Monoubiquitylation of histone H2B on Lys123 (H2BK123ub1) plays a multifaceted role in diverse DNA-templated processes. Wozniak and Strahl show that H2BK123ub1 is regulated through the control of Bre1 protein stability. Bre1 stability is primarily controlled through its association with the PAF complex. Using a domain in Rtf1 that stabilizes Bre1, they show that inappropriate Bre1 levels lead to defects in gene regulation. These data uncover a novel quality control mechanism used by the cell to maintain proper Bre1 and H2BK123ub1 levels, thereby ensuring proper control of gene expression.
Keywords: chromatin, gene transcription, histones, ubiquitylation
Abstract
Monoubiquitylation of histone H2B on Lys123 (H2BK123ub1) plays a multifaceted role in diverse DNA-templated processes, yet the mechanistic details by which this modification is regulated are not fully elucidated. Here we show in yeast that H2BK123ub1 is regulated in part through the protein stability of the E3 ubiquitin ligase Bre1. We found that Bre1 stability is controlled by the Rtf1 subunit of the polymerase-associated factor (PAF) complex and through the ability of Bre1 to catalyze H2BK123ub1. Using a domain in Rtf1 that stabilizes Bre1, we show that inappropriate Bre1 levels lead to defects in gene regulation. Collectively, these data uncover a novel quality control mechanism used by the cell to maintain proper Bre1 and H2BK123ub1 levels, thereby ensuring proper control of gene expression.
Histone post-translational modifications (PTMs) play essential roles in the regulation of chromatin structure and function (Kouzarides 2007; Zentner and Henikoff 2013). One such histone PTM that has been well studied as a regulator of multiple DNA-templated processes is monoubiquitylation of histone H2B, which occurs at Lys123 (H2BK123ub1) in the budding yeast Saccharomyces cerevisiae (Robzyk et al. 2000). This PTM functions in the context of transcriptional regulation (both initiation and elongation) (Henry et al. 2003; Kao et al. 2004; Xiao et al. 2005; Pavri et al. 2006; Fleming et al. 2008; Chandrasekharan et al. 2009, 2010) but has also been linked to other processes, including DNA replication (Rizzardi et al. 2012; Trujillo and Osley 2012) and repair (Game and Chernikova 2009) and kinetochore function (Latham et al. 2011).
H2BK123ub1 functions in chromatin by several means. First, this mark physically alters chromatin compaction and nucleosome stability (Fleming et al. 2008; Chandrasekharan et al. 2009; Fierz et al. 2011). Another function of H2BK123ub1 is to promote histone H3 methylation at Lys4 (H3K4me) and Lys79 (H3K79me) in a mechanism of histone “cross-talk” referred to as trans-histone regulation (Briggs et al. 2002; Dover et al. 2002; Ng et al. 2002; Sun and Allis 2002). H3K4me and H3K79me, in conjunction with H2BK123ub1, serve as markers of euchromatin and act to facilitate transcription factor recruitment and prevent the binding of silencing factors (Wozniak and Strahl 2014). Accordingly, loss of these PTMs leads to aberrant gene regulation.
In yeast, H2BK123ub1 is catalyzed by the concerted efforts of the ubiquitin-conjugating enzyme (E2) Rad6 and the RING finger domain-containing ubiquitin ligase (E3) Bre1 (Robzyk et al. 2000; Hwang et al. 2003; Wood et al. 2003a). Similar to other E3 ligases, Bre1 serves as the substrate recognition module for the complex and is important for the recruitment of Rad6 to chromatin (Wood et al. 2003a). Studies have also found that the polymerase-associated factor (PAF) complex associates with Rad6 and facilitates its recruitment to gene bodies (Ng et al. 2003; Wood et al. 2003b; Xiao et al. 2005). Although the mechanistic underpinnings of this recruitment are not entirely clear, it is known that the Rtf1 subunit of the PAF complex plays a major role (Wood et al. 2003b; Xiao et al. 2005). In addition to Bre1 recruitment and catalysis, H2BK123ub1 levels are also controlled by the deubiquitylases Ubp8 and Ubp10 (Henry et al. 2003; Emre et al. 2005). Loss of Ubp8 or Ubp10 leads to phenotypes similar to the loss of H2BK123ub1, indicating that the levels of this PTM are carefully regulated in the cell.
In this study, we found that H2BK123ub1 is regulated through the control of Bre1 protein stability. Surprisingly, Bre1 stability is primarily controlled through its catalytic activity in addition to its association with the PAF complex that is likely responsible for its recruitment to chromatin. By taking advantage of a region in Rtf1 of the PAF complex that can stabilize Bre1, we found that inappropriate stabilization of Bre1 under normal conditions leads to defects in gene regulation. Our results suggest a “rheostat” control mechanism for H2BK123ub1 that contributes to proper transcriptional control.
Results and Discussion
Trans-histone regulatory pathways facilitate H2BK123ub1
A major mechanism by which histone modifications are regulated is via trans-histone pathways, which involve a histone region or histone PTM regulating the outcome of another histone modification in an intranucleosomal or internucleosomal manner. For example, methylation of H3 at Lys36 (H3K36me) is regulated by regions within H2A and H4, which form a nucleosomal surface that the responsible enzyme, Set2, binds to when catalyzing this mark (Du and Briggs 2010). H2BK123ub1, on the other hand, regulates the outcome of H3K4 and H3K79 methylation in a trans-histone pathway that controls the function of the Set1 and Dot1 enzymes, respectively. However, trans-histone pathways controlling the outcome of H2BK123ub1 have not been fully explored.
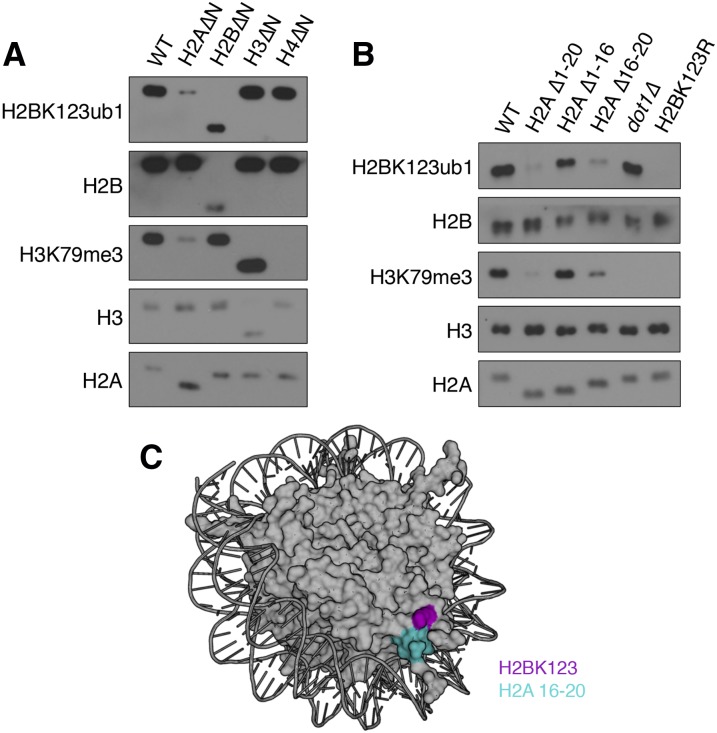
To identify regulatory mechanisms important for the outcome of H2K123ub1, we determined the levels of H2BK123ub1 as well as the downstream modification H3K79me3 in yeast strains lacking the N-terminal tails of histones H3, H4, H2A, and H2B. As expected, loss of the H4 tail eliminated H3K79me3 without affecting H2BK123ub1 (Fig. 1A; Fingerman et al. 2007). We also observed two opposing effects on H2BK123ub1. First, loss of the H2B tail reduced the levels of H2B without affecting H2BK123ub1, suggesting that a higher proportion of H2B in these cells is ubiquitylated (Fig. 1A). Second, and consistent with a previous report (Zheng et al. 2010), we found that loss of the H2A tail reduced H2BK123ub1 as well as H3K79me3 (Fig. 1A). Residues 16–20 of H2A (the HAR domain) primarily mediate this regulation, since deletion of this domain affects H2BK123ub1 and H3K79me3 to an extent similar to that of loss of the entire tail (Fig. 1B; Supplemental Fig. 1). Taken together, our data verify that multiple trans-histone pathways exist to regulate H2BK123ub1.
Figure 1.
The H2A N-terminal tail regulates H2BK123 ubiquitylation. (A) Shown is a Western screen of histone methylation and ubiquitylation states in wild-type (WT) and mutant strains lacking the N-terminal tails of each of the core histones. (H2A∆N) ∆1–20; (H2B∆N) ∆1–32; (H3∆N) ∆1–30; (H4∆N) ∆1–27. (B) Residues 16–20 of histone H2A (HAR domain) regulate H2BK123ub1 and H3K79me3. Strains harboring the indicated truncations of H2A were probed for ubiquitylation and methylation states as in A. (C) The HAR domain (cyan) is located next to H2BK123 (magenta) on the surface of the nucleosome (Protein Data Bank [PDB] ID: 1ID3).
The HAR domain regulates H2BK123ub1 by stabilizing Bre1
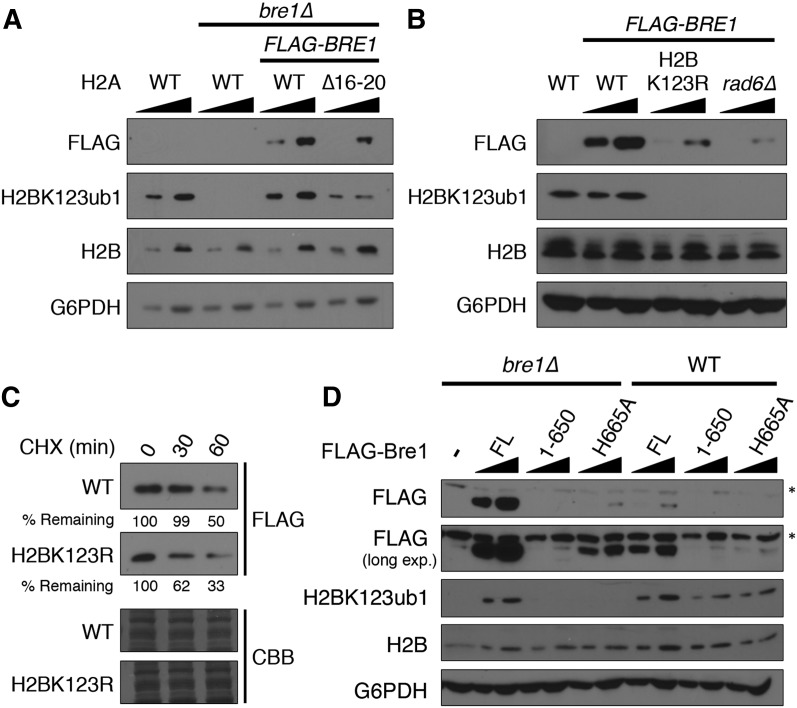
We next sought to uncover the mechanism by which the HAR domain regulates H2BK123ub1. Given the close physical proximity of the HAR domain to H2BK123 on the nucleosomal surface (Fig. 1C), we hypothesized that the HAR domain may play a role in the ubiquitylation reaction itself. Thus, we investigated whether loss of the HAR domain had any effect on the E2 or E3 ubiquitin ligases Rad6 and Bre1, respectively. Loss of the HAR domain did not alter either total or bulk chromatin-bound levels of Rad6 (Supplemental Fig. 2). To assess Bre1 levels, we transformed bre1∆ strains either containing or lacking the HAR domain with a low-copy plasmid expressing ADH1-driven, N-terminally Flag-tagged Bre1. Importantly, this expression construct restores H2BK123ub1 to wild-type levels in the bre1∆ strain and behaves similarly to a version containing the native BRE1 promoter (Fig. 2A; Supplemental Fig. 3). Surprisingly, the levels of Bre1 in the HAR deletion strain were reduced, matching the decrease in H2BK123ub1 (Fig. 2A). Moreover, this was not the result of decreased BRE1 transcription as measured by RT–PCR (Supplemental Fig. 4), indicating that the HAR domain regulates Bre1 levels through a mechanism that is post-transcriptional.
Figure 2.
Bre1 stability is dependent on the catalysis of H2BK123ub1. (A) The HAR domain is important for the stability of Bre1. The indicated mutant strains were transformed with empty vector or ADH1-driven Flag-BRE1 and subjected to immunoblot analysis with the indicated antibodies. G6PDH served as a loading control. Increasing amounts of extract were loaded for each sample, as indicated by solid black triangles. (B) Catalysis of H2BK123ub1 is required for Bre1 stability. The indicated strains were analyzed as in A. (C) Loss of H2BK123ub1 destabilizes Bre1. Wild-type (WT) and H2BK123R strains were treated with cyclohexamide (CHX) for the indicated amount of time. Samples taken at each time point were analyzed by immunoblot analysis and Coomassie Brilliant Blue (CBB) staining. The percentage of signal compared with the 0-min time point for each sample is indicated (% remaining). (D) The RING finger domain of Bre1 is required for stability. Wild-type or bre1∆ strains expressing empty vector (−), full-length (FL) Flag-Bre1, or mutant derivatives lacking the RING finger domain (1–650) or harboring an inactivating point mutation (H665A) were analyzed by immunoblot analysis. (*) Nonspecific band.
Bre1 stability is dependent on its ability to ubiquitylate H2BK123
Given the possibility that the HAR domain might regulate Bre1 stability through its contribution to a nucleosomal surface required by Bre1 to catalyze H2BK123ub1, we next asked whether the loss of H2BK123ub1 itself might also regulate Bre1 stability. Strikingly, we found that Bre1 protein levels were nearly abolished in strains harboring a point mutation at H2BK123 (H2BK123R) (Fig. 2B). As with the loss of the HAR domain, the H2BK123R mutation did not affect BRE1 expression, suggesting that the regulation occurs at the level of the protein stability (Supplemental Fig. 4). Consistent with this, a cyclohexamide (CHX) pulse-chase analysis revealed that Bre1 is more rapidly turned over in the H2BK123R strain (Fig. 2C, cf. wild-type and H2BK123R at 30 min after CHX treatment). Taken together, these data provide strong support that Bre1 in the HARΔ and H2BK123R strains is subject to post-transcriptional control. We note that Bre1 regulation does not appear to involve the proteasome, since MG132 treatment failed to stabilize Bre1 (Supplemental Fig. 5). This result is in agreement with another report showing that MG132 decreases H2BK123ub1 levels (Mimnaugh et al. 1997).
We next ascertained whether mutations in the ubiquitylation machinery would also affect Bre1 stability. We found that loss of Rad6, like the H2BK123R mutant, also decreased Bre1 levels (Fig. 2B). Moreover, both deletion of the catalytic RING finger domain of Bre1 (1–650) and a point mutation that disrupts its enzymatic function (H665A) (Wood et al. 2003a) destabilize Bre1 (Fig. 2D). Additionally, RING finger mutants of Bre1 also had a destabilizing effect on the protein when expressed in the context of wild-type endogenous Bre1, indicating that destabilization is not merely the consequence of a global loss of histone ubiquitylation (Fig. 2D). Thus, the ability of Bre1 to ubiquitylate chromatin is important for its stability.
The PAF complex contributes to Bre1 stability via a conserved domain in Rtf1
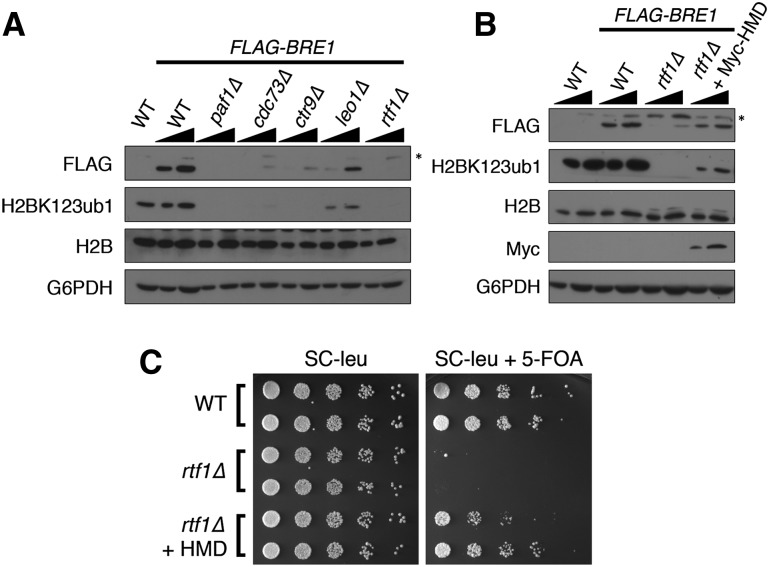
Given that Bre1 stability is dependent on catalysis, we next sought to determine whether other proteins that promote H2BK123ub1 also regulate Bre1 stability. We focused on the PAF complex, which has been well studied as a regulator of H2BK123ub1 (Jaehning 2010). As shown in Figure 3A, deletions of individual members of the complex have varying effects on H2BK123ub1, with the paf1∆ and rtf1∆ strains having the strongest effect. Significantly, we found that the loss of H2BK123ub1 correlates with the loss of Bre1 levels in these mutant strains, thereby linking the PAF complex to Bre1 stability and H2BK123ub1.
Figure 3.
The histone modification domain (HMD) of Rtf1 stabilizes Bre1. (A). The PAF complex regulates Bre1 stability. The indicated strains, transformed with empty vector or Flag-BRE1, were subjected to immunoblot analysis. Increasing amounts of extract were loaded for each sample, as indicated by solid black triangles. (*) Nonspecific band. (B) The HMD of Rtf1 stabilizes Bre1. The indicated strains were transformed with Flag-BRE1 and/or NLS-Myc-HMD (Myc-HMD, CEN) and subjected to immunoblot analysis as in A. (C) The HMD is sufficient for mediating the telomeric silencing function of Rtf1. Empty vector or a plasmid expressing NLS-Myc-HMD (HMD, 2µ) were transformed into wild-type (WT) or rtf1∆ telomeric silencing reporter strains harboring the URA3 gene inserted within a subtelomeric region of chromosome VII. Strains were plated on SC-leu medium with or without 5-FOA.
Rtf1 is the only subunit of the PAF complex that is absolutely required for H2BK123ub1 (Ng et al. 2003; Wood et al. 2003b; Xiao et al. 2005). This appears to be mediated by a small conserved domain of Rtf1 called the histone modification domain (HMD), which is capable of facilitating H2BK123ub1 independently of the PAF complex (Piro et al. 2012). Based on this finding, we hypothesized that the HMD promotes H2BK123ub1 by stabilizing Bre1. To test this idea, we coexpressed Myc-tagged HMD fused to a nuclear localization sequence (NLS-Myc-HMD) and Flag-Bre1 in the rtf1∆ strain. In agreement with published data (Piro et al. 2012), we found that the HMD could restore H2BK123ub1 in the rtf1∆ strain (Fig. 3B). Moreover, we found that expression of the HMD could also rescue Bre1 levels, indicating a critical role for the HMD in stabilizing Bre1.
To examine the functional relevance of HMD-mediated Bre1 stabilization, we investigated its role in telomeric silencing, a function linked to both Bre1 and Rtf1. We made use of a telomeric silencing reporter strain that has the URA3 gene inserted near the telomere of chromosome VII. Loss of Rtf1 in this strain shows a severe growth defect when grown on medium containing 5-FOA, indicating a loss of silencing (Fig. 3C). In line with the finding that the HMD could rescue Bre1 levels and H2BK123ub1, expression of the HMD was able to restore the silencing defect of the rtf1∆ strain (Fig. 3C). These data demonstrate that the HMD plays an important role in gene silencing by stabilizing Bre1.
Altering the balance of Bre1 leads to defects in gene regulation
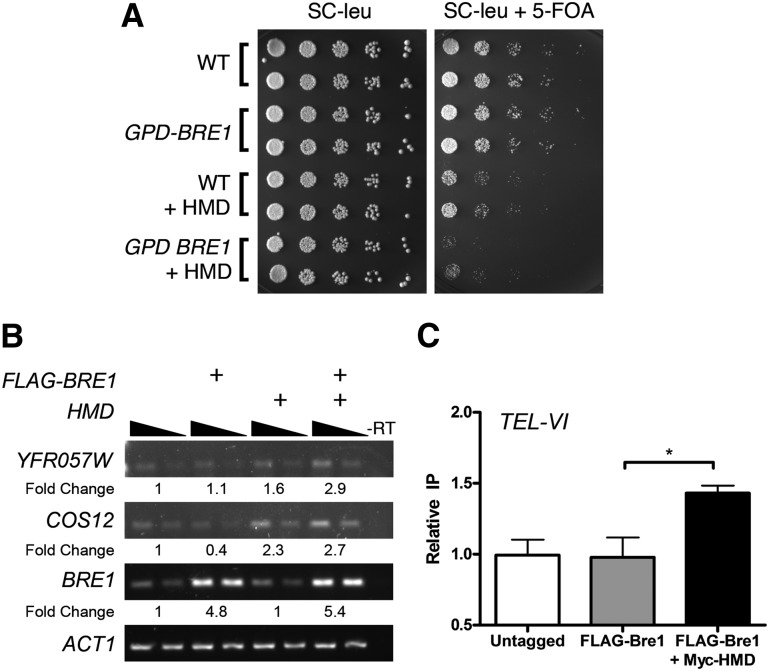
The ability of the HMD to stabilize Bre1 allowed us to use it as a tool to ask why Bre1 is under such careful regulation. To address this question, we again used the telomeric silencing reporter strain used above. In this strain, we overexpressed Bre1 from the highly expressed GPD promoter either alone or in combination with the HMD and measured growth on 5-FOA. Overexpression of Bre1 alone did not result in any growth defect on 5-FOA (Fig. 4A), consistent with inability of Bre1 overexpression to increase the levels of H2BK123ub1 (Fig. 2D). In contrast, we found that overexpression of the HMD resulted in reduced growth on 5-FOA, and this effect was exacerbated when Bre1 was also overexpressed, indicating loss of silencing of the URA3 reporter (Fig. 4A). In validation of the reporter strain, we also observed increased transcription of two naturally silenced subtelomeric genes (YFR057W [chromosome VI] and COS12 [chromosome VII]) with Bre1 stabilization, indicating that aberrant levels of Bre1 impact transcription of normally silenced telomere-proximal genes (Fig. 4B).
Figure 4.
Aberrant Bre1 stabilization disrupts gene silencing. (A) Stabilization of Bre1 causes defective silencing at telomeres. Telomeric silencing strains that overexpressed Bre1 from the GPD promoter (GPD-BRE) and/or the HMD (2μ) were used and analyzed as in Figure 3C. (B) Stabilized Bre1 alters the expression of naturally silenced telomeric genes. RT–PCR was performed with RNA isolated from strains expressing Flag-BRE1 (CEN) and/or the HMD (2μ) with primers directed toward the subtelomeric genes YFR057W (chromosome VI), COS12 (chromosome VII), and BRE1 or the housekeeping gene ACT1. Decreasing amounts of cDNA were used for each PCR, as indicated by solid black triangles. The expression of each target was normalized to ACT1, and the fold change versus wild-type (WT) was calculated and is shown below each strain. (C) The HMD recruits Bre1 to telomeres. ChIP was performed with M2 Flag agarose under each of the indicated conditions. ChIP and input DNA were used as template for PCR reactions containing primers specific to a subtelomeric region of chromosome VI (TEL-VI). Relative immunoprecipitation represents fold change enrichment versus untagged. See the Supplemental Material for further details. Data represent mean ± SEM (n = 3). (*) P < 0.04.
Last, we sought to determine whether the observed changes in gene expression were the result of HMD-mediated binding of Bre1 at telomeres. To determine this, we performed chromatin immunoprecipitation (ChIP) to measure Bre1 binding to a subtelomeric region of chromosome VI proximal to YFR057W, where the HMD has been previously shown to bind (Piro et al. 2012). In agreement with the up-regulation of YFR057W, we found increased Bre1 binding in this region in the presence of the HMD (Fig. 4C). Taken together, these observations demonstrate that aberrant stabilization of Bre1 at telomeres leads to defects in gene silencing. Given that loss of Ubp8 and Ubp10 also results in increased H2BK123ub1 levels at euchromatic and telomeric regions (Henry et al. 2003; Emre et al. 2005), the collective data support a model in which the ubiquitylation machinery is present across the genome but is kept in check by the opposing functions of RNA polymerase II (RNAPII)-dependent PAF recruitment and the deubiquitylating enzymes that reduce H2BK123ub1, both of which would control Bre1 stability and hence H2BK123ub1 levels genome-wide.
Concluding remarks
In this study, we uncover a novel pathway of H2BK123ub1 regulation that involves the precise control of Bre1 protein stability. Using mutants that disrupt (1) the nucleosomal surface targeted by Bre1, (2) Bre1 catalytic activity, or (3) proteins that aid in Bre1 catalysis (i.e., Rad6 and the PAF complex), we show that the ability to ubiquitylate H2B is critical for the stabilization of this E3 ligase. By expressing a domain in Rtf1 that couples the PAF complex with Bre1 and leads to its stabilization, we show that aberrant Bre1 levels result in adverse consequences for gene silencing. Taken together, these findings reveal a novel control mechanism for Bre1 that we suggest functions to fine-tune the appropriate levels of H2BK123ub1 genome-wide.
In addition to the regulation of Bre1, another mechanism that acts to fine-tune the levels of H2BK123ub1 across the genome is the deubiquitylating enzymes Ubp8 and Ubp10. A question remains as to why the cell would use two distinct mechanisms to control H2BK123ub1 levels. Perhaps, similar to histone acetylases and deacetylases, where the equilibrium of the “on” and “off” enzymes define the precise levels of histone acetylation at any given point across the genome, it may be that the level of H2BK123ub1 across the genome is similarly governed by the equilibrium of Rad6/Bre1 and Ubp8/Ubp10. Consistent with this idea, deletion of the heterochromatin-associated Ubp10 deubiquitylase results in increased levels of H2BK123ub1 in silenced regions of the genome (Emre et al. 2005). This finding implies that Bre1/Rad6 can localize to these regions but is prevented from functioning by the removal of H2BK123ub1. Notably, we were unable to detect Bre1 at a subtelomeric region of chromosome VI under normal conditions (Fig. 4C), suggesting that it may interact transiently with these regions. In contrast, within transcribed regions where Bre1 is stabilized by the PAF complex, the equilibrium shifts toward productive H2BK123ub1 (Fig. 5A). Thus, a possible surveillance mechanism comprising the deubquitylating enzymes ensures loss of Bre1 and erasure of H2BK123ub1 where it would otherwise drive inappropriate functions (Fig. 5B).
Figure 5.
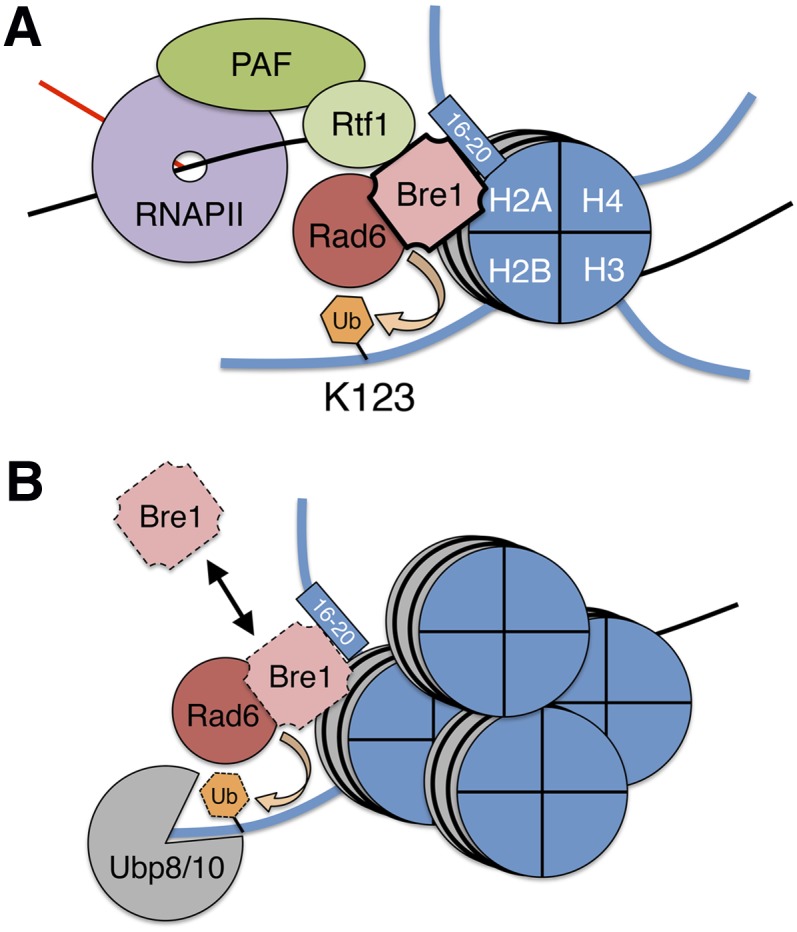
Transcription-coupled stabilization of Bre1 fine-tunes H2B ubiquitylation. We found that the Rtf1 subunit of the PAF complex is important for stabilizing Bre1 and promoting H2BK123ub1. Given the close association of the PAF complex with transcribing RNAPII, we propose that Rtf1, residues 16–20 of histone H2A, and perhaps other proteins associated with the transcriptional apparatus interact with and stabilize Bre1 (indicated by solid black outline) to promote H2BK123ub1 in active regions of the genome. Once transcription is complete or in repressed regions, the absence of the transcriptional machinery leads to Bre1 instability (indicated by dashed outline). Transient interactions of Bre1/Rad6 with chromatin in repressed regions catalyze short-lived H2BK123ub1 (dashed outline) that is rapidly removed by the deubiquitylating enzymes Ubp8/10.
Our observations also provide insight into the regulation of H2BK123ub1 by the PAF complex. Previous work has shown that Bre1 directly interacts with the PAF complex in vitro using purified recombinant proteins (Kim and Roeder 2009). In addition, we demonstrated that Rad6/Bre1 is associated with the PAF complex in yeast (Xiao et al. 2005). Given these observations, we propose that the PAF complex, through the HMD, stabilizes Bre1 in transcribed regions, which in turn promotes Rad6 recruitment (Wood et al. 2003a) and H2BK123ub1 (Fig. 5A). It is not entirely clear how a potential interaction with Rtf1 could stabilize Bre1, but the interaction may either mask specific degradation sequences within Bre1 or aid in the recruitment of Bre1 to its nucleosomal substrate, which may be the actual stabilizing interaction.
One of the important mechanistic functions of H2BK123ub1, in addition to promoting nucleosomal disruption and stability during transcription elongation, is the regulation of histone methylation at H3K4 and H3K79 (Briggs et al. 2002; Dover et al. 2002; Ng et al. 2002; Sun and Allis 2002; Chandrasekharan et al. 2010). This form of histone “cross-talk” has been the focus of numerous studies over the past decade, but the mechanism remains to be fully elucidated. Two primary models exist, which suggest that H2BK123ub1 acts as either a wedge in chromatin to facilitate enzyme access (Fierz et al. 2011) or a bridge to the histone methyltransferases (either directly [McGinty et al. 2008; Kim et al. 2013] or indirectly [Lee et al. 2007; Vitaliano-Prunier et al. 2008]). The indirect recruitment mechanism has been proposed to involve Cps35/Swd2, which is a subunit of the H3K4-methylating COMPASS complex and has been suggested to interact with the H3K79 methyltransferase Dot1 (Lee et al. 2007). However, both of these models share the common theme that the ubiquitin moiety itself at H2BK123 mediates the “cross-talk.” Intriguingly, our data demonstrate that the same mutations used to characterize H2BK123ub1-mediated “cross-talk” also disrupt the stability of Bre1. Thus, it will be intriguing to determine whether any aspect of the trans-histone pathway of H3K4 and H3K79 methylation might involve Bre1 itself independent of H2BK123ub1. In support of this idea, Bre1 has been shown to interact with Cps35/Swd2 in vivo (Vitaliano-Prunier et al. 2008), and, intriguingly, mutations that disrupt H2BK123ub1 (and hence Bre1 stability) also disrupt the ability of Cps35/Swd2 to facilitate COMPASS-mediated H3K4 methylation (Lee et al. 2007; Vitaliano-Prunier et al. 2008). Thus, Cps35/Swd2 may be a link between Bre1 and H3K4 methylation. Future studies will be required to revisit some of the basic assumptions of H2BK123ub1-mediated histone “cross-talk” and the details that underlie Bre1 regulation.
Materials and methods
Yeast strains and plasmids
Strains and plasmids used in this study are listed in Supplemental Tables 1 and 2. Gene disruptions and endogenous overexpression were performed as previously described (Janke et al. 2004) and verified by both PCR and immunoblotting.
Yeast whole-cell extracts and Western blot analysis
Yeast were grown in YPD or synthetic complete dropout (SC) medium at 30°C to mid-log phase, and extracts were prepared as previously described (Mehta et al. 2010). Antibodies and dilutions are listed in the Supplemental Material.
Phenotypic spotting assays
Fivefold serial dilutions of saturated overnight yeast cultures were plated on YPD or SC medium with or without the indicated drugs. Cells were plated at a starting OD600 of 0.5 on the appropriate medium and imaged after 2–4 d of growth at 30°C.
RNA isolation and RT–PCR
RNA was prepared from 10 OD600 units of mid-log phase cells using hot acid phenol-chloroform extraction followed by ethanol precipitation. Crude RNA was DNaseI-treated (Promega) and then purified using an RNeasy minikit (Qiagen). cDNA was synthesized using SuperScript II first strand synthesis system (Life Technologies) and diluted 1/10 prior to amplification by PCR. Primers are listed in Supplemental Table 3. Reactions were run on 2% agarose gels and visualized by UV with SYBR Safe DNA gel stain (Life Technologies). Bands were quantified using ImageJ software.
ChIP
ChIP was performed as described previously (Jha and Strahl 2014) with some exceptions. Sonication for each sample was performed for 20 min with alternating on/off cycles of 30 sec using a Bioruptor Standard (Diagenode). Immunoprecipitation was performed overnight with 1 mg of clarified, sonicated extract and 20 µL of equilibrated Flag M2 agarose (Sigma). Primer sequences are listed in Supplemental Table 3, and analysis methodologies are described further in the Supplemental Material.
Acknowledgments
We thank members of the Strahl laboratory for a critical reading of the manuscript. We also thank David Allis, Elizabeth Gjoneska, Judith Jaehning, Michael Parra, Ali Shilatifard, Jeff Smith, Zu-Wen Sun, and Fred Winston for yeast strains and plasmids. This work was supported by a National Science Foundation grant to B.D.S. (grant no. 1330320).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.243121.114.
References
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418: 498. [DOI] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW. 2009. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci 106: 16686–16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW. 2010. Histone H2B ubiquitination and beyond: regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics 5: 460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277: 28368–28371 [DOI] [PubMed] [Google Scholar]
- Du HN, Briggs SD. 2010. A nucleosome surface formed by histone H4, H2A, and H3 residues is needed for proper histone H3 Lys36 methylation, histone acetylation, and repression of cryptic transcription. J Biol Chem 285: 11704–11713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emre NC, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, Henry KW, Li K, Marmorstein R, Greenblatt JF, Shilatifard A, et al. 2005. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol Cell 17: 585–594 [DOI] [PubMed] [Google Scholar]
- Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. 2011. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol 7: 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerman IM, Li HC, Briggs SD. 2007. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev 21: 2018–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. 2008. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 31: 57–66 [DOI] [PubMed] [Google Scholar]
- Game JC, Chernikova SB. 2009. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair 8: 470–482 [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell 11: 261–266 [DOI] [PubMed] [Google Scholar]
- Jaehning JA 2010. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta 1799: 379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962 [DOI] [PubMed] [Google Scholar]
- Jha DK, Strahl BD. 2014. An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat Commun 5: 3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. 2004. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev 18: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Roeder RG. 2009. Direct Bre1–Paf1 complex interactions and RING finger-independent Bre1–Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem 284: 20582–20592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim JA, McGinty RK, Nguyen UT, Muir TW, Allis CD, Roeder RG. 2013. The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol Cell 49: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Latham JA, Chosed RJ, Wang S, Dent SY. 2011. Chromatin signaling to kinetochores: transregulation of Dam1 methylation by histone H2B ubiquitination. Cell 146: 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. 2007. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131: 1084–1096 [DOI] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. 2008. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453: 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta M, Braberg H, Wang S, Lozsa A, Shales M, Solache A, Krogan NJ, Keogh MC. 2010. Individual lysine acetylations on the N terminus of Saccharomyces cerevisiae H2A.Z are highly but not differentially regulated. J Biol Chem 285: 39855–39865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimnaugh EG, Chen HY, Davie JR, Celis JE, Neckers L. 1997. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry 36: 14418–14429 [DOI] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem 277: 34655–34657 [DOI] [PubMed] [Google Scholar]
- Ng HH, Dole S, Struhl K. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem 278: 33625–33628 [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717 [DOI] [PubMed] [Google Scholar]
- Piro AS, Mayekar MK, Warner MH, Davis CP, Arndt KM. 2012. Small region of Rtf1 protein can substitute for complete Paf1 complex in facilitating global histone H2B ubiquitylation in yeast. Proc Natl Acad Sci 109: 10837–10842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzardi LF, Dorn ES, Strahl BD, Cook JG. 2012. DNA replication origin function is promoted by H3K4 di-methylation in Saccharomyces cerevisiae. Genetics 192: 371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287: 501–504 [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108 [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Osley MA. 2012. A role for H2B ubiquitylation in DNA replication. Mol Cell 48: 734–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaliano-Prunier A, Menant A, Hobeika M, Geli V, Gwizdek C, Dargemont C. 2008. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol 10: 1365–1371 [DOI] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. 2003a. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 11: 267–274 [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. 2003b. The Paf1 complex is essential for histone monoubiquitination by the Rad6–Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem 278: 34739–34742 [DOI] [PubMed] [Google Scholar]
- Wozniak GG, Strahl BD. 2014. Hitting the ‘mark’: interpreting lysine methylation in the context of active transcription. Biochim Biophys Acta doi: 10.1016/j.bbagrm.2014.03.002 [DOI] [PubMed] [Google Scholar]
- Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol 25: 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Henikoff S. 2013. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol 20: 259–266 [DOI] [PubMed] [Google Scholar]
- Zheng S, Wyrick JJ, Reese JC. 2010. Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Mol Cell Biol 30: 3635–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]