Abstract
All-optical electrophysiology—spatially resolved simultaneous optical perturbation and measurement of membrane voltage—would open new vistas in neuroscience research. We evolved two archaerhodopsin-based voltage indicators, QuasAr1 and 2, which show improved brightness and voltage sensitivity, microsecond response times, and produce no photocurrent. We engineered a novel channelrhodopsin actuator, CheRiff, which shows improved light sensitivity and kinetics, and spectral orthogonality to the QuasArs. A co-expression vector, Optopatch, enabled crosstalk-free genetically targeted all-optical electrophysiology. In cultured neurons, we combined Optopatch with patterned optical excitation to probe back-propagating action potentials in dendritic spines, synaptic transmission, sub-cellular microsecond-timescale details of action potential propagation, and simultaneous firing of many neurons in a network. Optopatch measurements revealed homeostatic tuning of intrinsic excitability in human stem cell-derived neurons. In brain slice, Optopatch induced and reported action potentials and subthreshold events, with high signal-to-noise ratios. The Optopatch platform enables high-throughput, spatially resolved electrophysiology without use of conventional electrodes.
Introduction
To disentangle the complex interactions underlying neural dynamics, one would like to visualize membrane voltage across spatial scales, from single dendritic spines to large numbers of interacting neurons, while delivering spatially and temporally precise stimuli1, 2. Optical methods for simultaneous perturbation and measurement of membrane potential could achieve this goal3.
One would further like to target the stimulation and recording to genetically specified cells. Genetic targeting is particularly important in intact tissue where closely spaced cells often perform distinct functions. Genetic targeting is also valuable in vitro, for characterizing heterogeneous cultures that arise during stem cell differentiation to neurons4, or while studying neurons co-cultured with other cell types.
Optical stimulation has been demonstrated with glutamate uncaging5, photoactivated ion channel agonists6, and microbial rhodopsin actuators7. Genetically encoded functional readouts include reporters of intracellular Ca2+ and membrane voltage8. Voltage-sensitive dyes offer good speed, sensitivity, and spectral tuning9, 10, but cannot be delivered to a genetically specified subset of cells and often suffer from phototoxicity.
Optical stimulation has been paired with voltage imaging11–14. However, robust and crosstalk-free genetically targeted all-optical electrophysiology has not been achieved due to limitations on the speed and sensitivity of genetically encoded voltage indicators (GEVIs), and spectral overlap between existing GEVIs and optogenetic actuators. GFP-based GEVIs experience severe optical crosstalk with even the most red-shifted channelrhodopsins, which retain ∼20% activation with blue light excitation15. There remains a need for sensitive, fast, and spectrally orthogonal tools for genetically targeted simultaneous optical perturbation and measurement of membrane voltage.
Here we introduce variants of a near infrared archaerhodopsin-based voltage indicator and a blue light gated channelrhodopsin actuator which individually show greatly improved performance relative to published optogenetic tools and which together constitute a tool for all-optical electrophysiology. First, we characterize the optical and electrophysiological properties of the indicator, the actuator, and the co-expressed pair (Optopatch) and compare against published tools. Second, we use Optopatch to probe neuronal excitation across spatial and temporal scales: from single dendritic spines to fields containing dozens of neurons measured in parallel, and from microsecond delays associated with action potential propagation to days-long changes in excitability. Third, we apply Optopatch to study excitability in human induced pluripotent stem cell (hiPSC)-derived neurons. These measurements revealed the first evidence for homeostatic plasticity of intrinsic excitability in hiPSC-derived neurons. Fourth, we apply Optopatch in tissue. In organotypic brain slice, Optopatch initiates and reports action potentials and subthreshold dynamics with higher signal-to-noise ratios, better photostability, and better time resolution than ArcLight, a recently introduced GFP-based GEVI16.
Results
Directed evolution of an Arch-based voltage indicator
We previously showed that Archaerhodopsin 3 (Arch) functions as a fast and sensitive voltage indicator17. Arch has the furthest red-shifted spectrum of any GEVI, giving it the unique property of little spectral overlap with channelrhodopsin actuators and GFP-based reporters. Thus it is natural to pair Arch-based indicators with optogenetic actuators for crosstalk-free all-optical electrophysiology.
However, wild-type Arch had some undesirable attributes for a reporter: it was very dim, and the brightness was a nonlinear function of illumination intensity18. Illumination for imaging generated a hyperpolarizing photocurrent, which partially suppressed neural firing. The mutant Arch(D95N) did not pump, but its step response was dominated by a 41 ms time constant, too slow to resolve action potential (AP) waveforms. Other non-pumping mutants improved speed relative to Arch(D95N) but did not reach the speed of wild-type Arch and did not address the low brightness14.
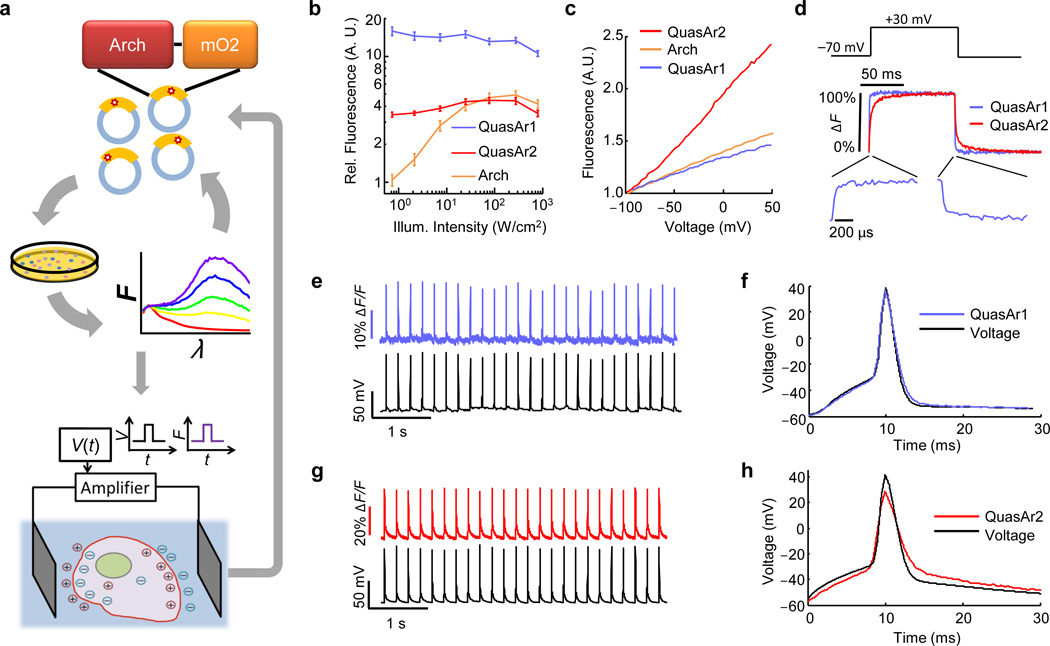
We sought to repair these defects in engineered mutants of Arch. To accommodate the multiple selection criteria, we adopted a hierarchical screen (Fig. 1a). Five rounds of brightness screening in E. coli and random mutagenesis on a library of > 104 Arch mutants resulted in a brighter Arch variant, containing 5 point-mutations (Methods). Further site-directed mutagenesis at known key residues improved voltage sensitivity and speed (Supplementary Fig. 1), while addition of an endoplasmic reticulum export motif and a trafficking sequence improved trafficking (Methods). The two most promising mutants were named QuasArs (Quality superior to Arch). QuasAr1 comprised mutations P60S, T80S, D95H, D106H, F161V and QuasAr2 comprised QuasAr1(H95Q). Both proteins had fluorescence excitation maxima at 590 nm and emission maxima at 715 nm (Supplementary Fig. 2). The fluorescence quantum yields of solubilized QuasAr1 and 2 were 19- and 10-fold enhanced, respectively, relative to the non-pumping voltage indicator Arch(D95N) (Supplementary Table 1). All fluorescence microscopy of QuasArs used 640 nm excitation.
Figure 1. Non-pumping Arch-derived voltage indicators with improved speed, sensitivity, and brightness.
A) Hierarchical screen to select improved Arch mutants. Five rounds of random mutagenesis and screening for brightness were performed in E. coli. The brightest mutants were subjected to targeted mutagenesis and screening for speed and voltage sensitivity in HeLa cells via induced transient voltage (Supplementary Fig. 1). B) Fluorescence of Arch mutants fused to eGFP and expressed in HEK cells, as a function of illumination intensity. The plot shows Arch fluorescence normalized by 640 nm illumination intensity and by eGFP fluorescence (488 nm exc., 525 – 575 nm em.) to control for cell-to-cell variations in expression. A linear fluorophore (i.e. brightness proportional to illumination intensity) would appear as a horizontal line. Error bars represent s.e.m. (n = 7 cells for each mutant). C) Fluorescence vs. membrane voltage for Arch, QuasAr1, and QuasAr2 expressed in HEK cells. D) Fluorescence responses to a step in membrane voltage from −70 to +30 mV. E) Simultaneous optical and electrical recording of APs in a rat hippocampal neuron expressing QuasAr1. Frame rate 1 kHz. F) Overlay of mean optically and electrically recorded AP waveforms. Frame rate 2 kHz. G, H) Same as E, F in neurons expressing QuasAr2. Data in B – H acquired on a 128 × 128 pixel EMCCD camera (Methods).
We compared the fluorescence, voltage sensitivity, and speed of the QuasArs to wild-type Arch in HEK cells, using epifluorescence microscopy and whole-cell patch clamp electrophysiology. Under low intensity illumination (500 mW/cm2), QuasAr1 was 15-fold brighter than wild-type Arch, and QuasAr2 was 3.3-fold brighter (Fig. 1b; Methods). Neither mutant showed the optical nonlinearity seen in the wild-type protein, implying that fluorescence was a 1-photon process with the voltage-sensitive transition occurring from the ground state. At high intensity (> 100 W/cm2) QuasAr1 was 2.5-fold brighter than wild-type Arch, while the brightness of QuasAr2 and of wild-type Arch were comparable.
Fluorescence of Arch, QuasAr1, and QuasAr2 increased nearly linearly with membrane voltage between −100 mV and +50 mV (Fig. 1c, Methods). Sensitivities were (ΔF/F per 100 mV): 32 ± 3% for QuasAr1 (n = 5 cells; all statistics are mean ± s.e.m. unless specified) and 90 ± 2% for QuasAr2 (n = 6 cells). The sensitivity of QuasAr2 is a substantial improvement over both Arch (40% per 100 mV) and Arch(D95N) (60% per 100 mV).
Steps in membrane voltage (−70 mV to +30 mV) induced rapid fluorescence responses in both mutants, which we quantified on a fast photomultiplier (Fig. 1d). At room temperature (23 °C) QuasAr1 had a step response time constant of 0.053 ± 0.002 ms (n = 6 cells), close to the 0.05 ms time resolution of the electronics and substantially faster than the 0.6 ms step response of wild-type Arch18. QuasAr2 had a bi-exponential step response with time constants of 1.2 ± 0.1 ms (68%) and 11.8 ± 1.5 ms (32%) (n = 6 cells). At 34 °C, the apparent speed of QuasAr1 remained at the 0.05 ms resolution of the electronics, and the time constants of QuasAr2 decreased to 0.30 ± 0.05 ms (62%) and 3.2 ± 0.4 ms (38%) (n = 7 cells). Both mutants had similar response times on rising and falling edges (Supplementary Table 2). Neither QuasAr1 nor QuasAr2 generated detectable photocurrent under red light (tested up to 900 W/cm2) or blue light (Supplementary Fig. 3).
We expressed QuasArs in cultured rat hippocampal neurons, evoked APs via current injection from a patch pipette and recorded the fluorescence responses from the soma and proximal dendrites (1 kHz frame rate, Fig. 1e–h, Supplementary Fig. 4). Single APs produced fluorescence transients with amplitude ΔF/F = 21 ± 2% for QuasAr1 (n = 11 cells) and ΔF/F = 48 ± 3% for QuasAr2 (n = 24 cells). Signal-to-noise ratios (SNRs) for single APs increased with illumination intensity. For QuasAr1, SNR values were 21 ± 2 (300 W/cm2, n = 6 cells) and 32 ± 4 (800 W/cm2, n = 6 cells). For QuasAr2, SNR values were 41 ± 4 (300 W/cm2, n = 12 cells) and 70 ± 8 (800 W/cm2, n = 12 cells). These SNRs correspond to equivalent electrical noise levels of 3.0 to 4.3 mV (800 to 300 W/cm2) for QuasAr1, or 1.5 to 2.2 mV (800 to 300 W/cm2) for QuasAr2 (Methods).
QuasAr1 did not introduce detectable broadening in the optically recorded AP waveform, acquired at a 2 kHz frame rate (Fig. 1f). At room temperature, QuasAr2 broadened the optically recorded AP by 650 ± 150 µs relative to the simultaneously recorded electrical waveform at 70% maximum depolarization (n = 5 cells; mean ± s.d.) (Fig. 1h). At 34 °C, QuasAr2 broadened the optically recorded AP by 180 ± 120 µs (n = 5 cells; mean ± s.d). Both probes reported AP peak times with < 100 µs jitter relative to simultaneously acquired patch clamp recordings (Methods).
Photostability is a concern with any voltage indicator, so we quantified the stability of QuasArs under continuous illumination at standard imaging intensity (300 W/cm2). Photobleaching time constants were 440 s for QuasAr1 and 1020 s for QuasAr2. We further tested for red light-induced phototoxicity using QuasAr2 as the readout. Under continuous illumination at 300 W/cm2, QuasAr2 reported APs with 100% fidelity for the 30 min duration of the experiment, with no detectable change in AP width or waveform (Supplementary Fig. 5).
We compared the QuasArs to ArcLight A24216. Photophysical comparisons were performed in HEK cells (Supplementary Table 2), and action potential comparisons were performed in matched neuronal cultures (Methods). ArcLight can be imaged with ∼30-fold lower illumination intensity than is required for the QuasArs, facilitating measurements on readily available microscope systems. However, the QuasArs reported action potentials with 7 to 16-fold larger fractional fluorescence changes, 3 to 8-fold higher SNR, 30 to 1000-fold higher temporal resolution, and 6 to 15-fold greater photostability (Supplementary Fig. 6).
The QuasArs represent the fastest and most sensitive GEVIs reported to-date. The 50 µs response time of QuasAr1 is more than 10-fold faster than the fastest previously reported GEVIs18, 19 and is comparable to fast voltage-sensitive dyes. QuasAr1 opens the possibility of accurate mapping of AP waveforms for even the fastest-spiking neurons20. The QuasArs report voltage with greatly improved sensitivity and time resolution compared to the first generation of Arch-based GEVIs, despite requiring ∼5-fold lower illumination intensity17. From a signal-to-noise perspective, QuasAr2 is superior to QuasAr1: the greater voltage sensitivity of QuasAr2 outweighs the greater brightness of QuasAr1. From a temporal resolution perspective, QuasAr1 is superior. We recommend QuasAr2 for spike counting and measurement of sub-threshold events, and QuasAr1 for measurement of microsecond-precision AP waveforms and timing. Furthermore, the far red excitation of the QuasArs allows, in principle, combination with channelrhodopsin actuators or other GFP-based reporters.
CheRiff, a sensitive blue-shifted optogenetic actuator
We next sought to combine the QuasAr reporters with a blue light-activated channelrhodopsin. To achieve spatially precise optical excitation, the channelrhodopsin should trigger APs when only a subsection of a cell is excited. Existing optogenetic actuators have had only marginal success in achieving this goal21. To avoid optical crosstalk, the blue light intended for the channelrhodopsin should not perturb the GEVI fluorescence. Existing optogenetic actuators require blue light intensities that perturb QuasAr fluorescence (Supplementary Fig. 7). We thus sought a more sensitive channelrhodopsin that could reliably trigger APs with sub-cellular illumination and at lower light intensity, while maintaining fast opening and closing kinetics.
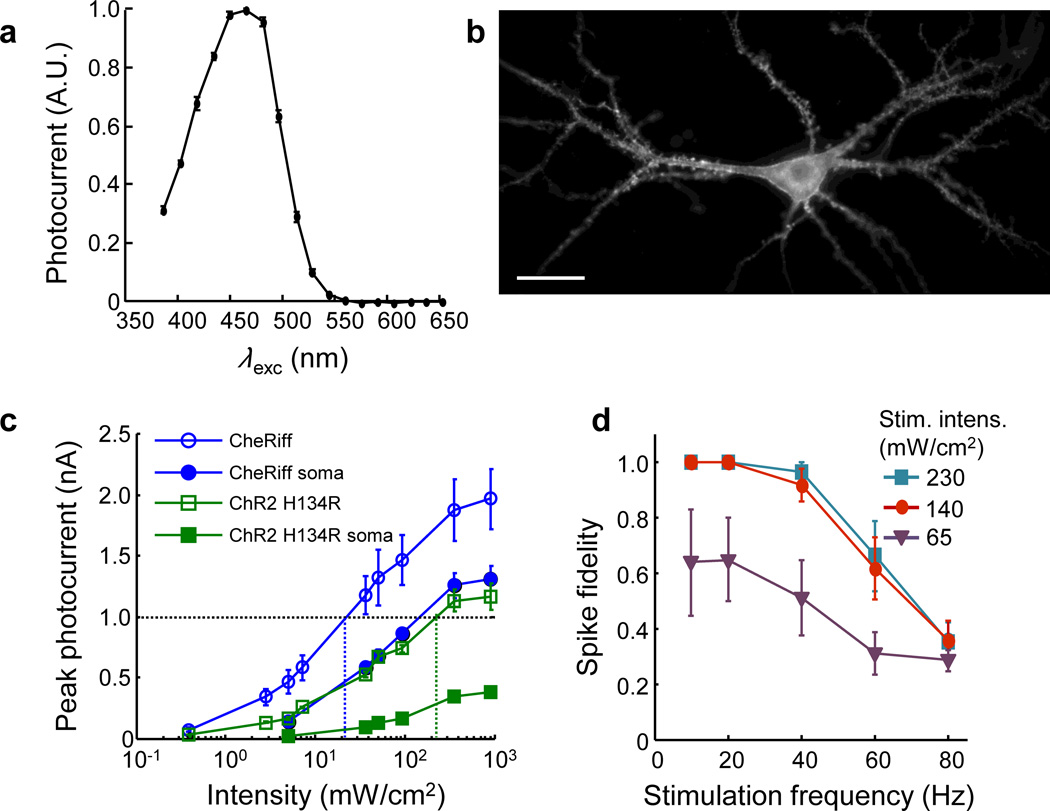
During a screen of plant genomes22 we identified a novel optogenetic actuator, Scherffelia dubia ChR (sdChR)15, derived from a fresh-water green alga first isolated from a small pond in Essex, England (Supplementary Fig. 8)23. SdChR had promising sensitivity and a blue action spectrum (λmax = 474 nm). Addition of a trafficking sequence improved membrane targeting (Supplementary Fig. 8). Introduction of the mutation E154A sped the kinetics and shifted the peak of the action spectrum to λmax = 460 nm (Fig. 2a), which decreased spurious channelrhodopsin activation by red light (Supplementary Fig. 9, Supplementary Table 3, Methods). We dubbed the final construct CheRiff in reference to Scherffelia, its genus of origin. CheRiff showed good expression and membrane trafficking in cultured rat hippocampal neurons (Fig. 2b).
Figure 2. CheRiff is a fast and sensitive blue-shifted channelrhodopsin.
A) Action spectrum acquired in HEK293T cells (n = 6 cells). CheRiff had a blue-shifted action spectrum with a peak at λmax ∼ 460 nm. B) Cultured rat hippocampal neuron expressing CheRiff-eGFP, imaged via eGFP fluorescence. Scale bar 25 µm. Image acquired on an sCMOS detector (Methods). C) Comparison of photocurrents as a function of illumination intensity in matched neuronal cultures expressing CheRiff (n = 5 cells) or ChR2 H134R (n = 5 cells). Illumination was either over the whole cell or confined to the soma. D) Spiking fidelity as a function of stimulation frequency and illumination intensity in neurons expressing CheRiff (n = 5 cells). Cells were stimulated with trains of 40 pulses (2 ms pulse width, 10 to 80 Hz) at three different blue light intensities. Error bars in C and D represent s.e.m.
Under typical neural culture conditions, rapid and robust AP initiation requires currents of approximately 1 nA (ref. 24). In a paired comparison, CheRiff passed a photocurrent of 1 nA at a whole-cell illumination intensity of 22 ± 10 mW/cm2 (n = 5 neurons), 9-fold lower than was required for ChR2 H134R (200 ± 80 mW/cm2, n = 5 neurons) (Fig. 2c). For stimulation localized to the soma, CheRiff passed a photocurrent of 1 nA under illumination at 100 mW/cm2, while ChR2 H134R did not achieve this photocurrent under any illumination intensity. Upon pulsed whole-cell illumination, CheRiff induced high-frequency and high-reliability spike trains (Fig. 2d) at five to ten-fold lower illumination intensities than have been reported for ChR2 H134R, ChIEF, or ChETA24 using the same protocol.
We measured the photophysical properties of CheRiff, ChR2 H134R and ChIEF25 in matched neuronal cultures (Supplementary Fig. 9, Supplementary Table 4). CheRiff showed 2-fold larger maximal photocurrents than ChR2 H134R or ChIEF. CheRiff had an opening rate 2-fold faster than ChR H134R and 4-fold faster than ChIEF. CheRiff had a similar closing rate to ChIEF, and was 1.5-fold faster than ChR H134R.
Finally, we tested for optical crosstalk between QuasArs and CheRiff in cultured neurons (Supplementary Table 5). Illumination sufficient to induce high-frequency trains of APs (488 nm, 140 mW/cm2) perturbed fluorescence of QuasArs by < 1% (Supplementary Fig. 7). Illumination with high intensity red light (640 nm, 900 W/cm2) induced an inward photocurrent through CheRiff of 14.3 ± 3.1 pA, which depolarized neurons by 3.1 ± 0.2 mV (n = 5 cells) (Supplementary Fig. 9). For most applications this level of optical crosstalk is acceptable.
Of the many attributes that determine channelrhodopsin function, the most important in CheRiff are its greatly increased sensitivity at low illumination intensity and its fast opening kinetics. These properties allow sub-cellular and low-intensity triggering of precisely timed APs.
Optopatch constructs
Optopatch1 and Optopatch2 consisted of bicistronic vectors for co-expression of CheRiff-eGFP and QuasAr1- or QuasAr2-mOrange2, respectively (Supplementary Fig. 10). We also made Optopatch variants which contained non-fluorescent eGFP and mOrange2 mutants, freeing the spectrum for other uses (Methods). The QuasAr and CheRiff constructs could also be delivered separately, but the bicistronic vector maintained a more uniform ratio of actuator to reporter expression levels.
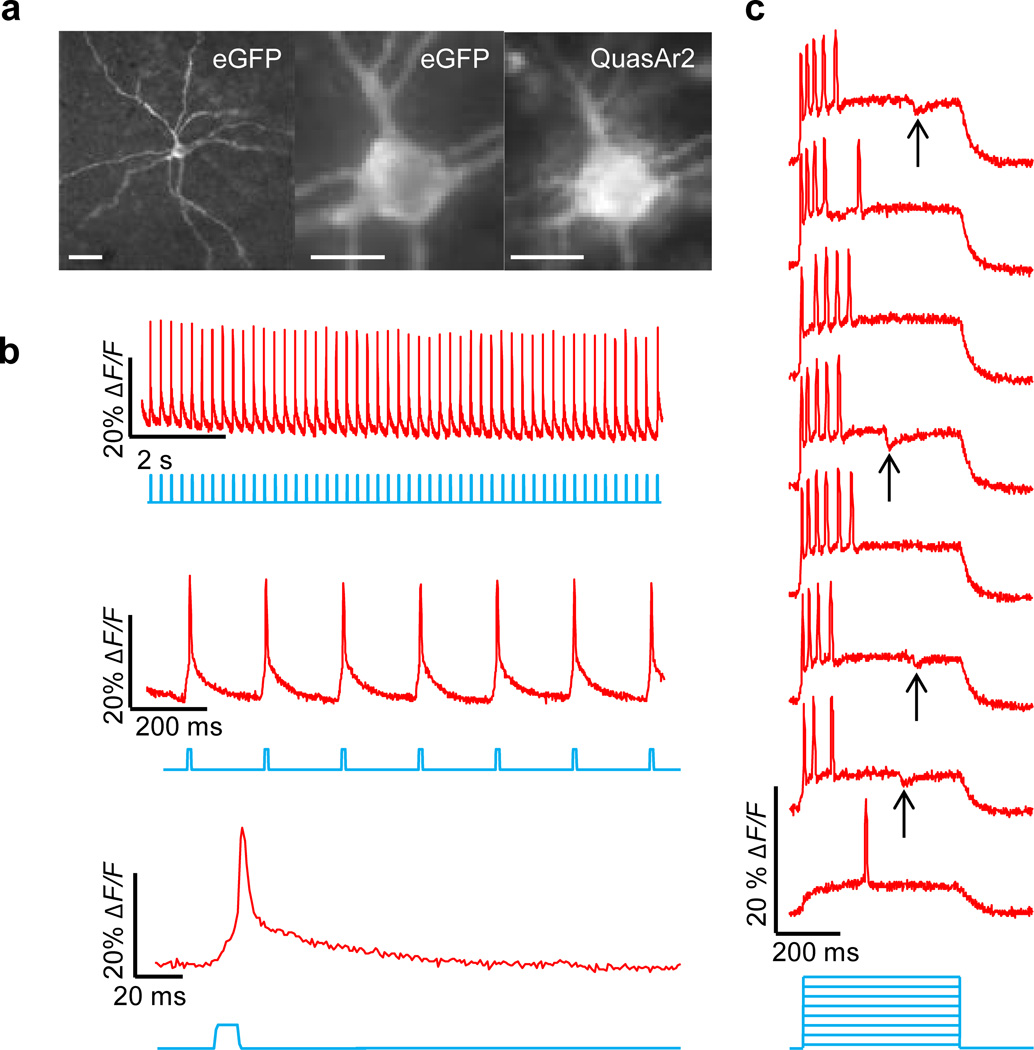
We characterized Optopatch2 in detail. When expressed under a CaMKIIα promoter in cultured rat hippocampal neurons (Methods), Optopatch2 showed high expression and good membrane trafficking of both CheRiff and QuasAr2 (Fig. 3a). Patch clamp measurements found no statistically significant effect of expression on membrane resistance (P = 0.72), membrane capacitance (P = 0.87), resting potential (P = 0.31), threshold current (P = 0.67), or threshold potential (P = 0.38), when compared to paired cultures transfected with cytoplasmic GFP driven by a CaMKIIα promoter (n = 8 cells for Optopatch2, n = 7 cells for GFP) (Supplementary Fig. 11).
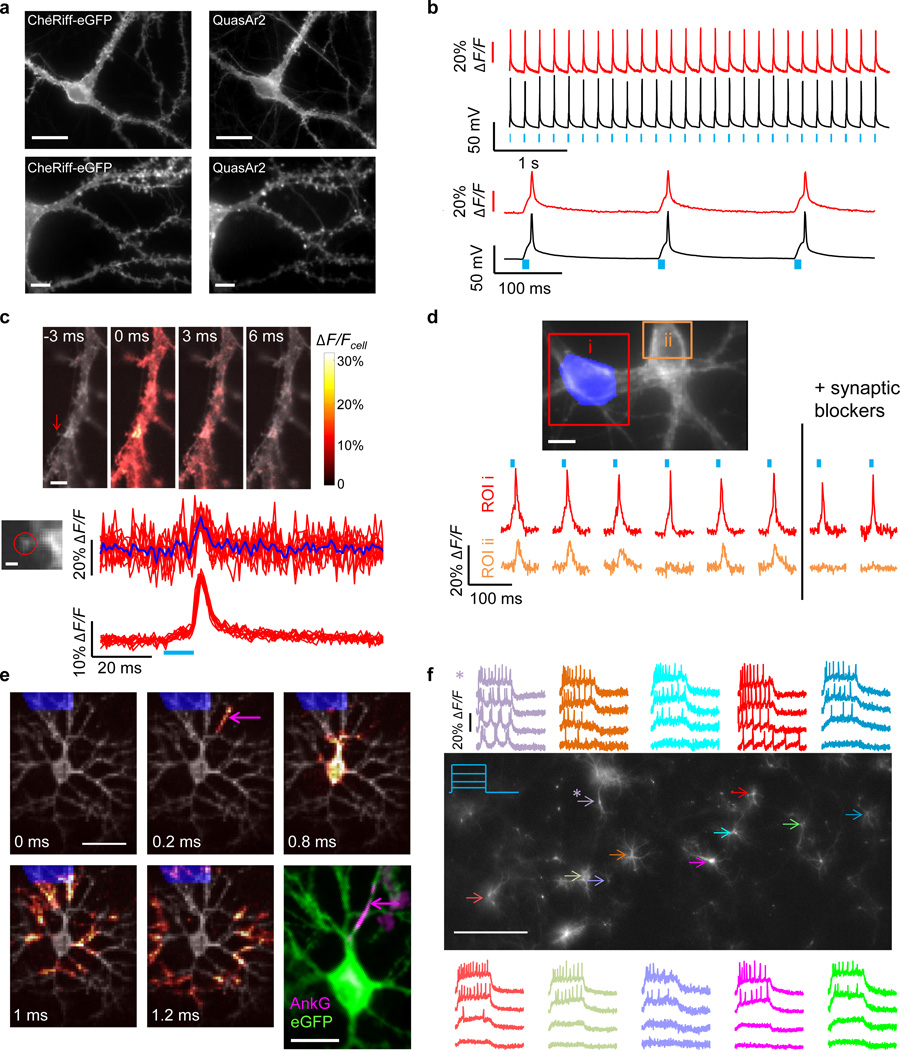
Figure 3. Optopatch enables high fidelity optical stimulation and recording in cultured neurons.
A) Trafficking of Optopatch components in cultured rat hippocampal neurons. Left: CheRiff-eGFP, measured via eGFP fluorescence. Right: QuasAr2, measured via retinal fluorescence. Scale bars: top 20 µm, bottom 5 µm. B) Temporally precise optical initiation and monitoring of single APs. Blue: illumination. Red: whole-cell single-trial unfiltered fluorescence. Black: patch clamp recording. C) Optical mapping of an AP induced via illumination of the soma. Top: Filmstrip showing average of n = 20 temporally registered APs. Fluorescence is normalized to mean fluorescence of the dendrite. Images are composite of mean fluorescence (gray) and changes in fluorescence (heat map). Arrow indicates dendritic spine. Scale bar 5 µm. Bottom: Single-trial detection of back-propagating APs in a single dendritic spine. Scale bar 1 µm. Top traces: ten single-trial recordings from the spine (red) and their average (blue). Bottom traces: ten single-trial recordings from the parent dendrite. D) Synaptic transmission. Optical stimulation of one soma (highlighted in blue) induced single APs in the stimulated cell (i) and EPSPs in the neighboring cell (ii). Synaptic blockers suppressed the response in the postsynaptic cell but not in the presynaptic cell. E) Sub-frame interpolation of AP propagation in a neuron expressing Optopatch1 (Supplementary Movie 4). Scale bar 50 µm. Bottom right: Immunostaining of the same cell with anti-eGFP and anti-AnkyrinG. Scale bar 25 µm. Magenta arrows: site of action potential initiation, distal end of the AIS. F) Parallel optical recording under increasingly strong 0.5 s optical step-stimuli. Asterisk indicates a cell that showed periodic bursts of 3–4 APs under weak stimulation. Scale bar 500 µm. Image is of eGFP fluorescence. Data in (A, C, F) acquired on an sCMOS detector; data in (B, D, E) acquired on an EMCCD (Methods). Detailed protocols for each panel are in Methods.
A neuron expressing Optopatch2 was exposed to whole-field illumination with pulses of blue light (10 ms, 25 mW/cm2) to stimulate CheRiff, and simultaneous constant illumination with red light (800 W/cm2) to excite fluorescence of QuasAr2. We imaged fluorescence of QuasAr2 at a 1 kHz frame rate and calculated fluorescence from whole-cell average intensity (Supplementary Fig. 4), while simultaneously recording membrane voltage via a patch pipette. The optical and electrical traces corresponded closely (Fig. 3b).
Raw movies acquired at 1 kHz clearly showed fluorescence changes due to optically triggered APs (Supplementary Movie 1). Averaging temporally registered AP movies over multiple trials improved the SNR for sub-cellular AP mapping (Supplementary Movie 2). Under focused red illumination (2200 W/cm2), back-propagating APs were detected in single dendritic spines on a single-trial basis, with an SNR of 2.5 (Fig. 3c).
Probing synaptic transmission
With Optopatch, one can stimulate and record from independently selected and rapidly reconfigured regions of a neural circuit. We implemented this capability in culture using a digital micromirror device (DMD) to pattern the blue CheRiff excitation26, and wide-field red illumination to monitor voltage throughout the field of view (Methods).
We probed synaptic transmission by optically stimulating the soma of single cells, and optically monitoring electrical responses in neighboring cells. Optically induced single APs in the presynaptic cell led to fluorescence transients indicative of excitatory post-synaptic potentials (EPSPs), as well as occasional failures of synaptic transmission, in the postsynaptic cell (Fig. 3d). The mean interval between the peak of the AP in the upstream cell and the onset of the optically detected EPSP in the downstream cell was < 2 ms, indicating a monosynaptic connection27. Addition of synaptic blockers (10 µM NBQX, 20 µM Gabazine, 25 µM AP-V) quieted the fluorescence response in the postsynaptic cell, without perturbing presynaptic activity. Validation measurements with patch clamp recordings showed millivolt-level correspondence of optically and electrically recorded postsynaptic potentials, as well as inhibition of these signal by synaptic blockers (Supplementary Fig. 12).
Probing AP propagation
We next sought to apply the extremely fast response of Optopatch1 (containing QuasAr1) to probe the microsecond-timescale dynamics of AP initiation and propagation within a single cell. We used the DMD to target optical stimulation to either a dendrite or the soma and recorded the fluorescence dynamics at a 1 kHz frame rate. To improve the SNR we averaged 100 to 400 temporally registered optically induced APs. These mean-AP movies showed spread of the subthreshold voltage outward from the stimulated region, followed by a sudden spike in cell-wide fluorescence that peaked within two frames (Supplementary Movie 3). Thus the native 1 kHz frame rate was insufficient to resolve the details of AP propagation.
To probe microsecond-level dynamics, we adapted the sub-frame interpolation approach of Foust28 and Popovic29 (Supplementary Fig. 13, Methods, Supplementary Software). By interpolating a smoothly varying spline to the fluorescence intensity time-trace at each pixel, we inferred the timing with which the fluorescence crossed a user-selected threshold (e.g. 50% maximum deviation) with a precision better than the exposure time of the camera. Mean deviation between optically inferred and electrically recorded AP timing at the soma was 40 – 70 µs (Supplementary Fig. 13). We then constructed a higher time-resolution movie highlighting each pixel at the sub-frame time of its wavefront crossing. This interpolation technique does not rely on an assumed AP waveform, nor does it assume wavelike propagation; it is compatible with APs that change shape within or between cells.
The visually striking propagation movies clearly showed AP initiation 30 – 50 µm from the soma in a single thin neurite, presumed to be the axon, in n = 8 of 8 measured cells (Supplementary Fig. 3e, Supplementary Movies 4–6), regardless of stimulus location. The AP then propagated down the putative axon and back into the soma and dendrites. Latencies between AP initiation and arrival in the soma were 320 ± 220 µs (n = 8 cells, mean ± s.d).
After acquiring Optopatch data, we fixed the cells and stained for ankyrin-G, a marker of the axon initial segment (AIS, Fig. 3e). The optically detected zones of AP initiation coincided with the distal end of the AIS, consistent with previous experiments using voltage-sensitive dyes30. Thus Optopatch can resolve functionally significant sub-cellular and microsecond timescale details of AP dynamics.
Parallel measurements in neuronal cultures
To achieve high-throughput functional characterization in neurons, one would like to apply the technique to many cells in parallel. We constructed a low-magnification, high-speed microscope (Supplementary Fig. 14, Methods) which robustly reported APs and subthreshold dynamics in up to ∼50 cells simultaneously (Supplementary Movie 7). We used a DMD to pattern the blue illumination for targeted CheRiff excitation in user-selected regions. Optical stimulation of a segment of a synaptically connected culture induced network activity which manifested as action potentials and sub-threshold dynamics in the cells not directly stimulated (Supplementary Fig. 15).
We developed an all-optical protocol to measure neuronal excitability. Synaptic blockers were added to suppress network activity. Cells were stimulated with pulses of blue light (500 ms at 6 s intervals) of increasing intensity (0 to 14 mW/cm2), while firing patterns were recorded under continuous red illumination (100 W/cm2). In wide-field measurements on n = 169 neurons expressing Optopatch2, we observed several distinct firing patterns, including fast-adapting and slow-adapting spike trains (Fig. 3f and Supplementary Movie 8). Two neurons showed intermittent bursting (one of these is indicated by a star in Fig. 3f). The comparatively high throughput of Optopatch measurements enables detection of rare electrophysiological phenotypes that might be missed in a manual patch clamp measurement on a smaller number of cells.
To test whether high-throughput Optopatch could quantify subtle electrophysiological shifts, we applied the all-optical excitability protocol to a model of homeostatic plasticity of intrinsic excitability (HPIE)31, in which neurons adapt their excitability to maintain an approximately constant level of activity. Observation of HPIE in culture32, 33 and in vivo34 has traditionally required laborious patch clamp measurements on many cells.
Neurons expressing Optopatch2 were incubated in tetrodotoxin (TTX, 1 µM) for 48 h and then measured in TTX-free medium in the presence of synaptic blockers32. Paired control cultures were incubated without TTX. Cells that had been treated with TTX (n = 94) subsequently showed significantly lower illumination threshold for spiking (P = 5×10−6), shorter interval between first and second spike (P < 0.001), and more total spikes (P < 0.01) than control cells (n = 75), but only a small change in time from light onset to first spike (Supplementary Fig. 16), consistent with previous reports32. Total data acquisition time was less than 1 h, a fraction of the time that would be required for manual measurements.
While the HPIE measurements showed population-level changes in excitability, a unique promise of Optopatch is the ability to measure the same cell over several days. In 8 of 10 trials, neurons measured with a 1 minute Optopatch protocol were identified and re-recorded 48 hrs later (Supplementary Fig. 17). This capability could be important in studying neuronal maturation or long-term effects of pharmacological, genetic, or environmental perturbations.
Probing excitability in hiPSC-derived neurons
Human-derived neurons show promise as a platform for studying human neurophysiology in health and disease35. However, variability arises at multiple levels in this approach4. Each patient’s genetic background modifies neuronal function, even for nominally monogenic diseases. Additional variability arises between iPSC clones from a single patient, between differentiations, and between cells cultured within a single well. Differing degrees of maturation are a particularly large source of cell-to-cell variability within a dish36. Furthermore, for diseasemodeling efforts, one may wish to test many differentiation protocols or pharmacological perturbations. Together, these factors motivate a need for accurate functional characterization with robust statistics. The low throughput and selection bias of manual electrophysiology present a serious bottleneck.
We first assessed whether Optopatch expression perturbed electrophysiology in hiPSC-derived neurons. Cells were transfected with Optopatch2 and cultured on a rat glial monolayer. The CamKIIα promoter genetically targeted the measurement to mature neurons within this highly heterogeneous culture36. Patch clamp measurements on cells transfected with Optopatch2 (n = 11) or with GFP (n = 11) showed no significant difference in membrane resistance (P = 0.82), membrane capacitance (P = 0.88), resting potential (P = 0.34), or action potential activation threshold (P = 0.78) (Supplementary Fig. 18). Optically evoked and optically monitored trains of APs showed the expected changes upon addition of ion-channel blockers lidocaine and tetraethylamine (Supplementary Fig. 19).
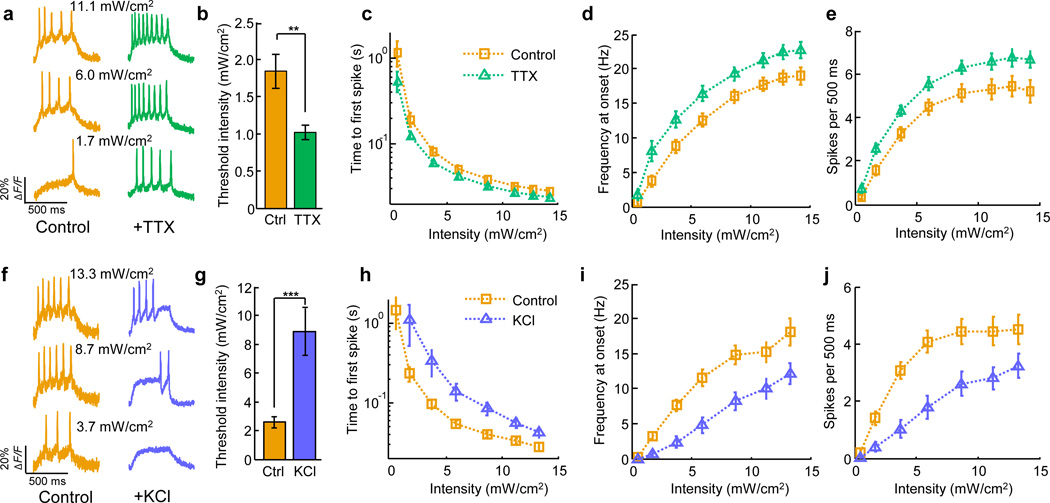
We then applied Optopatch to test for HPIE in hiPSC-derived neurons. This subtle electrophysiological effect is one of several forms of neural plasticity which have not previously been reported in human-derived neurons. HiPSC-derived neurons were incubated in 1 µM TTX for 48 h. Upon return to TTX-free medium, treated cells showed a substantial increase in subsequent optically measured excitability (n = 31 cells) relative to controls (n = 32 cells) (Fig. 4a – e), demonstrating positive HPIE.
Figure 4. Homeostatic plasticity of intrinsic excitability in human iPSC-derived neurons probed via Optopatch2.
A–E) Positive HPIE. Data from n = 32 control cells and n = 31 TTX-treated cells A) Representative optical recordings from single neurons after incubation in TTX and matched control cells. B) Threshold stimulation intensity (488 nm) to induce at least one spike in 500 ms. TTX treated cells had a significantly lower threshold than controls (P = 0.004). C) Time from onset of illumination to first spike. D) Spike frequency at onset (inverse time between first and second spike). E) Total spikes per 500 ms stimulus. Measures in D and E showed significantly increased excitability in TTX-treated cells relative to control cells (P < 0.05 for each stimulation intensity ≥ 1.7 mW/cm2). F–J) Negative HPIE. Data from n = 25 control cells and n = 28 KCl-treated cells. Panels are the same as A–E. KCl treated cells had a significantly higher stimulation intensity threshold than controls (P = 7×10−6). Measures in H – J showed significantly decreased excitability in KCl-treated cells relative to control cells (H: P < 0.01 for all stimulus intensities; I: P < 0.05 for stimulus intensities ≥ 1.7 mW/cm2; J: P < 0.05 for stimulus intensities ≤ 11.2 mW/cm2). For all experiments fluorescence was excited at 300 W/cm2, and collected at a 1 kHz frame rate on an EMCCD (Methods). All error bars represent s.e.m. Statistical significance determined by two-tailed student’s t-test or Mann–Whitney U test. ** P < 0.01; *** P < 0.001.
We next tested for negative HPIE: conditions that depolarize cells (e.g. high KCl) induce a gradual decrease in intrinsic excitability33. HiPSC-derived neurons were incubated in 15 mM KCl for 60 h. Upon return to baseline conditions (2.5 mM KCl), treated cells showed a substantial decrease in subsequent optically measured excitability (n = 28 cells) relative to controls (n = 25 cells) (Fig. 4f – j).
Post-measurement immunostaining with anti-human nuclear antigen 1 and anti-GFP antibodies confirmed that all neurons tested were of human origin (Supplementary Fig. 20). Validation measurements with manual patch clamp confirmed that the HPIE protocols did not change CheRiff photocurrents (Supplementary Fig. 21).
These experiments demonstrate that human iPSC-derived neurons undergo bidirectional HPIE, and that Optopatch measurements can non-perturbatively report subtle differences in electrophysiology in these cells. High-throughput Optopatch measurements promise to be a powerful tool for functional characterization of neuronal populations in hiPSC-based disease modeling efforts.
Imaging in organotypic slice culture
Voltage imaging with GEVIs in intact tissue would enable functional mapping of sub-cellular and circuit-level signal processing. Application of GEVIs to recording of APs in mammalian brain tissue has been limited by the low voltage sensitivity and slow response of existing indicators37, 38. No GEVI has been shown to report single-trial APs in tissue with high fidelity.
We expressed Optopatch2 in organotypic brain slice using biolistic gene delivery. Neurons that had taken up the gene were clearly visible via fluorescence of eGFP (indicating CheRiff) and QuasAr2 under wide-field epifluorescence imaging (20× N.A. 1.0 water immersion objective) (Fig. 5a). Upon illumination with pulses of blue light (10 ms, repeated at 5 Hz, 7.5 mW/cm2), the fluorescence under red excitation (1,200 W/cm2 nominal incident intensity, not corrected for light scatter) showed clearly resolved single-trial APs in the soma (Fig. 5b) and in dendrites (Supplementary Fig. 22). These traces represent raw fluorescence, without background subtraction or correction for photobleaching.
Figure 5. Optopatch2 in organotypic brain slice.
A) Left and middle: eGFP fluorescence, indicating CheRiff distribution. Right: QuasAr2 fluorescence. Scale bars from left to right: 50 µm, 20 µm, 20 µm. B) Single-trial optical recordings of APs initiated by pulses of blue illumination (10 ms, 7.5 mW/cm2). Signal represents whole-soma fluorescence without photobleaching correction or background subtraction. C) Bursts of APs triggered by steps of blue illumination (500 ms, 1–10 mW/cm2). Inhibitory potentials (arrows) were sometimes observed during the stimulation intervals, but not during rest intervals, suggesting inhibitory feedback arising from optically induced network activity. For B) and C), red illumination was 1,200 W/cm2 nominal incident intensity, not corrected for light scatter. Fluorescence collected at a frame rate of 1 kHz on an EMCCD camera (Methods).
We performed Optopatch measurements on n = 7 separately prepared organotypic brain slices (Supplementary Fig. 23). AP amplitudes ranged from ΔF/F = 7.2 to 26.1% (mean 15.9%, n = 7 cells), calculated without subtraction of background autofluorescence. Whole cell-body fluorescence reported APs with SNRs ranging from 7.8 to 65.6 (mean 31.9) in a 1 kHz bandwidth. At lower red excitation intensity (400 W/cm2 nominal incident intensity, not corrected for light scatter), whole cell-body fluorescence reported APs with SNRs ranging from 7.2 to 35.7 (mean 16.5) in a 1 kHz bandwidth.
We further tested the response of neurons in tissue to extended pulses of blue illumination (0.5 s, 1 to 10 mW/cm2). This stimulus elicited a variety of firing patterns, including single spikes, bursts (Fig. 5c) and sustained activation (Supplementary Fig. 23). Optically induced spike trains were often interrupted by hyperpolarizing fluorescence transients, which we provisionally ascribe to inhibitory feedback in the local micro-circuit. We did not observe these inhibitory potentials in the absence of optical stimulation. These results demonstrate the feasibility of optically measuring single-cell, single-trial AP waveforms, excitability, and subthreshold dynamics in tissue with high SNR.
Finally, we compared Optopatch to ArcLight in organotypic slice (Supplementary Fig. 23, Methods). Under manual patch clamp stimulation, ArcLight fluorescence resolved single action potentials with a mean value of ΔF/F = −1.5 ± 0.4% and an SNR of 7.1 ± 2.8 (n = 6 cells) (excitation at 488 nm, 50 W/cm2). The ArcLight fluorescence transients had a mean width (at 70% maximum deviation) of 21.5 ± 3.0 ms. Thus QuasAr2 reports action potentials in tissue with greater sensitivity, higher SNR, and better temporal resolution than ArcLight.
Discussion
The combination of improved reporter and improved actuator in the Optopatch constructs facilitates rapid, non-invasive characterization of genetically defined cells across spatial scales from microns to millimeters, and temporal scales from microseconds to days. Optopatch has not yet been implemented with real-time feedback on the illumination, so it is not suited for voltageclamp experiments. Nonetheless, with the assistance of computational modeling, open-loop voltage measurements can probe ionic conductances and membrane electrical properties39, 40. Absolute voltage measurements with GEVIs are challenging due to variations in expression level and membrane trafficking. A recent report demonstrated measurements of absolute voltage through its effect on photocycle dynamics in an Archaerhodopsin mutant41. A similar strategy may apply to the QuasArs.
Our discovery of homeostatic plasticity in intrinsic excitability of hiPSC-derived neurons serves as a paradigm for other Optopatch assays of neuronal excitability. Such assays may probe cell-autonomous functional phenotypes in hiPSC-based models of diseases such as ALS42, epilepsy43, and Parkinson’s disease44. Other neurodevelopmental and neuropsychiatric diseases arise primarily through synaptic dysfunction. These include Rett syndrome45, Fragile X46, and Phelan-McDermid syndrome36. Development of robust assays of synaptic function will likely benefit from incorporation of cell patterning techniques to control the type and location of synaptic connections. A merit of optical electrophysiology for disease modeling and drug screening is that it does not require mechanical access; thus it is compatible with microfluidic compound delivery and high throughput experimental formats. Optopatch measurements preserve the sterility of the sample and the integrity of the cell membrane, and thus are compatible with studies of long-term responses to chronic pharmacological perturbations.
We demonstrated that Optopatch functions in intact mammalian tissue. With extension to multiple genetically specified cells, Optopatch measurements in tissue may provide a useful tool for functional circuit mapping2. With improved optical sectioning capability, sub-cellular Optopatch measurements could enable inference of electrophysiological parameters in multicompartment neural models of single-cell information processing.
For applications in vivo, the new QuasAr reporters are likely to be appropriate for measurements on a few cells or small networks. The required laser power scales with the field of view, so tissue heating may be a concern for fields of view > ∼200 µm on a side. Applications to larger circuits will likely benefit from further improvements to the indicator, primarily increased brightness, and exploration of two-photon excitability or other contrast modalities. Small transparent organisms, e.g. zebrafish and C. elegans, are likely early targets as most illumination passes through the body without depositing heat.
Online Methods
Engineering of Arch
We adopted a hierarchical approach to screening that prioritized brightness over multiple secondary selection criteria. The brightness screen was conducted by examining the fluorescence of large libraries of variants expressed in bacterial colonies. Subsequent screens for trafficking, speed, and voltage sensitivity were performed in HeLa cells subjected to field stimulation and induced transmembrane voltages, and then in HEK cells with patch clamp.
Molecular biology procedure
Synthetic DNA oligonucleotides used for cloning and library construction were purchased from Integrated DNA Technologies (primers used for directed evolution of QuasArs are in Supplementary Table 6). Pfu polymerase (Fermentas) or AccuPrime Pfx SuperMix (Invitrogen) were used for high fidelity non-mutagenic PCR amplifications in the buffer supplied by the respective manufacturer. Taq polymerase (New England Biolabs) in the presence of MnCl2 (0.1 mM) was used for error-prone PCR. PCR products and products of restriction digests were routinely purified using preparative agarose gel electrophoresis followed by DNA isolation using the GeneJET gel extraction kit (Fermentas). Restriction endonucleases were purchased from Fermentas and used according to the manufacturer’s recommended protocol. Ligations were performed using T4 ligase (Invitrogen) or Gibson Assembly (New England Biolabs). Small-scale isolation of plasmid DNA was performed by GeneJET miniprep kit (Fermentas). The cDNA sequences for all Arch variants and fusion constructs were confirmed by dye terminator cycle sequencing using the BigDye Terminator Cycle Sequencing kit (Applied Biosystems). Site-directed mutagenesis and randomization of targeted codons was performed with either the QuikChange Lightning Single or Multi kit (Agilent Technologies).
Construction of Arch mutant libraries
A library of > 104 mutants was generated by error-prone PCR of the gene encoding Arch D95N. These variants were then joined with the gene encoding mOrange2 by a two-part overlap extension PCR. The 5’ piece used in the overlap extension was prepared by error-prone PCR of Arch D95N as template with a mixture of the forward primer (Fw_XbaI_Arch) and reverse primer (RV_Arch). Primer Fw_XbaI_Arch contains an XbaI site and primer RV_Arch contains an overlap region with primer FW_Arch_FP. The 3’ piece for use in the overlap extension was prepared by high fidelity PCR amplification of mOrange2 using a forward primer (FW_Arch_FP) and a reverse primer (RV_HindIII_FP). Primer RV_HindIII_FP contains a stop codon and a HindIII site. The full-length Arch-mOrange2 gene library was assembled by overlap extension PCR using an equimolar mixture of primers Fw_XbaI_Arch and RV_HindIII_FP together with a mixture of the 5’ and 3’ PCR fragments described above (50 ng each) as the template. In later rounds of directed evolution, error-prone PCR and StEP PCR DNA shuffling 47 were both used for construction of Arch-mOrange2 gene libraries.
The full-length PCR product (approximately 1,500 b.p.) was purified by agarose gel electrophoresis, doubly digested, and ligated between the XbaI and HindIII sites of a modified pBAD vector which was generated by deleting the ETorA tag between the NcoI and XbaI sites of the pTorPE vector 48 using Quikchange Lightning kit.
Following ligation, electrocompetent E. coli strain DH10B was transformed with the library of gene variants and cultured overnight at 37 °C on 10-cm Petri dishes of LB-agar supplemented with 100 µL of 4 mM retinal (Sigma), 100 µg/mL ampicillin (Sigma), and up to 0.0020% (wt/vol) l-arabinose (Alfa Aesar). The retinal solution was added on the surface of LB-agar plates evenly and air-dried prior to plating the cell suspension. At concentrations of l-arabinose higher than 0.0020% (wt/vol) we observed abnormal colony morphologies and reduced fluorescent brightness, presumably due to cytotoxicity caused by overexpression.
Screening of Arch mutants in E. coli
The imaging system used for library screening has previously been described in detail49. We screened 10,000–20,000 colonies (10–20 plates of bacterial colonies) per round of random mutagenesis. For libraries generated by randomization of one or more codons, we screened approximately 3-fold more colonies than the expected library diversity (e.g. 3,000 colonies for a 1,000-member library).
We acquired two images of colonies using filter sets for mOrange2 (exc. 540 – 580 nm, em. 600 – 660 nm) and Arch (exc. 604–640 nm and em. 660 – 700 nm). An image of the ratio of Arch: mOrange2 fluorescence was calculated, and the colonies with the top 0.01% to 0.1% highest ratios were manually picked. Picked clones were individually cultured in 2 mL liquid LB medium (200 µg/mL ampicillin) shaken (250 rpm) overnight at 37 °C.
Protein expression was induced by adding 2 mL of liquid LB medium containing 120 µM retinal, 200 µg/mL ampicillin and 0.2% l-arabinose to the overnight culture, followed by incubation at 37 °C for 3 hours. The cell pellets were collected by centrifugation, washed and resuspended in buffered M9 salt solution containing 7 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl and 1 g/L NH4Cl. The suspension was then diluted 5-fold prior to acquisition of its fluorescence spectrum in a Safire 2 fluorescence microplate reader (Tecan).
The emission profiles of each variant under excitation at 525 nm and 600 nm were acquired and normalized by the absorbance at 600 nm. The cell pellets of the three variants with the highest ratios of Arch to mOrange2 and the two variants with the brightest absolute Arch fluorescence were treated for plasmid DNA extraction, and the pooled genes were used as templates for construction of gene libraries in the next round of directed evolution.
After five iterations we arrived at a non-pumping variant of Arch with five mutations relative to wild-type (P60S, T80S, D95N, D106Y, and F161V) and substantially improved brightness under excitation with low illumination intensity. This variant, designated Arch 3.5, was used as the template for subsequent efforts to address the secondary selection criteria.
Random mutagenesis at positions Asp95 and Asp106
We next focused on tuning other properties of Arch including voltage sensitivity, response kinetics, membrane trafficking and the undesirable dependence of brightness on illumination intensity. Positions Asp95 and Asp106 of Arch are structurally aligned with positions Asp85 and Asp96 of bacteriorhodopsin, and have been reported to play key roles in proton translocation during the photocycle 50, 51. The voltage sensing mechanism of Arch is likely due to electric-field-dependent protonation of the retinal Schiff base 17,52, so we reasoned that perturbations of the proton translocation network around the Schiff base could potentially affect the voltage sensitivity, response kinetics, or complex photophysics18.
We constructed libraries in which Asp95 and Asp106 were randomized to a subset of all possible amino acid substitutions. First, we randomized position 95 using codon HVS (where H = A, C or T; V= A, C, or G; S = C or G), which encodes for all amino acids except Ala, Gly, Asp, Glu and Val. This library was screened by fluorescence imaging of E. coli colonies. Variants that retained a high ratio of Arch to mOrange2 fluorescence were picked and expressed in HeLa cells for screening via induced transmembrane voltage (see below).
The mutation N95H emerged as the best from the first round of screening in HeLa cells. We then constructed a second library by randomizing position 106 to a subset of amino acids with polar or charged side chains (codon NRC, where N = A, C, G, or T; R = A or G), and screened these in HeLa cells. The variant with histidine at position 106 proved most promising and was designated QuasAr1.
Solubilization and spectroscopic characterization of QuasAr1 and QuasAr2. E. coli
expressing QuasAr1 and QuasAr2 were grown in 12 mL liquid LB medium with 200 µg/ml ampicillin overnight. The next day, 12 mL of liquid LB medium containing 50 µM retinal, 200 µg/ml ampicillin and 0.1% arabinose was added into the overnight culture, followed by additional incubation at 37 °C for 4 hours. The cell pellets were collected by centrifugation and lysed by suspension in B-PER solution (Pierce). The cytoplasmic fraction was discarded after centrifugation and the colored insoluble fraction was resuspended in phosphate buffered saline (PBS) containing 1.5% n-dodecyl-β-D-maltopyranoside (Affymetrix, Inc.). The suspension was homogenized by an ultrasonic homogenizer and centrifuged (17,000 g for 15 mins, 4 °C). The solubilized protein in the supernatant was used for in vitro spectroscopic characterization.
Absorption spectra were recorded on a DU-800 UV-visible spectrophotometer (Beckman) and fluorescence spectra were recorded on a Safire2 plate reader (Tecan). Cy5 carboxylic acid (Cyandye) was used as the reference for quantum yield measurement. Quantum yield measurements were performed on a series of dilutions of each protein solution and standard, with absorbance values ranging from 0.01 to 0.05 at 600 nm. The fluorescence emission spectra of each dilution were recorded with excitation at 600 nm and the total fluorescence intensities obtained by integration. Integrated fluorescence intensity vs. absorbance was plotted for each protein and each standard. Quantum yields, Φ, were determined from the slopes (S) of each line using the equation: Φprotein = Φstandard × (Sprotein/Sstandard).
Expression vectors for HeLa cells
To express Arch-mOrange2 variants in HeLa cells, the gene in the pBAD vector was first amplified by PCR using primers Fw_BamHI_Kozak_Arch and RV_FP_ERex_stp_XbaI. This reverse primer encodes the endoplasmic reticulum (ER) export sequence from the inward-rectifier potassium channel Kir2.1 (FCYENE) 53, which has been reported to be effective for improving the membrane trafficking of Arch in mammalian cells54.
The purified DNA was digested with BamHI and XbaI restriction enzymes and ligated into a purified pcDNA3.1 plasmid that had been digested with the same two enzymes. The ligation reaction was used for transformation of electrocompetent E. coli strain DH10B cells. Cells were plated on LB/agar supplemented with ampicillin and individual colonies were picked into 4 mL of LB/ampicillin following overnight incubation at 37 °C. Liquid cultures were shaken at 250 rpm and 37 °C for 12–15 h and then a small scale isolation of plasmid DNA was performed. Each gene in pcDNA3.1 was fully sequenced using T7_FW, and BGH_RV primers. Plasmids were then used for cell transfection as described below.
Induced transmembrane voltage (ITV) in HeLa cells
We co-expressed prospective Arch variants in HeLa cells with the inward rectifier potassium channel, Kir2.1. Expression of Kir2.1 lowered the resting potential to approximately −60 mV, close to the resting potential of neurons55, 56. We reasoned that this effect would center the ITV close to the physiologically relevant range.
HeLa cells were grown to 40–60% confluence on home-made 35 mm glass bottom dishes or 24-well glass bottom plates. Cells were transfected with 1 µg of plasmid DNA comprising a 1:1 mixture of Arch variant and Kir2.1, using either 2 µL Turbofect (Thermo Scientific) or 2 µL Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 3 h incubation, the medium was exchanged to DMEM with 10% fetal bovine serum. Cells were incubated for an additional 24 h at 37 °C in a CO2 incubator. Immediately prior to imaging, cells were washed twice with Hanks balanced salt solution (HBSS) and then 1 mL of 20 mM HEPES buffered HBSS was added.
Cell imaging was performed with an inverted Eclipse Ti-E (Nikon) equipped with a Photometrics QuantEM 512SC camera, a 150 W mercury-xenon lamp (Hamamatsu), and a 10 mW 638 nm semiconductor diode laser (56ICS/S2669, Melles Griot CleanBeam) aligned just above the angle for total internal reflection. The filters were: 590–650 nm (excitation), 668–738 nm (emission), and 666 nm (dichroic). Movies were acquired at 10 ms/frame. The NIS-Elements Advanced Research software (Nikon) was used for microscope and camera control and data acquisition. A schematic of the setup is shown in Supplementary Fig. 1.
To probe the response kinetics and voltage sensitivity, we used a pair of parallel platinum electrodes to apply a reproducible electric field across the cell culture and induce transient asymmetries in the membrane voltage57. Platinum electrodes with a gap of 0.5 cm were mounted in a custom plastic support. The electrode pair was placed in the imaging dish or well, and voltage pulses from a 6824A 40V/25A DC Power Supply (HP / Agilent) were applied using waveforms generated by a pulse generator PG 58A (Gould Advance Ltd). The typical waveform had square-wave pulses lasting 20 ms, and pulse amplitudes from 25 – 35 V. Fluorescence was imaged at 100 Hz frame rate in 4×4 binning mode for 10 seconds. During each voltage pulse, opposite sides of the cell showed opposite fluorescence transients. Typical fluorescence traces are shown in Supplementary Fig. 1.
Raw fluorescence traces were corrected for background autofluorescence and photobleaching. The average voltage sensitivity (ΔF/Fmin) and signal-to-noise ratio of each Arch variant were compared to the best variant of the previous generation, and only the variants with equal or improved performance were chosen as templates for the next round of screening.
Expression vectors for HEK cells and neurons
To enable more accurate electrophysiological characterization via patch clamp in HEK cells and primary neuron cultures, we cloned QuasAr1 into the BamHI/EcoRI sites of lentivirus vector FCK-Arch-GFP (Addgene: 22217). This vector contains a CaMKIIα promoter and a Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element (WPRE) after the 3’ end of the open reading frame. The Arch cDNA was generated by PCR using forward primer FW_BamHI_Kozak_Arch_ValSer and overlapping reverse primers RV_FP_TS and RV_TS_ERex_ stp_EcoRI. These reverse primers introduce a trafficking signal (TS) motif and ER export signal peptide sequence at the C-terminus of the protein.
Simultaneous electrophysiology and fluorescence in HEK cells
HEK293T cells (ATCC CRL-11268) were cultured and transfected following standard protocols17. Cells tested negative for mycoplasma. Briefly, HEK-293 cells were grown at 37 °C, 5% CO2, in DMEM supplemented with 10% FBS and penicillin-streptomycin. 400 ng of plasmid DNA was transfected using Transit 293T (Mirus) following the manufacturer’s instructions, and assayed 48 hours later. The day before recording, cells were re-plated onto glass-bottom dishes (In Vitro Scientific) at a density of ∼10,000 cells/cm2.
Cells were supplemented with retinal by diluting stock retinal solutions (40 mM, DMSO) in growth medium to a final concentration of 5 µM, and then returning the cells to the incubator for 0.5 - 1 hour. All imaging and electrophysiology were performed in Tyrode buffer (containing, in mM: 125 NaCl, 2.5 KCl, 3 CaCl2, 1 MgCl2, 10 HEPES, 30 glucose pH 7.3, and adjusted to 305–310 mOsm with sucrose). A gap junction blocker, 2-aminoethoxydiphenyl borate (50 µM, Sigma), was added to eliminate electrical coupling between cells.
Filamented glass micropipettes (WPI) were pulled to a tip resistance of 5–10 MΩ, and filled with internal solution containing 125 mM potassium gluconate, 8 mM NaCl, 0.6 mM MgCl2, 0.1 mM CaCl2, 1 mM EGTA, 10 mM HEPES, 4 mM Mg-ATP, 0.4 mM Na-GTP (pH 7.3); adjusted to 295 mOsm with sucrose. Pipettes were positioned with a Sutter MP285 manipulator. Whole-cell, voltage and current clamp recordings were acquired using an Axopatch 700B amplifier (Molecular Devices), filtered at 2 kHz with the internal Bessel filter and digitized with a National Instruments PCIE-6323 acquisition board at 5–10 kHz. Data was only acquired from HEK cells having reversal potentials between −10 and −40 mV, access resistance < 25 MΩ and membrane resistance > 0.5 GΩ.
Simultaneous whole-cell patch clamp recordings and fluorescence recordings were acquired on a home-built, inverted epifluorescence microscope, described previously17 and described below in “Optopatch Apparatus”. For step response measurements, voltage clamp electronics were compensated 90–95%. We examined variants of QuasAr1 with mutations at position 95 (Asn, Cys, Gln, His and Tyr) and position 106 (Arg, Asp, Asn, Cys, Glu, His, Lys and Tyr). These experiments confirmed that histidine at position 106 provided undetectable photocurrent, and the best combination of improved voltage sensitivity, and fast kinetics. Mutants with Gln, Cys, or Asn at position 95 exhibited better voltage sensitivity compared to QuasAr1, while retaining fast kinetics. We designated the H95Q mutant QuasAr2.
Analysis of mutations in QuasAr1 and QuasAr2
We developed a structural model of Quasar1 (Supplementary Fig. 2) based on homologous protein Arch-2 (PDB: 2EI4, ref. 58). Mutations T80S and F161V are located in the periphery of the protein, while P60S is close to the Schiff base of the retinal chromophore. Given their location, we suspect that the T80S and F161V substitutions are unlikely to have a direct impact on the photophysical properties of the protein, and are more likely to have a role in improving the folding efficiency. In contrast, the close proximity of the P60S substitution to the Schiff base suggests that this mutation has a more direct influence on the photophysical properties.
We compared the Arch double mutants Arch(D95H, D106H) (termed “HH”) and Arch(D95Q, D106H) (termed “QH”) to the corresponding QuasAr1 and QuasAr2 mutants do determine whether the mutations in the proton-transport chain were sufficient to induce the improved sensor performance. QuasAr1 and QuasAr2 were both substantially brighter than the corresponding double mutants (Supplementary Fig 3). Furthermore, the voltage sensitivity of the HH, QH, QuasAr1 and wild-type protein were comparable, and three-fold less than the sensitivity of QuasAr2. The QuasArs were faster than their corresponding double mutants. Thus one or more of the three mutations outside the proton transport chain (P60S, T80S, F161V) plays an important role in the brightness, sensitivity, and speed of the QuasAr mutants.
The constructs described in this paper are available on Addgene.
Engineering of CheRiff
The gene for Scherffelia dubia Channelrhodopsin (sdChR) was synthesized with mouse codon optimization. This gene was selected from a screen of channelrhodopsins for its blue excitation peak (474 nm) and its large photocurrent relative to ChR2. However, the trafficking in neurons was poor (Supplementary Fig. 8). Addition of a trafficking sequence from Kir2.1 improved trafficking greatly,54. Addition of the ER2 export motif did not appear to further improve trafficking, so this motif was not included in the final construct.
The improved membrane trafficking led to large photocurrents in neurons under blue excitation (2470 ± 170 pA peak, 488 nm, 500 mW/cm2, n = 3 cells) but also led to unacceptably large photocurrents from the red laser used to illuminate the QuasArs (38 ± 4 pA, steady state, 640 nm, 300 W/cm2). Furthermore the off-time was undesirably slow (τoff = 26.0 ± 2.9 ms). Based on experience with mutations in other rhodopsins, we introduced the mutation E154A, which only slightly decreased the peak photocurrent under blue illumination (to 2030 ± 100 pA, n = 10), but decreased the crosstalk from red illumination (to 10.5 ± 2.8 pA) and shortened τoff to 16.0 ± 0.8 ms. This variant, termed CheRiff, showed excellent trafficking and neural activation at low illumination intensities.
The action spectrum of CheRiff was measured in HEK293 (n = 6 cells) with a monochromator (Till-Photonics Polychrome IV). 10 ms pulses of nearly equal photon fluxes (∼2.5 × 1021 photons/s/m2) were used across wavelengths. Small deviations in photon flux between wavelengths were corrected by dividing the recorded opsin response by the measured photon dose. For each cell, wavelengths were swept from blue-to-red and red-to-blue, and the responses were averaged.
Design of Optopatch
We tried several strategies to create an Optopatch construct. Kleinlogel and coworkers reported a gene fusion strategy for stoichiometric co-expression of two microbial rhodopsins as a single polypeptide chain59. We made fused constructs of several channelrhodopsins and Arch, but these showed poor trafficking.
We then explored 2A peptide ribosomal skip sequences as a means to achieve approximately stoichiometric co-expression. The porcine teschovirus-1 (P2A) sequence performed better than the Thosea asigna virus (T2A) sequence. Due to the greater need for high expression of the voltage indicator than the actuator, we placed the QuasAr gene before the P2A sequence and the CheRiff gene after (Supplementary Fig. 10).
In some applications one might wish to use the visible spectrum for other imaging modalities, e.g. for a reporter of Ca2+ or a GFP expression marker. In such cases, it is inconvenient to have GFP and mOrange2 fused to CheRiff and Arch, respectively. Removal of the eGFP tag from Arch resulted in poor membrane localization in neurons. To maintain the beneficial trafficking properties of the eGFP tag while eliminating the eGFP fluorescence, we mutated the eGFP chromophore from TYG to GGG using site-directed mutagenesis (Agilent). This mutation has been reported to preserve folding of eGFP60. We also made versions of Optopatch in which the mOrange2 was mutated to a non-fluorescent form by the mutation TYG to TAG.
Optopatch constructs were incorporated into lentiviral vectors under the CaMKIIα promoter, adapted from Addgene plasmid 22217.
Neuronal culture and gene delivery
All procedures involving animals were in accordance with the National Institutes of Health Guide for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committee at the institution at which they were carried out.
Primary neurons
Rat glial monolayers were prepared similarly to ref. 61. Briefly, 106 dissociated hippocampal cells from P0 rat pups (Sprague Dawley, Tocris)62 were plated on a 10 cm culture dish in glial medium GM, comprised of 15% FBS (Life), 0.4% (w/v) D-glucose, 1% glutamax (Life), 1% penicillin/streptomycin (Life) in MEM (Life). When the dish reached confluence (1–2 weeks), cells were split using trypsin onto glass-bottomed dishes (In Vitro Scientific, D35-20-1.5-N) coated with poly-D-lysine and matrigel (BD biosciences) at a density of (3500 cells/cm2). After ∼ 3–6 days, glial monolayers were at or near confluence and the medium was replaced by GM with 2 µM cytarabine (cytosine-β-arabinofuranoside, Sigma). Dishes were maintained in GM with 2 µM cytarabine until use. Dishes were discarded if microglia or neurons were identified on the monolayers.
Hippocampal neurons from P0 rat pups were dissected and cultured in neurobasal-based medium (NBActiv4, Brainbits llc.) at a density of 30,000–40,000 cm−2 on the pre-established glial monolayers62. At one day in vitro (DIV), cytarabine was added to the neuronal culture medium at a final concentration of 2 µM to inhibit further glial growth63.
Neurons were transfected on DIV 7 with the QuasArs or Optopatch plasmids via the calcium phosphate transfection method. Procedures closely followed ref. 64. Measurements on neurons were taken between DIV 13-18.
For comparisons between CheRiff, ChR2 H134R and ChIEF, neurons were plated on glass-bottom dishes coated with poly-d-lysine (Sigma P7205) and matrigel (BD biosciences 356234) without pre-established glial monolayers. On DIV 3 cytarabine (2 µM) was added. Cells were transfected on DIV 7 with channelrhodopsin-eGFP fusions, in identical lentiviral plasmids with a CaMKIIα promoter. All comparison measurements were taken between DIV14–15 at room temperature (23 °C).
For TTX-induced homeostatic plasticity, primary neurons were transfected via the calcium phosphate method on DIV7. TTX (1 µM) was added on DIV 16. Excitability was measured on DIV 18 in Tyrodes medium with synaptic blockers (10 µM NBQX, 25 µM AP-V, 20 µM Gabazine).
hiPSC-derived neurons
Human iPSC-derived iCell neurons were purchased from Cellular Dynamics Inc. Neurons were tested negative for mycoplasma by the manufacturer. Neurons were thawed and resuspended in complete iCell Neuron Maintenance Medium (CM) following manufacturer protocols. Cells were then plated at a density 125,000/cm2 on pre-established rat glial monolayers grown on glass-bottomed dishes. Medium was replaced 24 hours post plating with CM supplemented with 10 ng/mL BDNF (Peprotech). Thereafter, 50% media exchanges with CM were done every 5 days.
For TTX-induced homeostatic plasticity, hiPSC-derived neurons were transfected via the calcium phosphate method on DIV17. TTX (1 µM) was added on DIV 26. Excitability was measured on DIV 28 in Tyrodes medium with synaptic blockers (10 µM NBQX, 25 µM AP-V, 20 µM Gabazine).
For KCl-induced homeostatic plasticity, hiPSC-derived neurons were transfected on DIV 10. KCl (15 mM) was added from DIV 18 to DIV 21 (60 h). Excitability was measured on DIV 21 in Tyrodes medium with synaptic blockers (10 µM NBQX, 25 µM AP-V, 20 µM Gabazine).
Organotypic brain slice culture
Organotypic hippocampal slices cultures were prepared from postnatal day 6–8 Sprague-Dawley rats as described previously65. The brain was taken out and immediately placed in chilled dissection media. Transverse hippocampal slices were cut with 400 µm thickness and 4 to 6 slices were placed in a sterile culture plate insert (Millicell-CM, Millipore) in 6-well plates containing prewarmed media. Slices were biolistically transfected with a Helios Gene Gun (BioRad) at 2 days in vitro (DIV 2). Bullets were prepared using 12.5 µg of 1.6 µm gold particles and 80 − 100 µg of plasmid DNA. Slices were maintained until imaging at DIV 12–16.
Immediately prior to inverted imaging, slices were affixed to a nylon mesh weight and mounted upside down in a delta T brainslice adapter for inverted microscope imaging (Bioptechs). Artificial cerebrospinal fluid (ACSF) was bubbled with carbogen (95% O2, 5% CO2) and flowed over the slice at 1 mL/min at 23 °C.
Electrophysiology in neurons
Measurements were performed on primary cultures at 13 – 15 DIV. Experiments were conducted in Tyrode’s solution containing 125 mM NaCl, 2.5 mM KCl, 3 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 30 mM glucose (pH 7.3) and adjusted to 305–310 mOsm with sucrose. Prior to imaging, neurons were incubated with 5 µM all-trans retinal for 30 minutes and then washed with Tyrode’s solution.
Synaptic blockers were added to the imaging medium for measurements of single-cell electrophysiology. The blockers comprised NBQX (10 µM, Tocris), D(−)-2-amino-5-phosphonovaleric acid (AP5; 25 µM, Tocris), and gabazine (SR-95531, 20 µM, Tocris). For measurements of channelrhodopsin photocurrents in neurons, TTX (1 µM, Tocris) was included along with the synaptic blockers to prevent recruitment of voltage gated sodium channels. Patch clamp data was used if and only if access resistance was < 25 MΩ, and did not vary over the experiment. Recordings were terminated if membrane resistance changed by > 10%. Experiments were performed at 23 °C under ambient atmosphere unless otherwise noted.
Comparison of QuasArs to ArcLight A242
ArcLight A242 was prepared in an identical lentiviral plasmid driven by a CaMKIIαpromoter and was transfected (DIV 7) in parallel with the QuasAr plasmids in paired cultures. We used a standard ArcLight imaging intensity of 10 W/cm2 at 488 nm. QuasAr expressing neurons were imaged at two intensities (300 and 800 W/cm2). All recordings were made on the setup described below (“Optopatch apparatus”) at a 1 kHz frame rate and 60x magnification. Due to its slow kinetics at room temperature (Fig S6), ArcLight recordings were made at 34 °C to enhance SNR and to match previously published conditions 16. QuasAr2 reported APs with comparable SNR at 23 °C and 34 °C (41 ± 3, n = 8 cells, 300 W/cm2). For comparisons in organotypic brain slice, ArcLight was imaged at 50 W/ cm2 on an upright microscope to enable simultaneous patch clamp stimulation and recordings. Recordings were made at a 1 kHz frame rate as described below (“Optopatch apparatus”) and were acquired at 34 °C.
Immunostaining
Cultures were fixed immediately following data acquisition in a solution of 4% paraformaldehyde and 4% sucrose (w/v) in PBS, pH 7.4 at room temperature for 8 minutes. Fixed cultures were then washed three times in Dulbecco’s PBS supplemented with Ca2+ and Mg2+ (DPBS), pH 7.4, prior to permeabilization and blocking in a solution of 0.1% (w/v) gelatin and 0.3% Triton-X-100 (v/v) in PBS, pH 7.4 (GTB) for 12–48 hours at 4C.
For experiments using the sub-frame interpolation algorithm, primary cultures were fixed and stained using primary mouse monoclonal anti-ankyrin G (NeuroMab clone N106/36; 1:500), primary rabbit monoclonal anti-GFP (Abcam ab32146, lot YK011702CS, 1:1000), secondary goat anti-rabbit AlexaFluor 488 conjugated (Abcam ab150077, 1:500), and secondary goat anti-mouse AlexaFluor 647 conjugated (Abcam ab150115, 1:500) antibodies.
For experiments on human iPSC derived neurons, cultures were incubated with primary mouse anti-human nuclear antigen antibody (Millipore MAB1281 clone 235-1,1:500) in GTB overnight at 4 °C, then washed three times in DPBS, and incubated with rabbit anti-GFP AlexaFluor 488 conjugated (polyclonal, Life A21311, 1:300) and secondary antibody donkey anti-mouse AlexaFluor 647 (Life A31571, 1:300) in GTB overnight at 4C. Cultures were washed three times in DPBS prior to mounting in DAPI Fluoromount-G (Southern Biotech).
Optopatch measurements
Experiments were conducted on a home-built inverted fluorescence microscope, similar to the one described in the Supplementary Material to ref. 17. Briefly, illumination from a red laser 640 nm, 140 mW (Coherent Obis 637-140 LX), was expanded and focused onto the back-focal plane of a 60× oil immersion objective, numerical aperture 1.45 (Olympus 1-U2B616). Imaging of brain slices was performed with a 20× water-immersion objective, numerical aperture 1.0 (Zeiss W Plan-Apo).
Illumination from a blue laser 488 nm 50 mW (Omicron PhoxX) was sent through an acousto-optic modulator (AOM; Gooch and Housego 48058-2.5-.55-5W) for rapid control over the blue intensity. The beam was then expanded and modulated by a digital micromirror device (DMD) with 608×684 pixels (Texas Instruments LightCrafter). The DMD was controlled via custom software (Matlab) through a TCP/IP protocol. The DMD chip was re-imaged through the objective onto the sample, with the blue and red beams merging via a dichroic mirror. Each pixel of the DMD corresponded to 0.65 µm in the sample plane. A 532 nm laser was combined with the red and blue beams for imaging of mOrange2. We wrote software to map DMD coordinates to camera coordinates, enabling precise optical targeting of any point in the sample.
To achieve precise optical stimulation of user-defined regions of a neuron, it was necessary to determine the mapping from pixels on the DMD to pixels on the camera. A uniform fluorescent film (exc. 488 nm, em. 515 nm) was loaded into the microscope. The DMD projected an array of dots of known dimensions onto the sample. The camera acquired an image of the fluorescence. Custom software located the centers of the dots in the image, and created an affine transformation to map DMD coordinates onto camera pixel coordinates.
A dual-band dichroic (Chroma zt532/635rpc) separated fluorescence of mOrange2 and Arch from excitation light. A 531/40 nm bandpass filter (Semrock FF01–531/40-25) and 495 nm longpass dichroic (Semrock FF495-Di03) was used for eGFP imaging, a 710/100 nm bandpass filter (Chroma, HHQ710/100) was used for Arch imaging, and a quad-band emission filter (Chroma ZET405/488/532/642m) was used for mOrange2 imaging and pre-measurement calibrations. A variable-zoom camera lens (Sigma 18–200 mm f/3.5–6.3 II DC) was used to image the sample onto an EMCCD camera (Andor iXon+ DU-860), with 128 × 128 pixels. The variable zoom enabled imaging at a range of magnifications while maintaining the high light collection efficiency of the oil or water immersion objectives.
In a typical experimental run, images of mOrange2 and QuasAr fluorescence were first acquired at full resolution (128 × 128 pixels). Data was then acquired with 2 × 2 pixel binning to achieve a frame rate of 1,000 frames/s. For experiments with infrequent stimulation (once every 5 s), the red illumination was only on from 1 s before stimulation to 50 ms after stimulation to minimize photobleaching. Cumulative red light exposure was typically limited to < 5 min. per neuron, although continuous red light exposure for 30 minutes was well tolerated (Supplementary Fig. 5). Full resolution eGFP images were taken after functional recordings to prevent CheRiff excitation prior to the experiment.
Low magnification wide-field imaging was performed with a custom microscope system based around a 2×, NA 0.5 objective (Olympus MVX-2). Illumination was provided by six lasers 640 nm, 500 mW (Dragon Lasers 635M500), combined in three groups of two. Illumination was coupled into the sample using a custom fused silica prism, without passing through the objective. Fluorescence was collected by the objective, passed through an emission filter, and imaged onto a scientific CMOS camera (Hamamatsu Orca Flash 4.0). This microscope imaged a 1.2 × 3.3 mm field of view with 3.25 µm spatial resolution and 2 ms temporal resolution, or a 4 × 4 mm field of view with 10 ms temporal resolution.
Blue illumination for channelrhodopsin stimulation was provided by a 473 nm, 1 W laser (Dragon Lasers), modulated in intensity by an AOM and spatially by a DMD (Digital Light Innovations DLi4130 – ALP HS). The DMD was re-imaged onto the sample via the 2× objective. The DMD provided targeted stimulation with excitation with 3.5 µm spatial resolution and 0.1 ms temporal resolution.
During an experimental run, we first acquired an image of a neuron using wide-field illumination at 640 nm to probe Arch fluorescence, and/or 532 nm to probe mOrange2 fluorescence. A user then selected one or more regions of interest on the image of the neuron, and specified a timecourse for the illumination in each region. The software mapped the user-selected pixels onto DMD coordinates and delivered the illumination instructions to the DMD.
Illumination parameters for the data in Fig. 3 were:
B) Blue illumination: 488 nm, 25 mW/cm2, 10 ms pulses, repeated at 5 Hz.
C) Stimulation targeted to soma: 488 nm, 95 mW/cm2, 10 ms pulses, repeated at 5 Hz.
D) Stimulation targeted to soma in ROI 1: 488 nm, 140 mW/cm2, 10 ms pulses, repeated every 15 s.
E) Excitation targeted to dendrites: 488 nm, 100 mW/cm2, 50 ms pulses, repeated at 5 Hz. Movie constructed from an average of 203 temporally registered APs.
F) Whole-field stimulation: 488 nm, 0 to 10 mW/cm2, 500 ms pulses, repeated at 6 s intervals. Synaptic blockers were added to suppress network activity.
Data analysis
Statistics
All error ranges represent standard error of the mean, unless otherwise specified. For two-sample comparisons of a single variable, data was tested for normality using the Shapiro-Wilks test. If the data was detectably non-Gaussian, we performed a nonparametric Mann-Whitney U test. Otherwise we performed a two-tailed student’s t-test. Channelrhodopsin multi-way comparisons of a single variable were made using a one-way ANOVA with Dunnett’s post-hoc test, using CheRiff as a reference. No channelrhodopsin dataset was detectably non-Gaussian (Shapiro-Wilks). Probabilities of the null hypothesis P < 0.05 were judged to be statistically significant.
Extracting fluorescence from movies
Fluorescence values were extracted from raw movies in one of two ways. One approach used the maximum likelihood pixel weighting algorithm described in ref. 17. Briefly, the fluorescence at each pixel was correlated with the whole-field average fluorescence. Pixels that showed stronger correlation to the mean were preferentially weighted. This algorithm automatically found the pixels carrying the most information, and de-emphasized background pixels. This approach was used for all experiments in cultured neurons. In images containing multiple neurons, the segmentation was performed semi-automatically using the independent components-based approach of ref. 66.
Alternatively, a user defined a region comprising the cell body and adjacent processes, and calculated fluorescence from the unweighted mean of pixel values within this region. With the improved trafficking of the QuasAr mutants compared to Arch, the maximum likelihood pixel-weighting algorithm was only marginally superior to manual definition of an ROI (Supplementary Fig. 4). For calculations of ΔF/F in culture, background fluorescence from a cell-free region was subtracted from the baseline fluorescence of the cell. In measurements in brain slice, fluorescence was calculated from manually defined ROIs with equal pixel weighting and no background subtraction or correction for photobleaching.
Precision of optically recorded AP timing
To determine the temporal precision of the QuasAr indicators, we used the sub-frame interpolation algorithm of refs. 28, 29 to infer the timing with which the fluorescence reached 70% of maximum at each AP, and compared to simultaneously acquired high time-resolution patch clamp recordings. Root-mean-square (r.m.s.) temporal jitter was 44 µs for QuasAr1 (n = 97 APs) and 61 µs for QuasAr2 (n = 99 APs). This jitter reflects the combined errors in timing intrinsic to the optical measurement (shot-noise and distortion of the waveform by the reporter) and errors introduced by temporal discretization of the camera frames and the sub-frame interpolation. Thus optical recordings with QuasArs can determine spike timing with precision much greater than the camera exposure time.
Fitting channelrhodopsin photocurrents
Photocurrents of the channelrhodopsins were characterized following the protocols in ref. 24. Briefly, peak photocurrents in response to a light pulse (488 nm, 0.5W/cm2, 1 s duration) were identified by first smoothing the traces using robust Loess method with a filter width of 2 ms, and then finding the extremum of the filtered trace after laser onset and subtracting the baseline current. Time to peak (ton) was defined as the time between light onset and peak photocurrent of the filtered trace. The steady-state photocurrent was found by fitting a monoexponential curve to the filtered trace from 2 ms after the peak until laser offset. The offset of this fit was defined as the steady-state photocurrent. The time constant of this fit was defined as the desensitization rate (τdes).The channel closure rate (τoff) in response to a light pulse (488 nm, 0.5W/cm2, 5 ms duration) was measured by fitting a monoexponential to the decay of the photocurrent after light offset.
Illumination intensities for 50% effective light power density24 (EPD50, Supplementary Table 4) values were determined from measurements of peak photocurrents versus a series of whole field illumination intensities. For each cell, peak photocurrents at each intensity were first normalized by the maximum peak photocurrent. The resulting curves were then fit with a simple binding model (Y = Bmax × X /(EPD50 + X). The reported EPD50s are the average of the fit parameters from n = 5 cells.
Sub-frame interpolation of AP timing
The sub-frame interpolation algorithm consists of a series of computational image-processing steps (Supplementary Fig. 13). Each step may be modified to account for experiment-specific attributes of the data.
A neuron was induced to fire through repeated optical stimulation of a user-selected subcellular compartment (typically soma or dendrite). We typically observed 5% photobleaching over a 40 s acquisition. Photobleaching was typically dominated by non-specific background fluorescence, rather than by photobleaching of QuasAr, and often photobleaching did not follow a simple single-exponential decay. The photobleaching baseline was constructed from the whole-field intensity by a sliding minimum filter, followed by a sliding mean filter. Each frame of the movie was then corrected by dividing by this baseline.
QuasAr fluorescence intensity F(t) was determined either by the regression algorithm described in ref. 17 or by whole-field average intensity. Both procedures gave similar results, with slightly better signal-to-noise ratio returned by the regression algorithm (Supplementary Fig. 4).
Determination of spike times was performed iteratively. A simple threshold-and-maximum procedure was applied to F(t) to determine approximate spike times, {T0}. Waveforms in a brief window bracketing each spike were averaged together to produce a preliminary spike kernel K0(t). We then calculated the cross-correlation of K0(t) with the original intensity trace F(t). Whereas the timing of maxima in F(t) was subject to errors from single-frame noise, the peaks in the cross-correlation, located at times {T}, were a robust measure of spike timing. A movie showing the mean AP propagation was constructed by averaging movies in brief windows bracketing spike times {T}. Typically 100 – 400 APs were included in this average. The AP movie had high signal-to-noise ratio, but did not clearly show signal propagation.
We applied spatial and temporal linear filters to further decrease the noise in AP movie. The spatial filter consisted of convolution with a Gaussian kernel, typically with a standard deviation of 1 pixel. The temporal filter was based upon Principal Components Analysis (PCA) of the set of single-pixel time traces. The time trace at each pixel was expressed in the basis of PCA eigenvectors. Typically the first 5 eigenvectors were sufficient to account for >99% of the pixel-to-pixel variability in AP waveforms, and thus the PCA eigendecomposition was truncated after 5 terms. The remaining eigenvectors represented uncorrelated shot noise (Supplementary Fig. 13b). Projections of the movie onto the PCA eigenvectors only showed spatial features above noise for the first 5 eigenvectors (Supplementary Fig. 13c inset). To verify that the spatial and PCA filtering did not distort the underlying AP waveforms, we compared mean AP waveforms in subcellular compartments before and after the smoothing steps (Supplementary Fig. 13d). We observed no systematic deviations in the AP waveforms in the axon, soma, or dendrites.
The user then set a threshold depolarization to track (represented as a fraction of the maximum fluorescence transient), and a sign for dV/dt (indicating rising or falling edge). We chose 50% maximal depolarization on the rising edge. The filtered data was fit with a quadratic spline interpolation and the time of threshold crossing was calculated for each pixel to create a map of the AP delay (Supplementary Fig. 13e).
The sub-frame timing precision of the algorithm was calibrated by patch clamp measurements. Optically induced APs were recorded simultaneously via QuasAr1 fluorescence in the soma and by conventional patch clamp, also in the soma (Supplementary Fig. 13f). The r.m.s. error in timing was 54 µs in this instance, and did not show systematic bias at the frame boundaries.
The fits were converted into movies showing AP propagation as follows. Each pixel was kept dark except for a brief flash timed to coincide with the timing of the user-selected AP feature at that pixel. The flash followed a Gaussian timecourse, with amplitude equal to the local AP amplitude, and duration equal to the cell-average time resolution, σ. Frame times in the sub-frame interpolation movies were selected to be ∼2-fold shorter than σ.
Occasionally it was possible to enhance the spatial resolution of the high temporal resolution movies by mapping the timing data onto a higher spatial resolution static image of fluorescence of eGFP (from the CheRiff-eGFP fusion, Supplementary Fig. 13g) or of QuasAr1. The pixel matrix of the sub-frame interpolated movie was expanded to match the dimensions of the high resolution image and the amplitude at each pixel was then set equal to the mean brightness at that pixel. Selected frames from the resulting movie showed AP initiation at the axon initial segment in the first two frames (Supplementary Fig. 13h, Supplementary Movie 4). To assemble the color movies (Fig. 3e, Supplementary Fig. 13, Supplementary Movies 4–6), the timing signal was assigned to a colormap which was overlayed on a gray-scale image of mean QuasAr fluorescence. Optionally, the optically stimulated region of the cell was highlighted in blue.
Supplementary Material
Acknowledgments
We thank N. Anand, L. Rosenbaum, T. Shen, and V. Nathan for technical assistance, Y. Cho, A. Douglass, A. Ting, F. Zhang, L. Looger, and D. Kim for helpful discussions. We thank Cellular Dynamics Inc. for technical assistance with hiPSC neurons. This work was supported by the Harvard Center for Brain Science, PECASE award N00014-11-1-0549, US National Institutes of Health grants 1-R01-EB012498-01 and New Innovator grant 1-DP2-OD007428, the Harvard–Massachusetts Institute of Technology Joint Research Grants Program in Basic Neuroscience, a National Science Foundation Graduate Fellowship (D.R.H., S.L.F), the Natural Sciences and Engineering Research Council of Canada (Discovery grants to R.E.C. and D.J.H), the Canadian Institutes of Health Research (R.E.C.), and graduate scholarships from the University of Alberta and Alberta Innovates (Y.Z.). R.E.C. holds a Tier II Canada Research Chair. ESB was supported by DARPA Living Foundries HR0011-12-C-0068, the MIT Media Lab, McGovern Institute, and Synthetic Intelligence projects, NYSCF Robertson Neuroscience Investigator Award, IET A. F. Harvey Prize, Skolkovo Institute of Science and Technology, NIH grants 1R01NS075421, 1R01NS067199, 1DP2OD002002, and 1R01DA029639, Human Frontiers Science Program, and NSF CAREER Award CBET 1053233 and EFRI 0835878. Work in VNM’s lab was supported by NIH grants R01DC011291 and R01DC013329. The 1000 Plants (1KP) initiative, led by GKSW, is funded by the Alberta Ministry of Enterprise and Advanced Education, Alberta Innovates Technology Futures (AITF) Innovates Centre of Research Excellence (iCORE), Musea Ventures, and BGI-Shenzhen.
Footnotes
Author Contributions
DRH designed the Optopatch construct and system. YZ and DRH engineered the QuasArs. DRH, NK, and YC engineered CheRiff. DRH and SLF acquired the optical electrophysiology data. CAW developed the low-magnification imaging system. VK and JS designed and prepared the slice experiments. DM conceived the sub-frame interpolation algorithm. PZ assisted with measurements of rhodopsin photophysics. JMK screened ion-channel blockers on hiPSC-derived neurons. NSM performed cell culture and sample preparation. GLB performed immunostaining in hiPSC-derived neurons. CS assisted with measurements of ArcLight in slice. MM and GW provided the transcriptomic data from which SdChR was mined and phenotyped. DRH and AEC wrote the paper with input from SLF, YZ and REC. DJH, VM, BS, ESB, and REC supervised aspects of the research. Correspondence regarding directed evolution of Arch should be addressed to REC (robert.e.campbell@ualberta.ca). AEC conceived and oversaw the project.
Competing financial interests
AEC and JMK are founders of Q-State Biosciences, a company focused on combining all-optical electrophysiology with cell-based models of disease. DRH, YZ, SLF, NCK, JMK, DM, YKC, DJH, REC, ESB, and AEC have filed patents related to Optopatch.
References
- 1.Peron S, Svoboda K. From cudgel to scalpel: toward precise neural control with optogenetics. Nat. Meth. 2010;8:30–34. doi: 10.1038/nmeth.f.325. [DOI] [PubMed] [Google Scholar]
- 2.Petreanu L, Mao T, Sternson SM, Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scanziani M, Hausser M. Electrophysiology in the age of light. Nature. 2009;461:930–939. doi: 10.1038/nature08540. [DOI] [PubMed] [Google Scholar]
- 4.Boulting GL, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat. Biotechnol. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furuta T, et al. Brominated 7-hydroxycoumarin-4-ylmethyls: photolabile protecting groups with biologically useful cross-sections for two photon photolysis. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1193–1200. doi: 10.1073/pnas.96.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer RH, Fortin DL, Trauner D. New photochemical tools for controlling neuronal activity. Curr. Opin. Neurobiol. 2009;19:544–552. doi: 10.1016/j.conb.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 8.Looger LL, Griesbeck O. Genetically encoded neural activity indicators. Curr. Opin. Neurobiol. 2012;22:18–23. doi: 10.1016/j.conb.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 9.Miller EW, et al. Optically monitoring voltage in neurons by photo-induced electron transfer through molecular wires. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2114–2119. doi: 10.1073/pnas.1120694109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan P, et al. Palette of fluorinated voltage-sensitive hemicyanine dyes. Proc. Natl. Acad. Sci. U. S. A. 2012;109:20443–20448. doi: 10.1073/pnas.1214850109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogt KE, Gerharz S, Graham J, Canepari M. Combining membrane potential imaging with l-glutamate or GABA photorelease. PLoS One. 2011;6:e24911. doi: 10.1371/journal.pone.0024911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuda S, et al. Probing the function of neuronal populations: combining micromirror-based optogenetic photostimulation with voltage-sensitive dye imaging. Neurosci. Res. 2012;75:76–81. doi: 10.1016/j.neures.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim DH, et al. In vivo Large-Scale Cortical Mapping Using Channelrhodopsin-2 Stimulation in Transgenic Mice Reveals Asymmetric and Reciprocal Relationships between Cortical Areas. Front. Neural Circuits. 2012;6:11. doi: 10.3389/fncir.2012.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong Y, Li JZ, Schnitzer MJ. Enhanced Archaerhodopsin Fluorescent Protein Voltage Indicators. PLoS One. 2013;8:e66959. doi: 10.1371/journal.pone.0066959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat. Meth. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin L, et al. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 2012;75:779–785. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat. Meth. 2012;9:90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maclaurin D, Venkatachalam V, Lee H, Cohen AE. Mechanism of voltage-sensitive fluorescence in a microbial rhodopsin. Proc. Natl. Acad. Sci. USA. 2013;110:5939–5944. doi: 10.1073/pnas.1215595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakai R, Repunte-Canonigo V, Raj CD, Knopfel T. Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur. J. Neurosci. 2001;13:2314–2318. doi: 10.1046/j.0953-816x.2001.01617.x. [DOI] [PubMed] [Google Scholar]
- 20.Bean BP. The action potential in mammalian central neurons. Nature Reviews Neuroscience. 2007;8:451–465. doi: 10.1038/nrn2148. [DOI] [PubMed] [Google Scholar]
- 21.Schoenenberger P, Grunditz Å, Rose T, Oertner TG. Optimizing the spatial resolution of Channelrhodopsin-2 activation. Brain Cell. Biol. 2008;36:119–127. doi: 10.1007/s11068-008-9025-8. [DOI] [PubMed] [Google Scholar]
- 22.Johnson MTJ, et al. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLoS One. 2012;7:e50226. doi: 10.1371/journal.pone.0050226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melkonian M, Preisig HR. A light and electron microscopic study of Scherffelia dubia, a new member of the scaly green flagellates (Prasinophyceae) Nord. J. Bot. 1986;6:235–256. [Google Scholar]
- 24.Mattis J, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat. Meth. 2011;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys. J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi H, et al. Light-addressed single-neuron stimulation in dissociated neuronal cultures with sparse expression of ChR2. BioSystems. 2012;107:106–112. doi: 10.1016/j.biosystems.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Fitzsimonds RM, Song H, Poo M. Propagation of activity-dependent synaptic depression in simple neural networks. Nature. 1997;388:439–448. doi: 10.1038/41267. [DOI] [PubMed] [Google Scholar]
- 28.Foust A, Popovic M, Zecevic D, McCormick DA. Action potentials initiate in the axon initial segment and propagate through axon collaterals reliably in cerebellar Purkinje neurons. J. Neurosci. 2010;30:6891–6902. doi: 10.1523/JNEUROSCI.0552-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popovic MA, Foust AJ, McCormick DA, Zecevic D. The spatio-temporal characteristics of action potential initiation in layer 5 pyramidal neurons: a voltage imaging study. J. Physiol. 2011;589:4167–4187. doi: 10.1113/jphysiol.2011.209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kole MH, Stuart GJ. Signal processing in the axon initial segment. Neuron. 2012;73:235–247. doi: 10.1016/j.neuron.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Turrigiano G, Abbott L, Marder E. Activity-dependent changes in the intrinsic properties of cultured neurons. Science. 1994;264:974–976. doi: 10.1126/science.8178157. [DOI] [PubMed] [Google Scholar]
- 32.Desai NS, Rutherford LC, Turrigiano GG. Plasticity in the intrinsic excitability of cortical pyramidal neurons. Nat. Neurosci. 1999;2:515–520. doi: 10.1038/9165. [DOI] [PubMed] [Google Scholar]
- 33.Grubb MS, Burrone J. Activity-dependent relocation of the axon initial segment fine-tunes neuronal excitability. Nature. 2010;465:1070–1074. doi: 10.1038/nature09160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambo ME, Turrigiano GG. Synaptic and intrinsic homeostatic mechanisms cooperate to increase L2/3 pyramidal neuron excitability during a late phase of critical period plasticity. J. Neurosci. 2013;33:8810–8819. doi: 10.1523/JNEUROSCI.4502-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trounson A, Shepard KA, DeWitt ND. Human disease modeling with induced pluripotent stem cells. Curr. Opin. Genet. Dev. 2012;22:509–516. doi: 10.1016/j.gde.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Shcheglovitov A, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503:267–271. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akemann W, et al. Imaging neural circuit dynamics with a voltage-sensitive fluorescent protein. J. Neurophysiol. 2012;108:2323–2337. doi: 10.1152/jn.00452.2012. [DOI] [PubMed] [Google Scholar]
- 38.Cao G, et al. Genetically Targeted Optical Electrophysiology in Intact Neural Circuits. Cell. 2013;154:904–913. doi: 10.1016/j.cell.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huys QJ, Ahrens MB, Paninski L. Efficient estimation of detailed single-neuron models. J. Neurophysiol. 2006;96:872–890. doi: 10.1152/jn.00079.2006. [DOI] [PubMed] [Google Scholar]
- 40.Williams JC, et al. Computational optogenetics: empirically-derived voltage-and light-sensitive channelrhodopsin-2 model. PLoS Comp. Biol. 2013;9:e1003220. doi: 10.1371/journal.pcbi.1003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou JH, Venkatachalam V, Cohen AE. Temporal Dynamics of Microbial Rhodopsin Fluorescence Reports Absolute Membrane Voltage. Biophys. J. 2014;106:639–648. doi: 10.1016/j.bpj.2013.11.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wainger BJ, et al. Intrinsic Membrane Hyperexcitability of Amyotrophic Lateral Sclerosis Patient-Derived Motor Neurons. Cell Rep. 2014;7:1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higurashi N, et al. A human Dravet syndrome model from patient induced pluripotent stem cells. Molec. Brain. 2013;6:19. doi: 10.1186/1756-6606-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Badger J, Cordero-Llana O, Hartfield E, Wade-Martins R. Parkinson’s disease in a dish-Using stem cells as a molecular tool. Neuropharmacology. 2014;76:88–96. doi: 10.1016/j.neuropharm.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 45.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 47.Zhao H, Giver L, Shao Z, Affholter JA, Arnold FH. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat. Biotechnol. 1998;16:258–261. doi: 10.1038/nbt0398-258. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Y, et al. An expanded palette of genetically encoded Ca2+ indicators. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng Z, Campbell RE. Assessing the structural stability of designed β-hairpin peptides in the cytoplasm of live cells. ChemBioChem. 2006;7:1147–1150. doi: 10.1002/cbic.200500540. [DOI] [PubMed] [Google Scholar]
- 50.Lanyi JK. Proton translocation mechanism and energetics in the light-driven pump bacteriorhodopsin. Biochim. Biophys. Acta. 1993;1183:241–261. doi: 10.1016/0005-2728(93)90226-6. [DOI] [PubMed] [Google Scholar]
- 51.Lanyi JK. Bacteriorhodopsin. Annu. Rev. Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- 52.Kolodner P, Lukashev EP, Ching Y, Rousseau DL. Electric-field-induced Schiff-base deprotonation in D85N mutant bacteriorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11618–11621. doi: 10.1073/pnas.93.21.11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma D, et al. Role of ER export signals in controlling surface potassium channel numbers. Science. 2001;291:316–319. doi: 10.1126/science.291.5502.316. [DOI] [PubMed] [Google Scholar]
- 54.Gradinaru V, et al. Molecular and Cellular Approaches for Diversifying and Extending Optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirkton RD, Bursac N. Engineering biosynthetic excitable tissues from unexcitable cells for electrophysiological and cell therapy studies. Nat. Commun. 2011;2:300. doi: 10.1038/ncomms1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park J, et al. Screening fluorescent voltage indicators with spontaneously spiking HEK cells. PLoS One. 2013;8:e85221. doi: 10.1371/journal.pone.0085221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pucihar G, Kotnik T. Measuring the induced membrane voltage with di-8-ANEPPS. J. Vis. Exp. 2009;33:e1659. doi: 10.3791/1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Enami N, et al. Crystal structures of archaerhodopsin-1 and -2: Common structural motif in archaeal light-driven proton pumps. J. Mol. Biol. 2006;358:675–685. doi: 10.1016/j.jmb.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 59.Kleinlogel S, et al. A gene-fusion strategy for stoichiometric and co-localized expression of lightgated membrane proteins. Nat. Meth. 2011;8:1083–1088. doi: 10.1038/nmeth.1766. [DOI] [PubMed] [Google Scholar]
- 60.Barondeau DP, Putnam CD, Kassmann CJ, Tainer JA, Getzoff ED. Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12111–12116. doi: 10.1073/pnas.2133463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCarthy KD, De Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goslin K. In: Culturing nerve cells. Banker G, Goslin K, editors. Cambridge, MA: The MIT Press; 1998. [Google Scholar]
- 63.Chen G, Harata NC, Tsien RW. Paired-pulse depression of unitary quantal amplitude at single hippocampal synapses. Proc. Natl. Acad. Sci U.S.A. 2004;101:1063–1068. doi: 10.1073/pnas.0307149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat. Protocols. 2006;1:695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- 65.Stoppini L, Buchs P, Muller D. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 66.Mukamel EA, Nimmerjahn A, Schnitzer MJ. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron. 2009;63:747–760. doi: 10.1016/j.neuron.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.