Regeneration of injured adult skeletal muscle involves fusion of activated satellite cells to form new myofibers. Millay et al. find that the muscle-specific membrane protein myomaker is induced in satellite cells during acute and chronic muscle regeneration. Deletion of myomaker in adult satellite cells completely abolishes muscle regeneration, resulting in severe muscle destruction after injury. This study identifies myomaker as the only muscle-specific protein known to be absolutely essential for fusion of embryonic and adult myoblasts.
Keywords: myomaker, fusion, muscle regeneration, myogenesis, E-box elements
Abstract
Regeneration of injured adult skeletal muscle involves fusion of activated satellite cells to form new myofibers. Myomaker is a muscle-specific membrane protein required for fusion of embryonic myoblasts, but its potential involvement in adult muscle regeneration has not been explored. We show that myogenic basic helix–loop–helix (bHLH) transcription factors induce myomaker expression in satellite cells during acute and chronic muscle regeneration. Moreover, genetic deletion of myomaker in adult satellite cells completely abolishes muscle regeneration, resulting in severe muscle destruction after injury. Myomaker is the only muscle-specific protein known to be absolutely essential for fusion of embryonic and adult myoblasts.
During embryonic muscle development, myoblasts first proliferate to generate the appropriate number of muscle precursors and then differentiate and fuse to form multinucleated muscle fibers (Buckingham 2006). The muscle developmental process is recapitulated during adult muscle regeneration due to the presence of satellite cells, the skeletal muscle stem cells. After muscle injury, quiescent satellite cells proliferate, differentiate, and fuse to completely restore muscle architecture and function (Yin et al. 2013). Satellite cells reside within the basal lamina adjacent to mature muscle fibers but lack the ability to fuse until muscle becomes injured, at which time they become fusogenic and generate new myofibers. How satellite cells acquire fusogenic potential and the mechanism by which they selectively fuse to injured myofibers are unknown. It has also not been determined whether fusion during muscle development and regeneration involves common molecular pathways.
Numerous proteins have been implicated in myoblast fusion during mouse embryogenesis and adult regeneration; however, few have been investigated through genetic loss of function during both periods of muscle cell fusion, and none have been demonstrated to be absolutely essential for fusion at all stages (Hindi et al. 2013). Embryonic loss of proteins that regulate cytoskeletal dynamics, such as Rac1, cdc42, and N-WASP, result in early postnatal lethality due to a reduction of myoblast fusion (Vasyutina et al. 2009; Gruenbaum-Cohen et al. 2012). However, their roles in regeneration have not been explored. Similarly, many proteins have been implicated in fusion during regeneration but have not been investigated or have no obvious role in embryonic fusion, such as the tetraspanins CD9 and CD81, MOR23, and focal adhesion kinase (Griffin et al. 2009; Quach et al. 2009; Charrin et al. 2013). Myoferlin is one of the few proteins shown to modulate fusion during both development and regeneration, but myoferlin-null mice are viable and exhibit decreased muscle size attributed to a reduction in fusion during development (Doherty et al. 2005). Regeneration is also diminished after genetic loss of myoferlin; however, myoferlin-null myoblasts still maintain some fusion competency. Thus, it remains unclear whether the molecules that govern fusion during embryonic development are also required for fusion of satellite cells during regeneration.
Fusion of muscle precursor cells must be precisely controlled to prevent incorporation of nonmuscle nuclei into myofibers. Myomaker is a muscle-specific membrane protein that controls myoblast fusion during embryogenesis. Genetic loss of myomaker in mice results in a complete block of myoblast fusion, resulting in the absence of all skeletal muscle and early postnatal death (Millay et al. 2013). However, the regulation and function of this multipass membrane protein during regeneration have not been explored. Here, we show that myomaker is transiently expressed in adult satellite cells in response to muscle injury, concomitant with the acquisition of fusogenic activity. By analyzing the myomaker promoter, we demonstrate that the basic helix–loop–helix (bHLH) transcription factors MyoD and myogenin induce myomaker transcription. Additionally, inducible genetic deletion of myomaker in adult satellite cells reveals its essential role in muscle regeneration. Our results indicate that developmental and regenerative myoblasts deploy similar fusion mechanisms to accomplish muscle formation, and myomaker acts as a central and essential regulator of the fusion process.
Results and Discussion
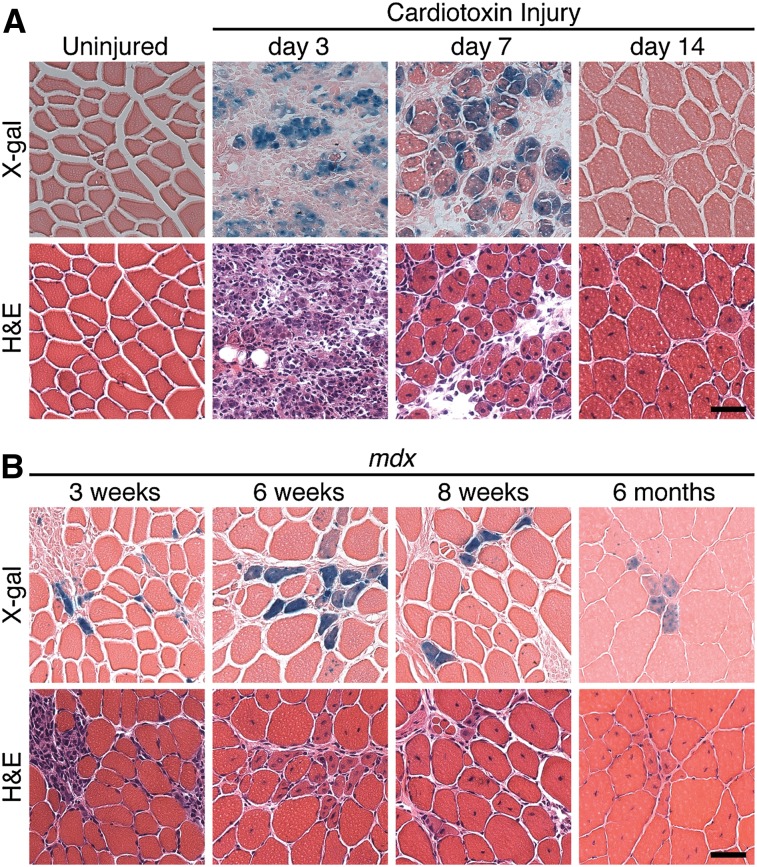
To begin to investigate the potential involvement of myomaker in muscle regeneration, we first examined its expression during acute and chronic muscle injury using mice harboring a LacZ cassette in intron 1 of the myomaker locus (myomakerLacZ/+) as a reporter for myomaker expression. Expression of this LacZ allele completely recapitulates endogenous myomaker expression during embryogenesis (Millay et al. 2013). To assess myomaker expression during acute muscle regeneration, the tibialis anterior (TA) muscles of 8-wk-old myomakerLacZ/+ mice were injured with cardiotoxin (CTX). Analysis of LacZ activity was assessed at multiple time points after CTX injury, revealing no myomaker expression in uninjured or day 1 CTX-injured TA (Fig. 1A). Myomaker was robustly expressed on day 3 and day 7 post-injury, when regeneration occurs, and dramatically down-regulated at day 14, when active myogenesis has ceased (Fig. 1A). Thus, myomaker is quickly induced after muscle injury to presumably perform an essential function for proper regeneration and is then deactivated once that function is no longer necessary.
Figure 1.
Transient expression of myomaker during acute and chronic muscle injury. (A) The TA muscle from myomakerLacZ/+ mice was injured with CTX, cryosectioned, and stained with X-gal. Myomaker-lacZ is expressed at days 3 and 7 post-injury and then down-regulated. (B) MyomakerLacZ/+ mice were crossed with mdx mice. TA muscles at the indicated time points were analyzed for myomaker expression through X-gal staining. For A and B, serial hematoxylin and eosin (H&E) sections accompany each X-gal image. Bar, 50 μm.
We also examined myomaker regulation during chronic muscle injury in the setting of muscular dystrophy, which is characterized by cycles of myofiber degeneration and regeneration (Durbeej and Campbell 2002). We interbred the dystrophin-deficient mdx mouse with myomakerLacZ/+ mice and analyzed myomaker expression in the TA at 3 wk, 6 wk, 8 wk, and 6 mo of age (Fig. 1B). We observed a relatively small and constant number of LacZ-positive myofibers present at each time point. These LacZ-positive myofibers likely represent the most recent fibers to have added new myonuclei. Given the transient activation of myomaker expression in response to injury, the LacZ-negative fibers containing centralized nuclei are likely to have already down-regulated myomaker expression. These data highlight the asynchronous regeneration of mdx myofibers. Moreover, our results show that activation of myomaker expression marks acute regeneration, as occurs in response to CTX injury, and chronic regeneration in mdx muscle.
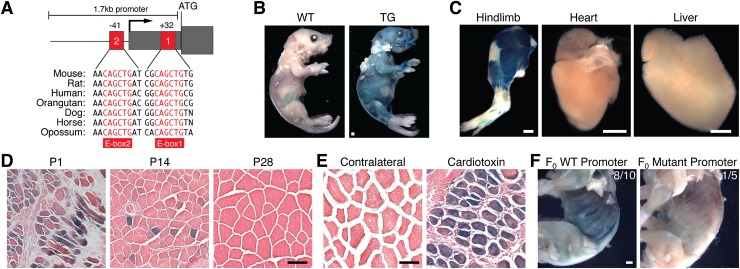
To define the mechanism that activates myomaker transcription, we analyzed the 5′ flanking region of the gene for regions of evolutionary sequence conservation likely to represent cis-regulatory elements. Indeed, we identified two highly conserved E-boxes (CANNTG) in close proximity to the myomaker transcription start site (Fig. 2A), conforming to the consensus binding site of the myogenic transcription factors MyoD and myogenin (Berkes and Tapscott 2005). We cloned a 1.7-kb genomic region containing these E-boxes, which also harbored three other E-box elements that are not conserved and thus is unlikely to be functional. Stable lines of transgenic mice bearing a reporter containing this 1.7-kb myomaker promoter upstream of LacZ exhibited β-galactosidase activity at postnatal day 3 (P3) in muscle (Fig. 2B). To be certain that the myomaker promoter element was expressed specifically in muscle, we stained various tissues at P3 with X-gal to detect β-galactosidase activity. Strong staining was observed in hindlimb skeletal muscle but not heart or liver (Fig. 2C). Compared with strong activity at P1, this promoter exhibited decreased activity at P14 and P28, when myogenesis is completed, overlapping with the expression of the myomaker transcript (Fig. 2D).
Figure 2.
Myomaker expression is controlled by promoter E-box elements. (A) Schematic of 1.7-kb myomaker promoter with proximal E-boxes notated. Mammalian conservation is shown below the schematic. The 1.7-kb promoter was cloned upstream of LacZ, and stable transgenic mice were generated. (B) Postnatal day 3 (P3) wild-type (WT) and transgenic mice stained with X-gal. Bar, 1 mm. (C) β-Galactosidase activity was detected in P3 hindlimbs but absent in heart and liver. Bar, 1 mm. (D) The myomaker promoter is expressed in muscle fibers at P1, is expressed to a lesser extent at P14, and is inactive at P28. Bar, 50 μm. (E) Transgenic mice harboring the myomaker promoter were subjected to CTX and stained with X-gal; the promoter is activated during regeneration. Bar, 50 μm. (F) The E-boxes (+32 base pairs [bp] and −41 bp) were mutated within the 1.7-kb promoter and assayed for the ability to drive LacZ expression in F0 transgenic mice. The wild-type promoter showed LacZ expression in eight of 10 P1 mice, but the mutant promoter lacked activity. Bar, 1 mm.
We also tested whether the 1.7-kb promoter element was reactivated upon muscle injury by subjecting the transgenic mice to CTX injection. Indeed, transgenic mice displayed robust LacZ expression after CTX injection but not in uninjured, contralateral muscle (Fig. 2E). To assess the potential contribution of the promoter-proximal E-boxes to myomaker expression, we mutated the E-boxes in the context of the 1.7-kb promoter. We observed LacZ staining in eight of 10 wild-type F0 transgenic mice (Fig. 2F). In contrast, only one of five F0 transgenic mice with the mutant promoter exhibited weak LacZ staining. While there are also conserved E-box elements in intron 1 of the myomaker locus that we did not investigate in this study, our results indicate a significant function for the two proximal E-box elements upstream of the myomaker start site.
The necessity of the proximal E-box elements suggests binding of the myogenic transcription factors MyoD and myogenin. We analyzed available chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) data sets from the ENCODE Project to explore the possibility that these transcription factors were detected at the proximal E-box sites within the myomaker promoter (The ENCODE Project Consortium 2012). This analysis revealed significant binding of both MyoD and myogenin at these sites during differentiation in the C2C12 muscle cell line (Supplemental Fig. 1). Prior studies have shown that CTX injury rapidly induces the expression of MyoD and myogenin in activated satellite cells (Mokalled et al. 2012). Thus, we propose that their acute induction leads to the rapid up-regulation of myomaker expression at the onset of muscle regeneration. The transient expression of myomaker after injury is potentially due to the lack of myocyte enhancer factor 2 (MEF2) sites in its promoter. MEF2 regulates differentiation during regeneration and persistent expression of structural and metabolic genes that allow muscle to function (Liu et al. 2014). Since there are extremely low levels of fusion in adult uninjured muscle, expression of myomaker is not required.
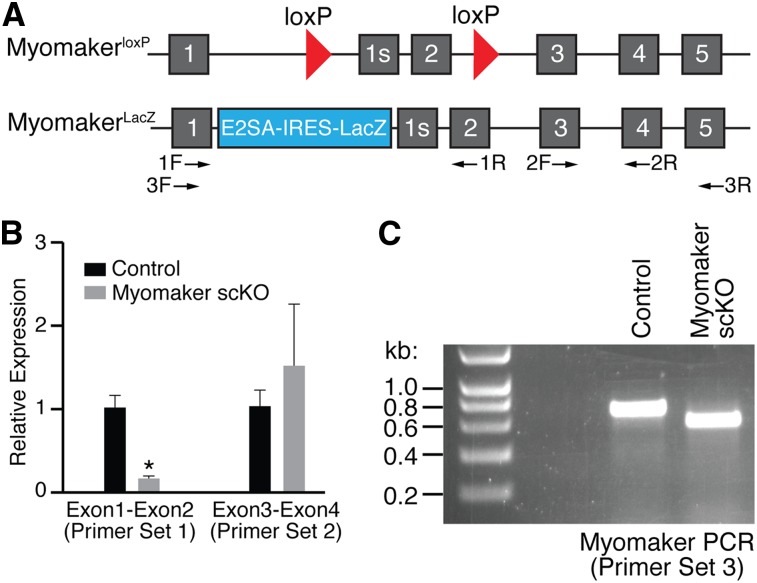
To investigate the potential function of myomaker in adult regeneration, we generated a myomaker conditionally targeted allele in which exon 2 was flanked by loxP sites (myomakerloxP/+) (Fig. 3A). To ensure efficient loss of myomaker during regeneration, we then generated mice that harbored one myomaker allele with loxP sites and one null allele with the LacZ cassette (myomakerLacZ/loxP) (Fig. 3A). The LacZ insertion blocks exon 1 from splicing with downstream exons and thus generates a null allele. Breeding of these mice with mice carrying the satellite cell-specific Cre recombinase (Pax7-CreERT2) allowed for deletion of myomaker in muscle progenitor cells upon tamoxifen treatment (Lepper et al. 2009). In the experiments below, we used both myomakerLacZ/loxP;Pax7-CreERT2 and myomakerloxP/loxP;Pax7-CreERT2 mice. We collectively refer to these lines as myomaker satellite cell knockout (scKO) mice. Controls were tamoxifen-treated myomakerLacZ/+;Pax7-CreERT2 or myomakerloxP/+;Pax7-CreERT2 mice.
Figure 3.
Analysis of myomaker transcript after tamoxifen-induced satellite cell deletion. (A) Diagram showing the myomaker targeted alleles used in this study. The myomakerloxP allele contains loxP sites flanking exon 2, and the myomakerLacZ/+ allele has a LacZ cassette in intron 1 that inhibits exon 1 splicing with exon 2. Exon 1s refers to the first exon of a short isoform of myomaker that is annotated but not highly conserved within mammalian species. The arrows below the schematic denote primer sets used in B and C. (B) Both control (n = 3, myomakerLacZ/+;Pax7-CreERT2 or myomakerloxP/+;Pax7-CreERT2) and myomakerscKO (n = 4, myomakerLacZ/loxP;Pax7-CreERT2) mice were treated with tamoxifen, injured with CTX, and analyzed by qPCR for the presence of exons 1 and 2 and exons 3 and 4 using the indicated primer sets. Data are presented as mean ± SEM. (*) P < 0.05. (C) PCR with primer set 3 shows a shorter amplicon in myomakerscKO muscle due to the Cre-mediated excision of exon 2.
To induce genetic deletion of myomaker in satellite cells, we injected myomakerscKO mice with tamoxifen daily for 5 d prior to injury of the TA muscle with CTX. We then injected tamoxifen every other day to ensure deletion of myomaker in the majority of satellite cells. Evaluation of myomaker RNA levels by quantitative PCR (qPCR) using a forward primer in exon 1 and a reverse primer in exon 2, which should be excised, revealed a decrease in myomaker expression (Fig. 3B). However, we did detect the presence of a transcript with primers that amplified exons 3 and 4 (Fig. 3B). To be certain that exon 2 was indeed deleted, we analyzed full-length myomaker transcript by PCR and observed the predicted smaller amplicon from myomakerscKO muscle (Fig. 3C). The myomaker transcript lacking exon 2 results in a frameshift and multiple premature stop codons and therefore does not generate a functional protein.
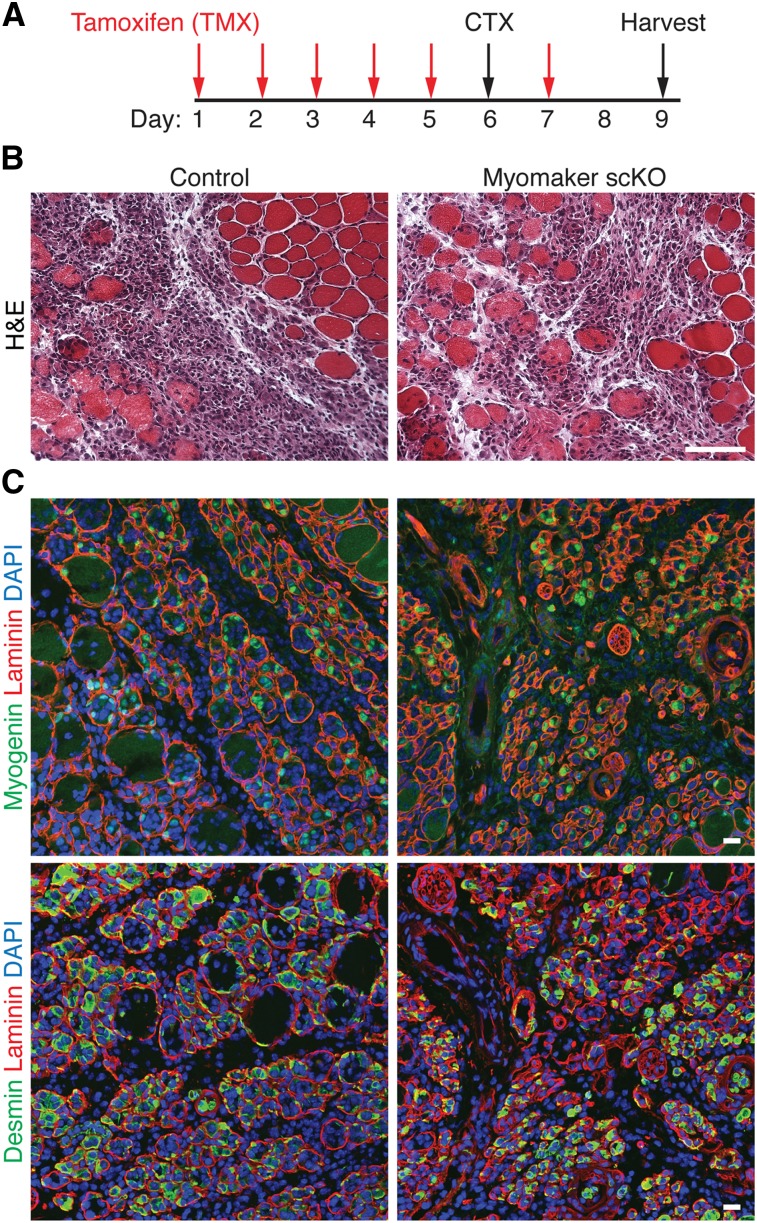
Our data indicate that myomaker is expressed in muscle after injury and not at appreciable levels in uninjured muscle. As a control, we analyzed uninjured muscle from myomakerscKO mice 14 d after the initial tamoxifen injection, and these muscles exhibited normal size and architecture (Supplemental Fig. 2). We then analyzed the consequences of loss of myomaker 3 d after CTX injury (Fig. 4A). Histological sections of control and myomakerscKO mice revealed an obvious presence of mononuclear cells, likely owing to both inflammatory infiltration and satellite cell activation (Fig. 4B). To evaluate the differentiation potential of the muscle progenitor pool after genetic deletion of myomaker, we analyzed expression of the muscle-restricted proteins myogenin and desmin. Cells positive for myogenin and desmin were clearly detected in myomakerscKO muscle, indicating that myomaker is not necessary to activate the myogenic program in satellite cells (Fig. 4C). Overall, control and myomakerscKO muscle were indistinguishable at 3 d post-injury, suggesting similar initial damage and proper activation of muscle precursors.
Figure 4.
Loss of myomaker in adult satellite cells does not alter the early response to muscle injury. (A) Schematic showing the timing of tamoxifen (TMX) and CTX. Injured muscle was analyzed 3 d after CTX injection. (B) Representative H&E-stained sections from control and myomakerscKO muscle demonstrate similar levels of damage. Bar, 50 μm. (C) Myogenin and desmin immunohistochemistry indicates the presence of myogenic cells in myomakerscKO muscle. Bar, 20 μm.
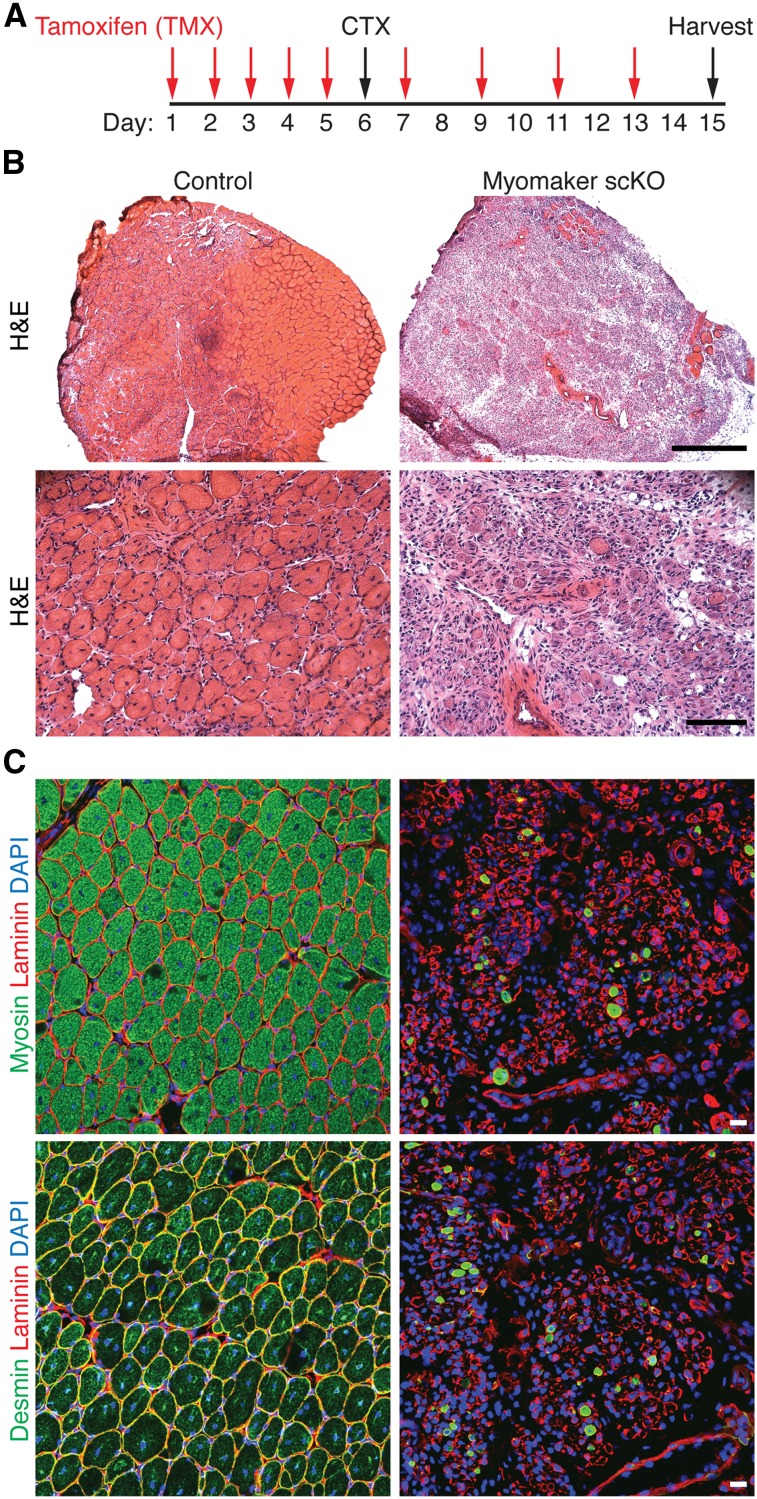
The complete regenerative capability of myomakerscKO mice was then assessed 9 d post-CTX (Fig. 5A). Analysis of hematoxylin and eosin (H&E)-stained histological sections showed that control mice had a robust regenerative response, indicated by large myofibers containing multiple centralized nuclei, a hallmark of regeneration (Fig. 5B). In contrast, we observed a dramatic absence of regenerative myofibers in myomakerscKO mice and a persistence of inflammatory infiltration (Fig. 5B). Furthermore, structures resembling muscle cells were present in myomakerscKO regenerating muscle; however, none were multinucleated. Staining with myosin and desmin 9 d after injury revealed a remarkable loss of muscle cells in myomakerscKO muscle, suggesting that activated myoblasts die if they do not undergo fusion (Fig. 5C). Taken together, these data demonstrate that myomaker is absolutely essential for adult muscle regeneration.
Figure 5.
Myomaker is necessary for muscle regeneration. (A) Time course of tamoxifen (TMX) and CTX treatment. TA muscles were analyzed 9 d post-injury. (B) H&E-stained sections from control and myomakerscKO mice show a complete lack of regeneration after genetic deletion of myomaker in satellite cells. Bars: top image, 500 μm; bottom image, 100 μm. (C) Myosin and desmin staining revealed a dramatic loss of muscle cells and no regenerated muscle fibers in myomakerscKO TA. Bar, 20 μm.
Myomaker is a muscle-specific plasma membrane protein that controls myoblast fusion during embryonic muscle development. The results of this study show that myomaker is transiently induced in activated satellite cells after both chemical and genetic injury to adult muscle. Up-regulation of myomaker expression during injury is mediated by E-box elements in the promoter, which bind myogenic regulatory factors and confer injury-dependent and muscle-specific expression to myomaker. Satellite cells that lack myomaker retain the ability to proliferate and differentiate but cannot fuse, resulting in a complete block of regeneration. Myomaker is the only muscle-specific protein shown to be absolutely necessary for the fusion of both embryonic and adult muscle precursors. The ability of myomaker to promote fusion to adult muscle fibers and muscle regeneration suggests opportunities to enhance muscle repair in the settings of acute and chronic injury through myomaker-directed cell–cell fusion.
Materials and methods
Mice
Both myomakerLacZ/+ and myomakerloxP/+ mouse strains used for this research project were created from C57BL/6 ES cells obtained from the KOMP Repository (http://www.komp.org) and generated by the Wellcome Trust Sanger Institute (Skarnes et al. 2011). MyomakerLacZ/+ was derived from Tmem8c clone EPD0626_5_C12 and was described previously (Millay et al. 2013). MyomakerloxP/+ was derived from clone DEPD00585_1_D04. Each of these clones contained a LacZ and NEO cassette in intron 1; however, clone EPD0626_5_C12 lacked the 3′ loxP site necessary for conditional deletion. Both clones were independently injected into 3.5-d-old C57BL/6 blastocysts by the Transgenic Technology Center at University of Texas Southwestern Medical Center. High-percentage chimeric male mice were crossed to C57BL/6 females to achieve germline transmission of the targeted allele. MyomakerloxP/+ mice were crossed with a mouse harboring a CAG-Flp allele to remove the NEO and LacZ cassettes and then intercrossed with myomakerLacZ/+ mice to generate myomakerLacZ/loxP mice. MyomakerLacZ/+ mice were genotyped with primers specific for LacZ (F, 5′-ATTGTCAGACATGTATACCCCGTACG-3′; and R, 5′-TTTTGACACCAGACCAACTGGTAATG-3′). MyomakerloxP/+ mice were genotyped for primers that amplify the 5′ loxP site (F, 5′-ACCCAAAGCTTGTCTTTTCCTGAGC-3′; and R, 5′-GCAAGTGCTGATTCTGACACACTGG-3′) and 3′ loxP site (F, 5′-GAGATGGCGCAACGCAATTAATG-3′; and R, 5′-TTCCTTCCCTTCCCTTTCTTTCCCC-3′). The Pax7-CreERT2 mice were kindly provided by Chen-Ming Fan (Carnegie Institution for Science, Baltimore, MD).
The myomaker promoter was cloned upstream of the HSP68 basal promoter and a lacZ reporter gene in the pGH-LacZ plasmid using KpnI (5′) and BamHI (3′). The mutant promoter was generated by independently cloning the regions immediately upstream (5′ PCR product) of and downstream (3′ PCR product) from the two proximal E-boxes and then using these products in a standard PCR sewing reaction followed by cloning into the pGH-LacZ vector. E-boxes were mutated from CANNTG to ACNNGT. Transgenic mice were generated by pronuclear injection of fertilized oocytes from B6C3F1 female mice and implanted into pseudopregnant ICR mice. Mice carrying LacZ were genotyped by PCR with primers specific for LacZ (described above). All experimental procedures involving animals in this study were reviewed and approved by the University of Texas Southwestern Medical Center’s Institutional Animal Care and Use Committee.
CTX injury
CTX from Naja mossambica mossambica (Sigma) was dissolved in sterile saline to a final concentration of 10 μM, aliquoted, and stored at −20°C. Mice were anesthetized by isoflurane inhalation, the legs were shaved and cleaned with alcohol, and TA muscles were injected with 50 μL of CTX with a 28-gauge needle.
X-gal staining
For whole-mount X-gal staining, mice, embryos, or tissues were fixed in 4% PFA/PBS (containing 0.01% deoxycholic acid, 0.02% Igepal) for 45 min at 4°C with gentle shaking and then rinsed twice with cold PBS. Samples were stained overnight in staining solution [5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCl2, 1mg/mL X-gal in PBS] followed by washing twice in PBS and post-fixing with 4% PFA/PBS. For X-gal staining of cryosections, the following procedure was employed: fixing with 2% glutaraldehyde/PBS; washing three times in 0.1% sodium deoxycholate, 0.2% NP40 substitute (Fluka), and PBS; and incubation in staining solution [4 mM K3Fe(CN)6, 4 mM K4Fe(CN)6, 0.4 mM MgCl2, 1mg/mL X-gal, 0.1% sodium deoxycholate, 0.2% NP40 substitute in PBS] overnight at 37°C in the dark. The samples were then rinsed in PBS and fixed in 4% PFA/PBS for at least 20 min. Tissue sections were costained with light eosin, dehydrated, and mounted with Permount (Fisher).
RNA analysis
Total RNA was extracted from mouse tissue with Trizol (Invitrogen), and cDNA was synthesized using SuperScript III reverse transcriptase with random hexamer primers (Invitrogen). Gene expression was assessed using standard qPCR approaches with Power SYBR Green. Analysis was performed on a 7900HT fast real-time PCR machine (Applied Biosystems) with the following myomaker SYBR primers: 1F, 5′-ATCGCTACCAAGAGGCGTT-3′; 1R, 5′-CACAGCACAGACAAACCAGG-3′; 2F, 5′-TATACTCCGGTCCCATAGGC-3′; and 2R, 5′-ATGCTCTTGTCGGGGTACAG-3′. Full-length myomaker cDNA was amplified using the following primers: 3F, 5′-ATGGGGACAGTTGTAGCCAAAC-3′; and 3R, 5′-TCAGACACAAGTGCAGCAGAGG-3′.
Histological analyses
For cryosections, skeletal muscle was dissected, embedded in gum tragacanth (1% in PBS), and frozen in 2-methylbutane cooled liquid nitrogen. Frozen sections were cut at 10 μm and stained with H&E using routine procedures. Immunohistochemistry was performed by fixation with 1% PFA/PBS; permeabilization with 0.2% Triton X-100 in PBS; blocking with PBS/1% BSA, 1% heat-inactivated goat serum, and 0.025% Tween20; incubation with primary antibody for at least 2 h; incubation with secondary Alexa-488 IgG1 antibody (Invitrogen) for 1 h; and mounting with VectaShield containing DAPI (Vector Laboratories). Anti-mouse myosin (my32, Sigma), desmin (DAKO), and myogenin (F5D, Developmental Studies Hybridoma Bank) antibodies were used at 1:100. Slides were visualized using a Zeiss LSM 780 confocal microscope.
Acknowledgments
We thank J. Cabrera for graphical assistance, the Transgenic Technology Center at University of Texas Southwestern Medical Center for embryonic stem cell injections, J. McAnally for blastocyst injections, and J. Shelton for microscopy assistance. D.P.M. was funded by a Development Grant from the Muscular Dystrophy Association. This work was supported by grants from the National Institutes of Health (HL-077439, HL-111665, HL-093039, 1R01-DK-099653, and U01-HL-100401), the Cancer Prevention and Research Institute of Texas, and the Robert A. Welch Foundation (grant 1-0025 to E.N.O.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.247205.114.
References
- Berkes CA, Tapscott SJ. 2005. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol 16: 585–595 [DOI] [PubMed] [Google Scholar]
- Buckingham M 2006. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev 16: 525–532 [DOI] [PubMed] [Google Scholar]
- Charrin S, Latil M, Soave S, Polesskaya A, Chretien F, Boucheix C, Rubinstein E. 2013. Normal muscle regeneration requires tight control of muscle cell fusion by tetraspanins CD9 and CD81. Nat Commun 4: 1674. [DOI] [PubMed] [Google Scholar]
- Doherty KR, Cave A, Davis DB, Delmonte AJ, Posey A, Earley JU, Hadhazy M, McNally EM. 2005. Normal myoblast fusion requires myoferlin. Development 132: 5565–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbeej M, Campbell KP. 2002. Muscular dystrophies involving the dystrophin-glycoprotein complex: an overview of current mouse models. Curr Opin Genet Dev 12: 349–361 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin CA, Kafadar KA, Pavlath GK. 2009. MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev Cell 17: 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum-Cohen Y, Harel I, Umansky KB, Tzahor E, Snapper SB, Shilo BZ, Schejter ED. 2012. The actin regulator N-WASp is required for muscle-cell fusion in mice. Proc Natl Acad Sci 109: 11211–11216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindi SM, Tajrishi MM, Kumar A. 2013. Signaling mechanisms in mammalian myoblast fusion. Sci Signal 6: re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan CM. 2009. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 460: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Nelson BR, Bezprozvannaya S, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. 2014. Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc Natl Acad Sci 111: 4109–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millay DP, O’Rourke JR, Sutherland LB, Bezprozvannaya S, Shelton JM, Bassel-Duby R, Olson EN. 2013. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499: 301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokalled MH, Johnson AN, Creemers EE, Olson EN. 2012. MASTR directs MyoD-dependent satellite cell differentiation during skeletal muscle regeneration. Genes Dev 26: 190–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach NL, Biressi S, Reichardt LF, Keller C, Rando TA. 2009. Focal adhesion kinase signaling regulates the expression of caveolin 3 and β1 integrin, genes essential for normal myoblast fusion. Mol Biol Cell 20: 3422–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasyutina E, Martarelli B, Brakebusch C, Wende H, Birchmeier C. 2009. The small G-proteins Rac1 and Cdc42 are essential for myoblast fusion in the mouse. Proc Natl Acad Sci 106: 8935–8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Price F, Rudnicki MA. 2013. Satellite cells and the muscle stem cell niche. Physiol Rev 93: 23–67 [DOI] [PMC free article] [PubMed] [Google Scholar]