Abstract
This review briefly addresses what has been learned about resistance durability in recent years, as well as the questions that still remain. Molecular analyses of major gene interactions have potential to contribute to both breeding for resistance and improved understanding of virulence impacts on pathogen fitness. Though the molecular basis of quantitative resistance is less clear substantial evidence has accumulated for the relative simplicity of inheritance. There is increasing evidence for specific interactions with quantitative resistance, though implications o this for durability are still unknown. Mechanisms by which resistance gene pyramids contribute to durability remain elusive, though ideas have been generated for identifying gene combinations that may be more durable. Though cultivar mixtures and related approaches have been used successfully, identifying the diseases and conditions that are most conducive to the use of diversity has been surprisingly difficult, and the selective influence of diversity on pathogen populations is complex. The importance of considering resistance durability in a landscape context has received increasing emphasis and is an important future area of research. Experimental systems are being developed to test resistance gene deployment strategies that previously could be addressed only with logic and observation. The value of molecular markers for identifying and pyramiding major genes is quite clear, but the successful use of quantitative trait loci (QTL) for marker-assisted selection of quantitative resistance will depend greatly on the degree to which the identified QTL are expressed in different genetic backgrounds. Transgenic approaches will likely provide opportunities for control of some recalcitrant pathogens, though issues of durability for transgenes are likely to be no different than other genes for resistance. The need for high quality phenotypic analysis and screening methodologies is a priority, and field-based studies are likely to remain of signal importance in the foreseeable future.
Keywords: durable resistance, host mixtures, landscape ecology, pathogen evolution, quantitative resistance, resistance gene pyramids
1. Introduction
A discussion of durable resistance must be considered in context of major global challenges. It has been predicted that a combination of changing diets and human population growth will result in an increased demand for agricultural production of 60-110% between the years 2005 and 2050 (Alexandratos, N. and J. Bruinsma, 2012; Tilman et al., 2011); increased demand for forest products could be even greater (WWF, 2012). Given the substantial losses caused by disease and pests globally (Oerke, 2006; Strange and Scott, 2005) and the increasing number of epidemic invasions resulting from globalization (Anderson et al., 2004; Crowl et al., 2008), meeting this demand will require an intense focus on disease and pest control. Further, these demands must be met while avoiding negative environmental impacts caused by current practices (Tilman et al., 2001) and in the face of significant global climate change (IPPC, 2007). Mean changes of temperature and precipitation can have positive, negative, or neutral impacts on specific diseases (Chakraborty, 2011; Garrett et al., 2006). Of greater concern may be the expected increase in climatic variability (IPPC, 2012), which could increase the number of diseases and pests of importance in a given locality, as well as the yearly fluctuations of their prevalence. Host plant resistance is generally the most favorable control method for environmental, economic, and social reasons. Thus, genes for resistance to diseases and pests can rightfully be considered one of the most important natural resources determining the survival of the human species (Mundt, 1994), while the evolutionary potential of plant pathogens to adapt to host resistance (McDonald and Linde, 2002) makes good stewardship essential to attain sustainable use of this precious resource.
The evolution of both organisms (Gould and Eldredge, 1977) and scientific thought (Kuhn, 1996) commonly experience periods of relative stasis punctuated by periods of rapid change. I suggest that the field of durable resistance had been in a period of relative stasis for some years, but recent information presented in this conference clearly suggests that the field is entering another period of significant advancement. This article will attempt to summarize what has been accomplished in this field of study and what remains to be done, with an emphasis on changes that have occurred since the last international conference on this topic held in 2000 (proceedings published in Vol. 124, Issue 2 of Euphytica). Throughout this short review, significant questions that remain to be answered will be listed as italicized “bullet points” in an attempt to frame future directions for the field, while recognizing that a summary by any individual is bound to contain gaps and shortcomings. I will focus primarily on genetic aspects of durability, though it is important to recall that other disease control practices can influence both the epidemiological impact and the durability of host plant resistance (Mundt et al., 2002).
2. Changes in outlook and approach
The field of durable resistance was once dominated by rigid dogma and competing views of both mechanism of resistance (e.g., horizontal versus vertical resistance) and resistance deployment strategies (e.g., pyramids versus mixtures). The field has largely moved beyond this outlook to a more mature one recognizing that all approaches of attaining durability have a potential value in different circumstances and, in fact, may complement each other when used in concert. The field of durable resistance also has broadened substantially in terms of host/pathogen systems under study. For many years, the field of durable resistance was largely dominated by studies of rusts and powdery mildews of small grain crops and of potato late blight. Over time, the field has expanded to a diversity of annual and perennial crops, to natural ecosystems, and to a wide range of fungi, oomycetes, bacteria, viruses, and nematodes (Zadoks, 2002), a healthy process that continued in the 2012 conference. This review will be dominated by plant pathogens, my area of familiarity, but it is very positive that the conference itself also included contributions regarding durability of host plant resistance to insect pests. Finally, the field of durable resistance has broadened in scope by more widely incorporating the information from the fields of molecular genetics/genomics, ecology, and population genetics.
3. Molecular mechanisms of host/pathogen interactions
A clear advance since 2000 has been exciting progress in understanding the elusive nature of gene-for-gene interactions in plant host/pathogen systems. Despite elucidation of the basic genetic system several decades ago (Ellingboe, 1976; Flor, 1971) and cloning of the first avirulence in the 1980s (Staskawicz et al., 1984), it had remained unclear why dominant genes conditioning avirulence would exist in pathogen populations. More recently, computational genomics has demonstrated that avirulence genes also serve as effectors of pathogen virulence, with substantial redundancy among effector genes (Cunnac et al., 2001; Jones and Dangl, 2006). These advances could have substantial relevance to understanding the dynamics of pathogens populations in response to resistance deployment (Michelmore et al., 2013). As one of many examples, it has often been observed that virulent races rarely revert to their initial frequencies after the end of a boom-and-bust cycle and the removal of the corresponding resistance gene (e.g., Fig. 1), an observation of relevance to understanding the potential success of deployment strategies such as resistance gene rotation or pyramiding of previously defeated resistance genes (see Section 5). Though it has long been suggested that compensatory mutations are crucial to the evolution of strains of both virulent plant pathogens (Parlevliet, 1981) and antiobiotic human pathogens (Björkman et al., 2000), recent understanding of the molecular basis of virulence in plant pathogens suggests that the process could in fact be determined, at least in part, by a “reshuffling” of effectors with differing impacts on pathogen virulence. We have barely scratched the surface in terms of applying molecular mechanism to resistance durability, and a frontier in coming years is to answer the question:
How can our knowledge of molecular host/pathogen interactions help us to better understand and attain durability of resistance?
Fig. 1.
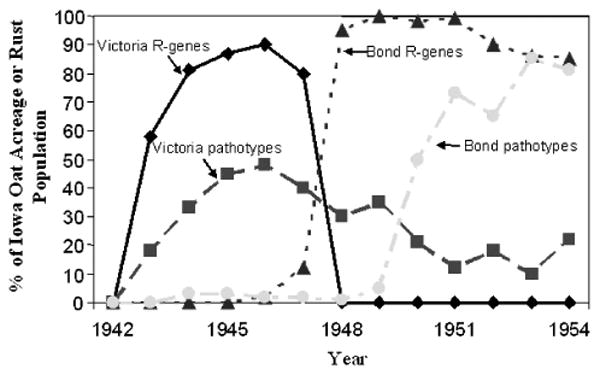
Example of a classic boom-and-bust cycle of major gene resistance to plant pathogens. Lines indicate the percentage of the Iowa oat area planted to cultivars possessing either the Victoria or the Bond major resistance and the percentage of the surveyed oat crown rust population virulent on cultivars carrying those resistance genes. Modified from McDonald (2004); used with permission. Original data from Browning and Frey (1969).
At this point in time, substantially less is known about the molecular basis of minor gene resistance. Whether quantitative resistance (QR) is fundamentally different from major gene resistance has been a point of debate for a long time (Nelson, 1978; Parlevliet and Zadoks, 1977; Vanderplank, 1982). More recent research suggests that multiple types of mechanisms potentially contribute to minor gene resistance (Poland et al., 2008), and the degree to which minor gene resistance conforms to these different mechanisms could have significant impact on durability of such resistance. For example, if minor genes function to impact morphological/developmental plant processes or influence basal host defense, QR would likely be more durable than if QR were instead due to weak major genes and thus subject to being overcome by lack-of-function mutations in the pathogen.
What is the mechanistic basis of quantitative resistance?
Is the mechanism underlying quantitative resistance relevant to its durability?
4. Use of quantitative resistance (QR)
4.1 Inheritance and selection
Substantial field experience suggests that QR often is much more available and easier to accumulate than originally expected, and simply purging the most susceptible lines each generation can provide substantial progress in accumulating QR (Parlevliet, 1989). Geiger and Heun (1989) reviewed the inheritance of QR from a biometric approach and concluded that the number of “effective factors” controlling QR ranges from 2 to 10, a range lower than had been predicted in earlier years. Similarly, a 1996 review of QR quantitative trait loci (QTL) concluded that the number of identified QTL associated with QR ranged from 2-11, with a median of 3.8 (Young, 1996). A more recent survey (Kover and Caicedo, 2001) included 85 QTL studies and found that the number of identified QTL ranged from 0 to 18, with a median of 4.2. A plethora of papers has been published subsequently with similar results, and Singh et al. (2008) recently suggested that 4-5 minor genes may be sufficient to keep wheat stem rust Ug99 “at negligible disease levels”, even under high disease pressure. The authors of the above reviews noted that estimates of QTL numbers are biased downward owing to small population sizes. Indeed, a resampling study showed that increasing sample size will result in identification of an increasing number of QTL with smaller effects (Vales et al., 2005). Nonetheless, it is clear that a relatively small number of genes can account for a large proportion of the QR trait. Researchers sometimes identify QR via components of resistance (latent period, infection efficiency, sporulation, etc.) (Parlevliet, 1979). These components are often highly correlated (Parlevliet, 1989), and evidence has been cited for pleitropic control (Parlevliet, 1986; Wang et al., 1994). More recently, development of near-isogenic barley lines containing different combinations of three QTL (Richardson et al., 2006) provided strong evidence for pleitropic control of latent period, infection efficiency, lesion size, and pustule density (a surrogate for sporulation) for stripe rust of barley (Fig. 2). If pleiotropic control is a general phenomenon, it would provide additional evidence for simplicity of the genetic control of QR and its use in breeding programs. On the other hand, pleiotropic control would reduce the number of genetic changes required for a pathogen to overcome such resistance.
Fig. 2.

Levels of four components of quantitative resistance regressed against number of quantitative trait loci (QTL) contained in eight different near-isogenic lines containing different combinations of three different QTL for resistance to barley stripe rust. Reproduced with permission from Richardson et al. (2006).
Despite the relative simplicity of inheritance of QR, there also appear to be an abundance of QR genes available, at least against some diseases. For example, Roswarne et al. (2013) recently cataloged 140 QTL for QR to stripe rust and assigned them to 49 different genomic regions of wheat. Relative simplicity and abundance of minor QR allow for rapid accumulation while also making progress for important agronomic traits, such as yield (Parlevliet, 1989). Though a tremendous amount has been learned, there still is a limited number of pathosystems for which QR has been thoroughly studied.
Is minor gene resistance available against all pathogens?
How many genes are sufficient?
Are components of resistance always pleiotropically controlled?
4.2 Mechanisms of durability
Extensive practical experience clearly demonstrates that quantitative resistance is more durable than major gene resistance on average (Parlevliet, 1989). The durability of QR is commonly assumed to be due to the number of genes controlling the trait, though arguments to the contrary have been made (Vanderplank, 1978, 1982). Certainly there are other mechanisms other than gene number that could contribute to durability of resistance of QR. For example, selection coefficients against individual genes controlling QR will be smaller than those against major gene resistance. The degree of host genotype x pathogen genotype specificity may be less for minor gene than major gene resistance (see below). Host x pathogen × genotype interactions (Kulkarni and Chopra, 1982) could also play an important role for genes of minor effect.
What determines the durability of QR?
4.3 Specificity of QR and potential ersosion
There have long been concerns that QR may select for pathogen virulence and/or aggressiveness. Parlevliet and Zadoks (1977) demonstrated through modeling that gene-for-gene interactions may occur in QR, but be difficult or impossible to detect with traditional analysis-of-variance approaches. Quantitative host genotype × pathogen genotype interactions have sometimes been detected experimentally (e.g., Latin et al., 1981; Lehman and Shaner, 1996; Parlevliet, 1977). Interactions with environment, however, may sometimes cause these interactions to be irrelevant to pathogen adaptation (Kulkarni and Chopra, 1982). For example, Leonards-Schippers et al. (1994) identified potato QTL that interacted with two races of P. infestans, though the results were not repeatable among trials.
Adaptation of pathogen populations to QR can be demonstrated in greenhouse and growth chamber evaluations, provided that the original source is a field-collected population and, hence, more heterogeneous or if sexual crosses have been made (Ahmed et al., 1995; Ahmed et al., 1996; Caten, 1974; Clifford and Clothier, 1974; Jeffrey et al., 1962; Kolmer and Leonard, 1986; Lehman and Shaner, 1996, 1997; Leonard, 1969). Such diverse populations would enable selection for more virulent types within the pathogen population. In some cases (Caten, 1974; Clifford and Clothier, 1974; Jeffrey et al., 1962), isolates have been found to be more virulent to the cultivar they were isolated from in the field, but subsequent cycling of single isolates failed to show increases in virulence. Such results could be expected, as large field populations may harbor significant variation, while individual isolates are likely to be invariant, or at least highly uniform, for pathogenicity. More recently, a serial passage experiment in the field demonstrated pathogen adaptation to QR for powdery mildew of barley (Villaréal and Lannou, 2000) and a serial passage experiment under controlled conditions resulted in complete erosion of the quantitative resistance to PVY in pepper (Montarry et al., 2012). Recent studies have also demonstrated pathogen adaptation to QR QTL under experimental conditions for apple scab (Caffier et al., this issue).
Pathogen adaptation to QR is more difficult to demonstrate in production situations. A wheat cultivar quantitatively resistant to Septoria tritici blotch eroded substantially over a 10-year period in the Willamette Valley of Oregon, as indicated by yearly comparison with a standard, susceptible cultivar (Mundt et al., 2002). This pathosystem represents the worst case scenario for pathogen adaptation, owing to yearly sexual recombination, unusually large effective population size, favorable environment, and lack of substantial immigration (Mundt et al., 1999). Thus, one might reasonably argue that these results are not relevant to the more common situation for plant pathogens (clonal reproduction, significant genetic bottlenecks, and variability of environment) and that the rate of pathogen adaptation would be very slow relative to the commercial life of a cultivar, at least for annual species. For potato late blight, arguments have been made for the stability of QR over both time (Vanderplank, 1978) and space (Forbes et al., 2005). However, an analysis of P. infestans populations from France versus Morocco (Andrivon et al., 2007) has raised questions about the potential of that pathogen to adapt to quantitative resistance. At the 2012 conference, Andrivon suggested that QR to potato late blight could be stable or unstable, depending upon the specific combinations of life history trade-offs, local adaptation, and gene flow.
In addition to specific adaptation discussed in the preceding paragraphs, QR may also select for increased pathogen aggressiveness. Here, I am using “aggressiveness” as originally defined by (Vanderplank 1968, 1978), i.e., genetic variation for pathogenicity among pathogen genotypes that does not interact differentially with host genotypes. Kolmer and Leonard (1986) found that both specific adaptation to host genotype as well as aggressiveness of Cochliobolus heterostrophus to maize genotypes could be increased through artificial selection over three sexual generations of the pathogen in the laboratory. There is evidence that selection pressure by both QR (Cowger and Mundt, 2002; Pink et al., 1992; Schouten and Beniers, 1997) and a protectant fungicide (Cowger and Mundt, 2002) selected for increased levels of pathogen aggressiveness.
As was recently discussed by Lannou (2012), interactions between the fields of plant pathology and evolutionary biology may be very helpful in understanding pathogen responses to QR.
Will QR select for increased pathogen adaptation and/or aggressiveness in the field?
Will QR erode against all pathogens?
How quickly will erosion occur?
Will QR erode to a practical level?
Are management strategies needed to increase the durability of QR?
5. Deployment strategies
Though deployment strategies are relevant to both major genes and to QR, they are more relevant to major gene resistance, given the long history for lack of durability of those genes when deployed singly.
5.1 One gene at a time
Although not often promoted by breeders or pathologists, there are cases in which deployment of a single major gene may make sense. Resistance is more likely to be durable in environments less conducive to the pathogen owing to smaller population sizes (Johnson, 1993). Thus, for environments in which a disease occurs at moderate or low severity, a single resistance gene may be adequate for a very long period of time, freeing resources to breed for durability to more serious/frequent diseases. A second example would be introductions of new pathogen species or new populations of a pathogen that are more highly aggressive. Though it is preferable to conserve resistance genes for use in some type of deployment scheme, it may be necessary to use single genes to keep an industry functional until such time that more durable strategies can be developed.
5.2 Gene rotation
Gene rotation involves deployment of an effective resistance gene, replacement with a different gene after appearance of a virulent race, and reuse of the original resistance in the future after the corresponding race has declined sufficiently in frequency (Crill, 1977). Though gene rotation schemes have been implemented against rice blast (Crill et al., 1981) and rice tungro disease (Manwan et al., 1985; Sama et al., 1991), success of these attempts is difficult to evaluate (Mundt, 1994). More generally, there are two substantial difficulties associated with gene rotation. The first is the very difficult logistical necessity of monitoring virulence accurately, having seed of replacement cultivars available in adequate quantities, and obtaining agreement among all farmers to simultaneously change cultivars. Perhaps more important, virulences may not decline to their original frequencies once the corresponding resistance gene is removed (e.g., Fig. 1).
Are there situations in which gene rotations will be an effective approach to durability?
5.3 Gene pyramids
There is broad agreement that combining genes for resistance (gene pyramids) is a useful approach for increasing durability, with many known successes. Perhaps the best success story, and certainly the best documented one, is for the control of stem and leaf rusts of wheat (Green and Campbell, 1979; McIntosh and Brown, 1997; Samborski, 1985; Schafer and Roelfs, 1985). For example, resistance gene combinations have kept wheat stem rust in check since the mid-1950s. The discovery of wheat stem rust race Ug99 in Uganda in the late 1990s now threatens these pyramids (Singh et al., 2011), and demonstrates that pyramids are not necessarily permanent. Nonetheless, controlling a globally-distributed pathogen of one the world's most important crops for over half of a century is an exceedingly impressive record of success befitting Johnson's (1981) definition of durability.
The mechanism(s) by which pyramids increase durability are not clear. The standard dogma has been that, if resistance genes have not previously been deployed singly or in less complex combinations, the probability of an asexual pathogen mutating to virulence against all resistance genes in the pyramid would be the product of the probabilities for each gene singly, thus making the probability of a virulent pathotype arising highly unlikely (e.g., Schafer and Roelfs, 1985; Wheeler and Diachun, 1983). It is very difficult to determine mechanism from available empirical data, however. There is not strong evidence for gene number per se as the dominant mechanism for the durability of pyramids (Johnson, 1981; Mundt, 1990, 1991; Vanderplank, 1978), and other mechanisms may be operative. For example, there may be a large fitness disadvantage associated with pathogen genotypes lacking avirulence against specific combinations of resistance genes (Green, 1975; Vanderplank, 1975). Johnson (1981) suggested that the slow rusting, adult plant resistance gene Sr2 may be strongly associated with durable resistance gene combinations against wheat stem rust, and combinations of Sr2 with other genes has played a crucial role in the international programs of CIMMYT (Rajaram et al., 1988; Singh et al., 2011), with stacking of major genes being a “supplementary strategy” (Rajaram et al., 1988). It would be interesting to test whether quantitative, adult plant resistance always contributes to durability of major genes in pyramids and, if so, whether it is the expression in the adult plant stage, the incomplete expression of resistance, both, or some other mechanism that contributes to this durability. Residual effects of defeated major genes (Pedersen and Leath, 1988) might also make a contribution to the durability of resistance gene combinations in some cases. Obviously, multiple mechanisms could operate simultaneously.
Elucidating the mechanisms by which pyramids provide durability is not merely an academic point, as mechanism may determine if one should focus on gene number per se or to finding favorable resistance gene combinations, or to combining adult plant genes with major genes, etc. For example, if the genetic probabilities hypothesis is not the main mechanism imparting durability, then a disappointing outcome could potentially result from a significant effort in building complex pyramids of major, race-specific genes.
What determines the durability of resistance gene pyramids?
Regardless of mechanism, it is reasonable to assume that some resistance gene combinations will be more durable than others, and methods have been proposed for identifying the most durable combinations. One such approach is through evaluation of fitness effects of individual genes (Fabre et al., 2009; Janzac et al., 2009; Khatabi et al., 2013; Leach et al., 2001; Vera Cruz 2000). If avirulence genes also function as virulence effectors (Cunnac et al., 2001; Jones and Dangl, 2006), then durability might reasonably be expected to be correlated with the fitness reduction associated with loss of the effector, and combining genes with large fitness losses would be expected to be more durable than combining genes with little or no fitness reduction. For clonal pathogens, another method is the lineage exclusion approach (Zeigler et al., 1994, 1995). It has been suggested that loss of an avirulence gene occurs more frequently in some clonal lineages of a pathogen than in others. Thus, it has been hypothesized that durability of pyramids can be increased by choosing combinations of resistance genes such that avirulence mutations occur infrequently against at least one resistance in each clonal lineage. Finally, I believe that the accumulated wisdom of plant breeders has often been underestimated. Genes that contribute to durable pyramids, such as Sr2 against wheat stem rust and Lr13 and Lr34 against wheat leaf rust, were uncovered through the experience of breeders working in the field, and many more such combinations are sure to be found.
Can we do a better job of predicting durable resistance gene combinations?
5.4 Mixtures
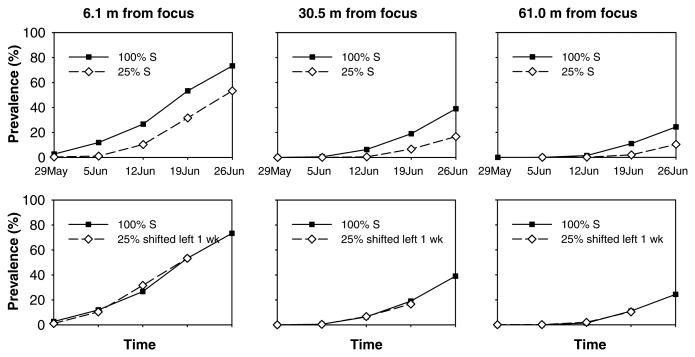
Resistance genes can also be mixed within a field, e.g., as multiline cultivars or cultivar mixtures. A significant degree of information has accumulated regarding the use of resistance gene mixtures for disease control, as well as a significant number of examples of successful implementation (Garrett and Mundt, 1999; Finckh et al, 2000; Mundt, 2002). As with most areas of durable resistance, much of the initial work was on rusts and powdery mildews of small grains. In more recent years, this work has expanded to a much wider diversity of pathosystems (Mundt, 2002). Despite attempts at prediction based on epidemiological principles (Garrett and Mundt, 1999), it has been very difficult to determine the types of pathosystem for which diversity provides a large degree of disease control. There are examples of both positive and negative effects of mixtures on disease control for most any type of pathosystem, e.g., small plant versus large plant, foliar versus soil-borne disease, specialized versus non-specialized pathogen, etc. (Mundt, 2002). Indeed, sometimes even long-held views on mixture mechanisms controlling rusts and powdery mildews can be brought into question. For example, it has long been assumed that rusts and mildews of cereals are controlled through dilution of inoculum (Chin and Wolfe, 1984a; Wolfe, 1985) and local reduction of intrinsic rates of disease increase (Browning and Frey, 1969; Mundt and Browning, 1985). However, recent studies suggest that perhaps diversity instead sometimes functions to reduce the number of new founders, with local infection rates being relatively unaffected (Fig. 3), a result consistent with the observation that mixtures are particularly vulnerable to the influence of outside inoculum (Mundt, 2002; Wolfe, 1985).
Fig. 3.
Temporal increase of stripe rust on a susceptible wheat genotype in monoculture or in a mixture of 25% susceptible/75% resistant plants at different distances from the initially inoculated focus. In bottom three panels, curves for the mixture are shifted left one week for comparison with the monoculture.
Under what conditions will mixtures provide a substantial epidemiological impact?
Given constant crop area, it is logical that a resistance gene will last longer in mixture than in pure stand simply owing to reduced exposure to the pathogen, and limited field observations support this expectation (Mundt, 1994). A more relevant question may be whether a given number of genes will last longer in mixtures than by sequential use in pure stand. In this regard, an issue has been whether use of mixtures will select for complex races (sometimes called “super-races”) that accumulate many or all resistance genes in a mixture or, alternatively, if a stable polymorphism will develop among races of varying complexity. Field observations and experiments to date do not suggest rapid dominance of host mixtures by highly fit, complex pathogen races, though available evidence is limited (Mundt, 2002). Mathematical models (summarized in Kiyosawa, 1989; Leonard and Czochor, 1980; Marshall, 1989; Mundt, 2002), generally suggest that complex races will eventually dominate the pathogen population, though the rate of evolution may be sufficiently slow to be manageable. Most of these models assume static costs associated with loss of avirulence genes to be the dominant mechanism countering evolution of pathogen complexity. There are many other potential mechanisms that could counter pathogen evolution towards complexity, however (Mundt, 2002). For example, quantitative adaptation of the pathogen to host genetic background in a three-way barley cultivar mixture apparently resulted in disruptive selection on the pathogen population and reduced fitness of the most complex race (Chin and Wolfe, 1984b).
How do pathogen populations evolve in diverse host populations?
5.5 Landscape approaches
Relatively little is known about the effects of landscape-level processes on epidemics or on pathogen evolution (Plantegenest et al., 2007; Real and Biek, 2007). A number of earlier studies were reported (Browning at al., 1969; Mundt and Browning, 1985), including suggestions for the regional deployment of resistance genes against rust and other pathogens that move on a continental scale during the course of a season (Browning et al., 1969; Knott, 1972; Reddy and Rao, 1979). Though such deployment strategies obviously cannot be studied experimentally, there have been situations in which regional deployment has been “tested” unintentionally, and suggest that such approaches may be highly effective (Browning et al., 1969), though not necessarily easy to implement. There has been a recent resurgence of interest in landscape issues influencing host plant resistance, including the effects of landscape structure and heterogeneity on epidemics (Fabre et al., 2012; Meentemeyer et al., 2011; Mundt et al., 2011; Skelsey et al., 2011). Recent studies with leaf rust of wheat (Papaïx et al., 2011; Lannou et al., this issue) have described an association between observed resistance levels of QR wheat cultivars and the cultivar composition on a national scale. Wingen et al. (2013) recently demonstrated via modeling how the nature of long-distance dispersal can influence the probability that virulent pathogen mutants will successfully colonize a resistant host in heterogeneous plant populations.
How do landscape factors influence population biology of plant pathogens and disease spread?
Will it be effective and feasible to manage landscapes for resistance durability?
6. Experimental Tests of Durability
Conclusions regarding durability strategies are often based solely on logic or observation, leaving many questions about causal relationships. Many important questions involve infrequent events that require long time-frames and/or large amounts of space to test using traditional approaches and, thus, experimental data have been very scarce. Fortunately, this situation is beginning to change. Experimental systems in both the laboratory and in plastic field tunnels demonstrated that the presence of minor QTL conditioning QR increased durability of a major gene for resistance to potato virus Y (PVY) in pepper (Palloix et al., 2009). The same result was obtained in a 5 yr, field-based system with Leptosphaeria maculans in Brassica napus (Brun et al., 2010; Delourme et al., this issue). In a combination field/greenhouse system, durability of a major gene for resistance to root knot nematode depended on host genetic background, perhaps owing to differences in quantitative resistance among potato genotypes (Fournet et al., 2013). Several other recent experimental systems to test different resistance gene deployment strategies were reported at the 2012 conference.
Can we further develop experimental tests of resistance durability?
7. Changing technologies
It is likely no accident that Nobel Laureate plant scientists Norman Borlaug and Barbara McClintock were known for spending countless hours in the field. There still is no substitute for spending the time to become intimately associated with the organism you are studying in its natural environment. In other words, if you want to become outstanding in your field, it can help to spend time standing in the fields. For those of us working with durable resistance, it is especially important to interact with farmers and other practitioners, as they observe their ecosystem on a daily basis and can provide perspectives that we cannot. One of many examples of this importance is a highly successful cultivar mixture program with against rice blast in China. Though scientists had the general idea of using host diversity for disease control, the specific spatial patterning of cultivars in the field that turned out to be a key to success of the program was suggested by a local farmer who was already using the practice (Zhu et al., 2000).
Can we maintain our “field wisdom” while adopting new technologies?
The importance of the field notwithstanding, molecular marker technologies have presented many new opportunities for achieving durable resistance. A clear application is in developing pyramids of major genes, as identifying these gene combinations is difficult or impossible based on phenotype alone. Scores, if not hundreds, of studies have been published identifying QTL for quantitative resistance to plant diseases, with the hope of using these QTL in marker-assisted selection (MAS). The “elephant in the room” regarding QTL is the degree to which they function in different genetic backgrounds; in absence of such transferability, markers are of limited use. At present, it appears that there will be many QR QTL that do not function in all genetic backgrounds, but this does not preclude using a combination of a limited number dependable markers as a pre-screen, and then using phenotypic analyses to chose the best progeny from among those. An important factor to consider in using MAS for QR is the potential danger for narrowing the genetic base for QR via wide-scale deployment of a limited number of QTL through use of MAS, though there may be ways to mitigate that danger (St. Clair, 2010).
How frequently will QR QTL function in multiple genetic backgrounds?
What is the most appropriate mix of markers versus phenotyping in selecting for QR?
Will deployment strategies be needed for QR QTL?
In the very near future, whole genome sequencing will likely make markers obsolete and greatly expand our options for studying disease resistance. As molecular technologies have expanded, there seems to be increasing agreement that the limitation to progress lies in improved phenotyping, especially for quantitative traits (Cobb, 2013; St. Clair, 2010). Along with this recognition have come calls for “next-generation phenotyping” via technologies such as field sensors, robots, and digital imaging (Cobb et al., 2013). Such technologies will certainly have their place and should be adopted when appropriate. However, there are many things that the human eye and brain can integrate while staring into a plant canopy that just simply will never be captured via these types of technology. It also is interesting that attempts to validate new marker technologies, for example, often result in field approaches that are faster, less expensive, and more accurate than the methodology they were attempting to validate. Thus, in addition to “next-generation phenotyping” at least an equal investment needs to be made in basic field biology, disease ecology, and experimental field design to improve “traditional” phenotyping methodologies.
What are the most effective ways to phenotype for disease resistance?
Transgenic technologies will likely provide opportunities for novel approaches to control of some recalcitrant pathogens, and to more quickly transfer currently available resistance genes within plant species (Dangl et al., 2013). We can hope for breakthroughs against plant diseases pathogens with outcomes that will be equivalent to the eradication of smallpox for humans.
More likely, however, transgenics will simply provide a greater diversity tools, but will be subject to the same issues of durability as traditional genetics.
How will transgenics contribute to resistance durability?
8. Closing comments
Kurt Leonard, my Ph.D. advisor and a significant contributor to the field of durable resistance, titled a recent summary of his career as “An Ideal Job” (Leonard, 2012). This is a wonderful reminder of how fortunate we are to study the dynamics of interacting organisms in a field that incorporates all aspects of biology - molecular biology, genetics, physiology, developmental biology, population biology, ecology, epidemiology environmental biology – and sometimes also a significant dose of the social and physical sciences as well. The excitement of this field fuels our energy, keeps us pursuing the next questions and, perhaps most importantly, provides the stimulating environment needed to attract the best and brightest young scientists into our field. However, another important reminder was delivered to me over 30 years ago while taking a course in “Tropical Plant Pathology” from H. David Thurston at Cornell University. During one class, Dr. Thurston was explaining how a very simple, low-cost project had an important impact on the well being of a rural community in the developing world. He then paused and said, “After all that's what plant pathology is all about – helping people”. This brings us back to the global issues mentioned in Introduction of this review, and a reminder that many people are depending on us to translate our science into workable solutions to achieve durability of resistance. We have a responsibility to do so.
Will we meet that challenge?
Highlights.
An expanded transcript of the Introductory Lecture given at the Plant Resistance Sustainability 2012 International Conference, Nice, France, 16-19 October 2012.
Achieving durable resistance is crucial to meeting global challenges of the next 50 years.
Accomplishments in the field are summarized.
Future directions are suggested.
Acknowledgments
Supported in part by NIH award R01GM96685 through the NSF/NIH Ecology and Evolution of Infectious Disease Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ahmed HU, Mundt CC, Coakley SM. Host-pathogen relationship of geographically diverse isolates of Septoria tritici and wheat cultivars. Plant Pathol. 1995;44:838–847. [Google Scholar]
- 2.Ahmed HU, Mundt CC, Hoffer ME, Coakley SM. Selective influence of wheat cultivars on pathogenicity of Mycosphaerella graminicola (anamorph Septoria tritici) Phytopathology. 1996;86:454–458. [Google Scholar]
- 3.Alexandratos N, Bruinsma J. ESA Working paper No 12-03. FAO; Rome: 2012. World Agriculture Towards 2030/2050: The 2012 Revision. [Google Scholar]
- 4.Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR, Daszak P. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol Evol. 2004;19:535–544. doi: 10.1016/j.tree.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Andrivon D, Pilet F, Montarry J, Hafidi M, Corbière R, Achbani EH, Pellé R, Ellissèche D. Adaptation of Phytophthora infestans to partial resistance in potato: Evidence from French and Moroccan populations. Phytopathology. 2007;97:338–343. doi: 10.1094/PHYTO-97-3-0338. [DOI] [PubMed] [Google Scholar]
- 6.Björkman J, Nagaev I, Berg OG, Hughes D, Andersson DI. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science. 2000;287:1479–1482. doi: 10.1126/science.287.5457.1479. [DOI] [PubMed] [Google Scholar]
- 7.Browning JA, Frey KJ. Multiline cultivars as a means of disease control. Annu Rev Phytopathol. 1969;14:355–382. [Google Scholar]
- 8.Browning JA, Simons MD, Frey KJ, Murphy HC. Regional deployment for conservation of oat crown rust resistance genes. Spec Rep—Iowa Agric Home Econ Exp Stn. 1969;64:49–56. [Google Scholar]
- 9.Brun H, Chèvre AM, Fitt BDL, Powers S, Besnard AL, Ermel M, Huteau V, Marquer B, Eber F, Renard M, Andrivon D. Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus. New Phytol. 2010;185:285–299. doi: 10.1111/j.1469-8137.2009.03049.x. [DOI] [PubMed] [Google Scholar]
- 10.Caten CE. Intra racial variation in Phytophthora infestans and adaptation to field resistance for potato late blight. Ann Appl Biol. 1974;77:259–270. [Google Scholar]
- 11.Chakraborty S, editor. Special issue: climate change and plant diseases. Plant Pathol. 2011;60:1–163. [Google Scholar]
- 12.Chin KM, Wolfe MS. The spread of Erysiphe graminis f.sp. hordei in mixtures of barley varieties. Plant Pathol. 1984a;33:89–100. [Google Scholar]
- 13.Chin KM, Wolfe MS. Selection on Erisyphe graminis in pure and mixed stands of barley. Plant Pathol. 1984b;33:535–546. [Google Scholar]
- 14.Clifford BC, Clothier RB. Physiologic specialization of Puccinia hordei on barley hosts with nonhypersensitive resistance. Trans Brit Mycol Soc. 1974;63:421–430. [Google Scholar]
- 15.Cobb JN, DeClerck G, Greenberg A, Clark R, McCouch S. Next-generation phenotyping: requirements and strategies for enhancing our understanding of genotype– phenotype relationships and its relevance to crop improvement. Theor Appl Genet. 2013;126:867–887. doi: 10.1007/s00122-013-2066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowger C, Mundt CC. Aggressiveness of Mycosphaerella graminicola isolates from susceptible and partially resistant wheat cultivars. Phytopathology. 2002;92:624–630. doi: 10.1094/PHYTO.2002.92.6.624. [DOI] [PubMed] [Google Scholar]
- 17.Crowl TA, Crist TO, Parmenter RR, Belovsky G, Lugo AE. The spread of invasive species and infectious disease as drivers of ecosystem change. Front Ecol Environ. 2008;6:238–246. [Google Scholar]
- 18.Crill P. An assessment of stabilizing selection in crop variety development. Annu Rev Phytopathol. 1977;15:185–202. [Google Scholar]
- 19.Crill P, Ham YS, Beachell HM. The rice blast disease in Korea and its control with race prediction and gene rotation. Korean J Breed. 1981;13:106–114. [Google Scholar]
- 20.Cunnac S, Chakravarthy S, Kvitko BH, Russell AB, Martin GB, Collmer A. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc Natl Acad Sci USA. 2001;108:2975–2980. doi: 10.1073/pnas.1013031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341:746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellingboe AH. Genetics of host-parasite interactions. In: Heitefuss R, Williams PH, editors. Physiological Plant Pathology Encyclopedia of Plant Physiology, New Series. Vol. 4. Springer; Berlin: 1976. pp. 761–778. [Google Scholar]
- 23.Fabre F, Bruchou C, Palloix A, Moury B. Key determinants of resistance durability to plant viruses: Insights from a model linking within- and between-host dynamics. Virus Res. 2009;141:140–149. doi: 10.1016/j.virusres.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 24.Fabre F, Rousseau E, Mailleret L, Moury B. Durable strategies to deploy plant resistance in agricultural landscapes. New Phytol. 2012;193:1064–1075. doi: 10.1111/j.1469-8137.2011.04019.x. [DOI] [PubMed] [Google Scholar]
- 25.Finckh MR, Gacek ES, Goyeau H, Lannou C, Merz U, Mundt CC, Munk L, Nadziak J, Newton AC, de Vallavieille-Pope C, Wolfe MS. Cereal variety and species mixtures in practice. Agronomie. 2000;20:813–837. [Google Scholar]
- 26.Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- 27.Forbes GA, Chacon MG, Kirk HG, Huarte MA, Van Damme M, Distel S, Mackay GR, Stewart HE, Lowe R, Duncan JM, Mayton HS, Fry WE, Andrivon D, Ellisseche D, Pelle R, Platt HW, MacKenzie G, Tarn TR, Colon LT, Budding DJ, Lozoya-Saldana H, Hernandez-Vilchis A, Capezio S. Stability of resistance to Phytophthora infestans in potato: an international evaluation. Plant Pathol. 2005;54:364–372. [Google Scholar]
- 28.Fournet S, Kerlan MC, Renault L, Dantec JP, Rouaux C, Montarry J. Selection of nematodes by resistant plants has implications for local adaptation and cross-virulence. Plant Pathol. 2013;62:184–193. [Google Scholar]
- 29.Garrett KA, Mundt CC. Epidemiology in mixed host populations. Phytopathology. 1999;89:984–990. doi: 10.1094/PHYTO.1999.89.11.984. [DOI] [PubMed] [Google Scholar]
- 30.Garrett KA, Dendy SP, Frank EE, Rouse MN, Travers SE. Climate change effects on plant disease: genomes to ecosystems. Annu Rev Phytopathol. 2006;44:489–509. doi: 10.1146/annurev.phyto.44.070505.143420. [DOI] [PubMed] [Google Scholar]
- 31.Geiger HH, Heun M. Genetics of quantitative resistance to fungal diseases. Annu Rev Phytopathol. 1989;27:317–341. [Google Scholar]
- 32.Gould SJ, Eldredge N. Punctuated equilibria: the tempo and mode of evolution reconsidered. Paleobiology. 1977;3:115–151. [Google Scholar]
- 33.Green GJ. Virulence changes in Puccinia graminis f. sp. tritici in Canada. Can J Bot. 1975;53:1377–1386. [Google Scholar]
- 34.Green GJ, Campbell AP. Wheat cultivars resistant to Puccinia graminis tritici in western Canada: Their development, performance, and economic value. Can J Plant Pathol. 1979;1:13–15. [Google Scholar]
- 35.Core Writing Team; Pachauri RK, Reisinger A, editors. IPCC (Intergovernmental Panel on Climate Change) Climate Change 2007: Synthesis Report, Contribution of Working Groups I, II and III to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. IPCC; Geneva: 2007. [Google Scholar]
- 36.IPCC (Intergovernmental Panel on Climate Change) Managing the Risks of Extreme Events and Disasters to Advance Climate Change Adaptation. In: Field CB, Barros V, Stocker TF, Qin D, Dokken DJ, Ebi KL, Mastrandrea MD, Mach KJ, Plattner GK, Allen SK, Tignor M, Midgley PM, editors. A Special Report of Working Groups I and II of the Intergovernmental Panel on Climate Change. Cambridge University Press; Cambridge and New York: 2012. pp. 3–21. Summary for policymakers. [Google Scholar]
- 37.Janzac B, Fabre F, Paloix A, Moury B. Constraints on evolution of virus avirulence factors predict the durability of corresponding plant resistances. Mol Plant Pathol. 2009;10:599–610. doi: 10.1111/j.1364-3703.2009.00554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeffrey SIB, Jinks JL, Grindle M. Intraracial variation in Phytophthora infestans and field resistance to potato late blight. Genetica. 1962;32:323–328. [Google Scholar]
- 39.Johnson R. Durable resistance: Definition of, genetic control, and attainment. Phytopathology. 1981;71:567–568. [Google Scholar]
- 40.Johnson R. Durability of disease resistance in crops: some closing remarks about the topic and the symposium. In: Jacobs T, Parlevliet JE, editors. Durability of Disease Resistance. Kluwer Academic Publishers; Dordrecht, Netherlands: 1993. pp. 283–300. [Google Scholar]
- 41.Jones JDJ, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 42.Khatabi B, Wen RH, Hajimorad MR. Fitness penalty in susceptible host is associated with virulence of soybean mosaic virus on Rsv1-genotype soybean: a consequence of perturbation of HC-Pro and not P3. Mol Plant Pathol. 2013 Jun 19; doi: 10.1111/mpp.12054. 2013. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiyosawa S. Breakdown of blast resistance in relation to general strategies of resistance gene deployment to prolong effectiveness of resistance in plants. In: Leonard KJ, Fry WE, editors. Plant Disease Epidemiology. Vol. 2. McGraw-Hill; New York: 1989. pp. 251–283. [Google Scholar]
- 44.Knott DR. Using race-specific resistance to manage the evolution of plant pathogens. J Environ Qual. 1972;1:227–231. [Google Scholar]
- 45.Kolmer JA, Leonard KJ. Genetic selection and adaptation of Cochliobolus heterostrophus to corn hosts with partial resistance. Phytopathology. 1986;76:774–777. [Google Scholar]
- 46.Kover PX, Caicedo AL. The genetic architecture of disease resistance in plants and the maintenance of recombination by parasites. Mol Ecol. 2001;10:1–16. doi: 10.1046/j.1365-294x.2001.01124.x. [DOI] [PubMed] [Google Scholar]
- 47.Kuhn TS. The Structure of Scientific Revolutions. third. University of Chicago Press; Chicago: 1996. [Google Scholar]
- 48.Kulkarni RN, Chopra VL. Environment as the cause of differential interaction between host cultivars and pathogen races. Phytopathology. 1982;72:1384–1386. [Google Scholar]
- 49.Lannou C. Variation and selection of quantitative traits in plant pathogens. Annu Rev Phytopathol. 2012;50:319–338. doi: 10.1146/annurev-phyto-081211-173031. 2012. [DOI] [PubMed] [Google Scholar]
- 50.Latin RX, MacKenzie DR, Cole HJ. The influence of host and pathogen genotypes on the apparent infection rates of potato late blight epidemics. Phytopathology. 1981;71:82–85. [Google Scholar]
- 51.Leach JE, Vera Cruz CM, Bai J, Leung H. Pathogen fitness penalty as a predictor of durability of disease resistance genes. Annu Rev Phytopathol. 2001;39:187–224. doi: 10.1146/annurev.phyto.39.1.187. [DOI] [PubMed] [Google Scholar]
- 52.Lehman JS, Shaner G. Genetic variation in latent period among isolates of Puccinia recondita f. sp. tritici on partially resistant wheat cultivars. Phytopathology. 1996;86:633–641. doi: 10.1094/PHYTO.1997.87.2.170. [DOI] [PubMed] [Google Scholar]
- 53.Lehman JS, Shaner G. Selection of populations of Puccinia recondita f. sp. tritici for shortened latent period on a partially resistant wheat cultivar. Phytopathology. 1997;87:170–176. doi: 10.1094/PHYTO.1997.87.2.170. [DOI] [PubMed] [Google Scholar]
- 54.Leonard KJ. Selection in heterogeneous populations of Puccinia graminis f. sp. avenae. Phytopathology. 1969;59:1851–1857. [Google Scholar]
- 55.Leonard KJ. An ideal job. Annu Rev Phytopathol. 2012;50:1–14. doi: 10.1146/annurev-phyto-081211-172957. [DOI] [PubMed] [Google Scholar]
- 56.Leonard KJ, Czochor RJ. Theory of genetic interactions among populations of plants and their pathogens. Annu Rev Phytopathol. 1980;18:237–258. [Google Scholar]
- 57.Leonards-Schippers C, Gieffers W, Pregl-Schafer R, Knapp SJ, Salamini F, Gebhardt C. Quantitative resistance to Phytophthora infestans in potato: a case study for QTL mapping in an allogamous plant species. Genetics. 1994;137:67–77. doi: 10.1093/genetics/137.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manwan I, Sama S, Rizvi SA. Use of varietal rotation in the management of rice tungro disease in Indonesia. Indonesian Res Dev J. 1985;7:43–48. [Google Scholar]
- 59.Marshall DR. Modeling the effects of multiline varieties on the population genetics of plant pathogens. In: Leonard KJ, Fry WE, editors. Plant Disease Epidemiology. Vol. 2. McGraw-Hill; New York: 1989. pp. 284–317. [Google Scholar]
- 60.McDonald BA. Population genetics of plant pathogens. The Plant Health Instructor. 2004 doi: 10.1094/PHI-A-2004-0524-01. [DOI] [Google Scholar]
- 61.McDonald BA, Linde C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol. 2002;40:349–379. doi: 10.1146/annurev.phyto.40.120501.101443. [DOI] [PubMed] [Google Scholar]
- 62.McIntosh RA, Brown GN. Anticipatory breeding for resistance to rust diseases in wheat. Annu Rev Phytopathol. 1997;35:311–326. doi: 10.1146/annurev.phyto.35.1.311. [DOI] [PubMed] [Google Scholar]
- 63.Meentemeyer RK, Cunniffe NJ, Cook AR, Filipe JAN, Hunter RD, Rizzo DM, Gilligan CA. Epidemiological modeling of invasion in heterogeneous landscapes: spread of sudden oak death in California (1990–2030) Ecosphere 2:art17. 2011 http://dx.doi.org/10.1890/ES10-00192.1.
- 64.Michelmore RW, Christopoulou M, Caldwell KS. Impacts of resistance gene genetics, function, and evolution on a durable future. Annu Rev Phytopathol. 2013;51:291–319. doi: 10.1146/annurev-phyto-082712-102334. [DOI] [PubMed] [Google Scholar]
- 65.Montarry J, Cartier E, Jacquemond M, Palloix A, Moury B. Virus adaptation to quantitative plant resistance: erosion or breakdown? J Evol Biol. 2012;25:2242–2252. doi: 10.1111/j.1420-9101.2012.02600.x. [DOI] [PubMed] [Google Scholar]
- 66.Mundt CC. Probability of mutation to multiple virulence and durability of resistance gene pyramids. Phytopathology. 1990;80:221–223. [Google Scholar]
- 67.Mundt CC. Probability of mutation to multiple virulence and durability of resistance gene pyramids: Further comments. Phytopathology. 1991;81:240–242. [Google Scholar]
- 68.Mundt CC. Techniques to manage pathogen co-evolution with host plants to prolong resistance. In: Teng PS, Heong KL, Moody K, editors. Rice Pest Science and Management. International Rice Research Institute; Los Baños: 1994. pp. 193–205. [Google Scholar]
- 69.Mundt CC. Use of multiline cultivars and cultivar mixtures for disease management. Annu Rev Phytopathol. 2002;40:381–410. doi: 10.1146/annurev.phyto.40.011402.113723. [DOI] [PubMed] [Google Scholar]
- 70.Mundt CC, Browning JA. Genetic diversity and cereal rust management. In: Roelfs AP, Bushnell WR, editors. The Cereal Rusts. Vol. 2. Academic Press; Orlando: 1985. pp. 527–560. [Google Scholar]
- 71.Mundt CC, Hoffer ME, Ahmed HU, Coakley SM, DiLeone JA, Cowger C. Population genetics and host resistance. In: Lucas JA, Bowyer P, Anderson HM, editors. Septoria on Cereals: A Study of Pathosystems. CAB International; Wallingford: 1999. pp. 115–130. [Google Scholar]
- 72.Mundt CC, Cowger C, Garrett KA. Relevance of integrated disease management to resistance durability. Euphytica. 2002;124:245–252. [Google Scholar]
- 73.Mundt CC, Sackett KE, Wallace LD. Landscape heterogeneity and disease spread: experimental approaches with a plant pathogen. Ecol Appl. 2011;21:321–328. doi: 10.1890/10-1004.1. [DOI] [PubMed] [Google Scholar]
- 74.Nelson RR. Genetics of horizontal resistance to plant diseases. Annu Rev Phytopathol. 1978;16:359–378. [Google Scholar]
- 75.Oerke EC. Crop losses to pests. J Agric Sci. 2006;144:31–43. [Google Scholar]
- 76.Palloix A, Ayme V, Moury B. Durability of plant major resistance genes to pathogens depends on the genetic background, experimental evidence and consequences for breeding strategies. New Phytol. 2009;183:190–199. doi: 10.1111/j.1469-8137.2009.02827.x. [DOI] [PubMed] [Google Scholar]
- 77.Papaïx J, Goyeau H, Du Cheyron P, Monod H, Lannou C. Influence of cultivated landscape composition on variety resistance: an assessment based on wheat leaf rust epidemics. New Phytol. 2011;191:1095–1107. doi: 10.1111/j.1469-8137.2011.03764.x. [DOI] [PubMed] [Google Scholar]
- 78.Parlevliet JE. Evidence of differential interaction in the polygenic Hordeum vulgare - Puccinia hordei relation during epidemic development. Phytopathology. 1977;67:776–778. [Google Scholar]
- 79.Parlevliet JE. Components of resistance that reduce the rate of epidemic development. Annu Rev Phytopathol. 1979;17:203–222. [Google Scholar]
- 80.Parlevliet JE. Stabilizing selection in crop pathosystems: An empty concept or a reality? Euphytica. 1981;30:259–269. [Google Scholar]
- 81.Parlevliet JE. Pleiotropic association of infection frequency and latent period of two barley cultivars partially resistant to barley leaf rust. Euphytica. 1986;35:267–272. [Google Scholar]
- 82.Parlevliet JE. Identification and evaluation of quantitative resistance. In: Leonard KJ, Fry WE, editors. Plant Disease Epidemiology. Vol. 2. McGraw-Hill; New York: 1989. pp. 215–248. [Google Scholar]
- 83.Parlevliet JE, Zadoks JC. The integrated concept of disease resistance; a new view including horizontal and vertical resistance in plants. Euphytica. 1977;26:5–21. [Google Scholar]
- 84.Pedersen WL, Leath S. Pyramiding major genes for resistance to maintain residual effects. Annu Rev Phytopathol. 1988;26:369–378. [Google Scholar]
- 85.Pink DAC, Lot H, Johnson R. Novel pathotypes of lettuce mosaic virus - Breakdown of a durable resistance? Euphytica. 1992;63:169–174. [Google Scholar]
- 86.Plantegenest M, Le May C, Fabre F. Landscape epidemiology of plant diseases. J R Soc Interface. 2007;4:963–972. doi: 10.1098/rsif.2007.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ. Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 2008;14:21–29. doi: 10.1016/j.tplants.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 88.Rajaram S, Singh RP, Torres E. Current CIMMYT approaches in breeding wheat for rust resistance. In: Simmonds NW, Rajaram S, editors. Breeding Strategies for Resistance to the Rusts of Wheat. CIMMYT; Mexico, D.F: 1988. pp. 101–108. [Google Scholar]
- 89.Real LA, Biek R. Spatial dynamics and genetics of infectious diseases on heterogeneous landscapes. J R Soc Interface. 2007;4:935–948. doi: 10.1098/rsif.2007.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reddy MSS, Rao MV. Resistance genes and their deployment for control of leaf rust of wheat. Indian J Genet Plant Breed. 1979;39:359–365. [Google Scholar]
- 91.Richardson KL, Vales MI, Kling JG, Mundt CC, Hayes PM. Pyramiding and dissecting disease resistance QTL to barley stripe rust. Theor Appl Genet. 2006;113:485–495. doi: 10.1007/s00122-006-0314-2. [DOI] [PubMed] [Google Scholar]
- 92.Rosewarne GM, Herrera-Foessel SA, Singh RP, Huerta-Espino J, Lan CX, He ZH. Quantitative trait loci of stripe rust resistance in wheat. Theor Appl Genet. 2013 doi: 10.1007/s00122-013-2159-9. Published online 17 August 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sama S, Hasanuddin A, Manwan L, Cabunagan RC, Hibino H. Integrated management of rice tungro disease in South Sulawesi, Indonesia. Crop Prot. 1991;10:34–40. [Google Scholar]
- 94.Samborski DJ. Wheat leaf rust. In: Roelfs AP, Bushnell WR, editors. The Cereal Rusts. Vol. 2. Academic Press; Orlando: 1985. pp. 39–59. [Google Scholar]
- 95.Schafer JF, Roelfs AP. Estimated relation between numbers of urediniospores of Puccinia graminis f. sp. tritici and rates of occurrence of virulence. Phytopathology. 1985;75:749–750. [Google Scholar]
- 96.Schouten HJ, Beniers JE. Durability of resistance to Globodera pallida I. Changes in pathogenicity, virulence, and aggressiveness during reproduction on partially resistant potato cultivars. Phytopathology. 1997;87:862–867. doi: 10.1094/PHYTO.1997.87.8.862. [DOI] [PubMed] [Google Scholar]
- 97.Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Njau P, Wanyera R, Herrera-Foessel SA, Ward RW. Will stem rust destroy the world's wheat crop? Adv Agron. 2008;98:271–309. [Google Scholar]
- 98.Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, Njau P, Herrera-Foessel S, Singh PK, Singh S, Govindan V. The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol. 2011;49:465–481. doi: 10.1146/annurev-phyto-072910-095423. [DOI] [PubMed] [Google Scholar]
- 99.Skelsey P, Rossing WAH, Kessel GJT, van der Werf W. Invasion of Phytophthora infestans at the landscape level: How do spatial scale and weather modulate the consequences of spatial heterogeneity in host resistance? Phytopathology. 2010;100:1146–1161. doi: 10.1094/PHYTO-06-09-0148. [DOI] [PubMed] [Google Scholar]
- 100.Staskawicz BJ, Dahlbeck D, Keen NT. Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race-specific incompatibility on Glycine max (L.) Merr Proc Natl Acad Sci USA. 1984;81:6024–6028. doi: 10.1073/pnas.81.19.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.St Clair DA. Quantitative disease resistance and quantitative resistance loci in breeding. Annu Rev Phytopathol. 2010;48:247–268. doi: 10.1146/annurev-phyto-080508-081904. [DOI] [PubMed] [Google Scholar]
- 102.Strange RN, Scott PR. Plant disease: a threat to global food security. Annu Rev Phytopathol. 2005;43:83–116. doi: 10.1146/annurev.phyto.43.113004.133839. [DOI] [PubMed] [Google Scholar]
- 103.Tilman D, Fargione J, Wolff B, D'Antonio Dobson A, Howarth R, Schindler D, Schlesinger WH, Simberloff D, Swackhamer D. Forecasting agriculturally driven global environmental change. Science. 2001;292:281–284. doi: 10.1126/science.1057544. [DOI] [PubMed] [Google Scholar]
- 104.Tilman D, Balzerb C, Hill J, Beforta BL. Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci USA. 2011;108:20260–20264. doi: 10.1073/pnas.1116437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vales MI, Capettini F, Chen X, Corey A, Hayes PM, Mather D, Mundt CC, Richardson K, Sandoval-Islas S, Schön CC, Utz HF. Effect of population size in the estimation of barley stripe rust QTL. Theor Appl Genet. 2005;111:1260–1270. doi: 10.1007/s00122-005-0043-y. [DOI] [PubMed] [Google Scholar]
- 106.Vanderplank JE. Disease Resistance in Plants. Academic Press; New York: 1968. [Google Scholar]
- 107.Vanderplank JE. Principles of Plant Infection. Academic Press; New York: 1975. [Google Scholar]
- 108.Vanderplank JE. Genetic and Molecular Basis of Plant Pathogenesis. Springer-Verlag; Berlin: 1978. [Google Scholar]
- 109.Vanderplank JE. Host Pathogen Interactions in Plant Disease. Academic Press; New York: 1982. [Google Scholar]
- 110.Vera Cruz CM, Bai J, Oña I, Leung H, Nelson RJ, Mew TW, Leach JE. Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. Proc Natl Acad Sci USA. 2000;97:13500–13505. doi: 10.1073/pnas.250271997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Villaréal LMMA, Lannou C. Selection for increased spore efficacy by host genetic background in a wheat powdery mildew population. Phytopathology. 2000;90:1300–1306. doi: 10.1094/PHYTO.2000.90.12.1300. [DOI] [PubMed] [Google Scholar]
- 112.Wang GL, Mackill DJ, Bonman JM, McCouch SR, Champoux MC, Nelson RJ. RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics. 1994;136:1421–1434. doi: 10.1093/genetics/136.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wheeler H, Diachun S. Mechanisms of pathogenesis. In: Kommedahl T, Williams PH, editors. Challenging Problems in Plant Health. American Phytopathological Society; St Paul: 1983. pp. 324–333. [Google Scholar]
- 114.Wingen LU, Shaw MW, Brown JKM. Long-distance dispersal and its influence on adaptation to host resistance in a heterogeneous landscape. Plant Pathol. 2013;62:9–20. [Google Scholar]
- 115.Wolfe MS. The current status and prospects of multiline cultivars and variety mixtures for disease resistance. Annu Rev Phytopathol. 1985;23:251–273. [Google Scholar]
- 116.WWF (World Wildlife Fund) Living Forests Report. World Wildlife Fund International, Gland; 2012. Forests and wood products. [Google Scholar]
- 117.Young ND. QTL mapping and quantitative disease resistance in plants. Annu Rev Phytopathol. 1996;34:479–501. doi: 10.1146/annurev.phyto.34.1.479. [DOI] [PubMed] [Google Scholar]
- 118.Zadoks JC. Epilogue: A summary with personal bias. Euphytica. 2002;124:259–264. [Google Scholar]
- 119.Zeigler RS, Tohme J, Nelson R, Levy M, Correa-Victoria FJ. Lineage-exclusion: a proposal for linking blast population analysis to resistance breeding. In: Zeigler RS, Leong SA, Teng P, editors. Rice Blast Disease. CAB International; Wallingford: 1994. pp. 267–292. [Google Scholar]
- 120.Zeigler RS, Cuoc LX, Scott RP, Bernardo MA, Chen DH, Valent B, Nelson RJ. The relationship between lineage and virulence in Pyricularia grisea in the Philippines. Phytopathology. 1995;85:443–451. [Google Scholar]
- 121.Zhu Y, Chen H, Fan J, Wang Y, Li Y, Chen J, Fan JX, Yang S, Hu L, Leung H, Mew TW, Teng PS, Wang Z, Mundt CC. Genetic diversity and disease control in rice. Nature. 2000;406:718–722. doi: 10.1038/35021046. [DOI] [PubMed] [Google Scholar]