Abstract
The relationship between synaptic excitation and inhibition (E/I ratio), two opposing forces in the mammalian cerebral cortex, affects many cortical functions like feature selectivity and gain1,2. Individual pyramidal cells show stable E/I ratios in time despite fluctuating cortical activity levels because when excitation increases, inhibition increases proportionally through the increased recruitment of inhibitory neurons, a phenomenon referred to as excitation-inhibition balance3–9. However, little is known about the distribution of E/I ratios across pyramidal cells. Through their highly divergent axons inhibitory neurons indiscriminately contact most neighboring pyramidal cells10,11. Is inhibition homogeneously distributed or is it individually matched to the different amounts of excitation received by distinct pyramidal cells? Here we discover that pyramidal cells in layer 2/3 of mouse primary visual cortex (V1) each receive inhibition in a similar proportion to their excitation. As a consequence E/I ratios are equalized across pyramidal cells. This matched inhibition is mediated by parvalbumin-expressing (PV) but not somatostatin-expressing (SOM) inhibitory neurons and results from the independent adjustment of synapses originating from the same PV cell but targeting different pyramidal cells. Furthermore, this match is activity-dependent as it is disrupted by perturbing pyramidal cell activity. Thus, the equalization of E/I ratios across pyramidal cells reveals an unexpected degree of order in the spatial distribution of synaptic strengths and indicates that the relationship between cortex’s two opposing forces is stabilized not only in time but also in space.
To determine the distribution of E/I ratios among layer 2/3 neighboring pyramidal cells (Fig. 1a), we used adeno-associated virus (AAV) to conditionally express Channelrhodopsin-2 (ChR2)12–14 in Scnn1a-Cre-Tg3 mice and photoactivated layer 4 excitatory neurons, one of the main sources of synaptic excitation to layer 2/3, in acute visual cortical slices (Extended Data Fig. 1). We compared the E/I ratios between 2–4 simultaneously recorded layer 2/3 pyramidal cells (inter-soma distance 39.4 ± 2.5 μm, mean ± s.e.m.; Extended Data Fig. 2) voltage clamped alternatively at the reversal potential for synaptic inhibition and excitation to isolate excitatory postsynaptic currents (EPSCs) and disynaptic inhibitory postsynaptic currents (IPSCs), respectively. EPSC amplitudes greatly varied between simultaneously recorded neurons and so did IPSC amplitudes (Fig. 1b). Despite the heterogeneous distributions of EPSC and IPSC amplitudes among pyramidal cells, however, we found a strong correlation between their amplitudes. That is, neurons with larger EPSCs also received larger IPSCs (Fig. 1c, e). As a consequence, the distribution of E/I ratios across pyramidal cells varied much less than the distributions of EPSC and IPSC amplitudes (Fig. 1d, f) and much less than if EPSCs and IPSCs were randomly paired between cells (Extended Data Fig. 2). These data indicate that E/I ratios are equalized across pyramidal cells.
Figure 1. Equalized E/I ratios across pyramidal cells.
(a) Two alternative models for the spatial distribution of E/I ratios across pyramidal cells. Top: the divergent axons of inhibitory neurons (IN) homogenously inhibit neighboring pyramidal cells. Pyramidal cells receiving more excitation have a larger E/I ratio. Bottom: despite divergent axons, inhibitory neurons generate larger inhibition in pyramidal cells receiving more excitation. Accordingly E/I ratios are equalized across pyramidal cells. (b) Left, schematic of experiments. Scnn1a-Cre-Tg3 mice with ChR2 in layer 4 excitatory neurons. Right, monosynaptic EPSCs and disynaptic IPSCs from four simultaneously recorded layer 2/3 pyramidal cells in response to layer 4 photoactivation. Note larger IPSCs in neurons receiving larger EPSCs. (c) EPSC amplitudes of the four neurons in (b) plotted against their IPSC amplitudes. Left: absolute amplitudes. Right, normalized amplitudes. EPSC (or IPSC) amplitudes are normalized by the mean of the simultaneously recorded EPSC (or IPSC) amplitudes. Lines: linear regression fits. (d) Distributions of normalized EPSC and IPSC amplitudes, and of normalized E/I ratios for the experiment in (b). E/I ratios are normalized by the mean of the simultaneously recorded ratios. Note narrower distribution of E/I ratios as compared to EPSCs or IPSCs. For computing average relative deviations, see Methods. (e) Summary graphs of normalized EPSCs and IPSCs from 20 similar experiments (n = 51 cells). Line: linear regression fit (R2 = 0.65, P < 0.0001). (f) Summary graphs of average relative deviations from 20 similar experiments. Bars: mean ± s.e.m. The average relative deviations of E/I ratios are 50% smaller than those of EPSCs (P < 0.0001) or IPSCs (P < 0.0001). (g) Left, schematic of experiments. Right, connectivity rates from PV and SOM cells to pyramidal cells.
This equalization could occur if distinct layer 2/3 pyramidal cells each receives inhibition from a “private” set of inhibitory neurons such that the excitatory afferents that more strongly excite a pyramidal cell also more strongly excite its private inhibitory neurons. However, the two classes of inhibitory neurons, PV and SOM cells, that provide most inhibition to layer 2/3 pyramidal cells showed broad connectivity with pyramidal cells (97% and 93%, respectively, Fig. 1g), as previously shown10,11, thus precluding the private connectivity.
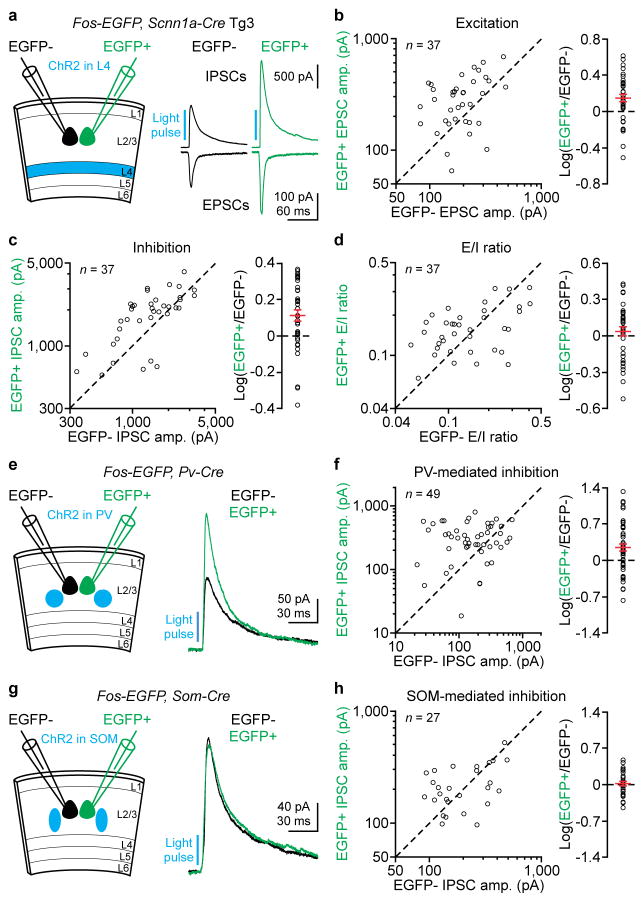
Alternatively, the correlation between excitation and inhibition could be an artifact of the slicing procedure, whereby damaged neurons receive less excitation and less inhibition. To address this possibility we used an independent marker to identify neurons receiving more excitation. We utilized Fos-EGFP mice in which the promoter of the activity-dependent immediate early gene Fos drives Fos-EGFP expression, as EGFP+ neurons receive more excitation than EGFP− neurons15. EGFP+ neurons were predominantly pyramidal cells (Extended Data Fig. 3). We photostimulated layer 4 in acute slices from Fos-EGFP, Scnn1a-Cre-Tg3 mice and simultaneously recorded pairs of EGFP+ and nearby EGFP− layer 2/3 pyramidal cells. Layer 4 activation generated larger EPSCs in EGFP+ neurons in 78% of all recorded pairs and EGFP+ neurons received, on average, 40% larger EPSCs (Fig. 2a, b, the average logarithm of EGFP+/EGFP− ratios was 0.15). Importantly, EGFP+ neurons also received larger disynaptic IPSCs (Fig. 2a, c). Consequently, the E/I ratios of EGFP+ and EGFP− neurons were similar (Fig. 2d).
Figure 2. PV cell-mediated inhibition matches layer 4-mediated excitation.
(a) Left, schematic of experiments. Fos-EGFP, Scnn1a-Cre-Tg3 mice with ChR2 in layer 4 excitatory neurons. Right, monosynaptic EPSCs and disynaptic IPSCs from simultaneously recorded EGFP− and EGFP+ neurons in response to layer 4 photoactivation. Note larger synaptic currents in EGFP+ neuron. (b–d) Summary graphs of 37 similar experiments. (b) Left, EPSC amplitudes in EGFP+ neurons plotted against those in EGFP− neurons. Right, logarithm of the ratio between EPSC amplitudes in EGFP+ and EGFP− neurons. Red: mean ± s.e.m. EPSC amplitudes are 40% larger in EGFP+ neurons (P = 0.0004). (c) As in (b), but for IPSCs. IPSC amplitudes are 30% larger in EGFP+ neurons (P = 0.001). (d) As in (b), but for E/I ratios. E/I ratios are similar between EGFP+ and EGFP− neurons (P = 0.7). (e) Left, schematic of experiments. Fos-EGFP, Pv-ires-Cre mice with ChR2 in PV cells. Right, IPSCs from simultaneously recorded EGFP− and EGFP+ neurons in response to PV cell photoactivation. Note larger IPSC in EGFP+ neuron. (f) Summary graph. Left, IPSC amplitudes in EGFP+ neurons plotted against those in EGFP− neurons. Right, logarithm of the ratio between IPSC amplitudes in EGFP+ and EGFP− neurons. Red: mean ± s.e.m. IPSC amplitudes are 77% larger in EGFP+ neurons (n = 49, P = 0.001). (g, h) As in (e, f), but for Fos-EGFP, Som-ires-Cre mice with ChR2 in SOM cells. IPSC amplitudes are similar between EGFP+ and EGFP− neurons (n = 27, P = 0.7).
Taken together, these results demonstrate that excitation and inhibition, despite varying in amplitudes between pyramidal cells, remain proportional to each other, thus equalizing E/I ratios.
Which type of interneuron provides the inhibition that matches layer 4-mediated excitation? We took advantage of the fact that EGFP+ neurons in Fos-EGFP mice receive larger excitation from layer 4 and crossed them to Pv-ires-Cre or Som-ires-Cre mice to conditionally express ChR2. Photoactivation of PV cells generated larger monosynaptic IPSCs in EGFP+ than in EGFP− neurons (Fig. 2e, f). In contrast, SOM cells generated similar IPSCs in EGFP+ and EGFP− neurons (Fig. 2g, h). These data indicate that PV cells, but not SOM cells, provide stronger inhibition onto neurons that receive stronger layer 4-mediated excitation, thereby contributing to the equalization of E/I ratios.
What mechanism regulates the strengths of excitation and/or of inhibition to achieve the observed proportionality? Excitation and inhibition may reach their specific ratio by using the pyramidal cell’s activity as a measure of their relative strengths. For example, the low activity caused by a strong PV cell-mediated inhibition or by a weak layer 4-mediated excitation could be the signal to increase layer 4-mediated excitation or to decrease PV cell-mediated inhibition, respectively, until a neuron’s specific higher set-point activity is reached. In both scenarios the initially small E/I ratio is increased by either increasing excitation to match the large inhibition or by decreasing inhibition to match the small excitation. Both scenarios are plausible since the activity of individual neurons can regulate the strengths of both excitatory and inhibitory synapses16–19. If this hypothesis is correct, perturbing the activity of pyramidal cells should disrupt the proportionality between excitation and inhibition. For example, reducing the excitability of a pyramidal cell should increase its E/I ratio by either increasing excitation (the first scenario), or decreasing inhibition (the second scenario), or both.
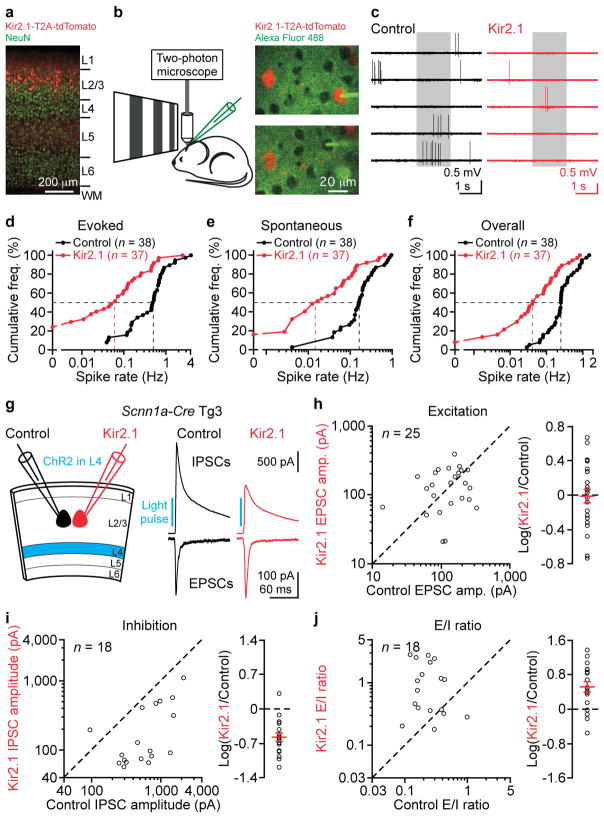
We reduced the excitability of a small, random subset of layer 2/3 pyramidal cells in V1 by overexpressing a Kir2.1 channel via in utero electroporation20–22 (Fig. 3a). Recordings in acute slices confirmed the reduced excitability in Kir2.1-overexpressing cells (Kir2.1 neurons) as compared to untransfected control pyramidal cells (Extended Data Fig. 4). In vivo targeted recordings from Kir2.1 and nearby control neurons (Fig. 3b, c) demonstrated that Kir2.1 overexpression drastically suppressed visual-evoked and spontaneous activity (Fig. 3d–f). We then examined the impact of this perturbation on excitation and inhibition. We photostimulated layer 4 and simultaneously recorded Kir2.1 and neighboring control neurons in the acute slices from Scnn1a-Cre-Tg3 mice. Surprisingly, layer 4-mediated excitation was not significantly different between these two groups (Fig. 3g, h), invalidating the first aforementioned possibility. In contrast, disynaptic inhibition was significantly smaller in Kir2.1 neurons (Fig. 3g, i), consistent with the second possibility. The effect on inhibition was due to the channel function of Kir2.1 because a non-conducting Kir2.1 mutant (Extended Data Fig 4) had no effect (Extended Data Fig. 5). Thus, perturbing layer 2/3 pyramidal cell excitability disrupts the proportionality between excitation and inhibition (Fig. 3j). These data indicate that pyramidal cell activity contributes to the equalization of E/I ratios across pyramidal cells.
Figure 3. Suppressing pyramidal cell activity reduces inhibition but not excitation.
(a) Fluorescent image of a V1 coronal section showing Kir2.1-T2A-tdTomato overexpression in a small, subset of layer 2/3 pyramidal cells (9 ± 1%, mean ± s.e.m., n = 12 sections from 6 mice). Cortical layers are identified by NeuN staining. L: layer, WM: white matter. (b) Left, schematic of in vivo experiments. Right, a Kir2.1 neuron (upper panel) and a control neuron (lower panel) were sequentially recorded with Alexa Fluor 488-filled pipettes. (c) Recordings from a control and a Kir2.1 neuron show spontaneous and visual-evoked spikes. Grey box: visual stimulation period. Note reduced spiking in Kir2.1 neuron. (d–f) Cumulative frequencies of evoked spike rate (d, median: control, 0.50 Hz; Kir2.1, 0.061 Hz; P < 0.0001), spontaneous spike rate (e, median: control, 0.16 Hz; Kir2.1, 0.017 Hz; P < 0.0001), and overall spike rate (f, median: control, 0.25 Hz; Kir2.1, 0.043 Hz; P < 0.0001) from 38 control neurons and 37 Kir2.1 neurons. (g) Left, schematic of slice experiments. Scnn1a-Cre-Tg3 mice with ChR2 in layer 4 excitatory neurons and Kir2.1 in a subset of layer 2/3 pyramidal cells. Right, monosynaptic EPSCs and disynaptic IPSCs from simultaneously recorded control and Kir2.1 neurons in response to layer 4 photoactivation. Note similar EPSC but smaller IPSC in Kir2.1 neuron as compared to control neuron. (h–j) Summary graphs. (h) Left, EPSC amplitudes in Kir2.1 neurons plotted against those in control neurons. Right, logarithm of the ratio between EPSC amplitudes in Kir2.1 and control neurons. Red: mean ± s.e.m. EPSC amplitudes are similar between Kir2.1 and control neurons (n = 25, P = 0.8). (i) As in (h), but for IPSCs. IPSC amplitudes in Kir2.1 neurons are 27% of those in control neurons (n = 18, P = 0.0003). (j) As in (h), but for E/I ratios. E/I ratios in Kir2.1 neurons are 3-fold of those in control neurons (n = 18, P = 0.004).
If pyramidal cell activity contributes to establishing the proportionality between layer 4-mediated excitation and PV cell-mediated inhibition, then the decrease in excitability should selectively decrease PV but not SOM cell-mediated inhibition. Conversely, an increase in excitability should selectively increase PV cell-mediated inhibition. Indeed, PV cell-mediated inhibition was significantly smaller in Kir2.1 than in control neurons, whereas SOM cell-mediated inhibition was similar (Fig. 4a–e). Overexpression of the non-conducting Kir2.1 mutant did not affect PV cell-mediated inhibition (Extended Data Fig. 5). We used a bacterial voltage-gated Na+ channel (mNaChBac) to enhance neuronal excitability23. Neurons expressing mNaChBac generate long-lasting action potentials and depolarization on the order of hundreds of milliseconds (Extended Data Fig. 6). Because constitutive expression of mNaChBac in cortical neurons from embryonic day 15.5 caused a neuronal migration defect (Extended Data Fig. 7), we devised a Flpo recombinase-mediated flip-excision strategy, F-FLEX switch (Extended Data Fig. 8), to conditionally express mNaChBac postnatally. We combined in utero electroporation of a Flpo-dependent mNaChBac-expressing plasmid, to randomly transfect a small subset of layer 2/3 pyramidal cells, with injection of an AAV expressing Flpo at postnatal day 1 (P1), to turn on mNaChBac expression. This allowed us to concurrently express ChR2 in PV or SOM cells, and mNaChBac in layer 2/3 pyramidal cells without affecting their migration (Extended Data Fig. 7). PV cell-mediated inhibition was significantly larger in mNaChBac neurons than in control neurons (Fig. 4f–h) and a non-conducting mNaChBac mutant (Extended Data Fig. 6) had no effect (Extended Data Fig. 5). mNaChBac expression did not alter SOM cell-mediated inhibition (Fig. 4i, j). To determine whether more acute perturbations of layer 2/3 pyramidal cell excitability also alter PV cell-mediated inhibition, we used Flpo and F-FLEX switch to express Kir2.1 or mNaChBac for only approximately one week starting around P12–P14. This acute decrease (Kir2.1) or increase (mNaChBac) in excitability caused a decrease or an increase in PV cell-mediated inhibition, respectively, similar to the changes caused by the chronic expression of Kir2.1 or mNaChBac (Fig. 4a–c, f–h). These data indicate that the proportionality between layer 4 mediated-excitation and PV cell-mediated inhibition is equalized across pyramidal cells through the bidirectional modulation of the strength of PV cell synapses.
Figure 4. Bidirectional regulation of PV but not SOM cell-mediated inhibition.
(a) Schematic of chronic and acute expression of Kir2.1. Red bars: approximate Kir2.1 expression time course. (b) Left, schematic of experiments. Pv-ires-Cre mice with ChR2 in PV cells and Kir2.1 in a subset of layer 2/3 pyramidal cells. Right, IPSCs from simultaneously recorded control neuron and neuron chronically or acutely expressing Kir2.1 in response to PV cell photoactivation. Note smaller IPSCs in Kir2.1 neuron. (c) Summary graphs. Left, IPSC amplitudes in Kir2.1 neurons plotted against those in control neurons. Right, logarithm of the ratio between IPSC amplitudes in Kir2.1 and control neurons. Red: mean ± s.e.m. IPSC amplitudes in Kir2.1 neurons are 23% (n = 36, P < 0.0001) and 31% (n = 16, P = 0.0005) of those in control neurons for chronic and acute conditions, respectively. (d, e) As in (b, c), but for Som-ires-Cre mice with ChR2 in SOM cells and Kir2.1 chronically in a subset of layer 2/3 pyramidal cells. IPSC amplitudes are similar between Kir2.1 and control neurons (n = 26, P = 0.3). (f) Schematic of chronic and acute expression of mNaChBac. Magenta bars: approximate mNaChBac expression time course. (g, h) As in (b, c), but for mNaChBac. IPSC amplitudes in mNaChBac neurons are 2.7-fold (n = 18, P = 0.001) and 2.2-fold (n = 24, P = 0.0003) of those in control neurons for chronic and acute conditions, respectively. (i, j) As in (d, e), but for mNaChBac. IPSC amplitudes are similar between mNaChBac and control neurons (n = 17, P = 0.7).
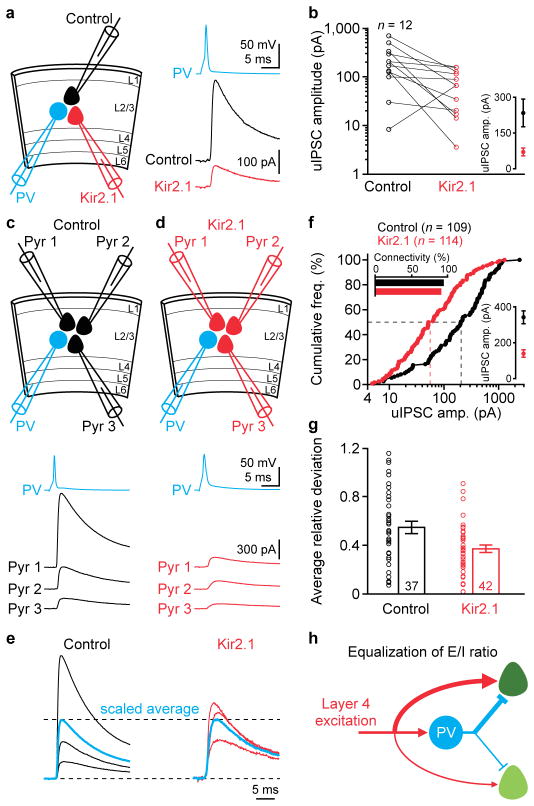
The above results show that the spatial heterogeneity of PV cell mediated-inhibition ensures the equalization of E/I ratios across pyramidal cells. Is a single PV cell-mediated inhibition also heterogeneous across its targeted pyramidal cells? We first determined whether the relative amplitudes of unitary IPSCs (uIPSCs) mediated by a PV cell onto its targets are predicted by the relative activity of these targets. We suppressed the activity of a small subset of layer 2/3 pyramidal cells by overexpressing Kir2.1 and simultaneously recorded from a layer 2/3 PV cell, a control and a Kir2.1 neuron (Fig. 5a). While the PV-to-pyramidal cell connectivity was similarly high regardless whether pyramidal cells overexpressed Kir2.1 (Fig. 5f), uIPSC amplitude was significantly smaller in Kir2.1 neurons than in control neurons (Fig. 5b, f). All properties of the unitary connections between PV cells and control neurons were similar to those recorded in mice that were not transfected with Kir2.1 (Extended Data Fig. 9), indicating a cell-autonomous effect of Kir2.1 overexpression. We assessed the variability of uIPSC amplitudes originating from a single PV cell and determined its dependency on the activity of the targeted pyramidal cells. We simultaneously recorded from a layer 2/3 PV cell and 2–3 nearby pyramidal cells that were either all control or all Kir2.1 neurons (Fig. 5c, d). uIPSC amplitudes varied greatly from one control neuron to another, but much less among Kir2.1 neurons (Fig. 5c–e, g), indicating reduced heterogeneity of uIPSC amplitudes among neurons with reduced activity (Extended Data Fig. 10). Thus, the inhibition generated by even an individual PV cell onto its targets is remarkably heterogeneous and this heterogeneity reflects in part the activity profile of the targeted pyramidal cell population. Hence, despite the indiscriminate connectivity of PV cells the amount of inhibition that they provide onto each of their targets is adjusted to equalize the E/I ratios (Fig. 5h).
Figure 5. Individual PV cell-mediated inhibition varies depending on targets’ activity.
(a) Left, schematic of experiments. Right, uIPSCs from simultaneously recorded control and Kir2.1 neuron in response to a PV cell action potential. Note smaller uIPSC in Kir2.1 neuron. (b) Summary graphs of uIPSC amplitudes from 12 similar experiments (P = 0.005). Inset: mean ± s.e.m. (c) Top, schematic of experiments. Bottom, uIPSCs from simultaneously recorded three control neurons in response to a PV cell action potential. (d) As in (c), but for three Kir2.1 neurons. (e) The uIPSCs from (c) and (d) were normalized by their respective mean amplitudes (scaled average). Note larger inter-cell variability of uIPSCs among control neurons. (f) Cumulative frequencies of uIPSC amplitudes (control: n = 109, median, 205.4 pA; Kir2.1: n = 114, 57.4 pA; P < 0.0001). Lower inset: mean ± s.e.m. of uIPSC amplitudes. Upper inset: unitary connectivity rates from PV cells to control (109 out of 116) and Kir2.1 (114 out of 126) neurons are similar (P = 0.3). (g) Summary graph for the average relative deviations of uIPSCs from 37 and 42 experiments as in (c) and (d). Bars: mean ± s.e.m. The average relative deviations for Kir2.1 neurons are 33% smaller than those for control neurons (P = 0.009). (h) Schematic of the equalized E/I ratios across cortical neurons.
Both theoretical and experimental evidence indicates that the relationship between synaptic excitation and inhibition in the cerebral cortex is fundamental for sensory processing1,2,24–27. Failure to establish or maintain this relationship may be the neural basis of neurological disorders like schizophrenia and autism28,29. We discover that E/I ratios are remarkably similar across different pyramidal cells despite large variations in the amplitudes of synaptic excitation and inhibition. The activity-dependent adjustment of inhibition to match excitation may result from activity-dependent gene expression18,19,30. Our study provides insight into how two opposing synaptic inputs, layer 4-mediated excitation and PV cell-mediated inhibition, remain proportional across a population of pyramidal cells and thus reveals an unanticipated degree of order in the distribution of synaptic strengths in cortical space.
METHODS
Mice
All procedures to maintain and use mice were approved by the Institutional Animal Care and Use Committee at the University of California, San Diego. Mice were maintained on a reverse 12-hour: 12-hour light: dark cycle with regular mouse chow and water ad libitum. CD-1 mice were purchased from Charles River Laboratories or Harlan Laboratories. Scnn1a-Cre-Tg331, Fos-EGFP32, Gad2-ires-Cre33, Pv-ires-Cre34, Som-ires-Cre33, and Rosa-CAG-LSL-tdTomato-WPRE31 mice were obtained from the Jackson Laboratory (stock number 009613, 014135, 010802, 008069, 013044, and 007909, respectively). Hemizygous transgenic mice and heterozygous knock-in mice of both sexes were used in the experiments.
DNA constructs and transfection of HEK cells
Two point mutations E224G and Y242F were introduced into mouse wild type Kir2.1 (Kcnj2) to enhance its ability to suppress neuronal activity. Mutation E224G attenuates the Mg2+ and polyamine block of Kir2.1 to reduce its inward rectification35. Mutation Y242F blocks tyrosine kinase phosphorylation of Kir2.1 at residue Y242 to enhance its plasma membrane surface expression36. Three point mutations G144A Y145A G146A were introduced to generate a non-conducting channel37. A Myc tag (EQKLISEEDL) was fused to the N-termini of Kir2.1 E224G Y242F and Kir2.1 E224G Y242F G144A Y145A G146A, and they were referred as Kir2.1 and Kir2.1Mut from now on, respectively. Both Kir2.1 and Kir2.1Mut were C-terminally fused with a T2A sequence (GSGEGRGSLLTCGDVEENPGP) followed by a tdTomato. The resulting constructs were then cloned into a plasmid containing a CAG promoter (pCAG) to generate pCAG-Kir2.1-T2A-tdTomato and pCAG-Kir2.1Mut-T2A-tdTomato.
The cDNA encoding a wild type bacterial Na+ channel NaChBac23 was synthesized de novo and codon-optimized for mammalian expression (referred as mNaChBac) by DNA2.0. A point mutation E191K was introduced to generate a non-conducting channel38, referred as mNaChBacMut. Both mNaChBac and mNaChBacMut were C-terminally fused with T2A-tdTomato and cloned into the pCAG plasmid to create pCAG-mNaChBac-T2A-tdTomato and pCAG-mNaChBacMut-T2A-tdTomato, respectively.
F-FLEX cassette using two wild type Frt sites and two F14 sites39 (Extended Data Fig. 8) was synthesized de novo and cloned into the plasmid pJ244 by DNA2.0 to generate pJ244-F-FLEX. mNaChBac-T2A-tdTomato and mNaChBacMut-T2A-tdTomato were subcloned into pJ244-F-FLEX in the inverted orientation. F-FLEX-mNaChBac-T2A-tdTomato and F-FLEX-mNaChBacMut-T2A-tdTomato cassettes were then subcloned into an AAV cis-plasmid containing an EF1α promoter to generate pAAV-EF1α-F-FLEX-mNaChBac-T2A-tdTomato and pAAV-EF1α-F-FLEX-mNaChBacMut-T2A-tdTomato, respectively. mNaChBac-T2A-tdTomato in pAAV-EF1α-F-FLEX-mNaChBac-T2A-tdTomato was replaced with inverted Kir2.1-T2A-tdTomato to generate pAAV-EF1α-F-FLEX- Kir2.1-T2A-tdTomato.
An improved version of Flp recombinase, Flpo40 was cloned into a pCAG plasmid and an AAV cis-plasmid containing a human Synapsin promoter to generate pCAG-Flpo and pAAV-hSynapsin-Flpo, respectively. pCAG-EGFP41, pCAG-mRFP42, and pCAG-Cre43 were obtained from Addgene (#11150, #28311, and #13775, respectively).
HEK-293FT cells (Life Technologies) were transfected with DNA constructs (0.1–0.5 μg) in 12-well plates using Lipofectamine 2000 (Life Technologies) to test their functionality. Constructs pCAG-EGFP, pCAG-mRFP, pCAG-Flpo, pCAG-Cre, pAAV-EF1α-F-FLEX-mNaChBac-T2A-tdTomato, and pAAV-EF1α-DIO-hChR2(H134R)-EYFP were used in Extended Data Fig. 8c, d.
In utero electroporation
Female CD-1 mice were crossed to male Scnn1a-Cre-Tg3, Pv-ires-Cre, or Som-ires-Cre mice to obtained timed-pregnancy. pCAG-Kir2.1-T2A-tdTomato, pCAG-Kir2.1Mut-T2A-tdTomato, pCAG-mNaChBac-T2A-tdTomato, and pCAG-mNaChBacMut-T2A-tdTomato were used at the final concentrations of 2–3 μg μL−1. pAAV-EF1α-F-FLEX-mNaChBac-T2A-tdTomato, pAAV-EF1α-F-FLEX-mNaChBacMut-T2A-tdTomato, or pAAV-EF1α-F-FLEX-Kir2.1-T2A-tdTomato (2–3 μg μL−1 final concentration) was mixed with pCAG-EGFP (0.2 μg μL−1 final concentration). Fast Green (Sigma-Aldrich, 0.01% final concentration) was added to the DNA solution. On embryonic day 14.5 or 15.5, female mice were anesthetized with 2.5% isoflurane in oxygen at the flow rate of 1 L min−1 and the body temperature was maintained by a feedback based DC temperature control system (FHC) at 37°C. Buprenorphine (3 μg, Reckitt Benckiser Healthcare) was administered subcutaneously along with 1 mL of Lactated Ringer’s Injection (Baxter Healthcare). The abdominal fur was shaved and the skin was cleaned with 70% alcohol and iodine. A sterile towel drape was laid on the mouse with only the abdomen exposed. Midline incisions (2 cm) were made on the abdominal skin and wall, and the uteruses were taken out of the abdominal cavity. A beveled glass micropipette (tip size, 100-μm outer diameter and 50-μm inner diameter) was used to penetrate the uterus and the embryo skull to inject about 1.5 μL of DNA solution into one lateral ventricle. 5 pulses of current (voltage, 39 V; duration, 50 ms) were delivered at 1 Hz with a Tweezertrode (5-mm diameter) and a square wave pulse generator (ECM 830, BTX Harvard Apparatus). The electrode paddles were positioned along the 70-degree angle to the embryo anteroposterior axis and the cathode was faced the caudal side of the injected ventricle to target the visual cortex. After electroporation, uteruses were put back into the abdominal cavity, and the abdominal wall and skin were sutured. Mice were returned to the home cage and recovered from anesthesia on a 37°C Deltaphase Isothermal Pad (Braintree Scientific). Another 3 μg buprenorphine was given subcutaneously on the next day. After birth, transfected pups were identified by the transcranial fluorescence of tdTomato or EGFP with a stereomicroscope (MVX10 Macroview, Olympus). Only the pups in which the majority of the transfection occurred in the primary visual cortex were used for experiments.
AAV production and injection
All recombinant AAV vectors were produced by the Penn Vector Core with the following titers: AAV2/9-CAGGS-Flex-ChR2-tdTomato44 (Addgene #18917, titer 1.15×1013 GC mL−1), AAV2/1-CAGGS-Flex-ChR2-tdTomato (titer 6.86×1012 or 1.22×1013 GC mL−1), AAV2/9-EF1α-DIO-hChR2(H134R)-EYFP45 (Addgene #20298, titer 6.24×1012 or 1.18×1013 GC mL−1), AAV2/1-EF1α-DIO-hChR2(H134R)-EYFP (titer 3.41×1013 GC mL−1), and AAV2/9-hSynapsin-Flpo (titer 1.57×1013 GC mL−1).
AAV was injected into the V1 of pups on postnatal days 0–3 using a Nanoject II nanoliter injector (Drummond Scientific Company) except for the acute expression of Kir2.1 and mNaChBac experiments (see below). Pups were anesthetized by hypothermia and secured on a custom-made plate. Fast Green (0.01% final concentration) was added to the virus solution for visualization. A beveled glass micropipette (tip size, 60-μm outer diameter and 30-μm inner diameter) was used to penetrate the scalp and skull, and to inject AAV at different depths (600, 500, 400, and 300 μm below the scalp) of one location (1.6 mm lateral, 0.3 mm caudal from the lambda). Total about 80–180 nL (adjusted based on the virus titers) of virus solution was injected over 60 s. After injection, the micropipette was kept in parenchyma at 300 μm depth for 30 s before being slowly withdrawn. Pups were placed on a 37°C Deltaphase Isothermal Pad to recover from anesthesia and then were returned to the dam. For chronic expression of mNaChBac or mNaChBacMut, AAV2/9-hSynapsin-Flpo (titer 1.57×1013 GC mL−1) was mixed with AAV2/9-EF1α-DIO-hChR2(H134R)-EYFP (titer 1.18×1013 GC mL−1) at 1:3 ratio for injection at postnatal day 1.
For acute expression of Kir2.1 or mNaChBac, pups that were injected with AAV2/1-EF1α-DIO-hChR2(H134R)-EYFP at postnatal days 0–2 were injected with AAV2/9-hSynapsin-Flpo at postnatal days 12–14. Pups were anesthetized with 2.5% isoflurane in oxygen at the flow rate of 1 L min−1 and the body temperature was maintained by a feedback based DC temperature control system at 37°C. Buprenorphine (1 μg) was administered subcutaneously along with 0.1 mL of Lactated Ringer’s Injection. Lubricant ophthalmic ointment (Artificial Tears Ointment, Rugby Laboratories) was applied to the corneas to prevent drying. The scalp fur was shaved and the skin was cleaned with 70% alcohol and iodine. A small incision (0.5 cm) was made on the skin medial to the visual cortex. The skull at the injection site (2.5-mm lateral to midline and 1-mm rostral to lambda suture) was thinned with a 0.3-mm diameter round bur (Busch Bur, Gesswein) on a high-speed rotary micromotor (Foredom) such that the injection glass micropipette (tip size, 50-μm outer diameter and 25-μm inner diameter) can penetrate the skull. Total 150 nL of virus solution was injected 450 μm below the skull at the speed of 20 nL min−1. After injection, the micropipette was kept in parenchyma for 5–10 min before being slowly withdrawn. The skin was sutured. Mice were returned to the home cage and recovered from anesthesia on a 37°C Deltaphase Isothermal Pad.
Immunocytochemistry
Mice were anesthetized by intraperitoneal injection of ketamine and xylazine mix (100 mg kg−1 and 10 mg kg−1, respectively), and were transcardial perfused with phosphate buffered saline (PBS, pH 7.4) followed by 4% paraformaldehyde in PBS (pH 7.4). Brains were removed, further fixed overnight in 4% paraformaldehyde, cryoprotected with 30% sucrose in PBS, and frozen in optimum cutting temperature medium (OCT) until sectioning. A HM 450 Sliding Microtome (Thermo Scientific) was used to section the brains to obtain 30–50 μm coronal slices. Slices were blocked with 1% bovine serum albumin (BSA), 2% normal goat serum (NGS), and 0.3% TritonX-100 in PBS at room temperature for 1 hour and incubated with primary antibodies in working buffer (0.1% BSA, 0.2% NGS, 0.3% TritonX-100 in PBS) at 4°C overnight. The following primary antibodies were used: rabbit anti-RFP (1:200, Abcam), rat anti-RFP (1:300, Chromotek), chicken anti-GFP (1:500, Aves Labs), rabbit anti-GFP (1:2000, Life Technologies), and mouse anti-NeuN (1:200, Millipore). The slices were washed 4 times with working buffer for 10 min each, incubated with secondary antibodies conjugated with Alexa Fluor 488, 594, or 647 (1:500 or 1:1000, Life Technologies) in working buffer for 1 hour at room temperature, and then washed 4 times with working buffer for 10 min each. NeuroTrace 435/455 blue fluorescent Nissl stain (1:200, Life Technologies) was used to label neurons after antibody staining. Slices were mounted in Vectashield Mounting Medium containing DAPI (Vector Laboratories) or ProLong Gold antifade reagent (Life Technologies). Images were acquired on an Olympus FV1000 Confocal, a Zeiss Axio Imager A1, or an Olympus MVX10 Macroview, and processed using NIH ImageJ. To estimate the fraction of layer 2/3 pyramidal cells that were transfected by in utero electroporation (Fig. 3a), transfected neurons (tdTomato+) and total neurons (NeuN+) in layer 2/3 were visually quantified. Assuming that 13.2% of layer 2/3 neurons were interneurons (Extended Data Fig. 3), transfected neurons were estimated to be 9 ± 1% (mean ± s.e.m, n = 12 sections from 6 mice) of layer 2/3 pyramidal cells.
In vitro physiology
Mice at postnatal days 14–23 were anesthetized by intraperitoneal injection of ketamine and xylazine mix (100 mg kg−1 and 10 mg kg−1, respectively), and were transcardial perfused with cold (0–4°C) slice cutting solution containing 80 mM NaCl, 2.5 mM KCl, 1.3 mM NaH2PO4, 26 mM NaHCO3, 4 mM MgCl2, 0.5 mM CaCl2, 20 mM D-glucose, 75 mM sucrose, and 0.5 mM sodium ascorbate (315 mOsm, pH 7.4, saturated with 95% O2/5% CO2). Brains were removed and a Super Microslicer Zero1 (D.S.K.) was used to section the brains to obtain 300 μm coronal slices in cutting solution. Slices were incubated in a custom-made interface holding chamber at 34°C for 30 min and then at room temperature for 20 min–8 hours until they were transferred to the recording chamber. During incubation, the holding chamber was saturated with 95% O2/5% CO2.
Recordings were performed on submerged slices in artificial cerebrospinal fluid (ACSF) containing 119 mM NaCl, 2.5 mM KCl, 1.3 mM NaH2PO4, 26 mM NaHCO3, 1.3 mM MgCl2, 2.5 mM CaCl2, 20 mM D-glucose, and 0.5 mM sodium ascorbate (300 mOsm, pH 7.4, saturated with 95% O2/5% CO2, perfused at 3 mL min−1) at 31–32°C. For whole-cell recordings, a K+-based pipette solution containing 142 mM K+-gluconate, 10 mM HEPES, 1 mM EGTA, 2.5 mM MgCl2, 4 mM ATP-Mg, 0.3 mM GTP-Na, 10 mM Na2-phosphocreatine (295 mOsm, pH 7.35) or a Cs+-based pipette solution containing 115 mM Cs+-methanesulfonate, 10 mM HEPES, 1 mM EGTA, 1.5 mM MgCl2, 4 mM ATP-Mg, 0.3 mM GTP-Na, 10 mM Na2-phosphocreatine, 2 mM QX 314-Cl, 10 mM BAPTA-tetracesium (295 mOsm, pH 7.35) was used. Membrane potentials were not corrected for experimentally measured liquid junction potential (11.4 mV for the K+-based pipette solution and 8.4 mV for the Cs+-based pipette solution).
Neurons were visualized with video-assisted infrared differential interference contrast imaging and fluorescent neurons were identified by epifluorescence imaging under a water immersion objective (40X, 0.8 NA) on an upright Olympus BX51WI microscope with an IR CCD camera (VX44, Till Photonics). For Fos-EGFP experiments, in a given field-of-view those pyramidal cells with the strongest EGFP fluorescence were visually identified as the EGFP+ neurons. The EGFP− neurons were those pyramidal cells whose fluorescence was equal to the slice background fluorescence level.
Data were low-pass filtered at 4 kHz and acquired at 10 kHz with an Axon Multiclamp 700A or 700B amplifier and an Axon Digidata 1440A Data Acquisition System under the control of Clampex 10.2 (Molecular Devices). Data were analyzed offline using AxoGraph X (AxoGraph Scientific). A collimated LED (470 nm) and a T-Cube LED Driver (Thorlabs) controlled by Axon Digidata 1440A Data Acquisition System and Clampex 10.2 were used to deliver blue light through the reflected light fluorescence illuminator port and the 40X objective to stimulate neurons via the activation of ChR2.
Synaptic currents were recorded in whole-cell voltage clamp mode with the Cs+-based patch pipette solution. Only recordings with series resistance below 20 MΩ were included. EPSCs and IPSCs were recorded at IPSC reversal potential (−60 mV) and EPSC reversal potential (+10 mV), respectively. For light pulse stimulation, pulse duration (0.5–5 ms) and intensity (1.1–5.5 mW mm−2) were adjusted for each recording to evoke small but reliable monosynaptic EPSCs or IPSCs to minimize voltage-clamp error. Disynaptic IPSCs were evoked using the same light pulses that were used for evoking the corresponding monosynaptic EPSCs. Light pulses were delivered at 30-s interstimulus interval.
To quantify the inter-cell variability of EPSCs (Fig. 1d, f), the EPSC amplitudes (EPSCi) of the pyramidal cells from one given experiment were first averaged to obtain the mean EPSC amplitude (EPSCmean). The average relative deviation of EPSCs was calculated as (N, the number of pyramidal cells in one given experiment). The average relative deviation of IPSCs or E/I ratios was obtained similarly for each experiment (Fig. 1d, f).
For the unitary connections from inhibitory neurons to pyramidal cells, PV and SOM cells were identified by the Cre-dependent expression of ChR2-tdTomato or hChR2(H134R)-EYFP in Pv-ires-Cre and Som-ires-Cre mice, respectively. Pyramidal cells were first recorded in whole-cell voltage clamp mode (+10 mV) with the Cs+-based patch pipette solution, and then a nearby PV or SOM cell was recorded in whole-cell current clamp mode with the K+-based patch pipette solution. Action potentials were elicited in PV or SOM cells by a 2-ms depolarizing current step (1–2 nA) at 15-s interstimulus interval. Unitary IPSCs were measured from the average of 10–50 sweeps. A connection was considered exist when the average IPSC amplitude was at least 3 times of the baseline standard deviation. For the average relative deviation of uIPSCs originating from one PV cell (Fig. 5 and Extended Data Fig. 9), the uIPSC amplitudes (uIPSCi) from all connected pyramidal cells in one given experiment were first averaged to obtain the mean uIPSC amplitude (uIPSCmean). The average relative deviation was calculated as (N, the number of pyramidal cells in one given experiment).
Neuronal intrinsic excitability was examined with the K+-based pipette solution in the presence of AMPA receptor blocker NBQX (10 μM), NMDA receptor blocker (RS)-CPP (10 μM), and GABAA receptor blocker SR 95531 (10 μM). After whole-cell current clamp mode was achieved and the bridge was balanced, resting membrane potentials were recorded within the first minute after break-in. Input resistances were measured by injecting a 500-ms hyperpolarizing current (10–100 pA) to generate a small membrane potential hyperpolarization (2–10 mV) from the resting membrane potentials. Depolarizing currents were increased at 5- or 10-pA steps to identify rheobase currents.
Ba2+-sensitive currents were measured with the K+-based pipette solution in the presence of NBQX (10 μM), (RS)-CPP (10 μM), SR 95531 (10 μM), and Na+ channel blocker TTX (1 μM). Only recordings with series resistance below 20 MΩ were included. Neurons were clamped at −25 mV and then membrane potential was ramped to −125 mV at the rate of 20 mV s−1. The membrane currents recorded in the presence of BaCl2 (50 μM) were subtracted from the ones recorded in the absence of BaCl2 to obtain the Ba2+-sensitive currents, which were then divided by the whole-cell membrane capacitances to calculate the current densities.
In vivo physiology
Mice at postnatal days 17–23 were anesthetized by intraperitoneal injection of chlorprothixene (5 mg kg−1) followed by urethane (1.2 g kg−1). Oxygen was given at the flow rate of 1 L min−1 during the experiments and isoflurane (< 0.5%) was supplemented if necessary. The body temperature was maintained by a feedback based DC temperature control system at 37°C. Dexamethasone sodium phosphate (2 mg kg−1) and Lactated Ringer’s Injection (3 mL kg−1 every 2 hours) were administered subcutaneously. Whiskers and eyelashes were trimmed, and a thin layer of silicone oil (viscosity 30,000 cSt, Sigma-Aldrich) was applied to the corneas to prevent drying. The scalp and periosteum were removed. Vetbond tissue adhesive (3M) was applied to all sutures to stabilize them. A custom-made recording chamber with a 3-mm diameter hole in the center was attached to the skull over V1 by Vetbond tissue adhesive and dental cement (Ortho-Jet BCA, Lang Dental). The recording chamber was then secured on a custom-made holder. A craniotomy (1.5–2 mm diameter, centered at 2.5-mm lateral to midline and 1-mm rostral to lambda suture) was performed with a 0.3-mm diameter round bur on a high-speed rotary micromotor. Dura was left intact and craniotomy was covered by a thin layer of 1.5% type III-A agarose in HEPES-ACSF containing 142 mM NaCl, 5 mM KCl, 10 HEPES-Na, 1.3 mM MgCl2, 3.1 mM CaCl2, 10 mM D-glucose (310 mOsm, pH 7.4). HEPES-ACSF was added into the recording chamber.
Targeted loose-patch recordings were performed under the guidance of two-photon laser scanning microscopy. Two-photon imaging was performed with a water immersion objective (40X, 0.8 NA, Olympus) on a Moveable Objective Microscope (Sutter Instruments) coupled with a Ti:Saphire laser (Chameleon Ultra II, Coherent) under the control of ScanImage 3.6 (Janelia Farm Research Campus, HHMI)46. Laser wavelength was tuned to 950 nm for two-photon excitation of tdTomato and Alexa Fluor 488. Laser average power was 25–50 mW after the objective.
An Axon Multiclamp 700B amplifier was used for extracellular recording of spikes. Fluorescent neurons located at 150–250 μm below dura were identified. A patch pipette containing HEPES-ACSF and 10–20 μM of Alexa Fluor 488 hydrazide (Life Technologies) was advanced along its axis towards the neuron with minimal lateral movements. A small positive pressure was applied to the patch pipette to avoid the clogging of the pipette tip and to inject small amount of fluorescent dye to stain the extracellular space. Non-fluorescent neurons were visualized as the negative images47. The pipette resistance was constantly monitored in voltage-clamp mode. The concurrence of pipette tip contacting the neuron and an increase in pipette resistance indicated a successful targeting, which was further confirmed post hoc (see below). Upon the release of positive pressure, a small negative pressure was applied to form a loose seal (10–30 MΩ). The amplifier was then switched to current-clamp mode with zero current injection to record voltage. Data were low-pass filtered at 10 kHz and acquired at 32 kHz with a NI-DAQ board (NI PCIe-6259, National Instruments) under the control of a custom-written program running in Matlab (Mathworks). Within a local region (< 50 μm), neighboring tdTomato+ and tdTomato− neurons were sequentially targeted for recording, but the order of recordings from tdTomato+ and tdTomato− was alternated. The correct targeting of tdTomato+ neurons was confirmed at the end of recordings by either filling the neuron with fluorescent dye via break-in or by the presence of partial fluorescent neuron in the recording pipette due to the suction.
Visual stimuli were generated in Matlab with Psychophysics Toolbox48 and displayed on a gamma-corrected LCD monitor (30 × 47.5 cm, 60 Hz fresh rate, mean luminance 50 cd m−2). The monitor was placed 25 cm away from the contralateral eye, covering 62° (vertical) ×87° (horizontal) of the visual space. For each local region in V1 (see above), the monitor was centered at the approximate retinotopic location that was mapped manually based on the single unit or multi-unit activity in response to a moving bar on the screen. During recordings, full field sinusoidal drifting gratings (temporal frequency 2 Hz, spatial frequency 0.04 cycles per degree, 100% contrast) were presented randomly at 12 different directions from 0° to 330° for 1.5 s, preceded and followed by the presentation of grey screen for 2 s and 1.5 s, respectively. The complete set of stimuli was repeated for 8–16 times.
Data analyses were performed offline using a custom-written program in Matlab. Voltage signals were high-pass filtered (125 Hz). Spikes were first detected as the events exceeding 5 time of the standard deviation of the noise, and then visually verified. Spontaneous spike rate was calculated as the average spike rate during the 2-s time window before the presentation of visual stimuli. Evoked spike rate was calculated as the average spike rate during the 1.5-s time window of visual stimulation. Overall spike rate was calculated as the average spike rate during the entire recording period.
Statistics
All reported sample numbers (n) represent biological replicates. Sample sizes were estimated to have 70–80% power to detect expected effect size using StatMate 2 (GraphPad Software). Statistical analyses were performed using Prism 5 (GraphPad Software) and Matlab. Linear regression with F test (two-sided) was used for Fig. 1e, Extended Data Fig. 2a, b. Bootstrapped distributions (Extended Data Fig. 2c) were used to determine the statistical significance for Fig. 1f. Wilcoxon matched-pairs signed rank test (two-sided) was used for Fig. 2, Fig. 3h–j, Fig. 4, Fig. 5b, Extended Data Figs. 1,5. Mann-Whitney test (two-sided) was used for Fig. 3d–f, uIPSC amplitudes in Fig. 5f, Fig. 5g, Extended Data Figs. 4,6, Extended Data Fig. 9c, d. Fisher’s exact test (two-sided) was used for connectivity rates in Fig. 5f, Extended Data Fig. 9b.
Extended Data
Extended Data Figure 1. Cre recombinase-expressing cells in Scnn1a-Cre-Tg3 mice are layer 4 excitatory neurons.

AAV-CAGGS-Flex-ChR2-tdTomato that expresses ChR2-tdTomato fusion protein in a Cre-dependent manner was injected into Scnn1a-Cre-Tg3 mice. (a) Representative fluorescent images of a coronal section of V1 showed that the ChR2-tdTomato expressing cells resided primarily in layer 4 (n = 11 mice). Cortical layers are indicated on the right based on the DAPI staining pattern. L: layer, WM: white matter. (b) Left, schematic of experiments. Right, a layer 2/3 pyramidal cell was voltage clamped at the reversal potential for excitation (+10 mV). Activation of ChR2-expressing neurons in layer 4 elicited GABAergic IPSC (black trace), which was abolished by the glutamatergic receptor blockers NBQX and CPP (red trace), indicating the disynaptic nature of the IPSC. (c) Summary data showed that NBQX and CPP reduced IPSC amplitudes by 98.0 ± 0.6% (mean ± s.e.m., n = 8, P = 0.008). The lack of blue light-evoked monosynaptic IPSC indicates that ChR2-expressing neurons are excitatory neurons.
Extended Data Figure 2. Characterization of the inter-cell variability of EPSCs, IPSCs, and E/I ratios.
(a, b) The inter-cell variability of EPSCs, IPSCs, and E/I ratios among neighboring pyramidal cells does not correlate with their inter-soma distances. (a) The average relative deviations of EPSCs, IPSCs, and E/I ratios from each experiment in Fig. 1f are plotted against the average inter-soma distance from the same experiment. The average inter-soma distance is the mean of the distances between each pair of pyramidal cells. For the experiments in which only two pyramidal cells were recorded, the inter-soma distance between the two pyramidal cells was used. Lines: linear regression fits. (b) The ratio of the EPSC amplitudes of two simultaneously recorded pyramidal cells was first calculated, and then the logarithm of this ratio was computed. The absolute value of the logarithm was plotted against the inter-soma distance between the two pyramidal cells. Similarly, the data for IPSCs and E/I ratios were plotted against the inter-soma distances. Lines: linear regression fits. (c) The distribution of the E/I ratios across the pyramidal cells varies less than if EPSCs and IPSCs were randomly paired between cells and less than the distributions of EPSC and IPSC amplitudes. To determine whether the precise E/I ratio recorded within each pyramidal cell minimizes the average relative deviation, we computed the E/I ratios from randomly but uniquely paired EPSCs and IPSCs within each of the 20 experiments from Fig. 1f. By randomizing within each experiment we ensure that the average relative deviation is only modified due to the pairing of EPSCs to IPSCs. Note, for an experiment with N pyramidal cells, there are N! possible randomized pairings of EPSCs and IPSCs, and hence N! possible E/I ratio average relative deviations (referred as random-E/I ratio average relative deviation). The distribution of the means of the random-E/I ratio average relative deviations (grey histogram) was constructed from the means of 10,000 samples. Each sample consisted of 20 random-E/I ratio average relative deviations, each of which was randomly chosen from the N! possible random-E/I ratio average relative deviations of each experiment. The grey vertical line represents the mean of the distribution. The distribution of the means of the E/I ratio average relative deviations (black histogram) was generated by bootstrapping (i.e. resampling 10,000 times with replacement). Each resample consisted of 20 randomly chosen E/I ratio average relative deviations from the 20 experiments in Fig. 1f and an E/I ratio average relative deviation was allowed to be repeated within one resample (i.e. sampling with replacement). The black vertical line represents the mean of the experimentally obtained E/I ratio average relative deviations. The E/I ratio average relative deviations are smaller than the random-E/I ratio average relative deviations (P < 0.0001). The distributions of the means of the EPSC average relative deviations (red histogram) and the means of the IPSC average relative deviations (blue histogram) were generated by similar bootstrapping to the E/I ratio average relative deviations. The red and blue vertical lines represent the mean of the experimentally obtained EPSC average relative deviations and IPSC average relative deviations, respectively. The E/I ratio average relative deviations are smaller than the EPSC average relative deviations (P < 0.0001) and the IPSC average relative deviations (P < 0.0001).
Extended Data Figure 3. Majority of layer 2/3 Fos-EGFP+ neurons in V1 are pyramidal cells.
Fos-EGFP mice were crossed to Gad2-ires-Cre and Rosa-CAG-LSL-tdTomato-WPRE mice to generate Fos-EGFP, Gad2-ires-Cre, Rosa-CAG-LSL-tdTomato-WPRE mice. (a) Representative fluorescent images showed a coronal section of V1. All neurons were visualized by NeuroTrace 435/455 blue fluorescent Nissl stain and GABAergic interneurons were labeled by tdTomato. EGFP was stained with an antibody against GFP and visualized with a secondary antibody conjugated with Alexa Fluor 647. Cortical layers are indicated on the left based on the Nissl staining pattern. L: layer. (b) The enlarged view of the boxed region in (a). In layer 2/3 of V1, only 5.3 ± 0.9% (mean ± s.e.m., n = 10 sections from 2 mice) of EGFP+ neurons were GABAergic interneurons (two examples are indicated by arrow heads). GABAergic interneurons constitute 13.2 ± 0.6% (mean ± s.e.m., n = 14 sections from 3 mice including one Gad2-ires-Cre, Rosa-CAG-LSL-tdTomato-WPRE mouse) of all layer 2/3 neurons.
Extended Data Figure 4. Overexpression of Kir2.1 increases a Ba2+-sensitive K+ current and decreases neuronal excitability.

(a) Schematics of experiments. Kir2.1 or a non-conducting mutant Kir2.1 (Kir2.1Mut) was overexpressed in a subset of layer 2/3 pyramidal cells by in utero electroporation. (b) Membrane currents in response to a 5-s membrane potential ramp from −25 mV to −125 mV from an untransfected control pyramidal cell, a pyramidal cell overexpressing Kir2.1, and a pyramidal cell overexpressing Kir2.1Mut. The purple traces were recorded in control condition and the grey traces were recorded in the presence of 50 μM BaCl2. The blue traces were obtained by subtracting the grey traces from the purple traces, representing the Ba2+-blocked K+ currents. At the concentration of 50 μM, Ba2+ primarily blocks the K+ channels of the Kir2 subfamily49. (c) The exogenously overexpressed Kir2.1 increased not only the Ba2+-blocked inward current density at −125 mV (P = 0.01), but also the outward current density at −45 mV (P = 0.001) due to its reduced inward rectification (see Methods). (d) Kir2.1Mut can bind to the endogenous Kir2.1 to form non-conducting channels49, acting as a dominant negative to decrease the inward current density at −125 mV (P = 0.004) but without affecting the outward current density at −45 mV (P = 0.2). (e) Membrane potentials (upper panels) in response to current injections (lower panels) from an untransfected control pyramidal cell, a pyramidal cell overexpressing Kir2.1, and a pyramidal cell overexpressing Kir2.1Mut. (f–h) Overexpression of Kir2.1 hyperpolarized the resting membrane potential (f, P = 0.0003), decreased the resting input resistance (g, P < 0.0001), and increased the rheobase current (h, P < 0.0001). (i–k) Overexpression of Kir2.1Mut increased the resting input resistance (j, P = 0.0002), but had no effects on the resting membrane potential (i, P = 0.5) and the rheobase current (k, P = 0.9). The numbers of recorded neurons are indicated on the bars. All data are expressed as mean ± s.e.m.
Extended Data Figure 5. Overexpression of Kir2.1Mut or mNaChBacMut in layer 2/3 pyramidal cells does not affect inhibition.
(a) Left, schematic of experiments. Scnn1a-Cre-Tg3 mice with ChR2 in layer 4 excitatory neurons and Kir2.1Mut in a subset of layer 2/3 pyramidal cells. Right, monosynaptic EPSCs and disynaptic IPSCs from simultaneously recorded control and Kir2.1Mut neurons in response to layer 4 photoactivation. (b–d) Summary graphs. (b) Left, EPSC amplitudes in Kir2.1Mut neurons plotted against those in control neurons. Right, logarithm of the ratio between EPSC amplitudes in Kir2.1Mut and control neurons. Red: mean ± s.e.m. EPSC amplitudes are similar between Kir2.1Mut and control neurons (n = 23, P = 0.7). (c) As in (b), but for IPSCs. IPSC amplitudes are similar between Kir2.1Mut and control neurons (n = 22, P = 0.6). (d) As in (b), but for E/I ratios. E/I ratios are similar between Kir2.1 and control neurons (n = 22, P = 0.6). (e) Left, schematic of experiments. Pv-ires-Cre mice with ChR2 in PV cells and Kir2.1Mut in a subset of layer 2/3 pyramidal cells. Right, IPSCs from simultaneously recorded control and Kir2.1Mut neurons in response to PV cell photoactivation. (f) Summary graphs. Left, IPSC amplitudes in Kir2.1Mut neurons plotted against those in control neurons. Right, logarithm of the ratio between IPSC amplitudes in Kir2.1Mut and control neurons. Red: mean ± s.e.m. IPSC amplitudes are similar between Kir2.1Mut and control neurons (n = 14, P = 0.8). (g, h) As in (e, f), but for a non-conducting mutant mNaChBac (mNaChBacMut). IPSC amplitudes are similar between mNaChBacMut and control neurons (n = 16, P = 0.9).
Extended Data Figure 6. Overexpression of mNaChBac increases neuronal excitability.

(a) Schematics of experiments. mNaChBac or a non-conducting mutant mNaChBac (mNaChBacMut) was overexpressed in a subset of layer 2/3 pyramidal cells by in utero electroporation. (b) Membrane currents (upper and middle panels) in response to voltage steps (lower panels) from an untransfected control pyramidal cell, a pyramidal cell overexpressing mNaChBac, and a pyramidal cell overexpressing mNaChBacMut. The endogenous voltage-gated inward Na+ current was fast inactivating and was blocked by tetrodotoxin (TTX, 1 μM), whereas the mNaChBac-mediated inward current was slow inactivating and insensitive to TTX. Inset: overlay of the two dashed boxes. Note that the fast component of the inward current representing the endogenous Na+ current was blocked by TTX. (c) Membrane potentials (upper panels) in response to current injections (lower panels) from a control neuron, a mNaChBac neuron, and a mNaChBacMut neuron. The mNaChBac neuron generated long-lasting action potentials and depolarization, whereas the mNaChBacMut neuron generated action potentials similar to the control neuron. (d, e) Overexpression of mNaChBac lowered the action potential threshold that was defined as the membrane potential when its derivative reached 2 V s−1 (d, P = 0.004), and decreased the rheobase current (e, P = 0.03). (f, g) Overexpression of mNaChBacMut did not alter the action potential threshold (f, P = 0.9) and the rheobase current (g, P = 0.8). The numbers of recorded neurons are indicated on the bars. All data are expressed as mean ± s.e.m.
Extended Data Figure 7. Postnatal expression of mNaChBac and Kir2.1 using Flpo and F-FLEX switch.
(a) Constitutive overexpression of mNaChBac causes a neuronal migration defect. mNaChBac or mNaChBacMut was overexpressed in a subset of pyramidal cells by in utero electroporation of pCAG-mNaChBac-T2A-tdTomato or pCAG-mNaChBacMut-T2A-tdTomato, respectively, on embryonic day 15.5 (E15.5). Representative fluorescent images of coronal sections of V1 showed that mNaChBac-expressing neurons (left panels) resided not only in layer 2/3, but also in layers 4–6 (n = 7 mice), whereas mNaChBacMut-expressing neurons (right panels) are all located in layer 2/3 (n = 5 mice). Cortical layers are indicated on the right based on the DAPI staining pattern. L: layer, WM: white matter. (b) Experimental procedures for conditional expression of mNaChBac or Kir2.1 in a subset of layer 2/3 pyramidal cells. Left: plasmids pAAV-EF1α-F-FLEX-mNaChBac-T2A-tdTomato or pAAV-EF1α-F-FLEX-Kir2.1-T2A-tdTomato together with pCAG-EGFP were in utero electroporated into V1 on E15.5. Successful transfection was indicated by the expression of EGFP. Middle: AAV-hSynapsin-Flpo was injected postnatally into V1. Right: Only those neurons that were transfected with pAAV-EF1α-F-FLEX-mNaChBac-T2A-tdTomato or pAAV-EF1α-F-FLEX-Kir2.1-T2A-tdTomato and at the same time infected by AAV-hSynapsin-Flpo will express mNaChBac-T2A-tdTomato or Kir2.1-T2A-tdTomato, respectively. (c) Representative fluorescent images of coronal sections of V1 showed that without injection of AAV-hSynapsin-Flpo, the transfected neurons did not express mNaChBac-T2A-tdTomato (left panels, n = 2 mice). The expression of mNaChBac-T2A-tdTomato in the transfected neurons was turned on by injection of AAV-hSynapsin-Flpo, and these neurons were all properly located in layer 2/3 (right panels, n = 7 mice). Cortical layers are indicated on the right based on the DAPI staining pattern. L: layer, WM: white matter. (d) Schematics of concurrent expression of mNaChBac or Kir2.1 in layer 2/3 pyramidal cells and ChR2 in PV or SOM cells. Plasmids pAAV-EF1α-F-FLEX-mNaChBac-T2A-tdTomato or pAAV-EF1α-F-FLEX-Kir2.1-T2A-tdTomato and together with pCAG-EGFP were in utero electroporated into V1 of Pv-ires-Cre or Som-ires-Cre mice on E15.5. AAV-EF1α-DIO-hChR2(H134R)-EYFP and AAV-hSynapsin-Flpo were injected into V1. ChR2 was conditionally expressed in PV or SOM cells, whereas mNaChBac or Kir2.1 was conditionally expressed in a subset of layer 2/3 pyramidal cells that were transfected by pAAV-EF1α-F-FLEX-mNaChBac-T2A-tdTomato or pAAV-EF1α-F-FLEX-Kir2.1-T2A-tdTomato, respectively.
Extended Data Figure 8. A Flpo recombinase-mediated FLEX (F-FLEX) switch for conditional gene expression.
(a) DNA sequence of the F-FLEX switch cassette. The first F14 site and Frt site were constructed in the forward direction and were separated by a 50-base pair linker. The second F14 site and Frt site were constructed in the reverse direction and were separated by another 50-base pair linker. Multiple cloning sites were inserted between the first Frt site and the second F14 site. (b) Principle of F-FLEX switch. The gene of interest is inserted between the first Frt site and the second F14 site of the F-FLEX switch cassette in an inverted orientation, and is driven by an EF1α promoter. Flpo recombinase-mediated recombination first occurs between the two F14 sites or the two Frt sites that are in the opposite direction, leading to a reversible inversion of the inverted gene of interest. Flpo-mediated recombination then occurs between the two F14 sites or the two Frt sites that are now in the same direction, excising the Frt site or the F14 site between them, respectively. The resulting construct contains only one F14 site and one Frt site, and the gene of interest is permanently locked in the forward orientation. (c) Flpo turns on F-FLEX switch. HEK cells were transfected with 1) Flpo, 2) F-FLEX-mNaChBac-T2A-tdTomato, 3) Flpo and F-FLEX-mNaChBac-T2A-tdTomato, or 4) Cre and F-FLEX-mNaChBac-T2A-tdTomato. EGFP was co-transfected to monitor the transfection. There was no leaky expression of mNaChBac-T2A-tdTomato in the absence of Flpo. mNaChBac-T2A-tdTomato expression was switched on by the expression of Flpo, but not by Cre. Similar results were obtained with other F-FLEX constructs (n = 5). (d) Flpo does not turn on Cre-dependent DIO switch45. HEK cells were transfected with 1) Cre, 2) DIO-hChR2(H134R)-EYFP, 3) Cre and DIO-hChR2(H134R)-EYFP, or 4) Flpo and DIO-hChR2(H134R)-EYFP. mRFP was co-transfected to monitor the transfection. There was no leaky expression of hChR2(H134R)-EYFP in the absence of Cre. hChR2(H134R)-EYFP expression was switched on by the expression of Cre, but not by Flpo. Similar results were obtained with other DIO constructs (n = 2).
Extended Data Figure 9. Overexpression of Kir2.1 in a small subset of layer 2/3 pyramidal cells does not affect PV-mediated inhibition onto untransfected pyramidal cells.
(a) Schematic of experiments. Left, unitary connection from a PV cell to nearby layer 2/3 pyramidal cells in control mice that were not electroporated with pCAG-Kir2.1-T2A-tdTomato. Right, unitary connection from a PV cell to nearby layer 2/3 untransfected pyramidal cells in mice that were in utero electroporated with pCAG-Kir2.1-T2A-tdTomato. (b) Connectivity rates from PV cells to layer 2/3 pyramidal cells in control mice (95%, 57 out of 60) and to untransfected pyramidal cells in electroporated mice (93%, 52 out of 56) are similar (P = 0.7). (c) Cumulative frequencies for uIPSC amplitudes (control: n = 57, median, 224.0 pA; untransfected: n = 52, median, 190.4 pA; P = 0.5). Inset: mean ± s.e.m. (d) Summary graph for the average relative deviations of uIPSCs from 20 and 17 similar experiment as in (a). Bars: mean ± s.e.m. (P = 0.6).
Extended Data Figure 10. A model for target activity-dependent inter-cell variability of PV cell-mediated inhibition.
Schematic illustration of how pyramidal cell activity regulates the inter-cell variability of PV cell-mediated inhibition. Left, pyramidal cells with different activity (dark and light colors indicate high and low activity, respectively) receive different amounts of PV cell-mediated inhibition (long and short bars indicate large and small inhibition, respectively), which consists of an activity-dependent component (green bars) and an activity-independent component (blue bars). The activity-dependent components are positively regulated by the pyramidal cell activity and are highly variable across neurons, whereas the activity-independent components are less variable. Right, when the activity of pyramidal cells is suppressed by overexpression of Kir2.1, the activity-dependent components are diminished and the remaining inhibition is largely the activity-independent component, hence less variable across neurons.
Acknowledgments
We thank M. Chan, J. Evora, A. Linder, and P. Abelkop for technical assistance; M.S. Caudill and S.R. Olsen for help with in vivo physiology recording program; E. Kim and A. Ghosh for pCAG-Kir2.1-T2A-tdTomato plasmid; J. Isaacson and H.Y. Zoghbi for comments on earlier versions of the manuscript; D.N. Hill and G.I. Allen for advise on statistical analysis; the members of the Scanziani and Isaacson laboratories for suggestions; and University of California, San Diego Neuroscience Microscopy Facility (P30 NS047101) for imaging equipments. M.X. was supported by a fellowship from Jane Coffin Childs Memorial Fund for Medical Research. M.S. is an investigator of the Howard Hughes Medical Institute. This work was also supported by the Gatsby Charitable Foundation.
Footnotes
Author Contributions
M.X. and M.S. designed the study. M.X. performed all experiments and data analysis. B.V.A contributed to data analysis. M.X. and M.S. wrote the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Haider B, McCormick DA. Rapid neocortical dynamics: cellular and network mechanisms. Neuron. 2009;62:171–189. doi: 10.1016/j.neuron.2009.04.008. S0896-6273(09)00289-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isaacson JS, Scanziani M. How inhibition shapes cortical activity. Neuron. 2011;72:231–243. doi: 10.1016/j.neuron.2011.09.027. S0896-6273(11)00879-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson JS, Carandini M, Ferster D. Orientation tuning of input conductance, excitation, and inhibition in cat primary visual cortex. J Neurophysiol. 2000;84:909–926. doi: 10.1152/jn.2000.84.2.909. [DOI] [PubMed] [Google Scholar]
- 4.Shu Y, Hasenstaub A, McCormick DA. Turning on and off recurrent balanced cortical activity. Nature. 2003;423:288–293. doi: 10.1038/nature01616. nature01616 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Wehr M, Zador AM. Balanced inhibition underlies tuning and sharpens spike timing in auditory cortex. Nature. 2003;426:442–446. doi: 10.1038/nature02116. nature02116 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Wilent WB, Contreras D. Synaptic responses to whisker deflections in rat barrel cortex as a function of cortical layer and stimulus intensity. J Neurosci. 2004;24:3985–3998. doi: 10.1523/JNEUROSCI.5782-03.200424/16/3985. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haider B, Duque A, Hasenstaub AR, McCormick DA. Neocortical network activity in vivo is generated through a dynamic balance of excitation and inhibition. J Neurosci. 2006;26:4535–4545. doi: 10.1523/JNEUROSCI.5297-05.2006. 26/17/4535 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okun M, Lampl I. Instantaneous correlation of excitation and inhibition during ongoing and sensory-evoked activities. Nat Neurosci. 2008;11:535–537. doi: 10.1038/nn.2105. nn.2105 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Atallah BV, Scanziani M. Instantaneous modulation of gamma oscillation frequency by balancing excitation with inhibition. Neuron. 2009;62:566–577. doi: 10.1016/j.neuron.2009.04.027. S0896-6273(09)00351-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fino E, Yuste R. Dense inhibitory connectivity in neocortex. Neuron. 2011;69:1188–1203. doi: 10.1016/j.neuron.2011.02.025. S0896-6273(11)00123-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Packer AM, Yuste R. Dense, unspecific connectivity of neocortical parvalbumin-positive interneurons: a canonical microcircuit for inhibition? J Neurosci. 2011;31:13260–13271. doi: 10.1523/JNEUROSCI.3131-11.2011. 31/37/13260 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–13945. doi: 10.1073/pnas.19361921001936192100. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. nn1525 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Li X, et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci U S A. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. 0509030102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benedetti BL, Takashima Y, Wen JA, Urban-Ciecko J, Barth AL. Differential Wiring of Layer 2/3 Neurons Drives Sparse and Reliable Firing During Neocortical Development. Cereb Cortex. 2012 doi: 10.1093/cercor/bhs257. bhs257 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burrone J, O’Byrne M, Murthy VN. Multiple forms of synaptic plasticity triggered by selective suppression of activity in individual neurons. Nature. 2002;420:414–418. doi: 10.1038/nature01242. nature01242 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Ibata K, Sun Q, Turrigiano GG. Rapid synaptic scaling induced by changes in postsynaptic firing. Neuron. 2008;57:819–826. doi: 10.1016/j.neuron.2008.02.031. S0896-6273(08)00213-4 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Peng YR, et al. Postsynaptic spiking homeostatically induces cell-autonomous regulation of inhibitory inputs via retrograde signaling. J Neurosci. 2010;30:16220–16231. doi: 10.1523/JNEUROSCI.3085-10.2010. 30/48/16220 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sim S, Antolin S, Lin CW, Lin YX, Lois C. Increased cell-intrinsic excitability induces synaptic changes in new neurons in the adult dentate gyrus that require npas4. J Neurosci. 2013;33:7928–7940. doi: 10.1523/JNEUROSCI.1571-12.2013. 33/18/7928 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. S0012-1606(01)90439-7 [pii] [DOI] [PubMed] [Google Scholar]
- 21.Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–872. doi: 10.1016/s0306-4522(01)00016-1. S0306452201000161 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Hatanaka Y, Hisanaga S, Heizmann CW, Murakami F. Distinct migratory behavior of early- and late-born neurons derived from the cortical ventricular zone. J Comp Neurol. 2004;479:1–14. doi: 10.1002/cne.20256. [DOI] [PubMed] [Google Scholar]
- 23.Ren D, et al. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635294/5550/2372. [pii] [DOI] [PubMed] [Google Scholar]
- 24.van Vreeswijk C, Sompolinsky H. Chaos in neuronal networks with balanced excitatory and inhibitory activity. Science. 1996;274:1724–1726. doi: 10.1126/science.274.5293.1724. [DOI] [PubMed] [Google Scholar]
- 25.Vogels TP, Abbott LF. Gating multiple signals through detailed balance of excitation and inhibition in spiking networks. Nat Neurosci. 2009;12:483–491. doi: 10.1038/nn.2276. nn.2276 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renart A, et al. The asynchronous state in cortical circuits. Science. 2010;327:587–590. doi: 10.1126/science.1179850. 327/5965/587 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogels TP, Sprekeler H, Zenke F, Clopath C, Gerstner W. Inhibitory plasticity balances excitation and inhibition in sensory pathways and memory networks. Science. 2011;334:1569–1573. doi: 10.1126/science.1211095. science.1211095 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012;35:57–67. doi: 10.1016/j.tins.2011.10.004. S0166-2236(11)00175-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 2013;503:121–125. doi: 10.1038/nature12743. nature12743 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. nn.2467 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barth AL, Gerkin RC, Dean KL. Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. J Neurosci. 2004;24:6466–6475. doi: 10.1523/JNEUROSCI.4737-03.200424/29/6466. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniguchi H, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. S0896-6273(11)00679-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hippenmeyer S, et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. 04-PLBI-RA-0875R3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Jan YN, Jan LY. Control of rectification and permeation by residues in two distinct domains in an inward rectifier K+ channel. Neuron. 1995;14:1047–1054. doi: 10.1016/0896-6273(95)90343-7. 0896-6273(95)90343-7 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Tong Y, et al. Tyrosine decaging leads to substantial membrane trafficking during modulation of an inward rectifier potassium channel. J Gen Physiol. 2001;117:103–118. doi: 10.1085/jgp.117.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tinker A, Jan YN, Jan LY. Regions responsible for the assembly of inwardly rectifying potassium channels. Cell. 1996;87:857–868. doi: 10.1016/s0092-8674(00)81993-5. S0092-8674(00)81993-5 [pii] [DOI] [PubMed] [Google Scholar]
- 38.Yue L, Navarro B, Ren D, Ramos A, Clapham DE. The cation selectivity filter of the bacterial sodium channel, NaChBac. J Gen Physiol. 2002;120:845–853. doi: 10.1085/jgp.20028699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turan S, Kuehle J, Schambach A, Baum C, Bode J. Multiplexing RMCE: versatile extensions of the Flp-recombinase-mediated cassette-exchange technology. J Mol Biol. 2010;402:52–69. doi: 10.1016/j.jmb.2010.07.015. S0022-2836(10)00768-0 [pii] [DOI] [PubMed] [Google Scholar]
- 40.Kranz A, et al. An improved Flp deleter mouse in C57Bl/6 based on Flpo recombinase. Genesis. 2010;48:512–520. doi: 10.1002/dvg.20641. [DOI] [PubMed] [Google Scholar]
- 41.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.22356881002235688100. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manent JB, Wang Y, Chang Y, Paramasivam M, LoTurco JJ. Dcx reexpression reduces subcortical band heterotopia and seizure threshold in an animal model of neuronal migration disorder. Nat Med. 2009;15:84–90. doi: 10.1038/nm.1897. nm.1897 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. 0610155104 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pologruto TA, Sabatini BL, Svoboda K. ScanImage: flexible software for operating laser scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-131475-925X-2-13. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitamura K, Judkewitz B, Kano M, Denk W, Hausser M. Targeted patch-clamp recordings and single-cell electroporation of unlabeled neurons in vivo. Nat Methods. 2008;5:61–67. doi: 10.1038/nmeth1150. nmeth1150 [pii] [DOI] [PubMed] [Google Scholar]
- 48.Brainard DH. The Psychophysics Toolbox. Spatial vision. 1997;10:433–436. [PubMed] [Google Scholar]
- 49.Hibino H, et al. Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol Rev. 2010;90:291–366. doi: 10.1152/physrev.00021.2009. 90/1/291 [pii] [DOI] [PubMed] [Google Scholar]