, regardless of source, typically goes through four steps to reach a useful outcome: (1) collection, (2) preparation, (3) processing, and (4) providing an output (Fig. 1). While most micro- and nanofluidic research implies practical utility, a significant portion focuses more on the latter parts of these steps while neglecting the importance and challenges of collection and preparation. Traditionally, having material to be processed in sufficient purity, concentration, and condition has been assured. In that paradigm micro- and nanofluidic efforts focused on developing smaller reactions (e.g. PCR and sequencing), better separation (e.g. capillary electrophoresis), enhanced sensitivity (e.g. immunoassays), or more physiologic cell culture. Appropriately prepared materials went in and information or processed material came out. As we send more of our labs-on-a-chip out from the lab into hands of users with real-world applications or as these devices are made to accommodate raw clinical or environmental samples, former assumptions regarding ready access to clean materials no longer hold. DNA samples may have impurities and too much salt. Proteins may be partially degraded. And unlike cell lines comprised predominantly from one type of reasonably well-characterized cell, patient samples may be mixtures of various ill-characterized cells. Under such circumstances, even if the micro- and nanofluidic processing step works well, the output results can be inferior, wrong, or difficult to reproduce. Such challenges and difficulties are not unique to the field of micro- and nanofluidics. Indeed, the National Institutes of Health (NIH) and others have highlighted the tremendous challenges in ensuring reproducibility of biomedical research in general.2,3
Figure 1.
This perspective focuses on collection and preparation of materials for introduction into micro- and nano-fluidic devices. We collectively call these steps “sampling”. Much effort in the field of lab on a chip has been devoted to processing and output steps, but less attention has traditionally been paid to collection and preparation. In real world applications however, effective processing and good output depends critically on these earlier steps.
Challenges may also present opportunities. Accordingly, some recent advances in micro- and nanofluidics have made significant impact because they contribute to the domains of collection and preparation, rather than downstream analysis or processing steps. A prominent example is the field of circulating tumor cell (CTC) capture devices pioneered by Toner, Nagrath, and others.4,5 The functional goal of these devices is to purify CTC targets from large volumes (at least relative to typical microfluidic devices) of whole blood and enrich them in a solid state capture scheme after which the cells are simply counted by microscopy or lysed for off-chip analysis. This application, now considered part of microfluidic canon, was somewhat counter-intuitive as a “micro” fluidic project. By focusing on sampling rather than processing steps, these devices broke new ground and highlighted the tremendous (and largely unrealized) opportunities available in the field.
Many of us have been taught that not all problems in collection and sampling should or even can be addressed by micro- and nanofluidics. For example, many chemical analysis textbooks caution that micro- and nanochannel methods are generally ill-suited for tackling issues requiring exceptionally high throughput or the detection of remarkably sparse targets. Consider a target present at a concentration of 10 particles/mL in a sample (not an unreasonable number for CTCs). Simple logic dictates that in order to have a reasonable chance of detecting the target, one would need to examine at least 100 µL of sample. Further, since sampling is governed by Poisson statistics, the probability of finding n target particles in a sample voxel containing an average of s target particles is given by (Eqn. 1):6
The probability of a false negative within the 100 µL of the sample (that is finding no target particle, n=0, when s=1) is 37 percent! Although P(0,s) decreases exponentially with increasing number of target particles, s, this treatment assumes an ideal system. To accommodate imperfect handling and device operation, one must thus process an even larger volume. Previously, the message of such analysis was interpreted to be that microfluidic sampling for targets present in low concentration is challenging and that such projects are best avoided. Overcoming such challenges and bias, however, especially when coupled with a compelling need, presents many opportunities as exemplified by work in CTC capture.
Challenges and Opportunities
Challenges can arise before the sample ever reaches the lab. What is collected and how the sample is stored can have enormous impact on downstream preparation and processing steps. Even for patients with the same disease, blood sample properties as well as target purity and concentration can vary considerably depending on treatments, therapies, and procedures patients may have undergone prior to collection. Storage of the sample can further aggravate these differences, or in some cases, introduce heterogeneity by itself. One biosample preservation study attributed the “majority” of errors in biobank analysis to preanalytical variables such as storage stability, type of anticoagulant used, or the collection technique.7 A second report put that number at 60–90 percent.8 Micro- and nanofluidics may address these issues by either enabling point of care (POC) devices that can collect, prepare, process, and output biosamples in the field and on demand. In cases where storage cannot be avoided or POC analysis is not possible a variety of micro- and nanofluidic technologies may be able to circumvent or temper the problems posed by sample heterogeneity resulting from collection and storage. Clearly, careful consideration of such factors will be fundamental to the success of any sampling effort.
Once a sample safely reaches the lab, it must undergo various sample preparation processes. We categorize the various steps that may be needed into four categories: (1) isolation and cleanup such as dissolution, cell lysis, filtration, centrifugation, (2) concentrating (or diluting) to an appropriate concentration, (3) conditioning of the sample by buffer exchange, adjusting pH, chelating, reacting with labels, etc., and (4) introduction to the device. Since the volume of sample required by micro- and nanofluidic analysis systems is typically small, and sample can be precious and not available in large volumes (e.g. banked patient blood or rodent blood samples), it would be ideal if sample preparation were also micro-scale. Over a half-century later we have not yet realized the “hundred tiny hands” Feynman contemplated1 to efficiently perform these steps to prepare materials for micro- and nanofluidic processing. Due to general sample complexity and the amount of work required, sample preparation is still considered the “weak link” in practical microfluidic applications.6 Conventional techniques (such as centrifugation and turbulent mixing) often prove difficult to miniaturize or, once miniaturized, encounter problems not normally apparent at larger scales (like filter clogging or small volumes of sample evaporating). Developing miniaturized sample preparation techniques can be further complicated by the need for compatibility with other aspects of the micro- or nanofluidic device (assay chemistry, detection methods, etc.) and the wide range of sample characteristics (viscosities, particulate content, etc) that may need to be accommodated.
Although responsible for some of the “problems” associated with small length scales, micro- and nano-scale phenomena have also been leveraged to accomplish sample preparation tasks in ways not possible at the macro-scale. For example, rather than lament inefficient turbulent mixing, laminar flow can be taken advantage of to perform filtering operations (the so-called H-filter that is described in more detail below). Problems of fast evaporation associated with high surface area to volume ratios can be utilized for sample concentration.9 Once the appropriate sample volume is identified and prepared, that voxel must be introduced into the device, flowed, metered, and otherwise controlled for appropriate preparation, processing, and readout – all preferably without the use of bulky external equipment. Unfortunately, small channel dimensions mean clogging can become a substantial issue. Voxels consisting of either large particles or dense suspensions can pose problems to physical filters, valves, and other elements. Even materials normally considered small – such as chromatin – can frustrate studies being conducted in sufficiently miniaturized (i.e. nano-scale) dimensions.
The nature of the target materials within a sample can also present obstacles. Cells, proteins, or other delicate materials need to be protected from certain environmental conditions (e.g. pH or temperature), agents present in the sampled milieu (e.g. enzymes that may degrade a biomarker), chemicals and processes used for preparation and interrogation on-chip (e.g. lytic agents), or even general sample handling (e.g. channel-induced shear). The fine control afforded by micro- and nano-scale tools can often be used to circumvent these sample introduction or conditioning issues; if not, further engineering strategies must be considered. These targets, delicate or not, can be found in a variety of suspensions. Variations in fluid viscosity, density, and even viscous behavior (whole blood, for example, is non-Newtonian) must be considered during system design and accounted for during conditioning steps. Finally, the target species in a sampled volume is rarely present at appropriate purity and concentration for downstream interrogation and analysis. Therefore, much attention must be devoted to discarding extraneous elements of the sample that may interfere with the ability to detect and probe the target. Even after such procedures are complete, buffer changes and similar operations may be necessary to prepare the voxel for successive processing or output steps.
Examples of Specific Workarounds
In seeking to address these shortcomings, the greatest advantage of micro- and nanofluidics remains the unique benefits afforded by operating at such small scales. This offers inspiration not just for augmenting strengths, but also for addressing weaknesses; a list of some specific workarounds can be found in Table 1. To improve sampling, researchers should look beyond miniaturizing conventional macro-scale systems (e.g. filters) and instead focus on leveraging unique phenomena of the micro-scale. A classic example of such an approach is the H-filter which takes advantage of laminar flow and short diffusion distances to create a fluidic element capable of separating particles by size.10 Remarkably the filter does not clog (so long as the particles are not larger than the channel itself) and can be dynamically tuned to sort particles of different sizes by adjusting the flow rate.
Table 1.
Common challenges and examples of some available workarounds for sampling.
| Problem | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Collection | Storage | Purification | Concentration | Conditioning | Introduction | Clogging | Retrieval | ||
| Solutions | Nano-scavengers11,12 | ✓ | ✓ | ✓ | ✓ | ||||
| Paper μ-fluidics14,16 | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| Electrophoresis23 | ✓ | ✓ | ✓ | ✓ | |||||
| H-filter10 | ✓ | ✓ | |||||||
| ATPS19 | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| μ -Centrifuges20–22 | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| Nano-channels23–25 | ✓ | ✓ | |||||||
| Tunable channels25 | ✓ | ✓ | ✓ | ✓ | |||||
| Oil dehydration32 | ✓ | ||||||||
| Droplet μ-fluidics17 | ✓ | ✓ | ✓ | ||||||
| EWOD18 | ✓ | ✓ | ✓ | ||||||
| μ-fluidic dispenser26 | ✓ | ||||||||
| Valving30,31 | ✓ | ||||||||
| Tapering24 | ✓ | ✓ | |||||||
| Biological sampling | ✓ | ✓ | ✓ | ||||||
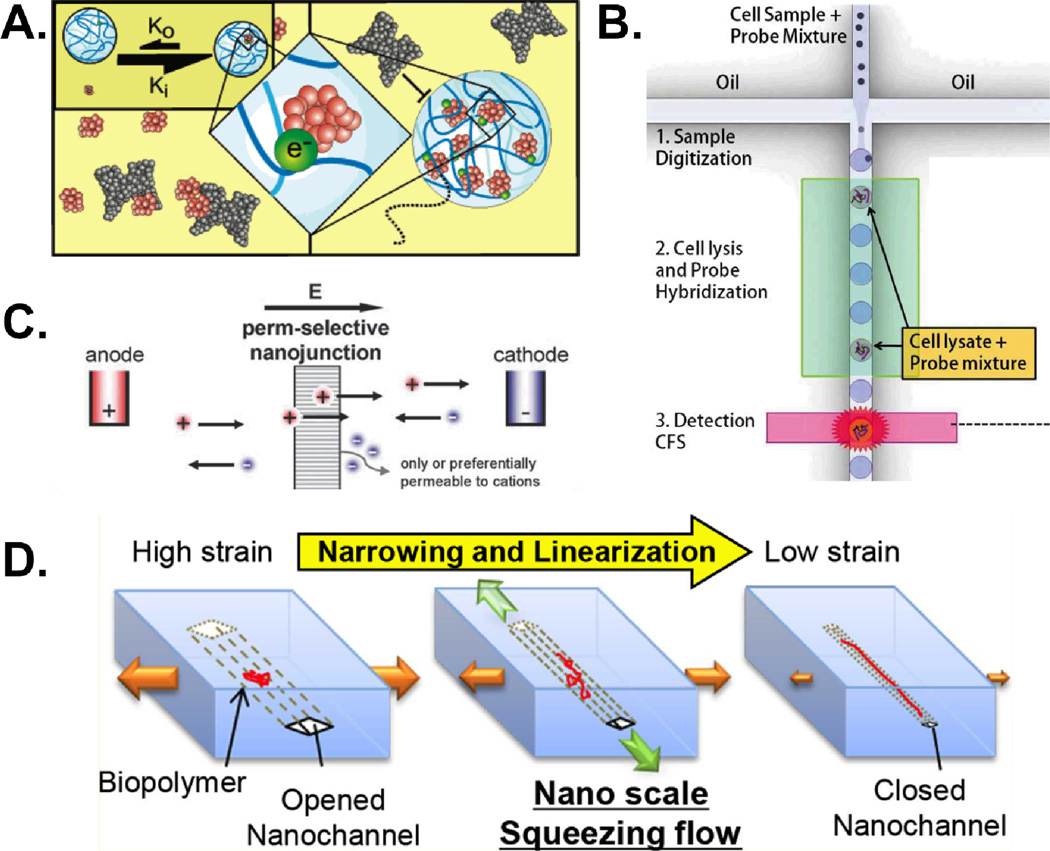
Nanotechnology also has been used to improve on-chip sampling. Luchini et al developed a new class of core-shell nano-particle capable of selectively sequestering, protecting, and subsequently eluting a target analyte from a complex sample. The porous shell is capable of excluding high molecular weight species – such as albumin – while allowing smaller molecules to diffuse through. The core region consists of a “bait” material meant to attract and sequester a specific class of analyte. Essentially, these “nano-scavengers” serve as a two-dimensional purification process – selecting based on size as well as affinity (Fig. 2A).11 Capture and subsequent elution were found to enrich proteins such as PDGF from spiked biosamples (here serum) by orders of magnitude. Sequestration by these small molecule-scavenging nano-particles even protected target molecules from enzymatic degradation.12 Greater functionality would increase this technology’s utility even further. Combined with existing micro- and nanofluidic devices, these nano-particles could open up new avenues in sampling, offering increased specificity, selectivity, and efficiency of separation.
Figure 2.
Some examples of nano-fludic workarounds for common sampling challenges. (A) Core-shell nanoparticles are capable of selecting targets based on size and affinity. When trapped, the nanoparticle protects the target from possible enzymatic degradation. Reprinted with permission from ref. 11. Copyright 2008 American Chemical Society (B) By creating discrete water droplets in an oil-carrier phase, droplet microfluidics increases local concentrations and allows for digitized operations. Adapted from ref 17 with permission from The Royal Society. (C) Nanofluidic enrichment/depletion takes advantage of EDL’s on the order of channel dimension to use a porous, polar membrane for charge-based particle selection. Reproduced from ref. 23 with permission from The Royal Society of Chemistry. (D) Size adjustable nanochannels can address issues with sample introduction and clogging. Reprinted with permission from ref. 25. Copyright 2012 American Chemical Society.
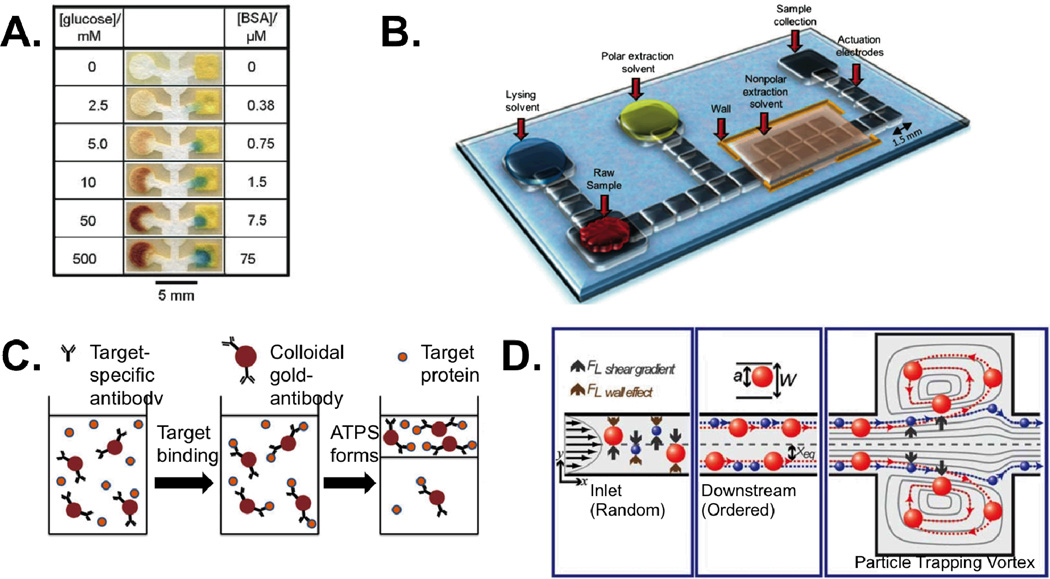
New methods for sampling are being developed in paper devices. Although paper microfluidics is not new – laminar flow test strips were introduced in the 1980’s – this technology may be the most commercially successful of all microfluidic efforts to date.13 Therefore efforts to revisit paper as a substrate are certainly warranted. Temporal and spatial flow control as well as traditional microfluidic schemes can now be realized in paper.14 Since flow in these devices is self-driven, paper microfludic sampling systems generally require fewer support systems. Integrated dipstick tests that simply wick sample from a reservoir through an assay region are already common in commercial applications and new designs that passively control flow rate and cutoff times are being incorporated into next-generation techniques.15 Increasingly, these assays also incorporate multiplexing – analyzing a single voxel for several biomarkers simultaneously (Fig. 3A).16 By probing multiple parameters of a sample, multiplexing reduces sample consumption and parallelizes experiments – further improving areas of micro- and nanofluidic sampling that are already seen as strengths.
Figure 3.
Some examples of micro-fluidic workarounds for common sampling challenges. (A) Paper devices can be used to perform multiplexed sampling operations, here measuring glucose and protein content in urine. Reproduced from ref. 16 with permission. (B) Digital microfluidic manipulation of droplets is used to lyse and extract a small molecule analyte from ~1 µL of tissue homogenate. Reproduced from ref. 18 with permission. (C) An aqueous two-phase system is used in concert with immuno-active particles to purify a target. Adapted from ref. 19 with permission. (D) Shear gradient and wall effect forces can be balanced to selectively trap larger particles in microvortices. Adapted from ref. 21 with permission from The Royal Society of Chemistry.
Just as paper microfluidics challenges convention about device structure and composition, digital microfluidics – microfluidics that deal with discrete (digital) voxels or droplets) – are opening up a world of microfluidics both inside and outside of channels. Droplet microfluidics rely on oil-water biphasic systems to create aqueous droplets (often nL to pL in volume) in an oil carrier phase within a microchannel. Each droplet can then be treated as an independent reaction volume allowing researchers to increase local concentrations of analytes, increase throughput, or perform binary operations such as droplet mixing, spitting, etc. One outstanding example of this technology is a report by Rane et al of a droplet-based system capable of executing many important sampling operations. The device was shown to encapsulate single pathogenic cells, lyse them, and carry out hybridization to detect bacterial RNA without amplification and entirely on-chip (Fig. 2B).17 Other droplet techniques can be realized outside the constraints of a microchannel. Electrowetting on a dielectric (EWOD) uses electrode arrays to control tiny droplets of water, mixing, merging, splitting, and moving them with applied voltage. For example, Wheeler and colleagues used an EWOD system to perform an estrogen extraction and quantification starting from 1 µL of biopsied breast tissue homogenate (Fig. 3B), a volume several orders of magnitude smaller than is used in conventional sample preparation methods.18 Both of these “sample-to-answer” devices ably demonstrate the potential of digital microfluidic platforms to miniaturize and automate sample preparation procedures.
Aqueous two-phase systems (ATPSs) – methods involving two immiscible yet water-based solutions – are another concept that can be used without the constraint of channels and associated problems such as clogging. Well-chosen ATPSs can also take advantage of differences in target properties to induce preferential partitioning to one phase or the other in order to purify and concentrate an analyte (Fig. 3C). Because of its aqueous nature, this method is particularly well suited for handling delicate materials such as cells, organelles, and biomolecules. The Kamei group used a micellar ATPS along with functionalized nano-particles to concentrate a target protein (transferrin) prior to use in a lateral flow assay, improving the test’s limit of detection by a full order of magnitude.19
Alternative, on-chip strategies to combat clogging include two power-free centrifuge analogs developed by the Beebe and Di Carlo groups, respectively. Warrick et al developed a device for concentrating small numbers of cells in a settling chamber using surface tension mediated pumping.20 Mach et al balanced shear gradient and wall effect forces to create vortices capable of trapping large particles for washing or sorting (Fig. 3D).21 Similar balancing of shear and wall forces can be used to sort particles by size and with high resolution through a process called inertial focusing.22 Inertial systems also utilize high flow rates allowing high throughput; a critical requirement for many sample preparation applications. In each case, both groups were able to recapitulate critical sample preparation processes in ways that took advantage of micro-scale phenomena and without simply miniaturizing the conventional system. Future efforts pertaining to micro- and nano-sampling should seek similar inspiration.
The still-smaller dimensions specific to nanochannels can offer unique advantages. Nanofluidic ion depletion utilizes the fact that, in nanochannels, the thickness of the electric double layer (EDL) of ions lining a charged surface can approach the width of the channel itself. In such cases a depletion-enrichment effect can occur where counter-ions (those with opposite polarity to the surface) are allowed to pass through a porous membrane while similarly charged particles are sieved – even if otherwise small enough to pass (Fig. 2C).23 In other applications, nanochannels can be useful for linearizing single fibers of DNA or chromatin for imaging. While the narrower nanochannels provide the highest degree of linearization, their extremely small size also exacerbates the common problems of sample loading and clogging. One workaround is to use a gradually narrowing tapered nanochannel24. Another workaround is the use of elastomeric nanochannels where the cross-sectional dimensions can be widened and narrowed reversibly. The widened state allow for easier sample loading and less clogging. Subsequent gradual narrowing of the channel leads to linearization of the DNA or chromatin fibers through a combination of hydrodynamic and confining forces (Fig. 2D). Eventually, the biopolymers are trapped in their elongated state for imaging of the DNA or histone modifications using fluorescence microscopy.25
Sample introduction is often a non-trivial process, especially when multiple samples or reagents must be delivered. Rather than using microfluidics exclusively for processing, some groups have developed microspotting devices capable of reagent/sample delivery as well. One group of researchers led by Wang built a microfluidic device capable of delivering nanoliter volumes. They used this device, along with a custom microwell plate, to perform parallelized, low volume PCR.26 In some microfluidic applications, ensuring both gentle sample introduction and final sample retrieval is important. One example is the use of microfluidics for manipulation of precious cells for in vitro fertilization. Heo and colleagues used a microfluidic funnel design to culture embryos in vitro under static and pulsatile conditions. Under pulsatile flow, embryo development approached in vivo levels in terms of development and cell number. Importantly, the micro-funnel design allowed for easy device output via cell retrieval. Embryos developed on chip were subsequently implanted and showed improved pregnancy rates compared to embryos from conventional static dish cultures.27
Some sampling challenges require more than clever devices and materials. Taking advantage of “biological” sampling or natural processes that collect, purify, or concentrate the target can be useful. CTCs and exosomes – cells or parts of cells from diseased tissue in the body – present an instructive example. Although the numbers of CTCs and exosomes in blood is scarce, that material, once obtained, is valuable as a high-fidelity representation of actual diseased tissue; thus sometimes the process is referred to as a non-invasive liquid biopsy. Isolated CTCs or exosomes can then serve as ample repositories that may reveal further information about the patient’s disease. While bloodborne DNA, RNA, or protein biomarkers released from diseased tissue may become too diluted or degraded, captured CTCs and exosomes offer opportunities to obtain these valuable diagnostic indicators in greater concentration and/or purity. Similarly, scientists may be able to apply these tactics to other targets and identify opportunities where nature has already performed a portion of the sampling work herself. Identifying areas where a “biological” sampling strategy would be feasible could lessen the burdens technologists must overcome and may lead to other interesting insights and opportunities.
Conclusion
Valuable technical and intellectual framework useful for target collection, purification, concentration, conditioning, and introduction already exist in many places. While some advances simply create miniaturized versions of macro-scale procedures, many of the most successful technologies are disruptive and leverage unique phenomena of the micro- and nano-scale. A most important need for accelerating this area is a better awareness and appreciation of the challenges and opportunities. Recent advances have shown sampling – the collection and preparation of a target– to be a useful and exciting goal in and of itself. Development of sampling technology is also a key to reaching the ultimate lab-on-a-chip goal of a “sample-to-answer” device or a system capable of real-time monitoring and environmental control.28,29 Micro- and nanofluidics is not a panacea but it can be an extremely powerful tool; one that may find ends more worthy than even Feynman’s miniaturized encyclopedia. We just need to be able to sample the world and introduce it into the devices more efficiently.
Acknowledgements
We thank the NIH (GM096040 and HG004653) for funding. JML gratefully acknowledges support from a US Department of Education GAANN fellowship and the Microfluidics in Biomedical Sciences Training Grant (NIH T32 EB005582).
Works cited
- 1.Feynman RP. Eng. Sci. 1960;23:22–36. [Google Scholar]
- 2.Francis Francis S., Tabak Lawrence A. Nature. 2014;505:612–613. doi: 10.1038/505612a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begley CG, Ellis LM. Nature. 2012;483:531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 4.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Engl N. N. Engl. J. Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 6.Mariella R. Biomed. Microdevices. 2008;10:777–784. doi: 10.1007/s10544-008-9190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson DH, Banks RE. PROTEOMICS - Clin. Appl. 2010;4:250–270. doi: 10.1002/prca.200900220. [DOI] [PubMed] [Google Scholar]
- 8.Hubel A, Aksan A, Skubitz APN, Wendt C, Zhong X. Biopreservation Biobanking. 2011;9:237–244. doi: 10.1089/bio.2010.0034. [DOI] [PubMed] [Google Scholar]
- 9.Walker GM, Beebe DJ. Lab. Chip. 2002;2:57–61. doi: 10.1039/b202473j. [DOI] [PubMed] [Google Scholar]
- 10.Brody JP, Osborn TD, Forster FK, Yager P. Sens. Actuators Phys. 1996;54:704–708. [Google Scholar]
- 11.Luchini A, Geho DH, Bishop B, Tran D, Xia C, Dufour RL, Jones CD, Espina V, Patanarut A, Zhou W, Ross MM, Tessitore A, Petricoin EF, Liotta LA. Nano Lett. 2008;8:350–361. doi: 10.1021/nl072174l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longo C, Patanarut A, George T, Bishop B, Zhou W, Fredolini C, Ross MM, Espina V, Pellacani G, Petricoin EF, Liotta LA, Luchini A. PLoS ONE. 2009;4:e4763. doi: 10.1371/journal.pone.0004763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sin Mandy LY, Gao Jian, Liao Joseph C, Wong Pak Kin. J. Biol. Eng. 2011;5 doi: 10.1186/1754-1611-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osborn JL, Lutz B, Fu E, Kauffman P, Stevens DY, Yager P. Lab. Chip. 2010;10:2659. doi: 10.1039/c004821f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutz BR, Trinh P, Ball C, Fu E, Yager P. Lab. Chip. 2011;11:4274. doi: 10.1039/c1lc20758j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Angew. Chem. Int. Ed. 2007;46:1318–1320. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rane TD, Zec HC, Puleo C, Lee AP, Wang T-H. Lab. Chip. 2012;12:3341. doi: 10.1039/c2lc40537g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mousa NA, Jebrail MJ, Yang H, Abdelgawad M, Metalnikov P, Chen J, Wheeler AR, Casper RF. Sci. Transl. Med. 2009;1:1ra2–1ra2. doi: 10.1126/scitranslmed.3000105. [DOI] [PubMed] [Google Scholar]
- 19.Mashayekhi F, Le AM, Nafisi PM, Wu BM, Kamei DT. Anal. Bioanal. Chem. 2012;404:2057–2066. doi: 10.1007/s00216-012-6278-y. [DOI] [PubMed] [Google Scholar]
- 20.Warrick J, Casavant B, Frisk M, Beebe D. Anal. Chem. 2010;82:8320–8326. doi: 10.1021/ac101866p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mach AJ, Kim JH, Arshi A, Hur SC, Di Carlo D. Lab. Chip. 2011;11:2827. doi: 10.1039/c1lc20330d. [DOI] [PubMed] [Google Scholar]
- 22.Di Carlo D, Irimia D, Tompkins RG, Toner M. Proc. Natl. Acad. Sci. 2007;104:18892–18897. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SJ, Song Y-A, Han J. Chem. Soc. Rev. 2010;39:912. doi: 10.1039/b822556g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westerlund F, Persson F, Kristensen A, Tegenfeldt JO. Lab. Chip. 2010;10:2049. doi: 10.1039/c004878j. [DOI] [PubMed] [Google Scholar]
- 25.Matsuoka T, Kim BC, Huang J, Douville NJ, Thouless MD, Takayama S. Nano Lett. 2012;12:6480–6484. doi: 10.1021/nl304063f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Zhou Y, Qiu H, Huang H, Sun C, Xi J, Huang Y. Lab. Chip. 2009;9:1831. doi: 10.1039/b901635j. [DOI] [PubMed] [Google Scholar]
- 27.Heo YS, Cabrera LM, Bormann CL, Shah CT, Takayama S, Smith GD. Hum. Reprod. 2010;25:613–622. doi: 10.1093/humrep/dep449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovarik ML, Gach PC, Ornoff DM, Wang Y, Balowski J, Farrag L, Allbritton NL. Anal. Chem. 2012;84:516–540. doi: 10.1021/ac202611x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chin CD, Linder V, Sia SK. Lab. Chip. 2012;12:2118. doi: 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- 30.Unger MA, Chou H-P, Thorsen T, Scherer A, Quake SR. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 31.Gu W, Zhu X, Futai N, Cho BS, Takayama S. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15861–15866. doi: 10.1073/pnas.0404353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He M, Sun C, Chiu DT. Anal. Chem. 2004;76:1222–1227. doi: 10.1021/ac035196a. [DOI] [PubMed] [Google Scholar]