A key question in evolutionary biology is how molecular variation relates to phenotypic diversity. Arabidopsis FLOWERING LOCUS C (FLC) plays a key role in controlling vernalization—the acceleration of flowering by prolonged cold. Li et al. identify five functionally distinct FLC haplotypes, defined by noncoding sequence variation, which vary in FLC expression level and silencing. Allelic heterogeneity at this single locus accounts for a large proportion of variation in Arabidopsis vernalization. This study advances our understanding of adaptation and provides a new paradigm for analysis of complex traits.
Keywords: allelic heterogeneity, FLOWERING LOCUS C, noncoding polymorphism, vernalization, adaptation
Abstract
Relating molecular variation to phenotypic diversity is a central goal in evolutionary biology. In Arabidopsis thaliana, FLOWERING LOCUS C (FLC) is a major determinant of variation in vernalization—the acceleration of flowering by prolonged cold. Here, through analysis of 1307 A. thaliana accessions, we identify five predominant FLC haplotypes defined by noncoding sequence variation. Genetic and transgenic experiments show that they are functionally distinct, varying in FLC expression level and rate of epigenetic silencing. Allelic heterogeneity at this single locus accounts for a large proportion of natural variation in vernalization that contributes to adaptation of A. thaliana.
Reproductive timing is central to plant adaptation through its major effect on fitness (Mitchell-Olds and Schmitt 2006). Many plants overwinter before flowering, thus aligning the transition to reproductive development with spring. In Arabidopsis thaliana, this process, called vernalization, involves expression and then subsequent epigenetic silencing of a floral repressor gene named FLOWERING LOCUS C (FLC) (Sheldon et al. 2000; Michaels 2009). A. thaliana accessions show extensive natural variation in vernalization (Nordborg and Bergelson 1999; Shindo et al. 2006; Alonso-Blanco and Mendez-Vigo 2014). Relatively rapid vernalization is thought to facilitate adaptation to habitats with high summer drought or other high mortality situations, whereas slow vernalization is thought to be an important adaptation to northern latitudes (Stinchcombe et al. 2004). In some cases, this variation has been mapped to the FLC locus itself (Coustham et al. 2012), but the extent to which FLC variation accounts for natural variation in vernalization response and the importance of this natural variation to adaptation remain important unanswered questions (Satake 2010). We exploited the extensive collection of genotyped A. thaliana accessions to characterize the genetic architecture of the FLC locus in worldwide accessions. Here, we describe the phenotypic variation conferred by the different FLC haplotypes, their geographical distribution, and the extent to which variation at FLC accounts for the phenotype. These data reveal that the extensive allelic heterogeneity at this single floral repressor gene can account for a major fraction of the natural variation in vernalization rate that underpins life history diversity in A. thaliana.
Results and Discussion
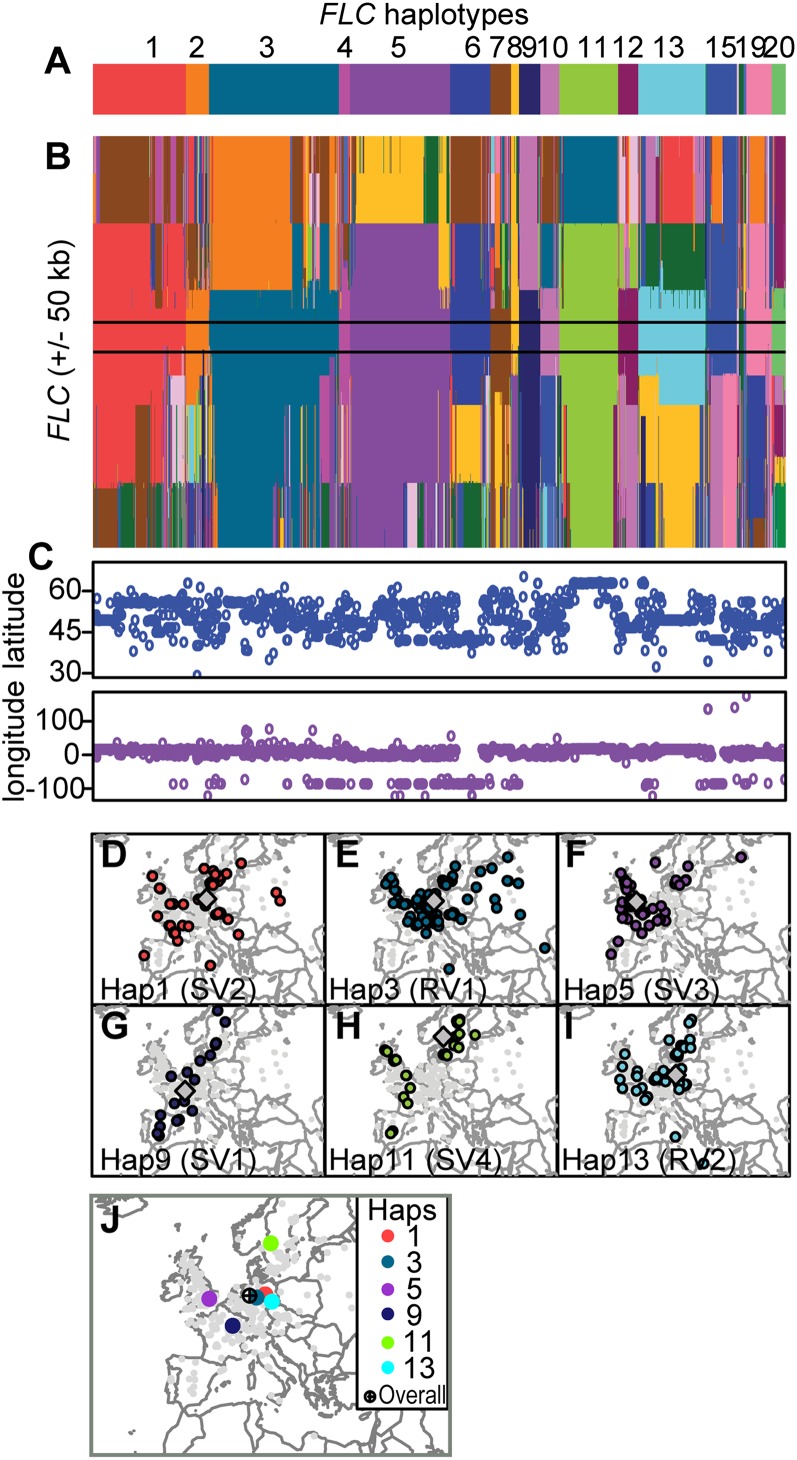
Single-nucleotide polymorphism (SNP) data from the Regional Mapping panel project (Horton et al. 2012) identified 20 FLC haplotypes across the 1307 accessions (Fig. 1A), with five major haplotypes predominating (Hap1, Hap3, Hap5, Hap11, and Hap13), numbered according to sequence similarity. A Northern Swedish accession, Lov-1, that we studied previously fell into Hap9, which showed intermediate frequency (Coustham et al. 2012). In the worldwide samples, the FLC region is characterized by high population structure and extensive haplotype sharing (Fig. 1B). Hap1, Hap3, and Hap13 have broad geographical distributions with centroids in central Europe, while Hap5 is predominantly in the United Kingdom, and Hap9 and Hap11 tend to be near coasts (Fig. 1C–J).
Figure 1.
Different FLC haplotypes in 1307 worldwide A. thaliana accessions. (A) Color-coded groups are indicated with numbers. The rare Hap14 and Hap16–18 are not marked. (B) Haplotype structure of the ±50-kb FLC region in 1307 global A. thaliana accessions. Each accession is represented in a column, and each SNP in the FLC ±50-kb window is represented in a row. At each SNP position, colors indicate the most likely haplotype membership for each accession as determined by fastPHASE analysis. Accessions are ordered by haplotype at the FLC locus itself, which is delineated by solid black horizontal lines. The prominent vertically oriented color blocks indicate that haplotype sharing among accessions often extends beyond the FLC locus itself. (C) Latitude and longitude of the collection site corresponding to the accessions above (Horton et al. 2012). (D–I) Geographical distribution of collection sites of A. thaliana accessions carrying the five predominant and one intermediate frequency FLC haplotypes. (J) Centroid distribution of the FLC haplotypes.
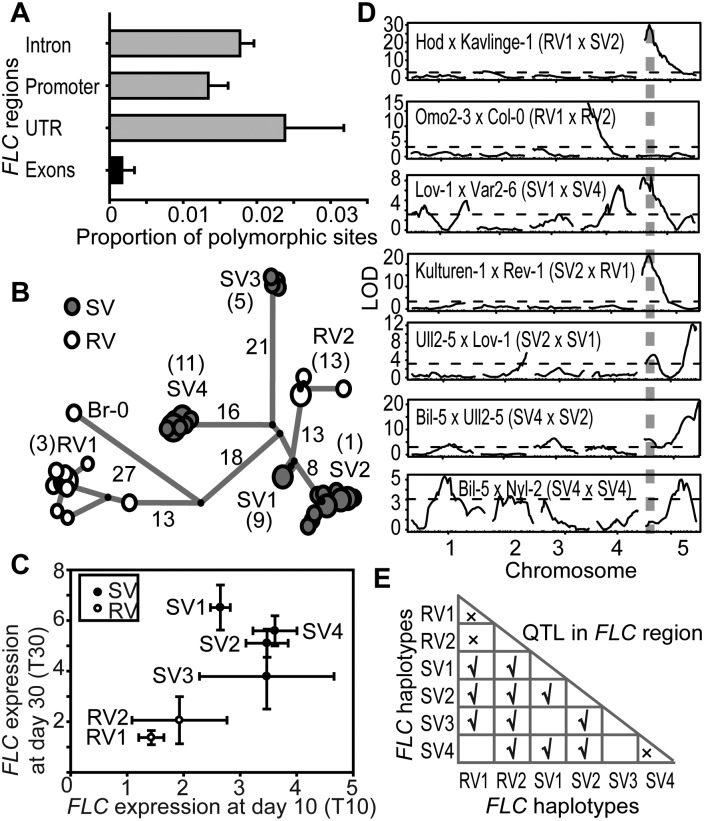
We selected 47 A. thaliana accessions that represented the major haplotypes and generated a high-quality FLC sequence for each accession (Supplemental Table S1). Remarkably, the predicted amino acid sequences from these FLC alleles were identical, with very low polymorphism in the coding sequences but considerable nucleotide sequence polymorphism in noncoding regions (Fig. 2A,B). The functional significance of this noncoding polymorphism was assessed through analysis of the vernalization response of the 47 accessions, using a partial vernalization treatment to enhance phenotypic diversity. The vernalization response—degree of reactivation of FLC expression after 4 wk of cold—varied among accessions and grouped with haplotype (Fig. 2). Two haplotypes (Hap3 and Hap13) showed relatively rapid vernalization (RV) response and so were named RV1 and RV2. Four others (Hap9, Hap1, Hap5, and Hap11) showed a relatively slow vernalization (SV) response and so were named SV1–4 (Fig. 2B,C; Supplemental Fig. S1A,B). The reactivation of FLC expression after transfer back to the warm differed for each haplotype and correlated with flowering time (Supplemental Fig. S1C,D). Additionally, 114 Swedish A. thaliana accessions (Supplemental Table S2), which had been genotyped using the 250K SNP array (Horton et al. 2012), were also phenotyped for their vernalization response. Similar groupings of haplotypes with phenotypes emerged from this analysis (Supplemental Fig. S2A,B). In general, therefore, accessions in the different FLC haplotype groups show distinct vernalization profiles that involve changes in silencing of FLC and altered flowering time.
Figure 2.
Functional differentiation of FLC haplotypes. (A) Distribution of genetic polymorphism (the mean [±SEM] proportion of polymorphic sites) across the 10-kb FLC region. (B) Haplotype analysis of 10-kb FLC genomic sequences from 47 A. thaliana accessions using FLUXUS network analysis. Circle size illustrates the frequency of the corresponding FLC haplotype. The number along the branch shows the number of nucleotide differences. Numbers in brackets indicate the corresponding haplotype numbers. (C) FLC expression of haplotypes containing FLC alleles with a slow vernalization (SV, black symbols) or rapid vernalization (RV, white symbols) response. Expression values shown are mean (±SEM, n = 2–13) from plants given 4 wk of cold followed by 10 d (T10) or 30 d (T30) of warm. Significant differences in vernalization response exist between haplotypes at 10 d (T10) and 30 d (T30) (Kruskal-Wallis test: H ≥ 18.42, d.f. = 5, P ≤ 0.002 in all tests). (D) QTL analysis on populations generated from crosses between accessions containing different FLC haplotypes. Dashed horizontal lines show the significance thresholds. The FLC region is indicated by the vertical dashed lines. (E) Summary of the presence of a QTL in the FLC region from D and Supplemental Table S3. The “✓” symbol indicates that there is a QTL in the FLC genomic region in the F2 population; the “×” indicates that there is no QTL.
The functional diversification of the different FLC haplotypes is supported by previous quantitative trait locus (QTL) analyses (Supplemental Table S3; Alonso-Blanco et al. 1998; Shindo et al. 2006; O’Neill et al. 2008; Li et al. 2010; Salome et al. 2011). Strong-effect QTL for vernalization response has been mapped over the FLC genomic region in populations generated from parents with different haplotypes: Col-0 (RV2) crossed to Lov-1 (SV1), Ull2–5 (SV2), or Var2–6 (SV4) (Shindo et al. 2006). To further test the functional diversity of FLC haplotypes, we undertook additional QTL analyses using F2 populations generated from crosses between accessions containing FLC alleles within and between haplotypes. In all crosses between the RV × SV accessions, a QTL was detected over the FLC genomic region, reinforcing the view that the haplotypes confer functionally distinct responses. No FLC QTL was detected in the cross between the RV1 × RV2 accessions due to their very similar vernalization phenotypes (Fig. 2D,E). This association between FLC alleles and flowering time was significantly reduced after 12 wk of vernalization compared with 4 wk, suggesting that the functional difference between haplotypes is cold exposure-dependent (Supplemental Fig. S2C,D).
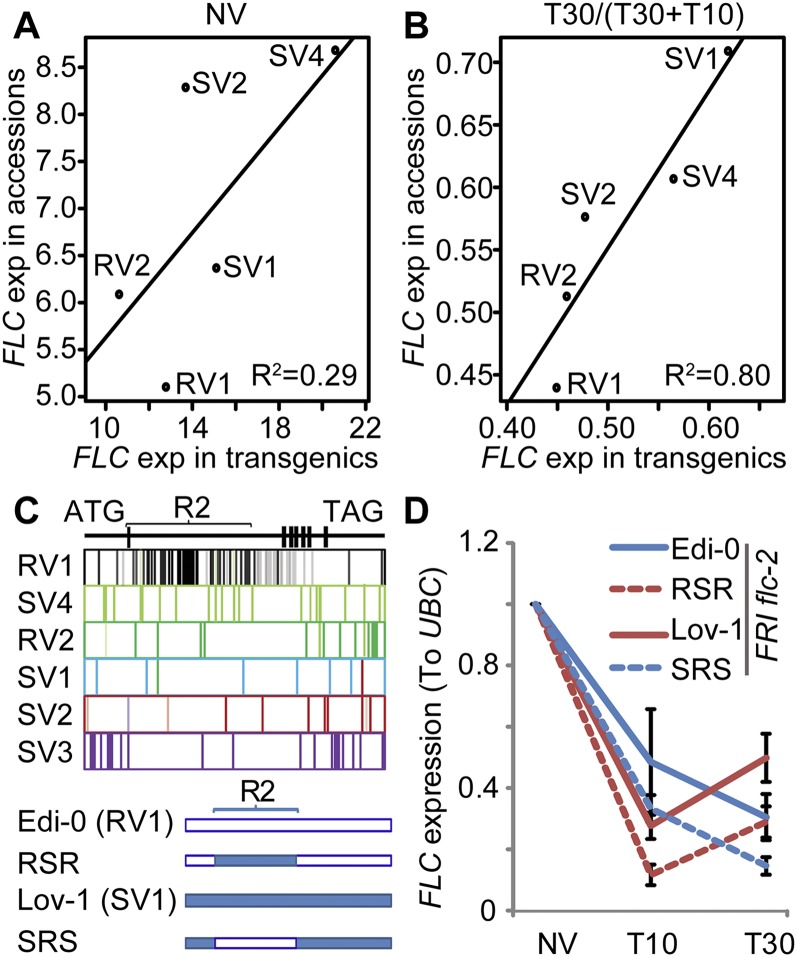
To conclusively demonstrate that FLC variation is the major contributor to the QTL, we generated transgenic lines differing only in their FLC allele. Twelve-kilobase FLC genomic clones from the A. thaliana accessions Edi-0, Col-0, Lov-1, Ull2-5, and Var2-6, representing haplotypes RV1, RV2, SV1, SV2, and SV4, respectively, were transformed individually into FRI flc-2 (Michaels and Amasino 2001). FLC expression in the transgenic lines was compared with that from the natural accessions both with and without vernalization. The differential response to vernalization shown by the FLC haplotypes was recapitulated in the transgenic plants (R2 = 0.80), whereas the correlation of prevernalization FLC expression between accessions and transgenic lines was weaker (R2 = 0.29) (Fig. 3A,B). The transgenic analysis therefore shows that the SNPs defining the FLC alleles confer the vernalization response characteristic of the parent A. thaliana accession, with trans-regulation playing a greater role in prevernalization FLC expression. These data enable us to conclude that noncoding cis variation within FLC is the major factor determining differential vernalization response in the characterized A. thaliana population.
Figure 3.
Transgenic analysis shows that noncoding FLC polymorphism plays a central role in vernalization response of natural Arabidopsis accessions. (A) Prevernalization FLC expression (nonvernalizaion [NV]) in transgenic lines correlates weakly with natural accessions. (B) Relative FLC expression [T30/(T30 + T10)] in transgenic lines correlates significantly with expression in natural accessions. R2 shows the explained amount of variation; the corresponding P-values are 0.201 and 0.025 in A and B. (C) Schematic illustration of FLC polymorphisms and the constructs of R2 chimeras. The SNPs defining the different FLC haplotypes are shown by the vertical bars. The bar transparency indicates the diversity of each SNP within each haplotype. The bars shared between haplotypes signify shared SNPs. The FLC gene structure is shown above: Filled black squares are exons, and horizontal lines are introns and untranslated regions. RSR and SRS chimeras show the exchange of R2 regions between Edi-0 and Lov-1 FLC alleles. (D) FLC expression analysis of transgenic FRI flc-2 plants containing chimeric FLC alleles. Plants were given either no cold (NV) or 4 wk of cold followed by 10 d (T10) or 30 d (T30) of warm. Values are means ±standard error from three biological repeats.
Analysis of chimeric FLC fusions previously showed that cis polymorphism quantitatively modulates a chromatin silencing mechanism underpinning the epigenetic regulation of FLC in one Northern Swedish accession (Coustham et al. 2012). To help define which of the many nucleotide differences between haplotypes determined variation in vernalization response, we first looked for natural recombination events across FLC genomic regions in accessions (Martin et al. 2010) and identified one that we called R2 (Supplemental Fig. S3A). Polymorphism in the R2 region showed the strongest association with the maintenance of FLC silencing after vernalization (Supplemental Fig. S3B). This region includes the nucleation region important for FLC silencing; the VRE (vernalization response element), defined as important for maintenance of silencing (Sheldon et al. 2002; Finnegan and Dennis 2007; Angel et al. 2011); and a large number of the SNPs defining the haplotype RV1 (Fig. 3C). We isolated and swapped the FLC R2 region reciprocally between Lov-1 (SV1) and Edi-0 (RV1) FLC alleles, which show slow and rapid silencing, respectively (Fig. 3C). The vernalization response of multiple transgenic lines was then compared. The SV1/RV1/SV1 (SRS) chimeric FLC shows stable FLC silencing after 4 wk of cold, which results in a rapid vernalization response compared with its SV1 FLC parent (Fig. 3D). Thus, the SRS vernalization response matches that of the RV1 accession rather than the SV1 accession. Similarly, the RV1/SV1/RV1 (RSR) chimera shows a vernalization response similar to the SV1 accession despite containing only a small part of the SV1 FLC allele (Fig. 3D). These results demonstrate that polymorphisms within R2 from RV1 and SV1 contribute to the differences in FLC silencing after 4 wk of vernalization, respectively. This region also contains the four single-nucleotide changes that cause the differential silencing of Lov-1 (SV1) and Col-0 (RV2) FLC alleles (Coustham et al. 2012). Remarkably, the Edi-0 (RV1) and Col-0 (RV2) FLC alleles do not share any polymorphisms that are not also found in other FLC haplotypes (Figs. 2B, 3C), demonstrating that the causative nucleotide changes that distinguish slow and rapid vernalization have evolved multiple times. The recombination event characterizing the RV1 haplotypes, the relative lack of variability of SNPs in the functional R2 region, and their relatively broad geographical distribution (Hap3/RV1) (Fig. 1E) suggest a relatively recent spread of RV1 accessions, potentially indicating a strong advantage of rapid vernalization in some locations.
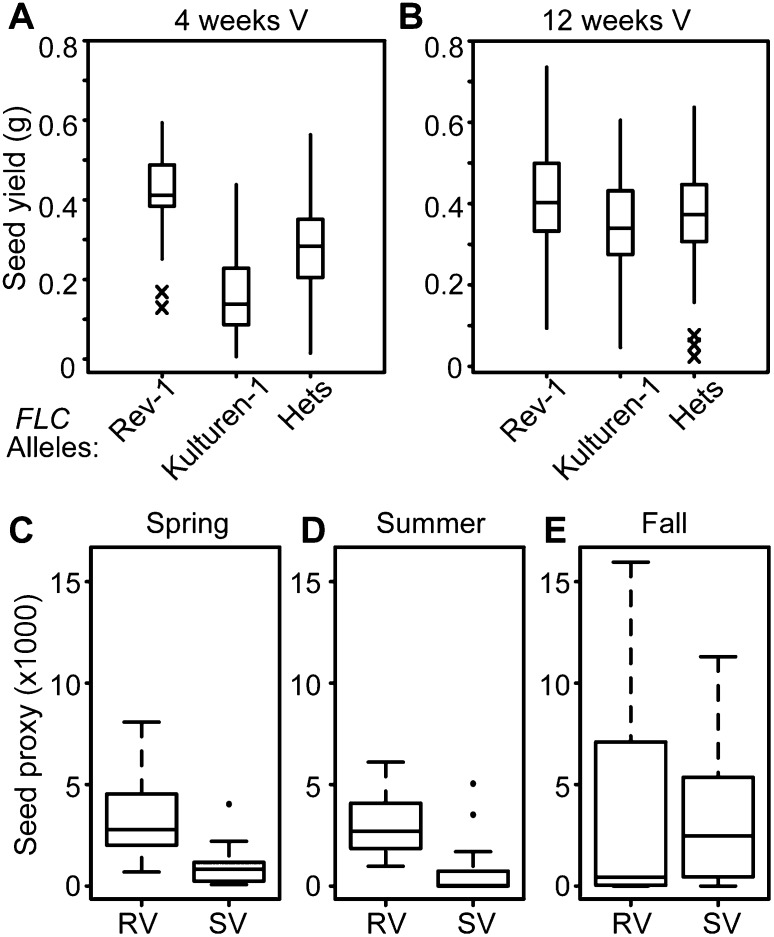
Consistent with this, faster vernalization has been predicted to confer a selective advantage to plants where avoidance of summer drought or other high risk is beneficial (Caicedo et al. 2004; Stinchcombe et al. 2004, 2005; Scarcelli et al. 2007; Satake 2010). We analyzed differences in fitness-related traits conferred by different FLC alleles in F2 populations (Fig. 4; Supplemental Fig. S4). After a relatively short vernalization (4 wk), the slow vernalization (Kulturen-1) FLC allele associated with lower seed yield than the rapid vernalization (Rev-1) FLC allele. After longer vernalization (12 wk), both alleles produced a similar seed yield (Fig. 4A,B; Supplemental Fig. S4A). Total seed weight represented an increased production of seed number, as no significant difference was found in the weight of each seed (Supplemental Fig. S4B). High fecundity has been associated with later flowering (Rédei 1962); however, for these rapid vernalization and slow vernalization alleles, an inverse relationship was found between flowering time and seed production after 4 wk of vernalization (Supplemental Fig. S4C). A similar relationship between vernalization response and fitness was also found for other rapid vernalization/slow vernalization allele combinations (Supplemental Fig.S4D). These data suggest that in some conditions, particularly after short cold periods, faster vernalization response can lead to enhanced fitness. To further explore the association of FLC haplotypes with fitness, we analyzed the field data of A. thaliana accessions grown under different natural seasonal conditions in Norwich (United Kingdom), where spring-, summer-, and fall-germinating cohorts are observed (Fournier-Level et al. 2011). Accessions with slow vernalization haplotypes produced less seed compared with rapid vernalization accessions in spring and summer sowings (which experienced only brief exposure to vernalizing temperatures), but fitness was similar between all groups in fall sowings (with exposure to extended cold such that all plants were fully vernalized) (Fig. 4C–E). These data encourage further extensive transplantation experiments that thoroughly investigate fitness differences between the haplotypes.
Figure 4.
Comparison of total seed production between plants with different vernalization responses. (A,B) Comparison of seed yield from F2 plants homozygous or heterozygous for the different FLC alleles after 4 wk (A; ANOVA, R2 = 0.41, F2227 = 80.43, P < 0.001) and 12 wk (B; ANOVA, R2 = 0.028, F2276 = 4.97, P = 0.008) of vernalization. Crosses indicate statistical outliers that fall more than three standard errors away from the mean. (C–E) Seed proxy values as a measure of fitness of field-grown accessions. The data set included only accessions with active FRIGIDA alleles (16 accessions from haplotypes RV1 and RV2 and 23 accessions from haplotype SV1-4). FLC haplotypes with rapid and slower vernalization response phenotypes were analyzed. Differences in yield were measured in field experiments in Norwich as in Fournier-Level et al. (2011). P-values are calculated from population structure-corrected ANOVA (see the Materials and Methods) (C; P = 0.0023), summer sowing (D; P = 0.00017), and fall sowing (E; P = 0.99).
The emerging picture is that functional differences in vernalization have evolved predominantly through cis polymorphism at FLC. Multiple alleles have arisen and been maintained in the A. thaliana population due to their contributions to life history diversity. Use of FLC genomic sequence from a close relative, Arabidopsis lyrata, did not help define which of the haplotypes are ancestral because of the high sequence divergence and large number of indels in the functionally important intronic regions. However, rapid vernalization might have arisen through selection pressure on the length of the plant life cycle due to high mortality from drought (Hoffmann 2005; Satake 2010), herbivory (Parachnowitsch and Lajeunesse 2012), pathogens (Kemen et al. 2011), agriculture, or human disturbance. The broader climate envelope of A. thaliana compared with its close relatives, which extends beyond the cool temperate regions into more Mediterranean climates, argues that faster vernalization could have increased the A. thaliana range (Hoffmann 2005). This and related studies should provide important knowledge needed to assess plant response to climate change.
Materials and methods
Plant materials and growth condition
Transgenic lines were obtained as described previously (Coustham et al. 2012). Plants were vernalized for 4 or 12 wk at 4°C and harvested for FLC expression analysis 10 d (T10) and 30 d (T30) after transfer from cold, along with nonvernalized plants (NV). Flowering time was assayed as days after bolting when stems reached 3 cm in height.
Plant fitness estimation
Controlled environment conditions were as follows: Plants were vernalized for 4 or 12 wk at 4°C under short-day conditions (8 h light) and moved to controlled environment conditions at 22°C under a long-day photoperiod (16 h light). Soil was from The Scotts Company Ltd., Sphagnum Moss peat (pH 5.5–6.0), with fertilizer added (milligram/liter): N150, P200, and K200. Twenty-five percent grit was added before use. Pot size was 5 × 5 cm. Plants were grown individually in each pot. In the greenhouse, total seed was collected from each plant after siliques had matured and dried naturally and was weighed.
In the field, plants of 39 accessions were grown in common gardens at the John Innes Center in spring, summer, and fall plantings as described previously (Fournier-Level et al. 2011). Fitness was estimated as the product of silique number at senescence and length of a representative silique (a proxy for seed number) for each individual (Fournier-Level et al. 2011).
FLC haplotype analysis using FLUXUS network analysis
FLC genome sequences were aligned using ClustalW with manual corrections in Mega5 (Tamura et al. 2011) and analyzed in FLUXUS network software using median joining setting (Bandelt et al. 1999).
FLC expression analysis
FLC expression data were obtained as described previously (Box et al. 2011). We generated 50 independent transgenic lines for each construct using the Agrobacterium tumefaciens floral dip technique. To overcome the variability in expression between transgenic lines, we pooled a random 10 seedlings from each of these 50 lines into a single sample. RNA was extracted from this pool and analyzed for FLC expression using quantitative RT–PCR as described in Box et al. (2011). For each time point, three independent pools of the 50 transgenic line pools were assayed.
Box plot and statistical analysis
Box plot and statistical analysis of flowering time and seed production were carried out in the GENESTAT package with the default setting (Ripatti et al. 2009).
Genotyping and QTL analysis of F2 populations
F2 populations were genotyped using a 384-SNP Illumina GoldenGate genotyping assay (http://www.illumina.com/technology/goldengate_genotyping_assay.ilmn). Genome Studio software (http://www.illumina.com/software/genomestudio_software.ilmn) and custom-made scripts in R software (http://www.R-project.org) were used for SNP calling.
Quantitative trait loci mapping was performed on raw phenotypic values without any transformation. QTL analyses were carried out using the default parameters of the scanone function implemented in the R/qtl package (Broman et al. 2003). The LOD significance threshold was determined by permutation test (1000 permutations, significance level of 5%) for each F2 population.
M&M test of the correlation between FLC polymorphism and gene expression
A general linear model (GLM) with T30/(T30 + T10) as response variable and SNP as the predictor was analyzed for each SNP separately. The F coefficient (adjusted MS/error variance) was calculated and plotted against the SNP position. The strength of the association of each SNP and the response variable was expressed by calculating the effect size estimator η2. This statistic expresses the ratio of variance explained in the dependent variable by a SNP. This analysis shows that SNPs in the R2 region (Supplemental Fig. S5B) explain ∼22.1% of the variation inT30/(T10 +T30) (η2 = 0.221), which is considered a large effect size.
fastPHASE haplotype analysis
SNPs in a window ±50 kb around FLC were extracted from a preimputation version of the Regional Mapping Project SNP panel described in Horton et al. (2012). These SNPs were used as the input into fastPHASE version 1.4.0 (Scheet and Stephens 2006), which was run using the default settings, with the exception of invoking the −Pzp option to output raw likelihood values. For each SNP in the analysis, the cluster membership with the highest likelihood was designated as the haplotype for that SNP.
Population structure-corrected ANOVAs for seed proxy analysis in accessions
A standard approach for accounting for such genomic background effects is linear mixed models (Vilhjálmsson and Nordborg 2013). We assessed the significance of phenotypic differences between haplotypes by carrying out an ANOVA on the residuals after subtracting the best linear unbiased predictors (BLUPs) of the effect explained by the genomic background. The BLUPs were estimated using EMMAX (Kang et al. 2010) with a kinship matrix estimated from the Regional Mapping Project data (Horton et al. 2012).
Acknowledgments
We thank all members of the Dean and Howard groups for discussions. This research was supported by UK Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/I007857/1, Institute Strategic Programme grant BB/J004588/1, European Research Council Advanced Investigator grant 233039 ENVGENE, and National Science Foundation Frontiers in Integrative Biological Research grant NSF EF-0425759.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.245993.114.
Freely available online through the Genes & Development Open Access option.
References
- Alonso-Blanco C, Mendez-Vigo B. 2014. Genetic architecture of naturally occurring quantitative traits in plants: an updated synthesis. Curr Opin Plant Biol 18C: 37–43 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, El-Assal SE, Coupland G, Koornneef M. 1998. Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149: 749–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel A, Song J, Dean C, Howard M. 2011. A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476: 105–108 [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48 [DOI] [PubMed] [Google Scholar]
- Box MS, Coustham V, Dean C, Mylne JS. 2011. Protocol: a simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Caicedo A, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD. 2004. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc Natl Acad Sci 101: 15670–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J, Dean C. 2012. Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science 337: 584–587 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. 2007. Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr Biol 17: 1978–1983 [DOI] [PubMed] [Google Scholar]
- Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM. 2011. A map of local adaptation in Arabidopsis thaliana. Science 334: 86–89 [DOI] [PubMed] [Google Scholar]
- Hoffmann MH 2005. Evolution of the realized climatic niche in the genus Arabidopsis (Brassicaceae). Evolution 59: 1425–1436 [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjalmsson BJ, et al. 2012. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet 44: 212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. 2010. Variance component model to account for sample structure in genome-wide association studies. Nat Genet 42: 348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemen E, Gardiner A, Schultz-Larsen T, Kemen AC, Balmuth AL, Robert-Seilaniantz A, Bailey K, Holub E, Studholme DJ, Maclean D, et al. 2011. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol 9: e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO. 2010. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci 107: 21199–21204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26: 2462–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD 2009. Flowering time regulation produces much fruit. Curr Opin Plant Biol 12: 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. 2001. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J. 2006. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441: 947–952 [DOI] [PubMed] [Google Scholar]
- Nordborg M, Bergelson J. 1999. The effect of seed and rosette cold treatment on germination and flowering time in some Arabidopsis thaliana (Brassicaceae) ecotypes. Am J Bot 86: 470–475 [PubMed] [Google Scholar]
- O’Neill CM, Morgan C, Kirby J, Tschoep H, Deng PX, Brennan M, Rosas U, Fraser F, Hall C, Gill S, et al. 2008. Six new recombinant inbred populations for the study of quantitative traits in Arabidopsis thaliana. Theor Appl Genet 116: 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parachnowitsch AL, Lajeunesse MJ. 2012. Adapting with the enemy: local adaptation in plant-herbivore interactions. New Phytol 193: 294–296 [DOI] [PubMed] [Google Scholar]
- Rédei GP 1962. Supervital mutants of Arabidopsis. Genetics 47: 443–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripatti S, Becker T, Bickeboller H, Dominicus A, Fischer C, Humphreys K, Jonasdottir G, Moreau Y, Olsson M, Ploner A, et al. 2009. GENESTAT: an information portal for design and analysis of genetic association studies. Eur J Hum Genet 17: 533–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salome PA, Bomblies K, Laitinen RA, Yant L, Mott R, Weigel D. 2011. Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics 188: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake A 2010. Diversity of plant life cycles is generated by dynamic epigenetic regulation in response to vernalization. J Theor Biol 266: 595–605 [DOI] [PubMed] [Google Scholar]
- Scarcelli N, Cheverud JM, Schaal BA, Kover PX. 2007. Antagonistic pleiotropic effects reduce the potential adaptive value of the FRIGIDA locus. Proc Natl Acad Sci 104: 16986–16991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P, Stephens M. 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 78: 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. 2000. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci 97: 3753–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Conn AB, Dennis ES, Peacock WJ. 2002. Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14: 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Lister C, Crevillen P, Nordborg M, Dean C. 2006. Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev 20: 3079–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, Purugganan MD, Schmitt J. 2004. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc Natl Acad Sci 101: 4712–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Caicedo AL, Hopkins R, Mays C, Boyd EW, Purugganan MD, Schmitt J. 2005. Vernalization sensitivity in Arabidopsis thaliana (Brassicaceae): the effects of latitude and FLC variation. Am J Bot 92: 1701–1707 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhjálmsson BJ, Nordborg M. 2013. The nature of confounding in genome-wide association studies. Nat Rev Genet 14: 1–2 [DOI] [PubMed] [Google Scholar]