piRNAs guide the repression of diverse transposable elements in metazoan germ cells. Le Thomas et al. show that piRNA biogenesis in Drosophila germ cells depends on the inheritance of homologous piRNAs from the previous generation. Transgenerationally inherited piRNAs trigger piRNA biogenesis in the progeny by two different mechanisms. First, inherited piRNAs guide post-transcriptional processing of precursors into mature piRNAs. Second, inherited piRNAs direct the modification of the chromatin state of cluster sequences. This study provides key insights into the transgenerational mechanism that specifies piRNA biogenesis in the germline.
Keywords: H3K9me3, epigenetics, piRNA
Abstract
Small noncoding RNAs that associate with Piwi proteins, called piRNAs, serve as guides for repression of diverse transposable elements in germ cells of metazoa. In Drosophila, the genomic regions that give rise to piRNAs, the so-called piRNA clusters, are transcribed to generate long precursor molecules that are processed into mature piRNAs. How genomic regions that give rise to piRNA precursor transcripts are differentiated from the rest of the genome and how these transcripts are specifically channeled into the piRNA biogenesis pathway are not known. We found that transgenerationally inherited piRNAs provide the critical trigger for piRNA production from homologous genomic regions in the next generation by two different mechanisms. First, inherited piRNAs enhance processing of homologous transcripts into mature piRNAs by initiating the ping-pong cycle in the cytoplasm. Second, inherited piRNAs induce installment of the histone 3 Lys9 trimethylation (H3K9me3) mark on genomic piRNA cluster sequences. The heterochromatin protein 1 (HP1) homolog Rhino binds to the H3K9me3 mark through its chromodomain and is enriched over piRNA clusters. Rhino recruits the piRNA biogenesis factor Cutoff to piRNA clusters and is required for efficient transcription of piRNA precursors. We propose that transgenerationally inherited piRNAs act as an epigenetic memory for identification of substrates for piRNA biogenesis on two levels: by inducing a permissive chromatin environment for piRNA precursor synthesis and by enhancing processing of these precursors.
piRNAs serve as guides for transcriptional and post-transcriptional repression of diverse transposable elements in germ cells of metazoa (Aravin et al. 2007a; Ghildiyal and Zamore 2009; Siomi et al. 2011). The sequences of the genomic regions that give rise to piRNAs, the piRNA clusters, define the repertoire of elements that are recognized and silenced. Therefore, proper selection of RNA molecules to be processed into mature piRNAs is critical for generating the diverse repertoire of piRNAs that guide effective silencing of selfish genomic elements while preventing degradation of functional RNAs. The precursors of other classes of small noncoding RNAs such as siRNAs and miRNAs are selected based on their unique structure: either long dsRNA or fold-back hairpins (Ghildiyal and Zamore 2009; Kim et al. 2009). However, no similar structural or sequence motifs have been identified in piRNA precursors in Drosophila or mammals. Furthermore, any sequence inserted into piRNA clusters can be efficiently converted into mature piRNAs (Todeschini et al. 2010; Muerdter et al. 2012), raising the question of how piRNA precursor transcripts are selected for processing.
In Drosophila, piRNAs are produced from extended (up to 200-kb) piRNA clusters that are strongly enriched in repetitive sequences, predominantly transposon remnants. piRNA clusters are located in pericentromeric and subtelomeric regions and are transcribed into long precursor molecules from both strands or just one strand (Aravin et al. 2007b; Brennecke et al. 2007). Unidirectional clusters are expressed predominantly in the somatic follicular cells of the Drosophila ovary, while bidirectional clusters are transcribed in germline-derived nurse cells (Brennecke et al. 2007; Lau et al. 2009). Interestingly, generation of piRNAs from unidirectional and bidirectional piRNA clusters is under the control of different sets of proteins (Malone et al. 2009).
Several studies suggested that piRNA clusters in Drosophila possess a unique chromatin structure that might play a role in piRNA biogenesis. Bidirectional piRNA clusters show characteristics of heterochromatin, with the enrichment of the repressive histone 3 Lys9 trimethylation (H3K9me3) mark and heterochromatin protein 1 (HP1) over clusters (Moshkovich and Lei 2010; Rangan et al. 2011). This heterochromatic environment seems to be required for proper expression of clusters and their processing into piRNAs. Indeed, the deficiency in histone methyltransferase Eggless/SETDB1 leads to loss of piRNAs (Rangan et al. 2011). A germline-specific homolog of HP1, Rhino, was shown to associate with double-stranded piRNA clusters, and Rhino deficiency also leads to loss of germline-specific piRNAs (Klattenhoff et al. 2009). In the nucleus, Rhino colocalizes in distinct areas with the nuclear DEAD-box RNA helicase protein UAP56 that binds to piRNA precursors (Zhang et al. 2012).
The recent study of de Vanssay et al. (2012) showed that a transgenic locus that generates piRNAs is able to induce piRNA production from another homologous locus that was originally incompetent for piRNA generation. Importantly, the study revealed that deposition of a cytoplasmic factor from the mother, which expresses the active locus, into the embryo is sufficient to activate piRNA generation from the recipient locus without the inheritance of the active locus itself. The investigators proposed that the epigenetic signal that activates piRNA biogenesis in the next generation is a pool of piRNAs that are generated from the active locus and inherited by the progeny. Indeed, Piwi proteins and the associated piRNAs that are expressed in the maternal germline during oogenesis are deposited into the developing egg and are present in the early embryo before the start of zygotic transcription (Harris and Macdonald 2001; Megosh et al. 2006; Brennecke et al. 2007). Furthermore, maternally inherited piRNAs are essential for effective piRNA-mediated silencing and the fertility of the progeny (Brennecke et al. 2008). Transgenerational inheritance of piRNAs that initiate piRNA biogenesis from homologous genomic regions in the progeny provides an attractive solution to the specification of piRNA precursors; however, the molecular mechanism for this process has remained unknown.
Here we use several genetic systems to dissect requirements for piRNA biogenesis. We found that the activity of double-stranded piRNA clusters in germ cells is greatly enhanced by the transgenerational inheritance of homologous piRNAs from the mothers. We show that, in the nucleus, inherited piRNAs modify the chromatin structure of homologous loci to recruit the Rhino/Cutoff complex, which is essential for transcription of piRNA precursors, while in the cytoplasm, they enhance processing of piRNA precursors.
Results
Activity of transgenic piRNA clusters correlates with the level of the H3K9me3 mark and the presence of transgenerationally inherited piRNAs
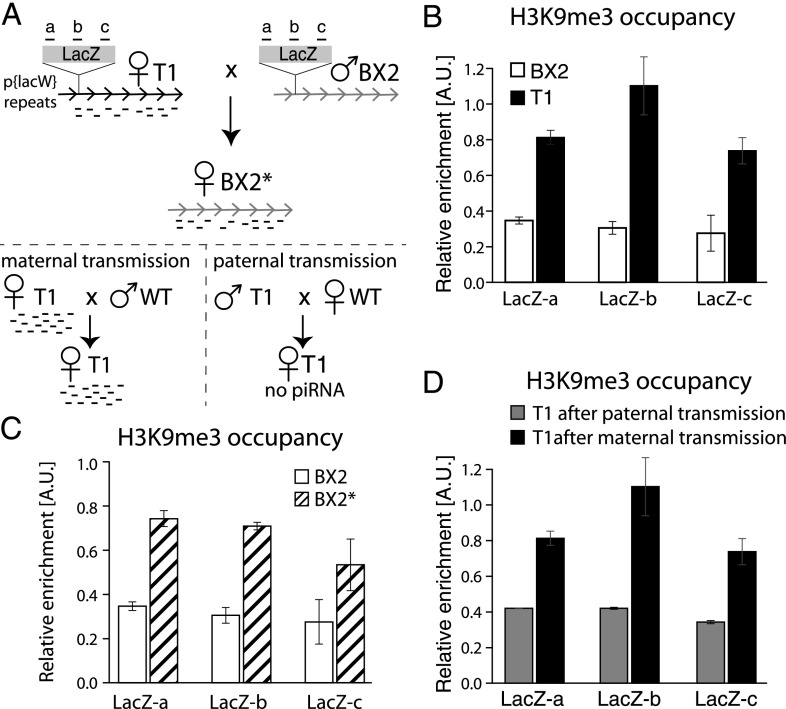
To identify the features that discriminate piRNA-generating regions from other genomic loci, we searched for examples in which the same locus is active in piRNA generation in one Drosophila strain but inactive in another. Two Drosophila melanogaster strains, T1 and BX2, contain an identical number of tandem repeats of the P-lacZ transgene inserted in the same locus in the middle of chromosome arm 2R, a genomic position that does not give rise to piRNAs in other strains (Dorer and Henikoff 1997; Ronsseray et al. 2001). In the T1 strain, the transgene gives rise to abundant piRNAs in germ cells, and these piRNAs are able to silence expression of another lacZ transgene in trans. In contrast, no piRNAs are generated from the same transgene in the BX2 strain (Fig. 1A; de Vanssay et al. 2012). As the sequence of the T1 and the BX2 transgenes is identical, the reason for their differential ability to generate piRNAs is unknown. Recent studies revealed that two chromatin factors, SETDB1 (a methyltransferase responsible for installation of the H3K9me3 mark) and the HP1 homolog Rhino, are required for piRNA biogenesis (Klattenhoff et al. 2009; Rangan et al. 2011). Based on this, we hypothesized that a difference in the chromatin state of the T1 and the BX2 transgenes explains their differential ability to produce piRNAs. To test this, we profiled the H3K9me3 mark on the transgene sequences in ovaries of flies from both strains using chromatin immunoprecipitation and quantitative PCR (ChIP-qPCR). The H3K9me3 mark was enriched by approximately fourfold over the transgene in the “active” T1 strain compared with the “inactive” BX2 strain (Fig. 1B). To rule out the possibility that the differences in the H3K9me3 mark between T1 and BX2 are caused by changes in nucleosome occupancy, we performed total H3 ChIP that showed no difference between the two strains (Supplemental Fig. S1). This result indicates that the ability of a genomic locus to generate piRNAs indeed correlates with its chromatin structure and in particular with a high level of the H3K9me3 mark.
Figure 1.
Activity of transgenic piRNA clusters correlates with the level of the H3K9me3 mark and the presence of transgenerationally inherited piRNAs. (A) Transgenic D. melanogaster strains used for the study. Both T1 and BX2 strains contain seven identical tandem copies of the P{LacW} transgene inserted at the cytological site 50C. Strain T1 was derived from BX2 by X-ray irradiation and carries large-scale chromosome rearrangements. Transgene-derived piRNAs are generated in T1 but not in BX2 females. Transgenerational inheritance of piRNAs from T1 females leads to activation of the BX2 locus in the progeny (indicated as BX2*), resulting in piRNA production. Note that inheritance of the T1 locus is not required for the activation of BX2. (Bottom) The ability to generate piRNAs from the T1 transgene is inherited through the female, but not the male, lineage. The locations of regions a, b, and c along the LacZ sequence, analyzed in the ChIP-qPCR experiments, are indicated. (B) piRNA production from transgenes correlates with enrichment of the H3K9me3 mark over the transgene. H3K9me3 signal on T1 and BX2 as measured by ChIP-qPCR on three different regions of the lacZ sequence shown in A. Signal was normalized to input and compared with signal at a control genomic region (chr2R: 2336913–2337023) highly enriched in the H3K9me3 mark. The ChIPs were performed on two independent biological samples followed by duplicate qPCRs on each sample. A two-sided t-test was used to calculate significance of the differences (P < 0.05). (C) Conversion of the inactive BX2 locus to a piRNA-producing BX2* locus correlates with an increase in the H3K9me3 signal, as measured by ChIP-qPCR on three different regions of the lacZ sequence. The ChIPs were performed on two independent biological samples followed by duplicate qPCRs on each sample. A two-sided t-test was used to calculate significance (P < 0.05). (D) Paternal transmission of the T1 transgene is accompanied by a loss of H3K9me3 signal, as measured by ChIP-qPCR. The ChIPs were performed on two independent biological samples followed by duplicate qPCRs on each sample. A two-sided t-test was used to calculate significance (P < 0.05).
In Drosophila, piRNAs expressed in germ cells during oogenesis are deposited into the embryo and are important for transposon silencing in the next generation (Brennecke et al. 2008). It was previously found that the BX2 locus, which is deficient in piRNA production, can be converted into an active locus (designated as BX2*) by exposure to maternally inherited cytoplasm carrying piRNAs homologous to the lacZ transgene (Fig. 1A; de Vanssay et al. 2012). To determine whether activation of the BX2 locus caused by transgenerationally inherited piRNAs was coupled with a change in the chromatin state of the locus, we analyzed this region for the presence of the H3K9me3 mark before and after conversion. Conversion of BX2 to BX2* was accompanied by an increase in the H3K9me3 mark on the transgene (Fig. 1C). The level of the H3K9me3 mark on the activated BX2* transgene was similar to the signal observed on the locus in the T1 strain. Therefore, exposure of an inactive locus to homologous piRNAs inherited from the previous generation leads to installment of the H3K9me3 mark and conversion of the locus to an active piRNA cluster.
The ability of the T1 and the activated BX2* loci to generate piRNAs was reported to be stable over many generations if the locus was inherited from the mother (Fig. 1A; de Vanssay et al. 2012). However, the transgenic loci lose their capacity to generate piRNAs after paternal transmission, suggesting that the presence of piRNAs is necessary to maintain the locus in an active state. We measured the enrichment of the H3K9me3 mark on the T1 transgene after maternal and paternal transmission. The paternal transmission reduced the level of the H3K9me3 mark approximately twofold, although levels were slightly higher than those on the nonactivated BX2 locus (Fig. 1D). Thus, lack of transgenerationally inherited piRNAs homologous to the transgene correlates with decreased levels of the H3K9me3 mark.
Taken together, our experiments demonstrate that the ability of a transgenic locus to generate piRNAs correlates with a high level of H3K9me3 over the transgene. Reciprocally, the high level of the H3K9me3 mark on the locus requires the presence of transgenerationally inherited piRNAs homologous to the locus. Specifically, an active locus loses its high H3K9me3 signal and becomes inactive if it is not exposed to homologous piRNAs, whereas an inactive locus with low H3K9me3 signal acquires both a higher level of the H3K9me3 chromatin mark and the ability to generate piRNAs after exposure to homologous piRNAs. Together, these observations suggest that piRNAs inherited from the previous generation induce a change in the chromatin state of homologous genomic regions in the progeny, and this change in chromatin is required to produce piRNAs from these regions.
Transgenerationally inherited piRNAs enhance the ping-pong processing of piRNAs
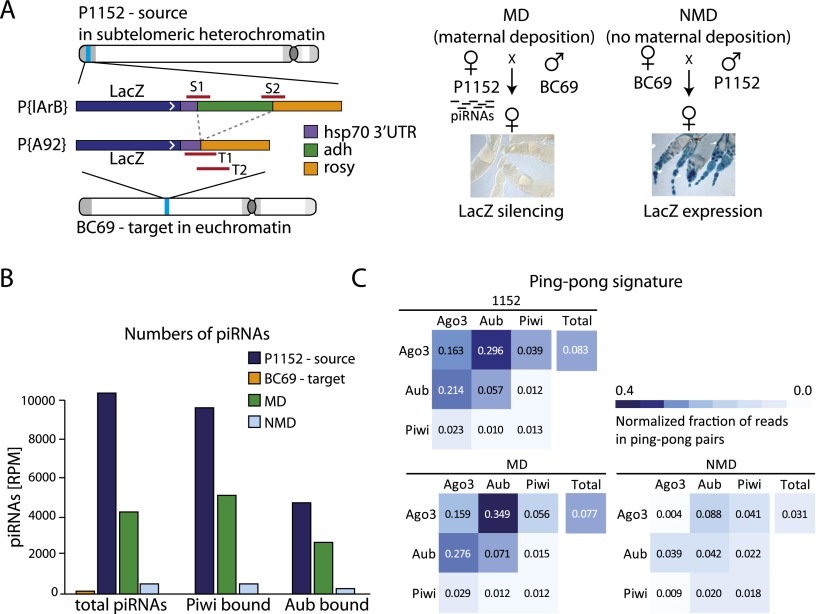
To test whether transgenerational inheritance of piRNAs is required for piRNA biogenesis from other loci, we used another transgenic system that contains single-copy unique sequences. The insertion of the P{lArB} construct into a subtelomeric piRNA cluster in strain P1152 leads to generation of abundant piRNAs from this transgene (Josse et al. 2007; Muerdter et al. 2012). Importantly, piRNAs generated from the lacZ sequence in P1152 are able to silence other lacZ transgenes located in euchromatic sites, such as the P{A92} in strain BC69, in trans (Ronsseray et al. 2003; Josse et al. 2007; de Vanssay et al. 2012).
We analyzed the generation of piRNAs from the P1152 locus in the parental strain and two reciprocal progenies of crosses between P1152 and BC69 stocks. Maternal deposition (MD) progeny inherit transgene-derived piRNAs from their P1152 mothers, while genetically identical no MD (NMD) progeny of the reciprocal cross do not inherit piRNAs (Fig. 2A). In addition to profiling total piRNA populations in the ovaries of each genotype, we immunoprecipitated the PIWI, AUB, and AGO3 proteins and defined piRNA populations associated with each protein. As reported before (de Vanssay et al. 2012; Muerdter et al. 2012), we detected a large number of piRNAs mapping to the P1152 transgene in the parental P1152 strain. Transgenic piRNAs are found in all three Piwi complexes (Fig. 2B). In contrast, practically no piRNAs were detected in the BC69 strain. The level of piRNAs mapped to the transgene drops twofold in the MD progeny compared with the parental P1152 flies, reflecting the fact that only one allele of the transgene is present in MD flies compared with two copies in the parental P1152 strain. The level of piRNAs decreases ∼10-fold in the NMD progeny compared with the genetically identical MD flies, indicating that inherited piRNAs are essential for piRNA generation (Fig. 2B).
Figure 2.
Inherited piRNAs activate the ping-pong cycle. (A) Transgenic D. melanogaster strains used for the study. The strain P1152 contains a single copy of the P{lArB} insertion at the telomeric X-TAS piRNA cluster on the X chromosome, which generates abundant piRNAs from both genomic strands (Josse et al. 2007; Muerdter et al. 2012). Strain BC69 contains a single copy of the P{A92} insertion at the euchromatic site 35B-C of chromosome 2 that does not generate piRNAs. The P{lArB} and P{A92} transgenes are identical in sequence, except for the Adh region that is specific to the P{lArB} construct in P1152. The bars (S1, S2, T1, and T2) indicate sequences used for ChIP-qPCR to measure the chromatin structure of the P1152 (source) and the BC69 (target) transgenes. BC69 females express lacZ from the transgene in the ovary; however, lacZ is repressed by lacZ piRNAs generated from the P1152 transgene if both transgenes are combined. The cross between the P1152 and BC69 strains was performed in two directions: The progeny designated as MD inherited P1152-derived piRNAs from their mothers. In contrast, the progeny of the reciprocal cross, designated as NMD, did not inherit these piRNAs. (B) piRNA production from the P1152 and the BC69 transgenes in the parental strains and the MD and NMD progenies. Shown are the numbers of piRNA reads normalized to library depth (reads per million [RPM]) mapped to the P{lArB} transgene in total RNA and in RNA purified from the PIWI and AUB complexes. Abundant piRNAs are present in the P1152 (piRNA source) but not in the BC69 (piRNA target) parental strain. piRNA levels in the MD progeny are twofold lower than in the P1152 strain, reflecting the heterozygous presence of the source locus in the progeny. The piRNA level is ∼10-fold lower in the NMD compared with the MD progeny in the total cellular piRNA population and in the PIWI and AUB complexes. (C) Analysis of ping-pong processing. An equal number of piRNA reads mapped to the P{lArB} transgene in each library was sampled 1000 times to calculate the fraction of reads in ping-pong pairs. Shown are the mean values of the normalized fraction of piRNAs that are in ping-pong pairs; the distributions of values from the 1000-times sampling are shown in box plots in Supplemental Fig. S1. Both the P1152 parental strain and the MD progeny have a high level of AUB- and AGO3-bound piRNAs generated by ping-pong processing. The fraction of piRNA generated by ping-pong processing drops fourfold to sevenfold in NMD progeny. PIWI-bound piRNAs are generated by a primary biogenesis mechanism and, accordingly, do not have a strong ping-pong signature.
One mechanism by which trangenerationally inherited piRNAs might boost production of new piRNAs in the progeny is processing of complementary transcripts through the ping-pong loop. In the ping-pong loop, piRNAs guide endonucleolytic cleavage of complementary transcript, leading to the generation of new piRNAs from the cleaved product (Brennecke et al. 2007; Gunawardane et al. 2007; Olovnikov and Kalmykova 2013). Indeed, piRNA clusters that are expressed in germ cells, including the telomeric cluster containing the P1152 locus, are transcribed from both genomic strands; therefore, transgenerationally inherited piRNAs can target transcripts from these regions to initiate their processing. To study the impact of inherited piRNAs on the ping-pong processing, we analyzed the characteristic feature of ping-pong: the number of complementary piRNA pairs with a 10-nucleotide (nt) overlap between their 5′ ends (so-called ping-pong pairs). To account for different numbers of sequences in each library, we sampled the same number of reads from each library 1000 times and calculated the fraction of piRNAs in ping-pong pairs. A large number of piRNAs derived from the transgene in the P1152 strain was generated by ping-pong processing (Fig. 2C; Supplemental Fig. S2). In agreement with previous studies (Brennecke et al. 2007; Li et al. 2009), the strongest ping-pong interaction was observed between AUB and AGO3, while PIWI participation in the ping-pong is negligible. The fraction of the ping-pong pairs in the total piRNA population as well as in AUB/AGO3 pairs is similar in the parental P1152 strain and MD progeny. However, the ping-pong processing dramatically decreased in the NMD progeny: The fraction of piRNAs in ping-pong pairs decreases fourfold to sevenfold in the most prominent partners, AUB and AGO3 (Fig. 2C; Supplemental Fig. S2). This result shows that inheritance of homologous piRNAs indeed boosts ping-pong processing in the progeny.
The level of PIWI-bound piRNAs mapped to the P1152 transgene is decreased 8.3-fold in the absence of inherited piRNAs in NMD progeny (Fig. 2B). However, analysis of piRNA sequences associated with PIWI showed that, in contrast to AUB and AGO3 piRNAs, they do not form significant ping-pong pairs (Fig. 2C; Supplemental Fig. S2), supporting previous studies that indicate that PIWI-bound piRNAs are generated by another biogenesis mechanism, so-called primary processing. Therefore, the boost in ping-pong processing is not sufficient to explain the whole impact of inherited piRNAs that also enhance primary piRNA biogenesis.
Inherited piRNAs trigger the deposition of the H3K9me3 mark and initiation of primary piRNA biogenesis from the same genomic sequence
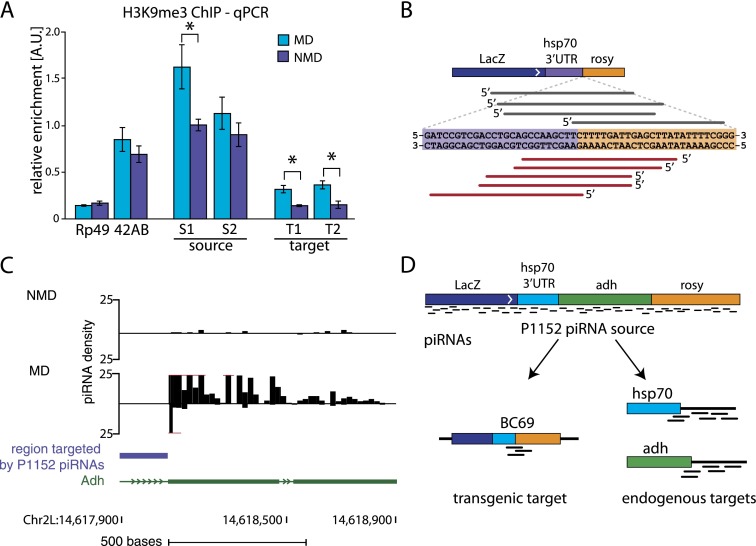
We analyzed chromatin states of both source (P1152) and target (BC69) loci in ovaries of MD and NMD progenies (Fig. 2A). Previously, we found that the presence of piRNAs generated from the P1152 source locus leads to an increase in the H3K9me3 mark and a decrease in the H3K4me mark on the BC69 target locus (Le Thomas et al. 2013). We confirmed accumulation of the H3K9me3 mark on the BC69 target locus and found that it occurs only in MD progeny that inherited homologous piRNAs from their mothers (Fig. 3A). Importantly, we also observe that, in the absence of inherited piRNAs, in NMD progeny, the level of H3K9me3 decreases on the P1152 source construct (Fig. 3A). These results showed that a high level of H3K9me3 on both the source and the target loci requires transgenerational piRNA inheritance.
Figure 3.
Inherited piRNAs activate piRNA biogenesis on target loci. (A) Repression of the BC69 target transgene by P1152 correlates with a twofold higher H3K9me3 signal in the MD progeny compared with the NMD progeny, as measured by ChIP-qPCR. The H3K9me3 mark also decreases at the P1152 locus in the NMD progeny, where cognate piRNAs are not inherited. Therefore, maternally deposited piRNAs are necessary to install the H3K9me3 mark on the piRNA target (BC69) and maintain the high level of the mark on the source of piRNAs (P1152). For this experiment, MD progeny was obtained by crossing P1152; BC69 females to wild-type males, and NMD progeny was obtained from the reciprocal cross. Signal was normalized to input and compared with signal at a control genomic region (chr2R: 2336913–2337023) highly enriched in the H3K9me3 mark. No H3K9me3 change was observed on the rp49 gene and the 42AB piRNA cluster. The ChIPs were performed on two independent biological samples followed by triplicate qPCRs on each sample. P-values were calculated with a two-sided t-test. (*)P < 0.05. (B) Primary piRNA processing is initiated on the BC69 target after exposure to homologous piRNAs in the MD progenies. Shown are piRNA sequences that mapped specifically to the BC69 target locus and not to P1152 (the junction between the hsp70 segment and the rosy segment, which is present in BC69 but not in P1152). Out of nine distinct piRNAs, two were detected in the total cellular piRNA library, and seven were detected in the Piwi-bound population in the MD progeny. None was detected in the parental strains or in the NMD progeny. BC69-derived piRNAs were also absent in the AUB and AGO3 complexes in the MD progeny, indicating that the target-derived piRNAs were generated by primary processing and not by ping-pong. The density of BC69 target-derived piRNAs in MD progeny seems to be ∼10-fold lower than that of P1152-derived piRNAs. (C) piRNAs are produced from the endogenous Adh locus targeted by the transgenic P1152 piRNAs in the MD progeny. The region targeted by the transgenic P1152 piRNAs is indicated by a blue bar. Shown are only piRNAs that uniquely mapped to the endogenous Adh locus. (D) Exposure to homologous piRNAs triggers piRNA generation from previously naïve loci. Note that piRNAs are generated from the regions that are not directly targeted by transgenic P1152 piRNA, indicating the spreading around the originally targeted sequence.
To test whether the deposition of H3K9me3 on the BC69 target also triggers initiation of piRNA biogenesis, we mapped piRNAs to the junction between the hsp70 and rosy sequence segments that is specific to this construct (Figs. 2A, 3B). As expected, no target-derived piRNAs were found in piRNA populations from the parental P1152 and BC69 strains. However, nine distinct sequences were detected in the MD progeny, while none was detected in the NMD progeny (Fig. 3B; Supplemental Fig. S3). This result shows that the same single-copy sequence acquires the H3K9me3 mark and becomes a source of new piRNA at the same time. Importantly, the target-derived piRNAs were detected in the MD progeny in the total piRNA population (two piRNA reads) and in complex with PIWI (seven reads), while they were absent in AUB and AGO3. This indicates that the new target-derived piRNAs are generated by primary processing and not by ping-pong. Overall, our data show that exposure to homologous piRNAs leads to an increase in the H3K9me3 mark and initiation of primary piRNA biogenesis on the target locus.
In addition to piRNAs that target the lacZ sequence, the P{lArB} transgene in the P1152 strain generates piRNAs that can potentially target three endogenous loci in the Drosophila genome, Adh, hsp70, and rosy. We tested whether exposure to homologous piRNAs initiated piRNA biogenesis at these genomic regions. Few piRNAs are generated from Adh, hsp70, and rosy loci in NMD progeny. In contrast, abundant piRNAs are generated from endogenous Adh and hsp70 sequences that flank the region targeted by transgene-derived piRNAs in MD progeny (Fig. 3C; data not shown). Importantly, the Adh sequences that generate piRNAs in MD flies are not directly targeted by transgene-derived piRNAs. This eliminates ping-pong processing initiated by transgene-derived piRNAs as a possible mechanism for biogenesis of piRNAs from the endogenous Adh locus. Instead, this result indicates that exposure to inherited piRNAs activates primary piRNA biogenesis at previously naïve genomic loci (Fig. 3D).
The H3K9me3 mark on piRNA clusters is recognized by the HP1 homolog Rhino
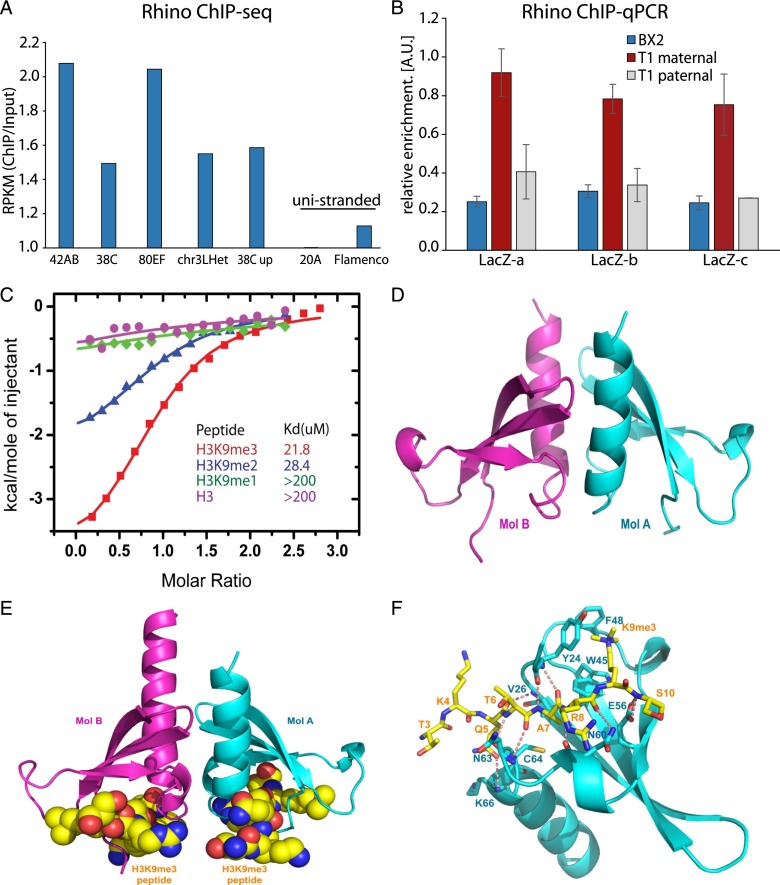
One possible reader of the H3K9me3 mark on piRNA cluster chromatin is Rhino, a germline-specific HP1 homolog that was previously shown to be enriched on double-stranded piRNA clusters and required for piRNA biogenesis (Klattenhoff et al. 2009). Rhino harbors a conserved chromodomain predicted to recognize methylated lysine residues (Supplemental Fig. S4). To get insights into the genome-wide distribution of Rhino on chromatin, we performed ChIP combined with deep sequencing (ChIP-seq) on ovaries of flies that expressed a BioTAP-tagged Rhino protein. Genome-wide analysis showed that Rhino is enriched on the regions that have high level of H3K9me3 (Supplemental Fig. S5). Analysis of the ChIP-seq data confirmed the previous observations of Rhino enrichment on chromatin of double-stranded, but not single-stranded, piRNA clusters (Fig. 4A).
Figure 4.
Chromodomain protein Rhino binds the H3K9me3 mark and is enriched on chromatin of piRNA clusters. (A) Rhino ChIP-seq signal is enriched at double-stranded piRNA clusters. No enrichment is seen on flamenco, a unistranded cluster that is active only in follicular cells, and on cluster 20A, a unistranded cluster that is expressed in germ cells. Shown are enrichments of ChIP-seq signal relative to input. Read counts are normalized to sequencing depth and region length (reads per kilobase per million [RPKM]). (B) Activity and H3K9me3 signal on the T1 and the BX2 transgenes correlates with Rhino binding. Rhino signal on T1 and BX2 was measured by ChIP-qPCR on three regions of lacZ (n = 2). Signal was normalized to input and compared with signal at a control genomic region (chr2R: 2336913–2337023). Error bars represent the standard error of the mean between two independent biological replicates. A two-sided t-test was used to calculate significance (P < 0.05). (C) Binding of Rhino chromodomain to different methylated states of the H3(1–15)K9me peptide measured by ITC. (D) Crystal structure of D. melanogaster Rhino chromodomain in the free state. The Rhino chromodomain forms a homodimer with each monomer, labeled as Mol A and Mol B. (E) Structure of the D. melanogaster Rhino chromodomain in complex with the H3(1–15)K9me3 peptide. Each monomer contains a bound H3K9me3 peptide. (F) Specific recognition of the H3K9me3 peptide by the Rhino chromodomain. Intermolecular hydrogen-bonding interactions are designated by dashed red lines.
Next, we tested whether the high level of the H3K9me3 mark on the transgenic T1 locus that was induced by transgenerationally inherited piRNAs leads to binding of Rhino protein. ChIP-qPCR showed that Rhino is enriched on the active T1 but not the inactive BX2 locus (Fig. 4B). Furthermore, Rhino localization on T1 chromatin is lost together with the H3K9me3 mark after paternal transmission of the locus.
To test whether the Rhino chromodomain directly recognizes the H3K9me3 mark, we monitored the binding affinity of the Rhino chromodomain as a function of the methylation state of the H3(1–15)K9me peptide using isothermal titration calorimetry (ITC). The H3K9me3 and H3K9me2 peptides showed similar binding affinities of 21.8 μM and 28.4 μM, respectively. In contrast, the H3K9me1 and unmodified H3 peptide showed no detectable binding affinity, establishing that recognition of the H3 tail by Rhino prefers high methylation states of Lys9 (Fig. 4C).
To further investigate the molecular mechanism of recognition of H3K9me3 by Rhino, we determined the crystal structures of the Drosophila Rhino chromodomain in the free form and in complex with the H3(1–15)K9me3 peptide at 1.5 Å and 2.5 Å resolution, respectively. The overall structure of the Rhino chromodomain (Fig. 4D) resembles that of the classic HP1 chromodomain, which has an N-terminal three-stranded twisted β sheet and a C-terminal short α helix, and forms a homodimer. The Rhino–H3(1–15)K9me3 complex also adopts a dimeric arrangement, similar to its free form (Fig. 4E; Supplemental Fig. S6A). We can trace and build the H3(1–15)K9me3 peptide from Thr3 to Ser10. The interaction between the peptide and the Rhino chromodomain consists of specific recognition of H3K9me3 by three conserved aromatic residues (Phe48, Try24, and Trp45), like that observed in the canonical recognition of H3K9me3 by the HP1 protein, complemented by hydrogen-bonding interactions mainly to the peptide backbone together with a few side chains (Fig. 4F; Supplemental Fig. S6B; Jacobs and Khorasanizadeh 2002). The dimer interface in the complex is stabilized by a network of hydrogen-bonding and hydrophobic interactions (Supplemental Fig. S7). Overall, our data suggest that the H3K9me3 mark installed on piRNA clusters in a manner that depends on transgenerationally inherited piRNAs is read by the Rhino chromodomain protein.
Rhino recruits the piRNA biogenesis factor Cutoff and promotes efficient transcription of piRNA clusters
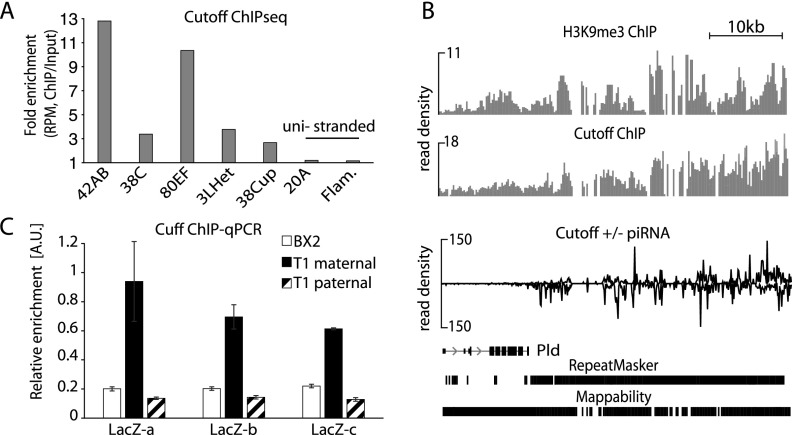
In the nucleus, Rhino colocalizes with Cutoff, a protein that plays an important but not well defined role in piRNA biogenesis (Pane et al. 2011). Cutoff is homologous to proteins that participate in RNA quality control and transcriptional termination in yeast and mammals (Xue et al. 2000; Kim et al. 2004; Jiao et al. 2010, 2013). Localization of Cutoff in specific nuclear foci was reported to be dependent on Rhino protein, suggesting that Cutoff might be recruited to chromatin through Rhino (Pane et al. 2011). We immunoprecipitated tagged Rhino and Cutoff proteins from Drosophila ovaries and confirmed that they form a complex (Supplemental Fig. S8). Next, we used genome-wide ChIP-seq to determine Cutoff localization on chromatin. In agreement with previous ChIP-PCR observations (Pane et al. 2011), we found that Cutoff localizes on double-stranded, but not single-stranded, piRNA clusters in perfect correlation with Rhino localization (Fig. 5A). The Cutoff profile at the border of piRNA clusters closely correlates with the H3K9me3 profile (Fig. 5B), suggesting that Cutoff is recruited to piRNA cluster chromatin through the interaction with Rhino that binds this mark. Interestingly, enrichment of Cutoff on piRNA cluster chromatin was abolished if, during the ChIP experiment, the cross-linked chromatin was treated with RNase prior to immunoprecipitation, indicating that Cutoff also interacts with nascent transcripts (Supplemental Fig. S9).
Figure 5.
Rhino recruits the piRNA biogenesis factor Cutoff to chromatin of piRNA clusters. (A) Cutoff ChIP-seq signal is enriched at double-stranded piRNA clusters. No enrichment is seen on unistranded flamenco and 20A clusters. Shown are enrichments of ChIP-seq signal relative to input. Read counts are normalized to sequencing depth and region length (RPKM). (B) Cutoff is enriched on chromatin of the active T1 but not the inactive BX2 or the paternally transmitted T1 transgene. Cutoff signal on T1 and BX2 as measured by ChIP-qPCR on three regions of lacZ (n = 2). Signal was normalized to input and compared with signal at a control genomic region (chr2R: 2336913–2337023). Error bars represent the standard error of the mean between two technical replicates. (C) Profiles of H3K9me3, Cutoff, and piRNAs along a fragment of the 42AB cluster (chromosome 2R: 2129920–2181079). The Cutoff ChIP-seq signal correlates with the H3K9me3 and piRNA profiles and progressively increases along the beginning of the 42AB cluster.
We tested whether the high level of the H3K9me3 mark and Rhino on the transgenic T1 locus leads to binding of the Cutoff protein. ChIP-qPCR showed that Cutoff is more enriched on the active T1 locus compared with the inactive BX2 locus (Fig. 5C). Furthermore, Cutoff localization on T1 chromatin is lost together with the H3K9me3 mark and Rhino after paternal transmission.
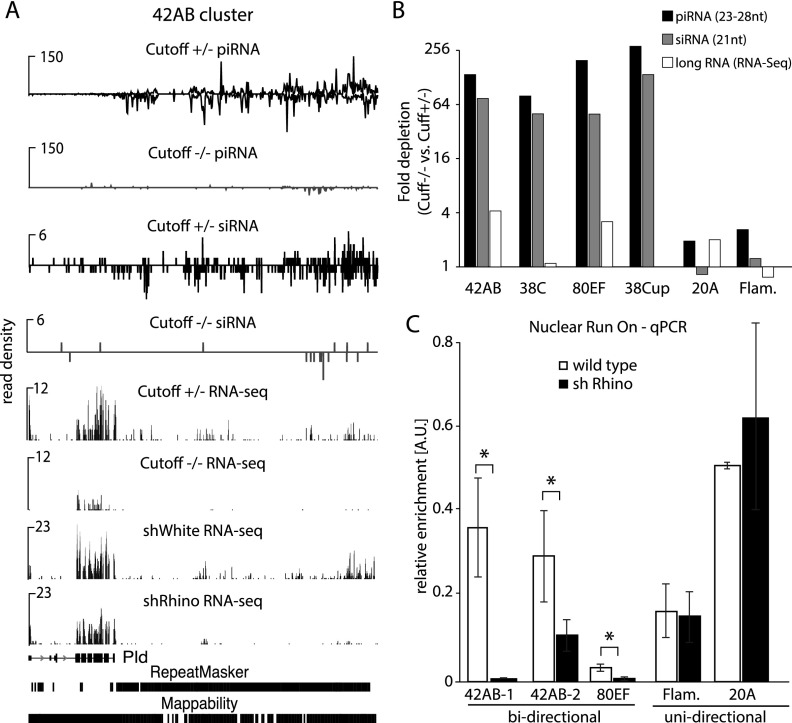
To study the functional importance of Cutoff localization on piRNA cluster chromatin, we profiled small and long RNA from ovaries of cutoff mutants and control flies. In agreement with previous observations (Pane et al. 2011), we found that piRNAs from double-stranded piRNA clusters are almost entirely eliminated in cutoff mutants (Fig. 6A,B). In contrast, cutoff mutation did not affect piRNA generation from flamenco, a unistranded cluster that is active only in follicular cells, and from 20A, a unistranded cluster that is expressed in germ cells but is not enriched in Rhino and Cutoff proteins. Double-stranded piRNA clusters also give rise to a significant fraction of the 21-nt siRNAs that are generated from dsRNAs by Dicer (Supplemental Fig. S10; Czech et al. 2008). Importantly, siRNAs from double-stranded clusters were also eliminated in the cutoff mutant (Fig. 6A,B), indicating that the function of Cutoff is broader than just targeting nascent transcripts to the piRNA processing machinery. Instead, this result suggests that Cutoff is required to generate precursor molecules for both siRNA and piRNA processing. Indeed, RNA sequencing (RNA-seq) of long rRNA-depleted RNA showed a decrease of transcripts from dual-stranded clusters upon depletion of Cutoff and Rhino (Fig. 6A,B). To study whether the Rhino/Cutoff complex is directly involved in transcription of piRNA precursor transcripts, we performed nuclear run-on/global run-on sequencing (GRO-seq) experiments on ovaries of control and Rhino-depleted flies. Quantification of nascent transcripts in two independent run-on experiments using two different methods—RT-qPCR and deep sequencing—showed that Rhino is indeed required for efficient transcription of dual-stranded piRNA clusters (Fig. 6C; Supplemental Fig. S11). Together, our data indicate that the H3K9me3 mark on piRNA clusters leads to recruitment of the Rhino/Cutoff complex, which is critical for transcription of piRNA precursors.
Figure 6.
Transcriptional output of piRNA clusters is decreased in the absence of Rhino and Cutoff. (A) Profiles of Cutoff, piRNA, siRNA, and long RNA (rRNA-minus RNA-seq) along a fragment of the 42AB cluster (chromosome 2R: 2129920–2181079). Both piRNAs and siRNAs as well as long RNA from the 42AB locus are almost entirely eliminated in cutoff mutant flies. A similar decrease in long RNA is seen in Rhino-depleted flies (shRhino). RNA-seq signal on the left corresponds to exons of the Pld gene. (B) The levels of piRNAs, siRNAs, and long RNAs (rRNA-minus RNA-seq) from double-stranded piRNA clusters are decreased in the Cutoff mutant. Shown are fold decrease in the normalized number of piRNA, siRNA, and long RNA reads generated from each piRNA cluster in the ovaries of cutoff homozygous mutant compared with control heterozygous animals. (C) Transcriptional output from double-stranded piRNA clusters is decreased upon Rhino depletion. Shown is RT-qPCR quantification of nascent transcripts from nuclear run-on performed on wild-type and rhino knockdown ovaries. The experiment was performed on two biological samples with three technical replicates each; P-values were calculated with a two-sided t-test. (*)P < 0.05.
Discussion
A previous study and our results reveal an essential role for a maternally transmitted transgenerationally inherited cytoplasmic factor in the generation of piRNAs. de Vanssay et al. (2012) showed that a maternal factor supplied to the progeny by females expressing piRNAs from the T1 locus activates piRNA generation from the homologous inactive BX2 locus (Fig. 1A). Furthermore, maintaining the activity of T1 in the subsequent generation also requires the maternal factor. We extended this observation to other systems and show that generation of piRNAs from a single-copy transgene inserted into a telomeric piRNA cluster also depends on a maternally transmitted cytoplasmic factor (Fig. 2B). This maternal factor also activates piRNA generation from a single-copy euchromatic sequence, which simultaneously becomes the target of repression and the source of new piRNAs (Figs. 2A, 3B). Finally, the activity of endogenous clusters in D. melanogaster also seems to require a maternally inherited factor: Our analysis of previously published piRNA profiles in interspecies hybrids between D. melanogaster females and Drosophila simulans males (Kelleher et al. 2012) showed that only D. melanogaster piRNA clusters generated piRNAs, while D. simulans piRNA clusters were inactive (Supplemental Fig. S12).
What is the nature of the maternally supplied epigenetic factor that triggers piRNA generation in the progeny? Multiple lines of evidence point to piRNAs themselves as the carriers of this epigenetic signal. First, as initially shown by de Vanssay et al. (2012), the epigenetic signal produced by the T1 locus does not require inheritance of the locus itself, indicating that the signal has a nonchromosomal nature (Fig. 1A). This eliminates the possibility that the signal is any kind of chromatin mark linked to the active locus. Second, the process of BX2 activation genetically depends on piRNA pathway genes but is independent of Dicer, which is required for siRNA biogenesis (de Vanssay et al. 2012). Third, both piRNAs and Piwi proteins are inherited from the maternal germline to the early embryos, while piRNAs are not transmitted through the sperm (Brennecke et al. 2008). Finally, piRNAs can be sequence-specific guides to identify and activate homologous loci. Importantly, recent studies have shown that piRNAs and the nuclear Piwi protein trigger installation of the H3K9me3 mark on homologous targets, providing a possible mechanism by which inherited piRNAs could lead to chromatin changes (Wang and Elgin 2011; Sienski et al. 2012; Le Thomas et al. 2013; Rozhkov et al. 2013). Together, these results strongly support the role of inherited piRNAs as a transgenerationally transmitted epigenetic signal that activates piRNA generation from homologous loci in the progeny.
How can transgenerationally inherited piRNAs activate piRNA generation from homologous loci? Our results imply two mechanisms that cooperate and work at different steps of piRNA biogenesis. In the cytoplasm, transgenerationally inherited piRNAs activate processing of complementary transcripts by the ping-pong amplification loop, as evidenced by a dramatic increase in piRNAs generated by the ping-pong processing upon MD of cognate piRNAs (Fig. 2C).
In the ping-pong processing, initial piRNAs guide generation of secondary piRNAs from complementary sequences. Previously, it was proposed that the ping-pong cycle requires two types of piRNA precursors: cluster transcripts and transcripts from active transposons provided in trans (Aravin et al. 2007a; Brennecke et al. 2007). Our results indicate that the ping-pong cycle can be activated by inherited piRNAs derived from the very same locus, provided that it is bidirectionally transcribed. Importantly, with the exception of one locus, all major piRNA clusters in the D. melanogaster germline are transcribed from both genomic strands, providing an abundant source of complementary transcripts to be used by the ping-pong process.
The major players in ping-pong processing in Drosophila are two Piwi proteins, AUB and AGO3, while the third Piwi protein, PIWI itself, is not involved in this process (Fig. 2C; Supplemental Fig. S2; Brennecke et al. 2007). AUB and AGO3 colocalize in cytoplasmic nuage granules, where the ping-pong processing is believed to take place (Brennecke et al. 2007). Therefore, the effect of inherited piRNAs on ping-pong processing impacts a late step of piRNA biogenesis after piRNA precursor transcripts are exported to the cytoplasm.
Although enhancing the ping-pong processing is clearly an important mechanism by which transgenerationally inherited piRNAs boost piRNA biogenesis, it cannot explain all of our observations, suggesting the existence of another mechanism. We found that maternal piRNAs are also required for the biogenesis of PIWI-associated piRNAs, although those are not generated by ping-pong processing (Fig. 2B,C). Using several genetic systems, we showed that inheritance of piRNAs leads to an increase of the H3K9me3 mark on regions homologous to the piRNAs. Importantly, acquisition of the H3K9me3 mark by genomic regions that did not previously produce piRNAs triggered piRNA generation in two transgenic systems (Figs. 1, 2). In contrast, the absence of inherited piRNAs led to a decreased H3K9me3 signal on homologous regions and a concomitant decrease of the corresponding piRNAs (Figs. 1, 2). These results suggest that modification of the chromatin structure of homologous genomic regions is the other mechanism by which transgenerationally inherited piRNAs turn on piRNA biogenesis in the progeny. Counterintuitively, we found that enrichment of the H3K9me3 mark, which is generally assumed to be repressive, strongly correlates with enhanced piRNA biogenesis. In agreement with our results, a previous study showed that biogenesis of piRNAs from double-stranded clusters requires Eggless/SETDB1, one of the methyltransferases responsible for installation of the H3K9me3 mark (Rangan et al. 2011).
Our analysis of several transgenic piRNA clusters revealed differences in the impact of inherited piRNAs on the level of the H3K9me3 mark. The inherited piRNAs seem indispensable to maintain high H3K9me3 signal on the transgenic T1 and BX2* loci (Fig. 1B,C). However, the absence of maternal piRNAs leads to a relatively mild decrease in H3K9me3 on the telomeric piRNA cluster in the P1152 strain (Fig. 3A). These results indicate that natural piRNA clusters are able to maintain a certain level of the H3K9me3 mark in a piRNA-independent fashion. This is not unexpected, as natural piRNA clusters are located close to heterochromatin, which is known to have a high level of H3K9me3 signal. In contrast, the T1 and BX2 transgenes are inserted in a euchromatic site that is normally lacking this mark. Overall, our data strongly support an essential role of the H3K9me3 mark in piRNA generation. They further reveal that enrichment of this mark on regions that generate piRNAs at least partially depends on the inheritance of homologous piRNAs from the previous generation. Finally, acquisition of the H3K9me3 mark by a naïve locus as a result of exposure to homologous piRNAs strongly correlates with the initiation of de novo primary piRNA biogenesis from such a locus. The exact mechanism for piRNA-dependent deposition of the H3K9me3 mark on piRNA regions remains to be elucidated; however, recent studies suggest that it might occur through recognition of nascent transcripts by the nuclear PIWI/piRNA complex, which is known to be deposited by the mother into the developing egg (Megosh et al. 2006; Brennecke et al. 2008) and has been shown to install H3K9me3 on its genomic targets (Kalmykova et al. 2005; Sienski et al. 2012; Le Thomas et al. 2013; Rozhkov et al. 2013).
The proposal that inherited piRNAs trigger piRNA biogenesis by changing the chromatin structure of homologous sequences raises the question of how the piRNAs distinguish a genuine transposon, a target that needs to be silenced, from a piRNA cluster that needs to be activated. Surprisingly, our results indicate that targeting by piRNAs leads to simultaneous repression of the target and activation of piRNA biogenesis from the same sequence. We found that targeting of a unique sequence by piRNAs triggers accumulation of the H3K9me3 mark, a decrease in target expression, and activation of piRNA biogenesis (Figs. 2, 3). Importantly, the target-derived piRNAs are not generated by the ping-pong mechanism (which would be a trivial explanation), as they are present in complex with PIWI, which does not participate in ping-pong processing. The similarity between transposon targets and piRNA-producing regions is supported by recent work that demonstrated that new transposon insertions in euchromatin start to generate piRNAs; i.e., they are becoming de novo piRNA clusters (Shpiz et al. 2014). Indeed, careful consideration suggests that the requirement to “silence” a genuine transposon target versus “activate’” a piRNA cluster is a false dichotomy: If nascent transcripts generated from piRNA target loci are channeled into the piRNA processing machinery instead of the standard mRNA processing pathway, the transcript will be effectively silenced, since no full-length mRNA will accumulate. The idea that the target of piRNA repression becomes a source of new piRNAs makes the distinction between piRNA clusters (source of piRNAs) and targets obsolete. Furthermore, our results expose a case in which the same genomic region is “silenced” and “activated” at the same time, depending on the exact output the researcher is looking at (generation of full-length mRNA or piRNAs). Similar phenomena might be more widespread than previously suspected, as studies in yeast suggest a very similar model in which centromeric repeats are “silenced” and generate siRNAs at the same time (Grewal and Jia 2007).
How can the high level of the allegedly repressive H3K9me3 mark enhance piRNA biogenesis? Our results show that the H3K9me3 mark provides a platform for the binding of Rhino, a chromodomain protein that shows specific enrichment over piRNA clusters (Fig. 4; Supplemental Fig. S5). As high levels of the H3K9me3 mark are also present in other genomic regions, it is possible that recognition of H3K9me3 is not sufficient for Rhino’s stable binding and that it interacts with other proteins to achieve its localization on chromatin of double-stranded clusters. Rhino forms a complex with Cutoff, a protein that is also required for piRNA biogenesis (Supplemental Fig. S8; Pane et al. 2011). The H3K9me3 mark, Rhino, and Cutoff colocalize at double-stranded piRNA clusters (Figs. 4A, 5A), and Cutoff is de-localized from nuclear foci in rhino mutants (Pane et al. 2011), suggesting that it is recruited to piRNA clusters through its interaction with Rhino. Taken together, our results suggest that Rhino and Cutoff, which were previously shown to be indispensable for piRNA generation from double-stranded piRNA clusters (Klattenhoff et al. 2009; Pane et al. 2011), are recruited to cluster chromatin through the H3K9me3 mark.
The exact molecular mechanism by which the Rhino/Cutoff complex activates piRNA biogenesis in the nucleus remains to be elucidated; however, two not necessarily mutually exclusive hypotheses can be proposed. First, Cutoff might bind and target nascent transcripts generated from piRNA clusters to the piRNA processing machinery instead of the normal pre-mRNA processing. In support of this idea, we found that the association of Cutoff with chromatin is RNA-dependent (Supplemental Fig. S9). We showed previously that inserting intron-containing heterologous gene sequences into piRNA clusters results in abundant piRNAs from both the exonic and intronic sequences, indicating that normal splicing is perturbed (Muerdter et al. 2012). According to the second hypothesis, the Rhino/Cutoff complex might enhance transcription of piRNA clusters, hence providing more precursors for piRNA biogenesis. Indeed, the run-on experiment showed that Rhino is required for efficient transcription of dual-stranded piRNA clusters. Furthermore, in agreement with an effect on transcription, we found that, in the cutoff mutant, both siRNAs and piRNAs as well as long RNAs are eliminated from double-stranded piRNA clusters (Fig. 6A,B), arguing against a role of Cutoff exclusive to piRNA processing.
The counterintuitive idea that the H3K9me3 mark might enhance rather than suppress transcription through binding of nonconventional epigenetic “readers” has interesting parallels in yeast. In Schizosaccharomyces pombe, H3K9 methylation induces binding of Swi6/HP1, which then recruits the Jumonji protein Epe1 that promotes nucleosome turnover, resulting in increased transcription of heterochromatic repeats and generation of siRNAs (Zofall and Grewal 2006; Grewal 2010). One possible mechanism by which Cutoff might enhance cluster transcription is by suppressing RNA polymerase II (Pol II) termination. Indeed, transgenic insertions that contain polyA cleavage/termination signals into piRNA clusters generate piRNAs downstream from the polyA signal, indicating that not only splicing but transcription termination is also suppressed in piRNA clusters (Muerdter et al. 2012). Ignoring transcription termination signals is likely an important feature of piRNA clusters, as otherwise, multiple signals within transposon sequences present in the clusters would terminate transcription and disrupt piRNA generation.
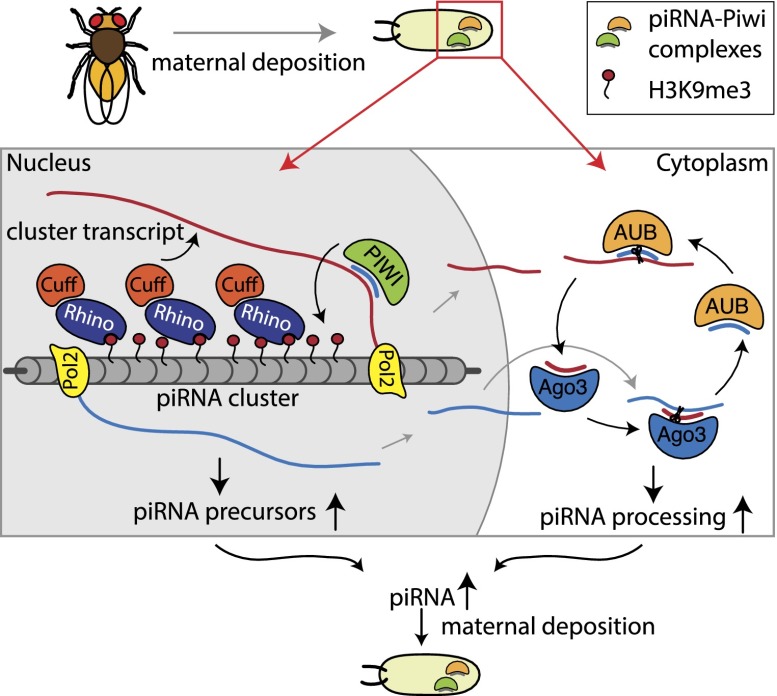
Overall, our data revealed that regions that produce piRNAs in Drosophila germ cells are defined by the epigenetic process of the transgenerational inheritance of cognate small RNAs. We found that inherited piRNAs trigger piRNA generation in the progeny by two mechanisms that seem to work simultaneously and cooperate to shape the final piRNA population (Fig. 7). In the nucleus, inherited piRNAs mark genomic regions that will give rise to new piRNAs and enhance early steps of piRNA biogenesis. In the cytoplasm, inherited piRNAs further trigger the post-transcriptional processing of cluster transcripts through the ping-pong amplification loop.
Figure 7.
A model for the role of transgenerationally inherited piRNAs in determining piRNA biogenesis in the next generation. The inherited piRNAs act in two ways: In the nucleus, piRNAs mediate installment of the H3K9me3 mark over piRNA-producing loci. An increased level of H3K9me3 is required for recruitment of the Rhino/Cutoff complex. Cutoff might target nascent transcripts to the piRNA processing machinery and/or enhance transcription. In the cytoplasm, piRNAs initiate the ping-pong amplification cycle, thereby boosting processing of piRNA cluster transcripts into mature piRNAs.
Materials and methods
Drosophila stocks
Strains T1 and BX2 were described in Dorer and Henikoff (1997) and de Vanssay et al. (2012). The strain P-1152, which carries the insertion of the P{lArB} construct in telomeric sequences of the X chromosome (site 1A), was described in Roche and Rio (1998). The strain BC69 that has the insertion of the P{A92} construct at a euchromatic location on chromosome 2L (site 35B10-35C1) was described in Lemaitre et al. (1993). Both stocks were a generous gift from S. Ronsseray. The Rhino-GFP fly line (GFP-tagged Rhino driven by the rhino promoter) was a generous gift from W. Theurkauff. The Cutoff-EGFP fly line (Nanos-GAL4/UASp-Cutoff-GFP), cuffwm25, and cuffqq37 were a generous gift from T. Schupbach. The Rhino-BioTAP flies were made by fusing the BioTAP tag (Alekseyenko et al. 2014) to the C-terminal region of the rhino gene under the UASp promoter. shRhino flies were obtained from the Bloomington Stock Center (stocks 34071 and 35171) and driven by Nos-Gal4 (stock 4937).
piRNA isolation, immunoprecipitation of PIWI proteins, and small RNA cloning
Immunoprecipitations of PIWI, AUB, and AGO3 proteins from D. melanogaster ovaries were carried out using rabbit polyclonal antisera directed against the N-terminal 14–16 amino acids according to previously described procedures (Brennecke et al. 2007). Small RNAs from immunoprecipitates and total RNA extracts were cloned as previously described in Brennecke et al. (2007) and Aravin et al. (2008). Briefly, small RNAs within a 19- to 29-nt window were isolated from 12% polyacrylamide gels. 3′ and 5′ linkers were ligated, and products were reverse-transcribed using SuperScript III (Invitrogen). Following PCR amplification, libraries were submitted for sequencing using the Illumina platform.
Analysis of the ping-pong processing
To determine the fraction of piRNAs that participate in ping-pong pairs, we counted uniquely mapped piRNA reads that map to opposite strands of each other and have a 10-base-pair (bp) distance between their 5′ ends. To account for variable sequencing depth, we carried out the analysis by sampling reads to ∼25% of the read counts in the library with the fewest reads in a region and repeating this 1000 times.
ChIP and ChIP-qPCR
ChIPs were carried out using commercially available antibodies (anti-H3K9me3 [ab8898], anti-RNA Pol II [ab5408], and anti-GFP [DHSB-4C9]) (Sanchez et al. 2014). ChIPs for Rhino and Cutoff were performed with anti-GFP antibodies on ovaries from transgenic animals expressing GFP-tagged proteins.
Prior to any ChIP experiment, 100 flies per immunoprecipitation condition were put on yeast for 2–3 d before ovary dissections. Ovaries were fixed using paraformaldehyde (PFA) at a final concentration of 1% and incubated for 10 min at room temperature. Samples were quenched by directly adding glycine (finale concentration 25 mM) for 5 min at room temperature and were then washed three times in PBS. Ovaries were afterward slightly dounced in Farnham buffer (5 mM HEPES at pH 8.0, 85 mM KCl, 0.5% NP-40/Igepal, protease inhibitor cocktail, 10 mM NaF, 0.2 mM Na3VO4) followed by strong douncing in RIPA buffer (20 mM Tris at pH 7.4, 150 mM NaCl, 1% NP-40/Igepal, 0.5% sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail, 10 mM NaF, 0.2 mM Na3VO4) prior to sonication. Sonication was done using a Bioruptor from Diagenode on medium power for 20 cycles (30 sec on, 30 sec off). Samples were centrifuged, and supernatant was collected and precleared for 2 h at 4°C using Dynabeads Protein G (Invitrogen) beads. If RNase treatment was required, half of the sample was incubated with 1 µL of 10 mg/mL RNase A during preclearing, and all samples were incubated for 2 h at room temperature. Meanwhile, in parallel, antibodies were conjugated to Dynabeads Protein G for 2 h at 4°C also. Five percent of precleared samples were saved for the Input fraction, and the rest was then transferred to the antibody-conjugated beads and incubated for 2 h at 4°C. Beads were then washed five times at 4°C using LiCl immunoprecipitation buffer (10 mM Tris at pH 7.5, 500 mM LiCl, 1% NP-40/Igepal, 1% sodium deoxycholate), rinsed in TE, and finally resuspended in Proteinase K buffer (200 mM Tris at pH 7.4, 25 mM EDTA, 300 mM NaCL, 2% SDS) with 100 µg of Proteinase K. Samples were incubated for 3 h at 55°C and then overnight at 65°C. DNA was then extracted following standard phenol-chloroform extraction, and concentration was measured by Qbit.
In the case of the Cutoff ChIP-seq, the experiment was performed using an altered protocol: Ovaries were treated with 2 mg/mL collagenase for 3 min at room temperature, washed once in PBS, and then fixed using fresh EGS (Thermo Scientific, 21565) solution (1.5 mg/mL final in PBS) for 30 min at room temperature. PFA was then added to the solution to a final concentration of 1% and incubated for 10 min at room temperature. Consecutive steps were as previously described. H3K9me3 and Rhino ChIP-qPCR experiments were performed on at least two biological replicates with three technical replicates each. Cutoff ChIP-qPCR was performed in two technical replicates. Values were normalized to respective inputs and to a genomic region known to be enriched in H3K9me3, Rhino, and Cutoff (chr2R: 2336913–2337023). Cutoff ChIP-qPCR, including RNase treatment, was normalized to respective inputs and to the control region RP49. For primers, see Supplemental Table S2. Error bars represent the standard error of the mean.
ChIP-seq and RNA-seq library construction and high-throughput data analysis
ChIP-seq and RNA-seq library construction and sequencing were carried out using standard protocols following the general principles described by Johnson et al. (2007) and Mortazavi et al. (2008), respectively. Libraries were sequenced on the Illumina HiSeq 2000 (50-bp reads) platform. Data sets for piRNAs were extracted from available Gene Expression Omnibus (GEO) data (Kelleher et al. 2012): D. melanogaster, SRX205641; D. simulans, SRX205642; and F1, SRX205643. The resulting sequencing reads were mapped against the genome using Bowtie 0.12.7 (Langmead et al. 2009) with the following settings: “-v 0–best–strata,” retaining only uniquely mappable reads with zero mismatches. Read mapping statistics for ChIP-seq data sets processed this way are presented in Supplemental Table S1. Data analysis was carried out using a combination of publicly available software tools and custom-written python scripts.
Rhino and Cutoff coimmunoprecipitation
Immunoprecipitation and Western blots were performed as previously described (Chen et al. 2007; Pane et al. 2011) on ovarian extracts from transgenic flies expressing Rhino-BioTAP and Cutoff-EGFP using anti-BioTAP antibody (Sigma, P1291) for immunoprecipitating Rhino-BioTAP. Western blots were carried out with anti-BioTAP antibody at 1:1000 and anti-GFP antibody at 1:1000 dilution in 5% nonfat milk.
Cloning, expression, and purification of Rhino
The D. melanogaster Rhino chromodomain (19–85) was inserted into the pETSumo expression vector (Invitrogen), which fuses a hexa-His tag plus a yeast sumo tag at the N terminus to the target protein. The plasmid was transformed into the Escherichia coli BL21(DE3) strain (Stratagene). The cells were cultured at 37°C until OD600 reached 0.8, and then the protein expression was induced with 0.2 mM IPTG overnight at 18°C. The hexa-His-Sumo tagged protein was purified using a HisTrap FF column (GE Healthcare). The tag was cleaved by Ulp1 protease and further removed by a second-step HisTrap FF column (GE Healthcare) purification. The target protein was further purified by a Hiload Superdex G200 16/60 column (GE Healthcare).
Crystallization and diffraction data collection
The purified Rhino protein was concentrated to ∼15 mg/mL in a storage buffer of 150 mM NaCl, 20 mM Tris (pH 7.5), and 5 mM DTT. Synthetic H3(1–15)K9me3 peptide was added at a threefold excess molar ratio to Rhino chromodomain and incubated for 30 min at 4°C. Crystallization was conducted at 20°C using the hanging drop vapor diffusion method by mixing 1 μL of protein solution and 1 μL of reservoir solution (0.2 M ammonium nitrate, 20% PEG3350) and was equilibrated against 0.5 mL of reservoir solution. The crystals were soaked into the reservoir solution supplemented with 15% glycerol before being flash-cooled in liquid nitrogen for diffraction data collection. The diffraction data were collected at beamline 24-ID-E at the Argonne National Laboratory, Chicago, and processed using the HKL2000 suite (Otwinowski and Minor 1997). The statistics of the diffraction data are summarized in Supplemental Table S4.
Structure determination and refinement
The structure of the Rhino chromodomain was solved using the molecular replacement method as implemented in Phaser (McCoy 2007) using the structure of the HP1 chromodomain as the search model (Protein Data Bank [PDB] ID: 1KNE). The model building was carried out using Coot (Emsley et al. 2010), and the structure refinement was conducted using Phenix (Adams et al. 2010). Throughout the refinement, a free R factor was calculated using 5% randomly chosen reflections. The stereochemistry of the structure model was analyzed using Procheck (Laskowski et al. 1993). The structure of the Rhino–H3(1–15)K9me3 complex was solved using the same protocol as the free-form structure. The statistics of the refinement and the structure model are summarized in Supplemental Table S4. All of the molecular graphics were generated using Pymol (DeLano Scientific, LLC).
ITC
All of the peptides, including H3(1–15), H3(1–15)K9me1, H3(1–15)K9me2, and H3(1–15)K9me3, were purchased from Tufts University Peptide Facility. Protein concentration was determined by absorbance spectroscopy. The Rhino chromodomain (23–85) was dialyzed against 25 mM sodium chloride, 20 mM Tris (pH 8.0), and 2 mM b-mercaptoethanol. The peptide was dissolved in the same buffer. ITC was conducted on a Microcal calorimeter ITC 200 instrument at 20°C. Binding curves were analyzed by nonlinear least-squares fitting of the data using Origin 7.0 software.
Nuclear run-on/GRO-seq
Nuclear run-on/GRO-seq experiments were performed on ovaries of shRhino flies (Bloomington Stock Center, 3407) driven by Nos-Gal4 (Bloomington Stock Center, 4937). Nos-Gal4 flies were used as a control. The nuclear run-on procedure was carried out as previously described (Shpiz et al. 2011) with slight modifications. BrUTP (5′-bromouridine-5′-triphosphate; Sigma, B7166)-labeled NRO-RNA was filtered through Illustra MicroSpin G25 columns (27-5325-01) twice to remove unincorporated BrUTP. The NRO-RNA was captured using the anti-BrdU antibody (Sigma, 032M 4753) for 1 h followed by incubation with Protein G beads (Dynabeads, Invitrogen, 1003D) for 1 h. The immunoprecipitation procedure was sequentially performed three times to yield highly enriched BrUTP RNA. As a negative control, the same procedure was performed on nonlabeled, total Drosophila ovary RNA. As a quality control, RT-qPCR was performed on 10% of the purified RNA with primers for Vasa, Rp49, and selected piRNA clusters. Libraries were cloned with the NEBNext Ultra Directional RNA library kit (E7420S) and sequenced on the Illumina HiSeq 2000 (50-bp reads) platform. Reads were mapped uniquely to the dm3 genome using Bowtie 0.12.7 (Langmead et al. 2009), allowing for no mismatches. The number of reads from the respective clusters are shown in Supplemental Table 5.
Accession codes
High-throughput sequencing data for ChIP-seq, RNA-seq, and piRNA-seq experiments are available through GEO (accession no. GSE59610). Coordinates and structure factors have been deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) PDB with the accession codes 4QUC for Rhino chromodomain in the free form and 4QUF for the Rhino chromodomain–H3K9me3 complex.
Acknowledgments
We thank members of the Aravin laboratory for discussion. We are grateful to the staff at beamline 24-ID-E at the Argonne National Laboratory and beamline X29A at the Brookhaven National Laboratory for support in diffraction data collection. E.S. is supported by a Ph.D. fellowship of the Boehringer Ingelheim Fonds. This work was supported by grants from the National Institutes of Health (R00 HD057233, R01 GM097363, and DP2 OD007371A) and by the Searle Scholar and the Packard Fellowship Awards to A.A.A., and by funds from the Abby Rockefeller Mauze Trust, Maloris Foundation, and Starr Foundation to D.P.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.245514.114.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Gorchakov AA, Kharchenko PV, Kuroda MI. 2014. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc Natl Acad Sci 111: 2488–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Hannon G, Brennecke J. 2007a. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 [DOI] [PubMed] [Google Scholar]
- Aravin A, Sachidanandam R, Girard A, Fejes-Toth K, Hannon G. 2007b. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin A, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon G. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone C, Aravin A, Sachidanandam R, Stark A, Hannon G. 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science, 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schüpbach T. 2007. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol 17: 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vanssay A, Bougé A-L, Boivin A, Hermant C, Teysset L, Delmarre V, Antoniewski C, Ronsseray S. 2012. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 490: 112–115 [DOI] [PubMed] [Google Scholar]
- Dorer DR, Henikoff S. 1997. Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics 147: 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore P. 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI 2010. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 20: 134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S, Jia S. 2007. Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. 2001. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128: 2823–2832 [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295: 2080–2083 [DOI] [PubMed] [Google Scholar]
- Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjian M. 2010. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Chang JH, Kilic T, Tong L, Kiledjian M. 2013. A mammalian pre-mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol Cell 50: 104–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. 2007. Genome-wide mapping of in vivo protein–DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Josse T, Teysset L, Todeschini A-L, Sidor C, Anxolabéhère D, Ronsseray S. 2007. Telomeric trans-silencing: an epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genet 3: 1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA. 2005. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res 33: 2052–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher E, Edelman N, Barbash D. 2012. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol 10: e1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. 2004. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522 [DOI] [PubMed] [Google Scholar]
- Kim V, Han J, Siomi M. 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana J, Zhang F, Schultz N, Koppetsch B, Nowosielska A, et al. 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. 1993. Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231: 1049–1067 [DOI] [PubMed] [Google Scholar]
- Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC. 2009. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res 19: 1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Ronsseray S, Coen D. 1993. Maternal repression of the P element promoter in the germline of Drosophila melanogaster: a model for the P cytotype. Genetics 135: 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27: 390–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63: 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megosh H, Cox D, Campbell C, Lin H. 2006. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol 16: 1884–1894 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Moshkovich N, Lei E. 2010. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet 6: e1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Olovnikov I, Molaro A, Rozhkov N, Czech B, Gordon A, Hannon G, Aravin A. 2012. Production of artificial piRNAs in flies and mice. RNA 18: 42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov IA, Kalmykova AI. 2013. piRNA clusters as a main source of small RNAs in the animal germline. Biochemistry (Mosc) 78: 572–584 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pane A, Jiang P, Zhao DY, Singh M, Schupbach T. 2011. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J 30: 4601–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Malone C, Navarro C, Newbold S, Hayes P, Sachidanandam R, Hannon G, Lehmann R. 2011. piRNA production requires heterochromatin formation in Drosophila. Curr Biol 21: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche SE, Rio DC. 1998. Trans-silencing by P elements inserted in subtelomeric heterochromatin involves the Drosophila Polycomb group gene, Enhancer of zeste. Genetics 149: 1839–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray S, Boivin A, Anxolabehere D. 2001. P-element repression in Drosophila melanogaster by variegating clusters of P-lacZ-white transgenes. Genetics 159: 1631–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray S, Josse T, Boivin A, Anxolabehere D. 2003. Telomeric transgenes and trans-silencing in Drosophila. Genetica 117: 327–335 [DOI] [PubMed] [Google Scholar]
- Rozhkov N, Hammell M, Hannon G. 2013. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev 27: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez P, Daniels KJ, Park YN, Soll DR. 2014. Generating a battery of monoclonal antibodies against native green fluorescent protein for immunostaining, FACS, IP, and ChIP using a unique adjuvant. Monoclon Antib Immunodiagn Immunother 33: 80–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Olovnikov I, Sergeeva A, Lavrov S, Abramov Y, Savitsky M, Kalmykova A. 2011. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res 39: 8703–8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Ryazansky S, Olovnikov I, Abramov Y, Kalmykova A. 2014. Euchromatic transposon insertions trigger production of novel Pi- and endo-siRNAs at the target sites in the Drosophila germline. PLoS Genet 10: e1004138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J. 2012. Transcriptional silencing of transposons by piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 [DOI] [PubMed] [Google Scholar]
- Todeschini A-LL, Teysset L, Delmarre V, Ronsseray S. 2010. The epigenetic trans-silencing effect in Drosophila involves maternally-transmitted small RNAs whose production depends on the piRNA pathway and HP1. PLoS ONE 5: e11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Elgin S. 2011. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci 108: 21164–21169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Bai X, Lee I, Kallstrom G, Ho J, Brown J, Stevens A, Johnson AW. 2000. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol Cell Biol 20: 4006–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch B, Schultz N, Vreven T, Meignin C, Davis I, Zamore P, et al. 2012. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 151: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Grewal SI. 2006. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell 22: 681–692 [DOI] [PubMed] [Google Scholar]