Abstract
Nemo-like kinase (Nlk) is related to the mitogen-activated protein (MAP) kinases and known to regulate signaling pathways involved in osteoblastogenesis. In vitro Nlk suppresses osteoblastogenesis, but the consequences of the Nlk inactivation in the skeleton in vivo are unknown. To study the function of Nlk, NlkloxP/loxP mice, where the Nlk exon2 is flanked by loxP sequences, were mated with mice expressing the Cre recombinase under the control of the paired-related homeobox gene 1 (Prx1) enhancer (Prx1-Cre), the Osterix (Osx-Cre) or the osteocalcin/bone gamma carboxyglutamate protein (Bglap-Cre) promoter. Prx1-Cre;NlkΔ/Δ mice did not exhibit a skeletal phenotype except for a modest increase in trabecular number and connectivity observed only in 3 month old male mice. Osx-Cre;NlkΔ/Δ male and female mice exhibited an increase in trabecular bone volume secondary to an increased trabecular number at 3 months of age. Bone histomorphometry revealed a decrease in osteoclast number and eroded surface in male mice, and decreased osteoblast number and function in female mice. Expression of osteoprotegerin mRNA was increased in calvarial extracts, explaining the decreased osteoclast and osteoblast number. The conditional deletion of Nlk in mature osteoblasts (Bglap-Cre;NlkΔ/Δ) resulted in no skeletal phenotype in 1 to 6 month old male or female mice. In conclusion, when expressed in undifferentiated osteoblasts, Nlk is a negative regulator of skeletal homeostasis possibly by targeting signals that regulate osteoclastogenesis and bone resorption.
Keywords: Nemo-like kinase, MAP kinases, bone remodeling, skeletal homeostasis, Notch
INTRODUCTION
The fate of mesenchymal cells and their differentiation toward cells of the osteoblastic lineage is tightly controlled by extracellular and intracellular signals. Some, such as bone morphogenetic proteins (BMPs) and Wnt, favor osteoblastogenesis (Canalis et al, 2003;Gazzerro and Canalis, 2006;Canalis, 2013;Monroe et al, 2012). Other signals, such as Notch, impair the differentiation of cells of the osteoblastic lineage (Deregowski et al, 2006;Zanotti and Canalis, 2010;Canalis et al, 2013;Zanotti and Canalis, 2013). The activity of these factors is tightly controlled by extracellular proteins that bind to the agent or to its receptors, and by intracellular proteins and regulatory kinases.
Nemo-like kinase (Nlk) is an evolutionary conserved kinase that phosphorylates serine or threonine residues immediately preceding a proline residue (Harada et al, 2002;Ishitani and Ishitani, 2013). Nlk is structurally related to mitogen activated protein (MAP) kinases and is ~42% identical to extracellular signal regulated kinase (ERK)2 (Brott et al, 1998). Recently, Nlk was found to play a critical role in the regulation of diverse signaling pathways, in particular the Wnt/β-catenin, BMP/Smad and the Notch signaling pathways (Ishitani et al, 2003;Ishitani et al, 2010;Zeng et al, 2007). Nlk phosphorylates T cell factor (TCF) and lymphoid enhancer factor (LEF) preventing TCF/LEF binding to DNA and gene transcription (Ishitani et al, 2003). As a consequence, Nlk downregulates the canonical Wnt/β-catenin signaling pathway acting as an intracellular Wnt antagonist. In addition, Nlk interacts and phosphorylates β-catenin in Drosophila, although it is not known whether this mechanism operates in mammalian cells (Mirkovic et al, 2011). Nlk phosphorylates Smad4 antagonizing BMP signaling (Shi et al, 2010;Zeng et al, 2007). In addition, the Notch intracellular domain (NICD) is a substrate of Nlk, and the phosphorylation of NICD impairs the formation of a Notch transcriptional complex and decreases canonical Notch signaling (Ishitani et al, 2010). Because BMP, Wnt and Notch play fundamental roles in osteoblastogenesis and osteoclastogenesis, Nlk has the potential to affect both processes of bone remodeling.
Nlk is expressed by calvarial osteoblasts, and forced expression of Nlk in ST-2 stromal cells and osteoblasts using retroviral vectors inhibits osteoblastic differentiation (Nifuji et al, 2010). Furthermore, we demonstrated that downregulation of Nlk enhanced osteoblastogenesis by augmenting BMP-2/Smad signaling and Wnt/β-catenin signaling (Zanotti and Canalis, 2012). However, the function of Nlk in the skeleton in vivo has not been defined. The targeted global disruption of Nlk in C57BL/6 mice causes embryonic lethality, although its disruption in 129SvJ/C57BL/6 mice leads to survival for up to 4 – 6 weeks (Kortenjann et al, 2001). These Nlk mutants exhibit a hematopoietic cell phenotype, decreased bone lining cells and increased bone marrow adipocytes, but a skeletal phenotype was not explored or reported (Kortenjann et al, 2001). The increased adipocytes are explained by an inhibitory effect of Nlk on adipogenesis secondary to a suppression of the transactivation of peroxisome proliferator-activated receptor (PPAR) γ (Takada et al, 2007). Since mesenchymal cells have the potential to differentiate into adipocytes and osteoblasts, this may serve as an additional regulatory control determined by Nlk (Pereira et al, 2002).
The intent of the present study was to define the function of Nlk in skeletal tissue in vivo. For this purpose, we created NlkloxP/loxP conditional mice. Nlk was inactivated by Cre recombination directed by either the paired-related homeobox gene 1 (Prx1) enhancer expressed in limb buds at day 10.5 of embryonic life (E10.5), the osterix (Osx) promoter expressed in osteoblast precursors or the osteocalcin/bone gamma carboxyglutamate protein (Bglap) promoter expressed in mature osteoblasts (Logan et al, 2002;Rodda and McMahon, 2006;Zhang et al, 2002). The skeletal phenotype of Nlk conditional null mice was determined by microcomputed tomography and by histomorphometric analysis.
MATERIALS AND METHODS
Generation of Nlk Null Mice
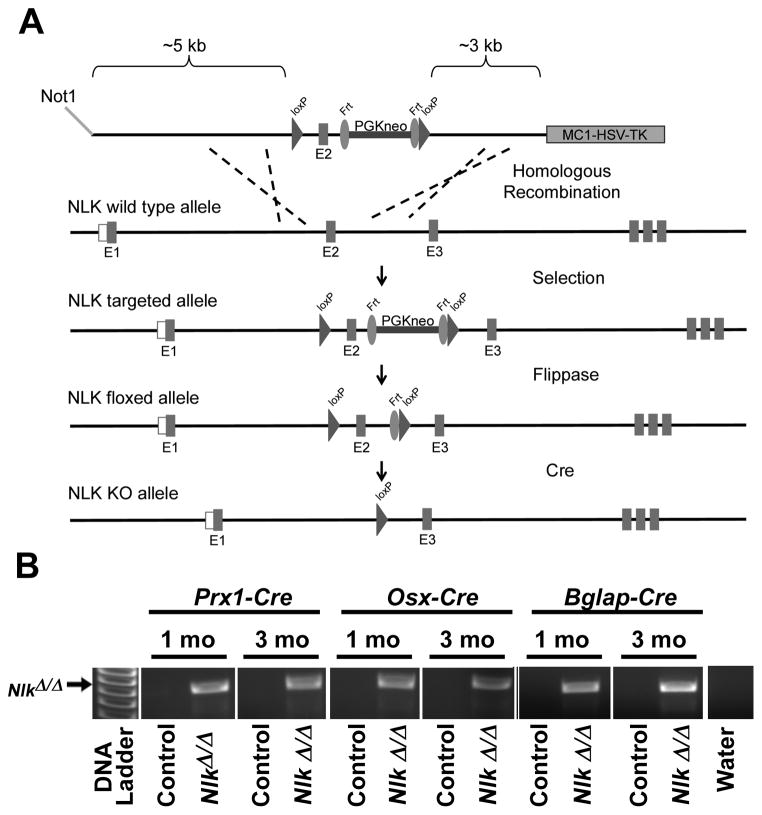
To generate a conditional allele of Nlk, approximately 10 kilobases (kb) of Nlk sequence containing exon 2 were selected from a bacterial artificial chromosome library of C57BL/6 mouse genomic DNA (id RP24-223B7), and retrieved into a PL253 vector. The vector contains an MC1 driven herpex simplex virus-thymidine kinase (MC1-HSV-TK) cassette for negative selection of embryonic stem (ES) cells (Liu et al, 2003;Mansour et al, 1988). A 5′ loxP site was introduced approximately 1.1 kb 5′ of exon 2, followed by the insertion of a phosphoglycerate kinase promoter-driven neomycin (neo) selection cassette flanked by flippase recognition target (Frt) sites and a 3′ loxP site (Frt-PGKneo-Frt-LoxP) approximately 500 bp 3′ of exon 2 (Figure 1A) (Buchholz et al, 1996). The targeting vector containing 5.5 kb of 5′ homology arm and 3.1 kb of 3′ homology arm was linearized by NotI digestion, and electroporated into ES cells derived from F1 (129Svj/C57BL/6) embryos. G418 and gancyclovir resistant colonies were selected and screened by long range nested polymerase chain reaction (PCR) using primers specific to the loxP sites and primers outside the 5′ and 3′ homology arms (Lay et al, 1998). Targeted ES clones were expanded and confirmed by long range PCR and used for aggregations to generate chimeric mice at the Gene Targeting and Transgenic Facility of the University of Connecticut Health Center (Farmington, CT). Chimeras were bred with mice expressing the Flp recombinase under the control of the Gt(ROSA)26Sor promoter (Jax Stock 003946; Jackson Laboratory, Bar Harbor, ME) for the removal of the PGKneo selection cassette, as previously reported (Canalis et al, 2010a;Canalis et al, 2010b;Farley et al, 2000). The excision of the selection cassette was confirmed by PCR, and the resulting Flp recombinase transgene was segregated by mating the mice with C57BL/6 wild type mice. Cre recombination mediates excision of exon 2, partially encoding for the kinase domain, and causes a frameshift so that the transcript is truncated upstream the kinase domain.
Figure 1.
A. Engineering of the Nlk conditional allele showing the wild type and Nlk targeted allele. In the conditional allele exon 2 is flanked by loxP sites upstream a PGK-neo selection cassette flanked by Frt sites. The Nlk targeted allele is shown before and after Flp and Cre recombination for the removal of the neo cassette and excision of exon 2, respectively. B. Cre-dependent recombination of NlkloxP/loxP sites following the crossing of NlkloxP/loxP mice with Prx1-Cre, Osx-Cre or Bglap-Cre transgenics.
As an initial step in the inactivation of Nlk, heterozygous transgenics expressing the Cre recombinase under the control of the Prx1 enhancer (Jackson Laboratory, Bar Harbor, ME), the osteocalcin or Bglap (T. Clemens, Baltimore, MD) or the osterix (Osx) (Jackson Laboratory) promoter were mated with NlkloxP/loxP to create Prx1-Cre+/−;NlkloxP/loxP, Bglap-Cre+/−;NlkloxP/loxP and Osx-Cre+/− ;NlkloxP/loxP mice (Logan et al, 2002;Rodda and McMahon, 2006;Zhang et al, 2002). These were mated with NlkloxP/loxP mice to generate an experimental cohort in which Nlk exon 2 is deleted by Cre in the limb bud (Prx1-Cre;NlkΔ/Δ), and in mature (Bglap-Cre;NlkΔ/Δ) or immature (Osx-Cre;NlkΔ/Δ) osteoblasts and to generate control littermates without Cre-mediated inactivation (NlkloxP/loxP). Prx1-Cre, Bglap-Cre and Osx-Cre transgenics were maintained in a C57BL/6 background and NlkloxP/loxP in a 129SvJ/C57BL/6 background. Because in Osx-Cre trangenics the expression of Cre is under the control of the tet-off cassette, pregnant dams were treated with a diet containing 625 mg of doxycycline hyclate/kg of chow to deliver 2–3 mg of doxycycline daily from the time of conception to delivery (Harlan Laboratories, Indianapolis, IN).
Genotyping of Prx1-Cre, Bglap-Cre, Osx-Cre, NlkloxP and NlkWT alleles was carried out by PCR in tail DNA extracts (Table 1A). Deletion of the neo cassette in NlkloxP mice by Flp recombination was determined by PCR in tail DNA and deletion of loxP flanked sequences in NlkloxP/loxP mice by Cre recombination was documented by PCR in DNA extracted from calvariae (Figure 1B). All animal experiments were approved by the Animal Care and Use Committee of Saint Francis Hospital and Medical Center.
Table 1.
| Table 1A. Primers used for allele identification by PCR. | |||
|---|---|---|---|
| Genotyping | |||
| Allele | Strand | Sequence 5′–3′ | Amplicon Size (bp) |
| Bglap-Cre transgene | Reverse Forward |
TGATACAAGGGACATCTTCC CAAATAGCCCTGGCAGAT |
300 |
| Osx-Cre transgene | Reverse Forward |
GTGAAACAGCATTGCTGTCACTT GCGGTCTGGCAGTAAAAACTATC |
100 |
| Prx1-Cre transgene | Reverse Forward |
GTGAAACAGCATTGCTGTCACTT GCGGTCTGGCAGTAAAAACTATC |
100 |
| Nlk | Reverse Forward |
AAAATCAAATGCCCACCAAA GTGGTGCACATCAGTCATCC |
NlkloxP = 400 NlkWT = 300 |
| Frt and LoxP recombination | Strand | Sequence 5′-3′ | Amplicon Size (bp) |
| Neo Cassette Frt recombination | Reverse Forward |
CAAGGGCCAATGATTCCTTA GGCTGTCCTGGAACTCAGAC |
Neo deleted = 373 Neo present = 507 |
| NlkloxP recombination | Reverse Forward |
CAAGGGCCAATGATTCCTTA GTGGTGCACATCAGTCATCC |
loxP recombination = 669 |
| Table 1B. Primers used for qRT-PCR determinations. | |||
|---|---|---|---|
| Gene | Strand | Sequence 5′–3′ | GenBank Accession Number* |
| Nlk | Forward Reverse |
AGTCCAGCAGCATACGTCTTC GCTGTTGTTGCCCAGGGTTT |
NM_008702 |
| Rpl38 | Forward Reverse |
AGAACAAGGATAATGTGAAGTTCAAGGTTC CTGCTTCAGCTTCTCTGCCTTT |
NM_001048057 NM_001048058 NM_023372 |
|
Tnfrsf11b (Opg) |
Forward Reverse |
CAGAAAGGAAATGCAACACATGACAAC GCCTCTTCACACAGGGTGACATC |
NM_008764 |
|
Tnfsf11 (Rankl) |
Forward Reverse |
TATAGAATCCTGAGACTCCATGAAAAC CCCTGAAAGGCTTGTTTCATCC |
NM_011613 |
GenBank accession number identifies transcript recognized by primer pairs.
Skeletal Staining
To assess the effect of Nlk on skeletal development, skeletons from newborns were fixed with ethanol. Cartilage was stained with Alcian blue and mineralized bone with alizarin red, followed by the addition of 1% potassium hydroxide to remove adjacent tissues, as described (O’Brien et al, 1996).
Microcomputed Tomography (μCT)
Bone microarchitecture of femurs from experimental and control mice was determined using a microcomputed tomography instrument (μCT 40; Scanco Medical AG, Bassersdorf, Switzerland), which was calibrated weekly using a phantom provided by the manufacturer (Bouxsein et al, 2010;Glatt et al, 2007). Femurs were scanned in 70% ethanol at high resolution, energy level of 55 kVp, intensity of 145 μA and integration time of 200 ms. A total of 100 slices at midshaft and 160 slices at the distal metaphysis were acquired at an isotropic voxel dimension of 6 μm or 216 μm3 and a slice thickness of 6 μm, and chosen for analysis. Trabecular bone volume fraction and microarchitecture were evaluated starting approximately 1.0 mm proximal from the femoral condyles. Contours were manually drawn every 10 slices a few voxels away from the endocortical boundary to define the region of interest for analysis. The remaining slice contours were iterated automatically. Trabecular regions were assessed for total volume, bone volume, bone volume fraction (bone volume/total volume, BV/TV), trabecular thickness, trabecular number, trabecular separation, connectivity density and structure model index (SMI), using a Gaussian filter (σ = 0.8) and user defined thresholds (Bouxsein et al, 2010). For analysis of femoral cortical bone, contours were iterated across 100 slices along the cortical shell of the femoral midshaft, excluding the marrow cavity. Analysis of BV/TV, porosity, cortical thickness, total cross sectional and cortical bone area, periosteal and endocortical perimeter and material density were performed using a Gaussian filter (σ = 0.8) and user defined thresholds. The terminology and units used are those recommended by a committee established by the Journal of Bone and Mineral Research (Bouxsein et al, 2010).
Bone Histomorphometry
Static and dynamic histomorphometry was carried out on experimental and control mice after they were injected with calcein, 20 mg/kg, and demeclocycline, 50 mg/kg, at an interval of 5 days. Five μm longitudinal sections of femurs and cross sections at the mid-femoral diaphysis were cut on a microtome (Microm, Richards-Allan Scientific, Kalamazoo, MI), and stained with 0.1% toluidine blue. Static parameters of bone formation and resorption were measured in a defined area between 360 μm and 2160 μm from the growth plate, using an OsteoMeasure morphometry system (Osteometrics, Atlanta, GA). For dynamic histomorphometry, mineralizing surface per bone surface and mineral apposition rate were measured on unstained sections under ultraviolet light using a triple diamidino-2-phenylindole/fluorescein/Texas red set long pass filter, and bone formation was calculated. The terminology and units used are those recommended by the Histomorphometry Nomenclature Committee of the American Society of Bone and Mineral Research (Dempster et al, 2013;Parfitt et al, 1987).
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from calvariae and mRNA levels determined by qRT-PCR (Nazarenko et al, 2002a;Nazarenko et al, 2002b). For this purpose, equal amounts of RNA were reverse-transcribed using iScript RT-PCR kit (BioRad, Hercules, CA), according to manufacturer’s instructions, and amplified in the presence of specific primers (Table 1B), and iQ SYBR Green Supermix (BioRad) at 60°C for 45 cycles. Transcript copy number was estimated by comparison with a serial dilution of cDNA for Nlk and Tnfrsf11b, encoding for Osteoprotegerin (Opg), both from American Type Tissue Culture Collection (ATCC; Manassas, VA), and Tnfsf11, encoding for Receptor activated of nuclear factor-kappa B ligand (Rankl) (BioSciences, Nottingham, UK). Reactions were conducted in a CFX96 qRT-PCR detection system (BioRad), and fluorescence was monitored during every PCR cycle at the annealing step. Data are expressed relative to ribosomal protein L38 (Rpl38) copy number, estimated by comparison with a serial dilution of cDNA for Rpl38 (ATCC).
Serum Osteoprotegerin
Murine Opg was measured by enzyme-linked immune-absorbent assay in serum from Osx-Cre;NlkΔ/Δ and control mice using a commercially available kit in accordance with manufacturer’s instructions (R and D Systems, Minneapolis, MN).
Statistical Analysis
Data are expressed as means ± SEM. Statistical differences were determined by unpaired Student’s t-test or ANOVA.
RESULTS
Conditional Inactivation of Nlk During Development
To induce the conditional inactivation of Nlk in the limb bud, Prx1-Cre+/−;NlkloxP/loxP mice were mated with NlkloxP/loxP mice to create limb bud specific Prx1-Cre;NlkΔ/Δ conditional null and NlkloxP/loxP littermate controls (Logan et al, 2002). Prx1-Cre;NlkΔ/Δ mice appeared normal and not different from wild type littermate controls. Alizarin red and alcian blue staining of the skeleton of newborn mice did not reveal apparent differences between Prx1-Cre;NlkΔ/Δ and control mice. At 1 and 3 months of age, the weight (not shown) and femoral length (Table 2) of Prx1-Cre;NlkΔ/Δ mice were not significantly different from controls. μCT revealed no differences between Prx1-Cre;NlkΔ/Δ and control mice at 1 month of age and only minor differences at 3 months of age, where Prx1-Cre;NlkΔ/Δ mice exhibited a ~15% increase in trabecular number and a ~75% increase in connectivity (Table 2).
Table 2.
Femoral microarchitecture assessed by μCT of 1 and 3 month old Prx1-Cre;NlkΔ/Δ conditional null mice and NlkloxP/loxP littermate controls.
| Males | 1 Month | 3 Month | ||
|---|---|---|---|---|
| Control | NlkΔ/Δ | Control | NlkΔ/Δ | |
| Femoral Length (mm) | 12.2± 0.4 | 11.6 ± 0.4 | 15.4 ± 0.2 | 15.7 ± 0.1 |
| Distal Femur Trabecular Bone | ||||
| Bone Volume/Total Volume (%) | 5.1± 0.8 | 4.6 ± 0.9 | 5.6 ± 0.7 | 8.0± 1.1 |
| Trabecular Number (1/mm) | 4.9± 0.7 | 3.9 ± 0.4 | 4.5 ± 0.1 | 5.1± 0.2* |
| Trabecular Thickness (μm) | 24± 1 | 24 ± 1 | 31 ± 1 | 32± 1 |
| Connectivity Density (1/mm3) | 182± 61 | 152 ± 38 | 135 ± 29 | 239± 26* |
| Structure Model Index | 2.8± 0.1 | 2.7 ± 0.1 | 2.9 ± 0.1 | 2.6± 0.1 |
| Femoral Midshaft Cortical Bone | ||||
| Bone Volume/Total Volume (%) | 90.1± 1.2 | 87.0 ± 1.0 | 92.1 ± 0.4 | 91.3± 0.5 |
| Cortical Thickness (μm) | 108± 5 | 88 ± 6 | 188 ± 5 | 185± 2 |
| Total Area (mm2) | 1.4± 0.1 | 1.5 ± 0.1 | 1.7 ± 0.1 | 1.9 ± 0.1 |
| Bone Area (mm2) | 0.49± 0.02 | 0.44 ± 0.04 | 0.89 ± 0.05 | 0.93 ± 0.02 |
| Periosteal Perimeter (mm) | 4.3± 0.1 | 4.3 ± 0.2 | 4.6 ± 0.1 | 4.8 ± 0.1 |
| Endocortical Perimeter (mm) | 3.4± 0.1 | 3.6 ± 0.2 | 3.2 ± 0.1 | 3.4 ± 0.1 |
| Females | 1 Month | 3 Month | ||
|---|---|---|---|---|
| Control | NlkΔ/Δ | Control | NlkΔ/Δ | |
| Femoral Length (mm) | 12.0± 0.1 | 11.9 ± 0.3 | 16.1 ± 0.1 | 16.3 ± 0.1 |
| Distal Femur Trabecular Bone | ||||
| Bone Volume/Total Volume (%) | 3.5± 0.2 | 4.2 ± 0.2 | 8.8 ± 0.8 | 9.7± 1.6 |
| Trabecular Number (1/mm) | 3.7± 0.1 | 3.8 ± 0.1 | 4.6 ± 0.1 | 4.5± 0.1 |
| Trabecular Thickness (μm) | 24± 0 | 24 ± 0 | 37 ± 1 | 38± 2 |
| Connectivity Density (1/mm3) | 84± 13 | 122 ± 16 | 168 ± 17 | 188± 25 |
| Structure Model Index | 2.8± 0.1 | 2.7 ± 0.1 | 2.4 ± 0.1 | 2.3± 0.2 |
| Femoral Midshaft Cortical Bone | ||||
| Bone Volume/Total Volume (%) | 88.5± 0.4 | 88.0 ± 1.6 | 91.0 ± 0.4 | 90.0± 0.3 |
| Cortical Thickness (μm) | 103± 3 | 98 ± 6 | 181 ± 5 | 169± 6 |
| Total Area (mm2) | 1.4± 0.1 | 1.5 ± 0.1 | 1.7± 0.1 | 1.8± 0.1 |
| Bone Area (mm2) | 0.48± 0.01 | 0.49 ± 0.03 | 0.87± 0.03 | 0.85± 0.02 |
| Periosteal Perimeter (mm) | 4.2± 0.1 | 4.3 ± 0.1 | 4.7± 0.1 | 4.7± 0.1 |
| Endocortical Perimeter (mm) | 3.4± 0.1 | 3.5 ± 0.1 | 3.3± 0.1 | 3.4± 0.1 |
μCT was performed in distal femurs for trabecular bone and midshaft for cortical bone from 1 and 3 month old male and female Prx1-Cre;NlkΔ/Δ conditional null mice (NlkΔ/Δ) and control NlkloxP/loxP littermates (Control). Values are means ± SEM; n = 3 to 7.
Significantly different from controls, p < 0.05 by unpaired t-test.
Conditional Inactivation of Nlk in Osteoblast Precursors
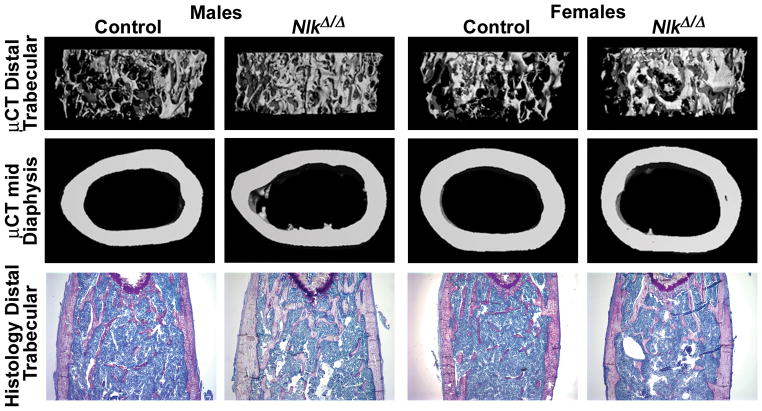
For the conditional inactivation of Nlk in osteoblast precursors, Osx-Cre+/−;NlkloxP/loxP mice were crossed with NlkloxP/loxP mice to create Osx-Cre;NlkΔ/Δ experimental and NlkloxP/loxP controls. Osx-Cre;NlkΔ/Δ mice appeared normal. Except for an unexplained 15% decrease in the weight of 6 month old female Osx-Cre;NlkΔ/Δ mice, the weight (not shown) and femoral length (Table 3) of experimental null mice were not different from littermate controls. μCT did not reveal a skeletal phenotype in 1 month old Osx-Cre;NlkΔ/Δ mice of either sex (Table 3). In contrast, at 3 months of age Osx-Cre;NlkΔ/Δ of both sexes exhibited a 1.5 to 1.8 fold increase in cancellous bone volume, secondary to an increase in trabecular number (Table 3, Figure 2). Connectivity density was increased by >2 fold, and SMI revealed a predominance of plate-like trabeculae. Minor changes in cortical microarchitecture, such as a ~10% decrease in cortical thickness, were observed in 3 month old female, but not in male, mice. The skeletal phenotype was transient and μCT revealed that the skeletal microarchitecture of 6 month old Osx-Cre;NlkΔ/Δ mice of both sexes was not different from controls, other than a modest decrease in trabecular thickness in male mice. Histomorphometric analysis of Osx-Cre;NlkΔ/Δ male 3 month old mice confirmed an increase in trabecular bone volume, albeit not statistically significant, and a decrease in the number of osteoclasts and in eroded surface, explaining the skeletal phenotype detected by μCT (Table 4). Three month old female Osx-Cre;NlkΔ/Δ mice exhibited an increase in trabecular bone volume, and a decrease in the number of osteoblasts but no changes in osteoclast number, or in parameters of bone resorption or dynamic parameters of bone formation (Table 4). However, osteoid surface/bone surface was decreased in female and male mice. Bone marrow adipocyte number was not changed in Osx-Cre;NlkΔ/Δ mice of either sex.
Table 3.
Femoral microarchitecture assessed by μCT of 1, 3 and 6 month old Osx-Cre;NlkΔ/Δ conditional null mice and NlkloxP/loxP littermate controls.
| Males | 1 Month | 3 Month | 6 Month | |||
|---|---|---|---|---|---|---|
| Control | NlkΔ/Δ | Control | NlkΔ/Δ | Control | NlkΔ/Δ | |
| Femoral Length (mm) | 12.3± 0.1 | 12.0 ± 0.2 | 15.5 ± 0.3 | 15.9 ± 0.2 | 16.1 ± 0.1 | 16.0± 0.1 |
| Distal Femur Trabecular Bone | ||||||
| Bone Volume/Total Volume (%) | 5.2± 0.4 | 5.2 ± 0.7 | 5.8 ± 0.6 | 10.7± 1.8* | 7.9 ± 0.9 | 7.1± 0.9 |
| Trabecular Number (1/mm) | 4.3± 0.2 | 4.4 ± 0.2 | 4.4 ± 0.1 | 5.4± 0.3* | 3.6 ± 0.2 | 4.0± 0.2 |
| Trabecular Thickness (μm) | 26± 1 | 25 ± 1 | 32 ± 1 | 34± 2 | 44 ± 2 | 36± 2* |
| Connectivity Density (1/mm3) | 142± 20 | 165 ± 58 | 145 ± 20 | 327± 65* | 101 ± 20 | 129± 24 |
| Structure Model Index | 2.8± 0.1 | 2.8 ± 0.1 | 2.8 ± 0.1 | 2.2± 0.2* | 2.3 ± 0.2 | 2.3± 0.1 |
| Femoral Midshaft Cortical Bone | ||||||
| Bone Volume/Total Volume (%) | 84.8± 1.1 | 83.3 ± 0.7 | 94.1 ± 0.1 | 94.0± 0.2 | 91.3 ± 0.4 | 91.6± 0.4 |
| Cortical Thickness (μm) | 102± 4 | 93 ± 3 | 189 ± 4 | 191± 9 | 187 ± 5 | 189± 9 |
| Total Area (mm2) | 1.3± 0.1 | 1.3 ± 0.1 | 1.6 ± 0.1 | 2.0± 0.2 | 2.1 ± 0.2 | 1.9± 0.1 |
| Bone Area (mm2) | 0.48± 0.02 | 0.47 ± 0.03 | 0.83 ± 0.05 | 0.97± 0.09 | 1.00 ± 0.07 | 0.97± 0.05 |
| Periosteal Perimeter (mm) | 4.0± 0.1 | 4.0 ± 0.1 | 4.5 ± 0.1 | 4.9± 0.2 | 5.1 ± 0.2 | 4.9± 0.1 |
| Endocortical Perimeter (mm) | 3.1± 0.1 | 3.1 ± 0.1 | 3.2 ± 0.1 | 3.5± 0.1 | 3.7 ± 0.2 | 3.4± 0.1 |
| Females | 1 Month | 3 Month | 6 Month | |||
|---|---|---|---|---|---|---|
| Control | NlkΔ/Δ | Control | NlkΔ/Δ | Control | NlkΔ/Δ | |
| Femoral Length (mm) | 12.3± 0.3 | 12.7 ± 0.2 | 15.5 ± 0.2 | 15.6 ± 0.1 | 16.4 ± 0.2 | 16.1± 0.1 |
| Distal Femur Trabecular Bone | ||||||
| Bone Volume/Total Volume (%) | 3.8± 0.4 | 4.1 ± 0.6 | 4.0 ± 0.8 | 6.1± 0.3* | 3.2 ± 0.5 | 3.2± 0.6 |
| Trabecular Number (1/mm) | 4.0± 0.2 | 4.0 ± 0.2 | 3.4 ± 0.2 | 3.8± 0.1+ | 2.8 ± 0.1 | 2.7± 0.1 |
| Trabecular Thickness (μm) | 23± 0 | 23 ± 1 | 36 ± 1 | 35± 0 | 40 ± 3 | 37± 2 |
| Connectivity Density (1/mm3) | 77± 16 | 124 ± 35 | 56 ± 16 | 130± 11* | 34 ± 7 | 38± 8 |
| Structure Model Index | 2.9± 0.1 | 2.9 ± 0.1 | 3.0 ± 0.2 | 2.5± 0.1* | 3.0 ± 0.2 | 2.8± 0.1 |
| Femoral Midshaft Cortical Bone | ||||||
| Bone Volume/Total Volume (%) | 85.1± 0.6 | 85.0 ± 1.2 | 94.3 ± 0.1 | 93.6± 0.2* | 91.8 ± 0.3 | 91.3± 0.3 |
| Cortical Thickness (μm) | 103± 2 | 100 ± 5 | 187 ± 4 | 171± 4* | 193 ± 4 | 187± 6 |
| Total Area (mm2) | 1.4± 0.1 | 1.5 ± 0.1 | 1.4 ± 0.1 | 1.5± 0.1 | 1.7± 0.1 | 1.7± 0.1 |
| Bone Area (mm2) | 0.53± 0.02 | 0.53 ± 0.03 | 0.75 ± 0.02 | 0.73± 0.01 | 0.87± 0.02 | 0.87± 0.02 |
| Periosteal Perimeter (mm) | 4.2± 0.1 | 4.3 ± 0.1 | 4.3 ± 0.1 | 4.4± 0.1 | 4.6± 0.1 | 4.7± 0.1 |
| Endocortical Perimeter (mm) | 3.3± 0.1 | 3.4 ± 0.1 | 3.0 ± 0.1 | 3.2± 0.1* | 3.2± 0.1 | 3.2± 0.1 |
μCT was performed in distal femurs for trabecular bone and midshaft for cortical bone from 1, 3 and 6 month old male and female Osx-Cre;NlkΔ/Δ conditional null mice (NlkΔ/Δ) and control NlkloxP/loxP littermates (Control). Values are means ± SEM; n = 5 to 6.
Significantly different from controls, p < 0.05 by unpaired t-test,
p < 0.051.
Figure 2.
Representative μCT of the distal femur (upper panel), cross sectional μCT images of the femoral midshaft (middle panel), and histological sections of the distal femur stained with toluidine blue (lower panel; magnification 40x), of 3 month old male and female Osx-Cre;NlkΔ/Δ and control NlkloxP/loxP littermates.
Table 4.
Femoral histomorphometry of 3 month old Osx-Cre;NlkΔ/Δ conditional null mice and NlkloxP/loxP controls.
| Males | ||
|---|---|---|
| Control | NlkΔ/Δ | |
| Distal Femur Trabecular Bone | ||
| Bone Volume/Tissue Volume (%) | 10.5 ± 1.7 | 14.1 ± 2.2 |
| Trabecular Number (1/mm) | 2.9 ± 0.2 | 3.4 ± 0.2 |
| Trabecular Thickness (μm) | 35 ± 3 | 40 ± 4 |
| Osteoblast Surface/Bone Surface (%) | 9.2 ± 1.3 | 8.8 ± 1.4 |
| Osteoblasts/Bone Perimeter (1/mm) | 11.1 ± 1.6 | 10.4 ± 1.5 |
| Osteoid Surface/Bone Surface (%) | 2.2 ± 0.4 | 1.0 ± 0.4* |
| Osteoclast Surface/Bone Surface (%) | 9.1 ± 0.8 | 6.2 ± 0.7* |
| Osteoclasts/Bone Perimeter (1/mm) | 5.7 ± 0.5 | 3.9 ± 0.4* |
| Eroded Surface/Bone Surface (%) | 15.0 ± 1.2 | 10.1 ± 1.0* |
| Mineral Apposition Rate (μm/day) | 1.1 ± 0.1 | 1.0 ± 0.1 |
| Mineralizing Surface/Bone Surface (%) | 14.6 ± 1.5 | 11.9 ± 1.7 |
| Bone Formation Rate (μm3/μm2/day) | 0.17 ± 0.03 | 0.12 ± 0.03 |
| Adipocytes (mm2) | 122 ± 19 | 94 ± 17 |
| Females | ||
|---|---|---|
| Control | NlkΔ/Δ | |
| Distal Femur Trabecular Bone | ||
| Bone Volume/Tissue Volume (%) | 4.7 ± 0.5 | 7.3 ± 0.5* |
| Trabecular Number (1/mm) | 1.7 ± 0.1 | 2.2 ± 0.1* |
| Trabecular Thickness (μm) | 27 ± 1 | 33 ± 1 |
| Osteoblast Surface/Bone Surface (%) | 18.5 ± 1.4 | 13.5 ± 1.6* |
| Osteoblasts/Bone Perimeter (1/mm) | 19.8 ± 1.3 | 14.5 ± 1.6* |
| Osteoid Surface/Bone Surface (%) | 4.5 ± 1.0 | 1.8 ± 0.6* |
| Osteoclast Surface/Bone Surface (%) | 11.7 ± 1.0 | 9.6 ± 0.7 |
| Osteoclasts/Bone Perimeter (1/mm) | 7.6 ± 0.6 | 6.5 ± 0.5 |
| Eroded Surface/Bone Surface (%) | 19.9 ± 1.7 | 16.9 ± 1.5 |
| Mineral Apposition Rate (μm/day) | 1.7 ± 0.1 | 1.7 ± 0.1 |
| Mineralizing Surface/Bone Surface (%) | 15.2 ± 3.9 | 13.3 ± 1.9 |
| Bone Formation Rate (μm3/μm2/day) | 0.25 ± 0.06 | 0.23 ± 0.03 |
| Adipocytes (mm2) | 215 ± 31 | 213 ± 37 |
Bone histomorphometry was performed in distal femurs from 3 month old male and female Osx-Cre;NlkΔ/Δ conditional null mice (NlkΔ/Δ) and control NlkloxP/loxP littermates (Control). Values are means ± SEM; n = 6 to 8.
Significantly different from controls, p < 0.05 by unpaired t-test.
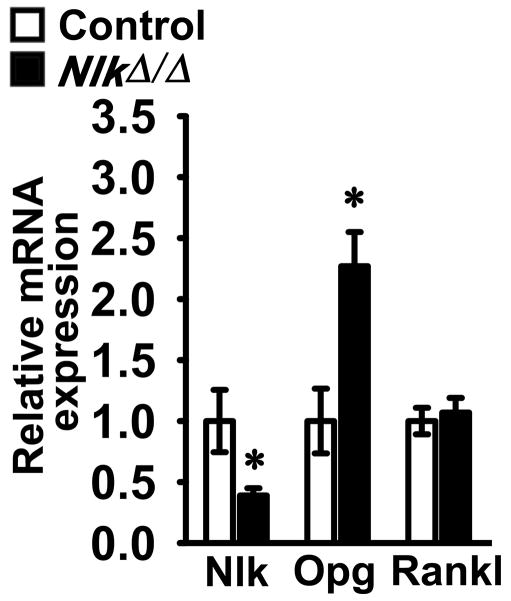
To determine possible mechanisms responsible for the phenotype observed, calvariae from Osx-Cre;NlkΔ/Δ and control mice were extracted and analyzed for changes in mRNA expression. The Nlk inactivation resulted in a ~2 fold increase in Opg mRNA expression without a significant change in Rankl mRNA, explaining the suppression of bone resorption (Figure 3). Immunoreactive serum Opg was not different between Osx-Cre;NlkΔ/Δ and control mice of either sex at 1 month (not shown) or 3 months of age. Serum Opg in 3 month old male control mice was (means ± SEM; n = 3 – 5) 3.6 ± 0.1 and in Osx-Cre;NlkΔ/Δ was 3.5 ± 0.4 μg/L; serum Opg in female control mice was 4.3 ± 0.2 and in Osx-Cre;NlkΔ/Δ was 4.5 ± 0.2 μg/L. This suggests that Opg acted locally in the bone microenvironment, and not systemically.
Figure 3.
Nlk, Opg and Rankl mRNA levels in calvarial extracts from 3 month Osx-Cre;NlkΔ/Δ mice (black bars) and control NlkloxP/loxP littermates (white bars) of both sexes. mRNA levels are expressed as copy number corrected for Rpl38 and controls normalized to 1. Values are means ± SEM; n = 7 – 8. *Significantly different from control mice, p < 0.05 by unpaired t test.
Conditional Inactivation of Nlk in Mature Osteoblasts
For the conditional deletion of Nlk in mature osteoblasts, Bglap-Cre+/−;NlkloxP/loxP were mated with NlkloxP/loxP mice to create Bglap-Cre;NlkΔ/Δ as an experimental group and NlkloxP/loxP as littermate controls. Bglap-Cre;NlkΔ/Δ conditional null mice appeared visually normal, had normal weight (not shown) and their femoral length was not affected (Table 5). μCT of trabecular and cortical bone of 1, 3 and 6 month old mice of both sexes did not reveal any differences between Bglap-Cre;NlkΔ/Δ mice and littermate controls, indicating that Nlk is dispensable for the function of mature osteoblasts (Table 5).
Table 5.
Femoral microarchitecture assessed by μCT of 1, 3 and 6 month old Bglap-Cre;NlkΔ/Δ conditional null mice and NlkloxP/loxP littermate controls.
| Males | 1 Month | 3 Month | 6 Month | |||
|---|---|---|---|---|---|---|
| Control | NlkΔ/Δ | Control | NlkΔ/Δ | Control | NlkΔ/Δ | |
| Femoral Length (mm) | 12.6± 0.1 | 12.6 ± 0.2 | 15.5 ± 0.3 | 15.5 ± 0.1 | 16.4 ± 0.1 | 16.6±0.2 |
| Distal Femur Trabecular Bone | ||||||
| Bone Volume/Total Volume (%) | 5.7± 0.3 | 6.3 ± 0.8 | 7.0 ± 1.2 | 7.1± 1.6 | 3.8 ± 0.8 | 4.8±0.8 |
| Trabecular Number (1/mm) | 4.8± 0.2 | 4.7 ± 0.3 | 4.5 ± 0.2 | 4.5± 0.3 | 3.5 ± 0.2 | 3.7±0.1 |
| Trabecular Thickness (μm) | 25± 0 | 26 ± 1 | 34 ± 3 | 33± 1 | 34 ± 2 | 33±2 |
| Connectivity Density (1/mm3) | 188± 25 | 249 ± 70 | 157 ± 28 | 155± 45 | 72 ± 19 | 116±31 |
| Structure Model Index | 2.8± 0.1 | 2.8 ± 0.1 | 2.7 ± 0.1 | 2.6± 0.2 | 3.0 ± 0.1 | 2.7±0.1 |
| Femoral Midshaft Cortical Bone | ||||||
| Bone Volume/Total Volume (%) | 87.0± 0.5 | 86.0 ± 0.5 | 94.0 ± 0.2 | 93.9± 0.1 | 92.4 ± 0.4 | 92.3±0.3 |
| Cortical Thickness (μm) | 107± 3 | 103 ± 2 | 187 ± 8 | 182± 3 | 199 ± 0 | 194±4 |
| Total Area (mm2) | 1.4± 0.1 | 1.3 ± 0.1 | 1.7 ± 0.1 | 1.7± 0.1 | 1.9 ± 0.1 | 2.0±0.1 |
| Bone Area (mm2) | 0.51± 0.01 | 0.49 ± 0.02 | 0.83 ± 0.04 | 0.80± 0.04 | 0.97 ± 0.04 | 0.99±0.05 |
| Periosteal Perimeter (mm) | 4.2± 0.1 | 4.1 ± 0.1 | 4.6 ± 0.1 | 4.6± 0.1 | 4.9 ± 0.1 | 5.1±0.1 |
| Endocortical Perimeter (mm) | 3.3± 0.1 | 3.3 ± 0.1 | 3.3 ± 0.1 | 3.3± 0.1 | 3.5 ± 0.1 | 3.6±0.1 |
| Females | 1 Month | 3 Month | 6 Month | |||
|---|---|---|---|---|---|---|
| Control | NlkΔ/Δ | Control | NlkΔ/Δ | Control | NlkΔ/Δ | |
| Femoral Length (mm) | 12.7± 0.1 | 12.8 ± 0.1 | 15.5 ± 0.2 | 15.6 ± 0.1 | 16.4 ± 0.1 | 16.5± 0.2 |
| Distal Femur Trabecular Bone | ||||||
| Bone Volume/Total Volume (%) | 5.2± 0.6 | 4.7 ± 0.5 | 3.8 ± 0.5 | 5.4± 0.8 | 2.1 ± 0.4 | 1.6± 0.3 |
| Trabecular Number (1/mm) | 4.6± 0.2 | 4.0 ± 0.2 | 3.6 ± 0.1 | 3.9± 0.2 | 2.7 ± 0.1 | 2.3± 0.2* |
| Trabecular Thickness (μm) | 24± 0 | 24 ± 0 | 31 ± 1 | 34± 0* | 34 ± 2 | 41± 3* |
| Connectivity Density (1/mm3) | 199± 68 | 147 ± 53 | 70 ± 16 | 101± 13 | 19 ± 7 | 12± 3 |
| Structure Model Index | 2.7± 0.1 | 2.8 ± 0.1 | 3.0 ± 0.1 | 2.7± 0.1 | 3.3 ± 0.1 | 3.4± 0.2 |
| Femoral Midshaft Cortical Bone | ||||||
| Bone Volume/Total Volume (%) | 88.0± 0.3 | 87.0 ± 0.5 | 93.8 ± 0.1 | 93.8± 0.1 | 92.3 ± 0.6 | 92.5± 0.2 |
| Cortical Thickness (μm) | 110± 3 | 109 ± 4 | 176 ± 2 | 179± 3 | 202 ± 4 | 198± 5 |
| Total Area (mm2) | 1.4± 0.1 | 1.4± 0.1 | 1.6 ± 0.1 | 1.7± 0.1 | 1.8± 0.1 | 1.8± 0.1 |
| Bone Area (mm2) | 0.50± 0.03 | 0.53± 0.02 | 0.77 ± 0.02 | 0.79± 0.02 | 0.89± 0.03 | 0.91± 0.03 |
| Periosteal Perimeter (mm) | 4.2± 0.1 | 4.2± 0.1 | 4.5 ± 0.1 | 4.6± 0.1 | 4.7± 0.1 | 4.7± 0.1 |
| Endocortical Perimeter (mm) | 3.4± 0.1 | 3.3± 0.1 | 3.3 ± 0.1 | 3.3± 0.1 | 3.3± 0.1 | 3.3± 0.1 |
μCT was performed in distal femurs for trabecular bone and midshaft for cortical bone from 1, 3 and 6 month old male and female Bglap-Cre;NlkΔ/Δ conditional null mice (NlkΔ/Δ) and control NlkloxP/loxP littermates (Control). Values are means ± SEM; n = 3 to 6.
Significantly different from controls, p < 0.05 by unpaired t-test.
DISCUSSION
Our findings demonstrate that the conditional inactivation of Nlk in Osx-expressing cells results in an increase in bone volume indicating that Nlk has a negative effect on bone homeostasis when expressed in undifferentiated cells of the osteoblastic lineage. In contrast, the conditional inactivation of Nlk in the limb bud or in mature osteoblasts, achieved by crossing Nlk conditional mice with transgenics expressing the Cre recombinase under the control of either the Prx1 enhancer or the Bglap or Osteocalcin promoter, did not cause an obvious skeletal phenotype. This would indicate that Nlk is dispensable for skeletal development or for the function of mature osteoblasts, but that it plays an important role in osteoblastogenesis and in the function of undifferentiated cells of the osteoblastic lineage. These findings are consistent with previous in vitro work demonstrating that Nlk modifies the activity of signaling molecules required for osteoblastic differentiation that target undifferentiated cells of the osteoblastic lineage (Zanotti and Canalis, 2012). It is also possible that the expression of MAP kinases by mature osteoblasts prevented the Nlk inactivation from affecting osteoblast function (Franceschi et al, 2007). The microarchitectural skeletal phenotype caused by the conditional Nlk inactivation in osteoblast precursors was not sexually dimorphic and was observed in male and female mice. However, it appeared to be somewhat more pronounced in male mice and it affected primarily cancellous and to a minor extent cortical bone. The phenotype was observed at 3 months of age, and not in older animals, possibly because Osx expressing cells differentiate into mature osteoblasts and osteocytes, a population of cells not influenced by Nlk inactivation (Liu et al, 2013;Maes et al, 2010). Osx-Cre;NlkΔ/Δ mice did not manifest a phenotype at 1 month of age, possibly because pregnant dams were treated with doxycycline preventing Cre activation during gestation, and in the progeny perinatally and at an early age.
The mechanism responsible for the increase in cancellous bone in male Osx-Cre;NlkΔ/Δ mice appeared to be a decrease in osteoclast number and an inhibition of bone resorption. However, the mechanism responsible for the increased bone volume in female mice was less clear. Indeed, a decrease in osteoblast number and osteoid surface was noted in female Osx-Cre;NlkΔ/Δ mice as well as a non-significant ~15% decrease in osteoclast number and eroded surface. It is possible that Nlk targets signals that suppress osteoclastogenesis and osteoblastogenesis, and its downregulation results in an enhanced activity of these inhibitory pathways. The inactivation of Nlk caused an inhibition of bone resorption, and the effect appeared related to an increase in the expression of Opg mRNA in bone extracts. Although Nlk is required for the induction of Opg following cellular mechanical loading, its downregulation did not preclude the induction of Opg mRNA (Yu et al, 2010). However, serum Opg levels were not affected suggesting that the impact of Nlk on Opg was limited to an effect within the skeletal microenvironment.
The mechanism of action of Nlk in vitro entails the inhibition of BMP-2 and Wnt signaling, but these mechanisms do not seem to explain the phenotype observed in vivo, since the Nlk inactivation did not result in increased osteoblast number or bone formation, as it would be expected from enhanced BMP or Wnt signaling (Canalis et al, 2003;Canalis, 2013;Zanotti and Canalis, 2012). If enhanced BMP and Wnt activity occurred, this could have been modest or transient. Nlk targets the Notch receptor, decreasing Notch signaling, and the inactivation of Nlk in osteoblast precursors could have caused enhanced Notch signaling explaining the phenotype observed in Osx-Cre;NlkΔ/Δ since Notch suppresses osteoclastogenesis and increases Opg expression (Bai et al, 2008;Hilton et al, 2008;Zanotti and Canalis, 2010). Enhanced Notch signaling may explain the decrease in osteoblast number observed in female mice, since Notch is an inhibitor of osteoblastic differentiation and function (Zanotti and Canalis, 2010;Zanotti and Canalis, 2013;Engin et al, 2008). However, changes in Notch signaling are of short duration and difficult to verify in in vivo models.
Nlk prevents the commitment of mesenchymal stem cells to the adipocytic lineage by modifying chromatin organization and suppressing PPAR-γ transactivation (Takada et al, 2007). Recent work has demonstrated that Osx-labeled cells generate various bone marrow stromal cells including adipocytes (Liu et al, 2013). However, adipocyte number was not affected in Osx-Cre;NlkΔ/Δ mice suggesting that adipocyte precursors were not targeted by Osx or that Nlk did not affect adipogenesis under the experimental conditions of this study.
In conclusion, our studies reveal that in osteoblast precursors Nlk has a negative effect on bone homeostasis, increasing bone resorption and decreasing cancellous bone volume, possibly by regulating selected signaling pathways in the skeleton.
Acknowledgments
National Institute of Arthritis and Musculoskeletal and Skin Diseases; AR063049 and National Institute of Diabetes and Digestive and Kidney Diseases; DK045227.
The authors thank T. Clemens for Bglap-Cre transgenics; Alison Kent and David Bridgewater for technical assistance, Mary Yurczak for secretarial help and Siu-Pok Yee for helpful discussions.
ABBREVIATIONS
- Bglap
bone gamma carboxyglutamate protein
- BMP
bone morphogenetic protein
- ERK
extracellular signal regulated kinase
- ES
embryonic stem
- FRT
Flp recognition target
- Id1
Inhibitor of DNA binding
- kb
kilobase
- LEF
lymphoid enhancer factor
- μCT
microcomputed tomography
- MAP
mitogen-activated protein
- neo
neomycin
- Nlk
Nemo-like kinase
- NICD
Notch intracellular domain
- Opg
Osteoprotegerin
- Osx
osterix
- PCR
polymerase chain reaction
- PPARγ
peroxisome proliferator-activated receptor γ
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- Prx
paired-related homeobox gene
- Rankl
Receptor activated of nuclear factor-kappa B ligand
- Rpl38
ribosomal protein L38
- SMI
structure model index
- TCF
T cell factor
Footnotes
Disclosure of potential conflict of interests: The authors declare that there are no conflicts of interest.
LITERATURE CITED
- Bai S, Kopan R, Zou W, Hilton MJ, Ong CT, Long F, Ross FP, Teitelbaum SL. NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J Biol Chem. 2008;283:6509–6518. doi: 10.1074/jbc.M707000200. [DOI] [PubMed] [Google Scholar]
- Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res. 2010;25:1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- Brott BK, Pinsky BA, Erikson RL. Nlk is a murine protein kinase related to Erk/MAP kinases and localized in the nucleus. Proc Natl Acad Sci U S A. 1998;95:963–968. doi: 10.1073/pnas.95.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F, Ringrose L, Angrand PO, Rossi F, Stewart AF. Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res. 1996;24:4256–4262. doi: 10.1093/nar/24.21.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E. Wnt signalling in osteoporosis: mechanisms and novel therapeutic approaches. Nat Rev Endocrinol. 2013;9:575–583. doi: 10.1038/nrendo.2013.154. [DOI] [PubMed] [Google Scholar]
- Canalis E, Economides AN, Gazzerro E. Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr Rev. 2003;24:218–235. doi: 10.1210/er.2002-0023. [DOI] [PubMed] [Google Scholar]
- Canalis E, Parker K, Feng JQ, Zanotti S. Osteoblast Lineage-specific Effects of Notch Activation in the Skeleton. Endocrinology. 2013;154:623–634. doi: 10.1210/en.2012-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E, Smerdel-Ramoya A, Durant D, Economides AN, Beamer WG, Zanotti S. Nephroblastoma Overexpressed (NOV) Inactivation Sensitizes Osteoblasts To Bone Morphogenetic Protein-2 But NOV Is Dispensable For Skeletal Homeostasis. Endocrinology. 2010a;151:221–233. doi: 10.1210/en.2009-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E, Zanotti S, Beamer WG, Economides AN, Smerdel-Ramoya A. Connective Tissue Growth Factor Is Required for Skeletal Development and Postnatal Skeletal Homeostasis in Male Mice. Endocrinology. 2010b;151:3490–3501. doi: 10.1210/en.2010-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR, Parfitt AM. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013;28:2–17. doi: 10.1002/jbmr.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deregowski V, Gazzerro E, Priest L, Rydziel S, Canalis E. Notch 1 Overexpression Inhibits Osteoblastogenesis by Suppressing Wnt/beta-Catenin but Not Bone Morphogenetic Protein Signaling. J Biol Chem. 2006;281:6203–6210. doi: 10.1074/jbc.M508370200. [DOI] [PubMed] [Google Scholar]
- Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, Chen Y, Wang L, Zheng H, Sutton RE, Boyce BF, Lee B. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med. 2008;14:299–305. doi: 10.1038/nm1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Franceschi RT, Ge C, Xiao G, Roca H, Jiang D. Transcriptional regulation of osteoblasts. Ann N Y Acad Sci. 2007;1116:196–207. doi: 10.1196/annals.1402.081. [DOI] [PubMed] [Google Scholar]
- Gazzerro E, Canalis E. Bone morphogenetic proteins and their antagonists. Rev Endocr Metab Disord. 2006;7:51–65. doi: 10.1007/s11154-006-9000-6. [DOI] [PubMed] [Google Scholar]
- Glatt V, Canalis E, Stadmeyer L, Bouxsein ML. Age-Related Changes in Trabecular Architecture Differ in Female and Male C57BL/6J Mice. J Bone Miner Res. 2007;22:1197–1207. doi: 10.1359/jbmr.070507. [DOI] [PubMed] [Google Scholar]
- Harada H, Yoshida S, Nobe Y, Ezura Y, Atake T, Koguchi T, Emi M. Genomic structure of the human NLK (nemo-like kinase) gene and analysis of its promoter region. Gene. 2002;285:175–182. doi: 10.1016/s0378-1119(02)00412-2. [DOI] [PubMed] [Google Scholar]
- Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, Long F. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T, Hirao T, Suzuki M, Isoda M, Ishitani S, Harigaya K, Kitagawa M, Matsumoto K, Itoh M. Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Nat Cell Biol. 2010;12:278–285. doi: 10.1038/ncb2028. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Ishitani S. Nemo-like kinase, a multifaceted cell signaling regulator. Cell Signal. 2013;25:190–197. doi: 10.1016/j.cellsig.2012.09.017. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Ninomiya-Tsuji J, Matsumoto K. Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23:1379–1389. doi: 10.1128/MCB.23.4.1379-1389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortenjann M, Nehls M, Smith AJ, Carsetti R, Schuler J, Kohler G, Boehm T. Abnormal bone marrow stroma in mice deficient for nemo-like kinase, Nlk. Eur J Immunol. 2001;31:3580–3587. doi: 10.1002/1521-4141(200112)31:12<3580::aid-immu3580>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Lay JM, Friis-Hansen L, Gillespie PJ, Samuelson LC. Rapid confirmation of gene targeting in embryonic stem cells using two long-range PCR techniques. Transgenic Res. 1998;7:135–140. doi: 10.1023/a:1008876526826. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Strecker S, Wang L, Kronenberg MS, Wang W, Rowe DW, Maye P. Osterix-cre labeled progenitor cells contribute to the formation and maintenance of the bone marrow stroma. PLoS One. 2013;8:e71318. doi: 10.1371/journal.pone.0071318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, Carmeliet G, Kronenberg HM. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Mirkovic I, Gault WJ, Rahnama M, Jenny A, Gaengel K, Bessette D, Gottardi CJ, Verheyen EM, Mlodzik M. Nemo kinase phosphorylates beta-catenin to promote ommatidial rotation and connects core PCP factors to E-cadherin-beta-catenin. Nat Struct Mol Biol. 2011;18:665–672. doi: 10.1038/nsmb.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe DG, McGee-Lawrence ME, Oursler MJ, Westendorf JJ. Update on Wnt signaling in bone cell biology and bone disease. Gene. 2012;492:1–18. doi: 10.1016/j.gene.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarenko I, Lowe B, Darfler M, Ikonomi P, Schuster D, Rashtchian A. Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore. Nucleic Acids Res. 2002a;30:e37. doi: 10.1093/nar/30.9.e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarenko I, Pires R, Lowe B, Obaidy M, Rashtchian A. Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Res. 2002b;30:2089–2195. doi: 10.1093/nar/30.9.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nifuji A, Ideno H, Ohyama Y, Takanabe R, Araki R, Abe M, Noda M, Shibuya H. Nemo-like kinase (NLK) expression in osteoblastic cells and suppression of osteoblastic differentiation. Exp Cell Res. 2010;316:1127–1136. doi: 10.1016/j.yexcr.2010.01.023. [DOI] [PubMed] [Google Scholar]
- O’Brien TP, Metallinos DL, Chen H, Shin MK, Tilghman SM. Complementation mapping of skeletal and central nervous system abnormalities in mice of the piebald deletion complex. Genetics. 1996;143:447–461. doi: 10.1093/genetics/143.1.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 1987;2:595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- Pereira RC, Delany AM, Canalis E. Effects of cortisol and bone morphogenetic protein-2 on stromal cell differentiation: correlation with CCAAT-enhancer binding protein expression. Bone. 2002;30:685–691. doi: 10.1016/s8756-3282(02)00687-7. [DOI] [PubMed] [Google Scholar]
- Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- Shi Y, Ye K, Wu H, Sun Y, Shi H, Huo K. Human SMAD4 is phosphorylated at Thr9 and Ser138 by interacting with NLK. Mol Cell Biochem. 2010;333:293–298. doi: 10.1007/s11010-009-0230-2. [DOI] [PubMed] [Google Scholar]
- Takada I, Mihara M, Suzawa M, Ohtake F, Kobayashi S, Igarashi M, Youn MY, Takeyama K, Nakamura T, Mezaki Y, Takezawa S, Yogiashi Y, Kitagawa H, Yamada G, Takada S, Minami Y, Shibuya H, Matsumoto K, Kato S. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat Cell Biol. 2007;9:1273–1285. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- Yu HC, Wu TC, Chen MR, Liu SW, Chen JH, Lin KM. Mechanical stretching induces osteoprotegerin in differentiating C2C12 precursor cells through noncanonical Wnt pathways. J Bone Miner Res. 2010;25:1128–1137. doi: 10.1002/jbmr.9. [DOI] [PubMed] [Google Scholar]
- Zanotti S, Canalis E. Notch and the Skeleton. Mol Cell Biol. 2010;30:886–896. doi: 10.1128/MCB.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, Canalis E. Nemo-like kinase inhibits osteoblastogenesis by suppressing bone morphogenetic protein and WNT canonical signaling. J Cell Biochem. 2012;113:449–456. doi: 10.1002/jcb.23365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, Canalis E. Notch signaling in skeletal health and disease. Eur J Endocrinol. 2013;168:R95–R103. doi: 10.1530/EJE-13-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng YA, Rahnama M, Wang S, Sosu-Sedzorme W, Verheyen EM. Drosophila Nemo antagonizes BMP signaling by phosphorylation of Mad and inhibition of its nuclear accumulation. Development. 2007;134:2061–2071. doi: 10.1242/dev.02853. [DOI] [PubMed] [Google Scholar]
- Zhang M, Xuan S, Bouxsein ML, von Stechow D, Akeno N, Faugere MC, Malluche H, Zhao G, Rosen CJ, Efstratiadis A, Clemens TL. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Chem. 2002;277:44005–44012. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]