In yeast, ribosome production is tightly controlled by transcriptional regulation of the 138 ribosomal protein genes (RPGs), yet the molecular basis for their coregulation is largely unknown. Here, a multifaceted analysis of RPG promoter architecture and function identifies two prevalent promoter types. Rapid depletion experiments establish a hierarchy of transcription factor (TF) binding in which Rap1 acts as a pioneer factor required for binding of all other TFs. Further studies suggest that direct interactions between specific TFs and unusually dynamic, promoter-proximal nucleosomes play a key role in RPG transcriptional regulation.
Keywords: transcription, ribosomal protein gene, yeast, Rap1, fragile nucleosome
Abstract
In yeast, ribosome production is controlled transcriptionally by tight coregulation of the 138 ribosomal protein genes (RPGs). RPG promoters display limited sequence homology, and the molecular basis for their coregulation remains largely unknown. Here we identify two prevalent RPG promoter types, both characterized by upstream binding of the general transcription factor (TF) Rap1 followed by the RPG-specific Fhl1/Ifh1 pair, with one type also binding the HMG-B protein Hmo1. We show that the regulatory properties of the two promoter types are remarkably similar, suggesting that they are determined to a large extent by Rap1 and the Fhl1/Ifh1 pair. Rapid depletion experiments allowed us to define a hierarchy of TF binding in which Rap1 acts as a pioneer factor required for binding of all other TFs. We also uncovered unexpected features underlying recruitment of Fhl1, whose forkhead DNA-binding domain is not required for binding at most promoters, and Hmo1, whose binding is supported by repeated motifs. Finally, we describe unusually micrococcal nuclease (MNase)-sensitive nucleosomes at all RPG promoters, located between the canonical +1 and −1 nucleosomes, which coincide with sites of Fhl1/Ifh1 and Hmo1 binding. We speculate that these “fragile” nucleosomes play an important role in regulating RPG transcriptional output.
In yeast cells growing under optimal nutrient conditions, ∼50% of all RNA polymerase II (RNAPII) initiation events occur at a ribosomal protein gene (RPG) promoter (Warner 1999). Consistent with this substantial energetic investment, the suite of 138 RPGs (59 of the 79 RPs are encoded by a pair of identical or very similar genes) is highly regulated in response to nutrient and stress conditions (DeRisi et al. 1997). Despite the high transcription rate and coregulation of RPGs, examination of their promoter sequences has not yielded clear insights into the relationship between promoter architecture and regulation. Early bioinformatic (Lascaris et al. 1999) and ChIP–chip (chromatin immunoprecipitation [ChIP] combined with microarray) analyses (Lieb et al. 2001) indicated that the transcription factor (TF) Rap1 binds to the promoters of many RPGs but also to a roughly equal number of other genes that are regulated differently. Subsequently, the forkhead (FH)-like DNA-binding protein Fhl1 was shown to localize nearly exclusively to RPGs (Lee et al. 2002), coincident with an essential interacting protein called Ifh1 (Jorgensen et al. 2004; Martin et al. 2004; Schawalder et al. 2004; Wade et al. 2004; Rudra et al. 2005). Perplexingly, a DNA sequence motif that determines the specificity of Fhl1 (and Ifh1) binding to RPG promoters has been surprisingly difficult to identify. One such potential motif, called IFHL, has instead been associated with binding of Hmo1 (Hall et al. 2006; Lavoie et al. 2010), an HMG-B-box protein found at many RPG promoters.
Here we present a multifaceted analysis of RPG promoter architecture and function. By combining ChIP-seq (ChIP with high-throughput DNA sequencing), MITOMI (mechanically induced trapping of molecular interactions), qPCR (quantitative PCR)-ChIP, and bioinformatic analysis, we show that the vast majority of RPG promoters can be classified into one of two distinct promoter architectures with respect to the localized binding of four TFs: Rap1, Fhl1/Ifh1, and Hmo1. Using both cis element mutations and strains that permit the rapid depletion of specific TFs, we establish a hierarchy of factor binding in which Rap1 is required for binding of the other three TFs, and Hmo1 is required, in addition, for Fhl1/Ifh1 binding at category I promoters. Surprisingly, we show that the FH DNA-binding domain of Fhl1 plays a major role in promoter binding at only a limited subset of RPGs, all of which lack Hmo1 binding. Furthermore, we present both in vitro and in vivo evidence that Hmo1 binding is indeed promoted by a specific DNA sequence found in multiple copies at its sites of binding. Finally, we show that the sites of Hmo1 and Fhl1/Ifh1 binding at category I promoters as well as Fhl1/Ifh1 binding at category II promoters are coincident with unusually micrococcal nuclease (MNase)-sensitive nucleosomes located in between the canonical +1 and −1 nucleosomes at these promoters in what have been previously characterized as nucleosome-free or nucleosome-depleted regions. We present evidence that the MNase sensitivity of these so-called “fragile” nucleosomes (FNs) depends to some extent on Rap1 binding but is likely to be driven by other trans-acting factors and promoter DNA sequences. Our data thus suggest that direct interactions between specific TFs and unusually dynamic promoter-proximal nucleosomes may play an important role in RPG transcriptional regulation.
Results
Identification of two predominant promoter architectures at RPGs
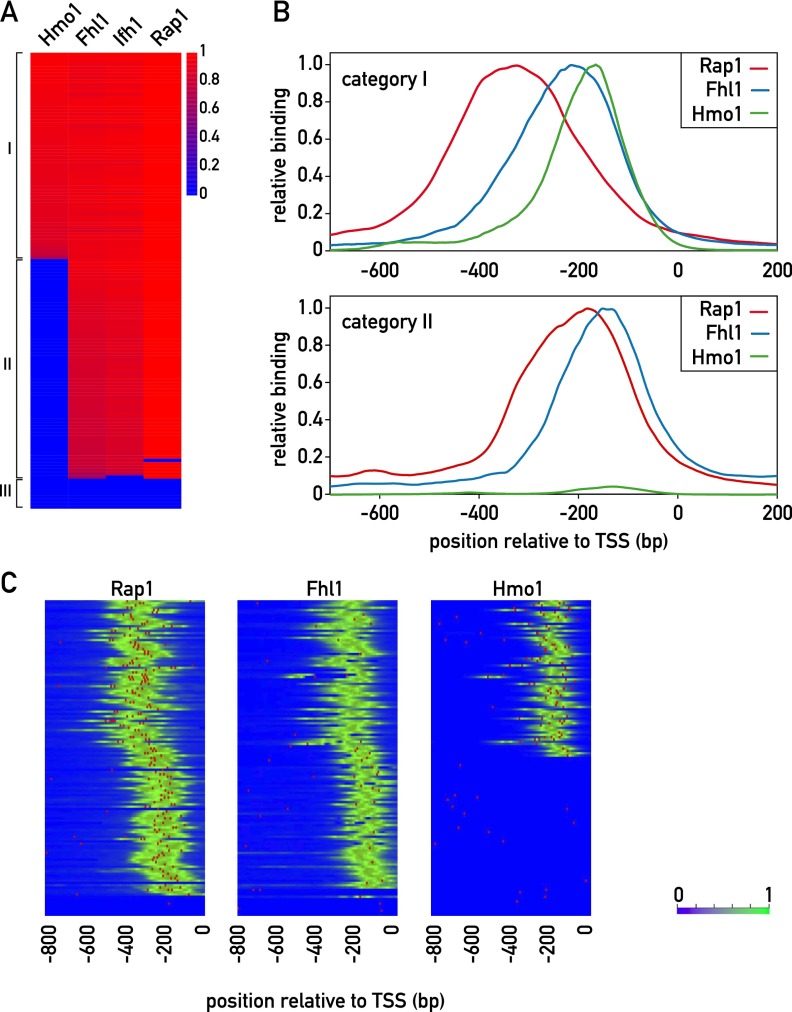
To gain insight into the organization of RPG regulatory regions, we generated ChIP-seq data sets for Rap1, Fhl1, Ifh1, and Hmo1. Based on the pattern of binding of these proteins (see the Materials and Methods), we identified two major categories of promoter architecture, both of which display highly stereotypical patterns of Rap1 and Fhl1/Ifh1 binding (Fig. 1). The first of these two groups (category I; 69 members) also displays Hmo1 binding at a position immediately downstream from Fhl1/Ifh1, whereas category II promoters (60 members) show no detectable Hmo1 binding (Supplemental Table S2). The remaining promoters (category III, nine members) did not show signals for any of the four TFs but instead showed evidence of Abf1 binding (Supplemental Fig. S1A). The cross-correlation between signals for all RPG promoters reveals that, on average, the Fhl1/Ifh1 pair and Hmo1 bind ∼100 and 150 base pairs (bp), respectively, downstream from Rap1 (Supplemental Fig. S1B). Examination of the average ChIP-seq signal in each of the categories shows that in category I promoters, Rap1 and Fhl1/Ifh1 bind further upstream of the transcription start site (TSS) than the category II promoters (Fig. 1B).
Figure 1.
Identification of distinct promoter architectures for RPG promoters. (A) ChIP-seq signals for the TFs Hmo1, Fhl1, Ifh1, and Rap1. The log-transformed signals (peak area) are color-coded as indicated. Signals were normalized to the maximum value over the whole set of RPG promoters for each TF. In the cases where no peak was found on the promoter, the value is given as 0. (B) Normalized average ChIP-seq profiles (Y-axis) for category I (top panel) and category II (bottom panel) promoters (X-axis; 0 indicates TSS). (Red) Rap1; (blue) Fhl1; (green) Hmo1. (C) Rap1, Fhl1, and Hmo1 ChIP-seq signals (green) and motifs (red) for all RPG promoters, ordered as in A. Motifs were defined according to the ChIP-seq-derived weight matrix in Supplemental Figure S2A. (Left) For Rap1, the threshold was 9.6 bits, and the maximum score was 23.5 bits. (Middle) For Fhl1, the threshold was 8.7 bits, and the maximum score was 12.5 bits. (Right) For Hmo1, the threshold was 10.6, and the maximum score was 15.2 bits. See also Supplemental Figures S1–S4 and Supplemental Tables S2 and S3.
The DNA sequences around peaks of Rap1, Fhl1, and Hmo1 binding were analyzed using MEME (Bailey et al. 2006) to identify conserved motifs. As expected, this identified a sequence highly related to motifs derived from PBM and MITOMI in vitro analyses of Rap1 (Supplemental Fig. S2A; Badis et al. 2008; Zhu et al. 2009) and previously associated with RPG promoters (Lascaris et al. 1999). We found that Rap1 ChIP-seq peaks at a large number of RPGs (75) were often coincident with two predicted binding sites, most of which (54) were arranged in a unique head-to-tail orientation with respect to the TSS, and had an end-to-end spacing between sites of 16–40 bp (Supplemental Fig. S1C). Of the 43 promoters predicted to contain only a single site, all displayed the same orientation with respect to the TSS (Supplemental Fig. S1C). In total, we identified five different groups with respect to predicted Rap1-binding site number and orientation. The average Rap1 ChIP-seq signals were similar for all five groups, although smaller peaks were observed in some cases where only one Rap1 motif is present (Supplemental Fig. S1D). Interestingly, while there were no major differences in the nucleotide composition of the sites from the five groups, the Rap1 motif from non-RP promoters showed a distinctly different consensus sequence that is much more similar to the telomeric Rap1 motif, which displays a strict CA/TG strand bias (Supplemental Fig. S2B). Although the presence of closely spaced multiple Rap1-binding sites at promoters has been described previously (Lascaris et al. 1999; De Sanctis et al. 2002), a recent report using a high-resolution ChIP method (ChIP-exo) (Rhee and Pugh 2011) has indicated that Rap1 binds to a unique site at most RPGs. We addressed this discrepancy through site mutagenesis experiments described below. Finally, we noted that there is a slight but statistically significant excess of category I genes in class 1 and, conversely, of category II genes in class 5 (overall P = 0.025 from Fisher’s exact test, with adjusted residual values of ±2 for class 1 and ±2.6 for class 5) (Supplemental Fig. S1C).
Previous motif searches using ChIP–chip Fhl1-binding data returned only the Rap1-binding motif (Harbison et al. 2004; Macisaac et al. 2006). By examining smaller regions (50 bp) around the centers of Fhl1 ChIP-seq peaks, we discovered a motif that matches a PBM-derived binding site for the FH domain of Fhl1 (Badis et al. 2008; Zhu et al. 2009) that is in turn similar to one that we obtained for this domain by MITOMI analysis (Supplemental Fig. S2A). Curiously, we observed that this Fhl1 motif overlapped with Fhl1 ChIP-seq signals in 55% of category II promoters but only 15% of category I promoters (Fig. 1C), thus raising questions about the role of the Fhl1 FH domain in promoter binding (see below).
Hmo1 was previously thought to bind nonspecifically to DNA (Kamau et al. 2004). Although a potential Hmo1-binding motif (referred to as the IFHL motif) has been described based on motif searches of genome-wide ChIP–chip binding data (Hall et al. 2006; Lavoie et al. 2010), Hmo1 was not thought to recognize this motif directly (Kasahara et al. 2007) and was reported to show no binding to multimerized motifs in vivo (Hall et al. 2006). However, using oligonucleotide pools constituting a De Bruijn representation of all possible 8-mer DNA sequences in a k-MITOMI (Geertz et al. 2012b) analysis of Hmo1 binding, we identified a sequence that is quite similar to the IFHL motif (Supplemental Fig. S2A). This finding raises the possibility that Hmo1 binding in vivo relies at least in part on sequence-specific DNA contacts (see below).
We next asked whether the precise location of TFs with respect to each other and to the TSS might have some relationship to TF-binding strength, as inferred by the measured number of tag counts underneath the respective ChIP-seq peaks. We noted that for category I promoters (Hmo1-bound), the Rap1 ChIP-seq peak intensities show a relatively weak but significant positive correlation (R = 0.46; P < 0.001) with the distances between Rap1 and Hmo1 peaks (Supplemental Fig. S3A). However, the predicted occupancies of these category I gene Rap1 sites, as estimated from their Rap1 motifs (see the Materials and Methods), do not show a significant correlation with the Rap1–Hmo1 distances (Supplemental Fig. S3B). Interestingly, the ChIP-seq signals for Fhl1 did not show significant correlation with Rap1–Hmo1 distance (Supplemental Fig. S3C). On the basis of these findings, we developed a thermodynamic model incorporating the DNA-binding energies and interaction energies between the TFs, which includes a distance-dependent interaction energy between Hmo1 and Rap1 bound at category I promoters (see the Supplemental Material; Supplemental Fig. S3D). One physical interpretation of this model is that Rap1 and Hmo1 contact each other and that this interaction requires DNA looping, which is more favorable at longer Rap1–Hmo1 spacing.
Relationship between RPG promoter architecture and expression
To investigate the effect of promoter architecture on transcriptional activity, we measured the activity of 119 RPG promoters using chromosomal reporter constructs. We created a library of diploid yeast strains, each of which expresses both cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) from identical RPG promoters integrated at the LEU2 locus on the two chromosome III homologs (Raser and O’Shea 2004). We grew these 119 yeast strains simultaneously in a microfluidic device that allowed for rapid perfusion of fresh medium, thereby permitting us to measure reporter output at the single-cell level during steady-state exponential growth (Denervaud et al. 2013). There was no significant difference between average promoter activities of category I and category II genes as measured by YFP fluorescence (Supplemental Fig. S4A; Supplemental Table S3). However, all of the highest-activity promoters were in category II, whereas category I promoters displayed less overall variation in expression level, as measured by the standard deviation of their activities, which was significantly smaller compared with those in category II (P < 0.0005 from F-test of equality of variances). This conclusion is consistent with another study that found that RPG promoters with intermediate activity are enriched for Hmo1 binding (Zeevi et al. 2011).
To identify which factors influence promoter activity, we used linear regression models that incorporate different features of our ChIP-seq data (peak area and distance between peaks) together with published nucleosome occupancy data (see the Materials and Methods). For category I promoters (Supplemental Fig. S4B,C, red), Hmo1 ChIP-seq signal intensity explains the largest fraction of the variance, although it does not quite reach statistical significance (P = 0.11) (Supplemental Fig. S4B,C). On the other hand, for category II promoters (Supplemental Fig. S4B,C, blue), Fhl1 ChIP-seq signal intensity together with nucleosome occupancy contributes significantly to the observed variance. This distinction between category I and category II promoters may arise from the fact that Hmo1 makes extensive, direct DNA contacts with the former class, whereas Fhl1 binds with high sequence selectivity only to the latter (see below). We also observed that for category II promoters, the promoter activities are negatively correlated with Rap1 signal strength in ChIP-seq, in contrast to a positive correlation for category I promoters (Supplemental Fig. S4B). Overall, the fraction of variance in promoter activities explained from the multilinear model for category I promoters is less (20%; P = 0.37 from F-test of the regression model) compared with that for the category II promoters (37%; P < 0.01 from F-test of the regression model) (Supplemental Fig. S4C).
To determine whether the different promoter architectures affect regulation, we compared their responses to various stress conditions using publically available microarray data. We identified only heat shock as a condition where the two categories displayed a small but significant difference, with repression of category I genes being slightly more pronounced than that of category II (P-value = 0.002 from two-sample t-test) (Supplemental Fig. S4D; Shivaswamy and Iyer 2008). The same trend was observed for RPG repression as cells enter into stationary phase (Shivaswamy and Iyer 2008), although the magnitude of the difference was minimal (Supplemental Fig. S4E). A similar tendency was observed following rapamycin inhibition of the TORC1 kinase, a key component of the signal transduction network that links cell growth with nutrient availability (De Virgilio and Loewith 2006), although in this case, the observed difference was not statistically significant (P = 0.1 from two sample t-test) (Supplemental Fig. S4F). Additional stresses were examined, including glucose limitation (Brauer et al. 2008), osmotic stress, and oxidative stress (Berry and Gasch 2008). However, under these stresses, no significant difference in RPG expression between categories I and II was observed (Supplemental Fig. S4G–I). On the whole, these observations indicate that the two promoter categories confer remarkably similar regulatory properties, consistent with the fact that they employ three common factors (Rap1, Fhl1, and Ifh1) in a stereotypical arrangement.
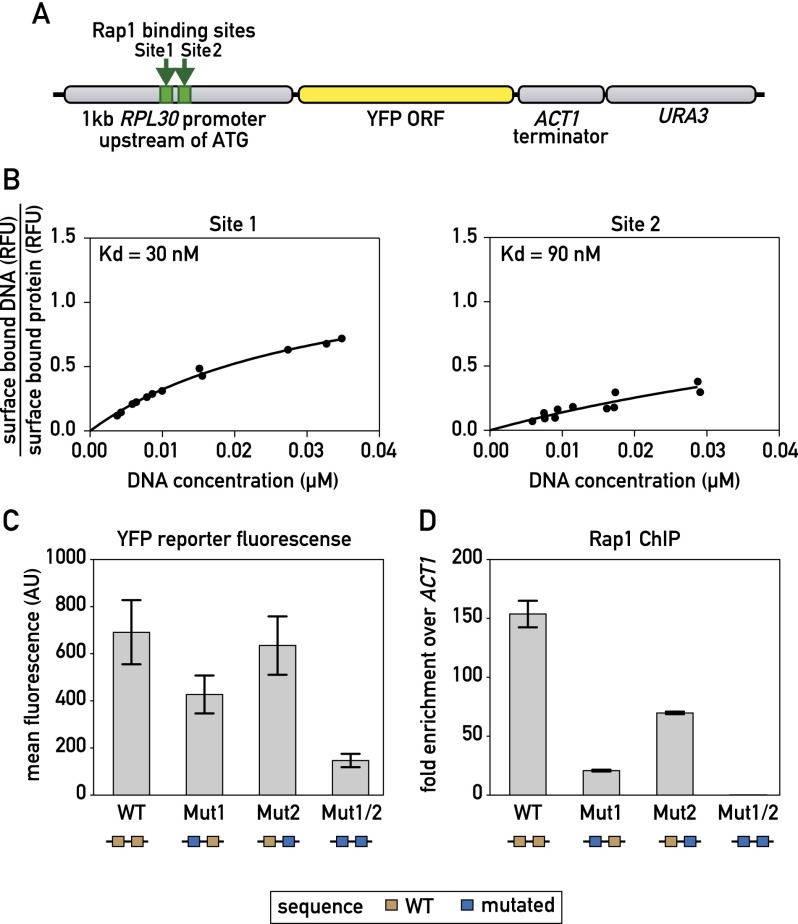
Tandem Rap1 sites make independent contributions to binding and activation
Our MEME analysis suggested that the majority of Rap1 ChIP-seq peaks are associated with two closely spaced motifs. However, a ChIP-exo study reporting binding at nucleotide resolution indicated that Rap1 binds to unique sites at most RPG promoters (Rhee and Pugh 2011). To address this discrepancy, we examined a 1-kb fragment containing the RPL30 promoter (Fig. 2A) where we had identified two Rap1 motifs underneath a ChIP-seq peak (Fig. 2B), but ChIP-exo revealed binding only to the more upstream site (site 1). We first used MITOMI to measure the binding affinity of the two sites separately and found site 1 binding to be slightly stronger than that of site 2 (Fig. 2B). We next generated mutations in the individual sites (Mut1 and Mut2) and showed that they each abolished Rap1 binding, as expected (Supplemental Fig. S5A,B). These mutations were then introduced into the RPL30 promoter-YFP construct individually and together. Interestingly, reporter fluorescence was decreased when site 1 was mutated and only slightly decreased when site 2 was mutated (Fig. 2C). However, when both Rap1 sites were mutated, transcriptional output was significantly decreased (Fig. 2C). Rap1 binding as measured by ChIP (using primer pairs specific to the mutated sites) was significantly decreased for both Mut1 and Mut2, with the effect of Mut1 more severe, in line with the MITOMI measurements of relative strength of the two sites (Fig. 2D). Only when both Rap1 sites were mutated, however, was Rap1 binding completely abolished in vivo (Fig. 2D). Even under these circumstances, YFP accumulation was reduced only to ∼25% of the wild-type level. This residual expression might result from a downstream poly(dA:dT) tract in the promoter (Goncalves et al. 1995; Iyer and Struhl 1995; Zhao et al. 2006) or through Rap1-binding events that are not detectable by the ChIP assay. In summary, these data indicate that closely spaced pairs of Rap1-binding sites are likely to cooperate in activation at most RPGs together with additional cis elements, including poly(dA:dT) tracts.
Figure 2.
Correlation between Rap1-binding constant and promoter output. (A) Schematic showing the RPL30 promoter-YFP reporter construct that was integrated at the LEU2 locus. (B) MITOMI measurements of the fraction of surface-bound target DNA are plotted against the concentration of target DNA in solution for the indicated binding site probes. Dissociation constants (Kd) were determined by performing a nonlinear regression fit using a one-site binding model. (C) YFP fluorescence measured by flow cytometry of exponentially growing cells containing the indicated RPL30 promoter-YFP reporter constructs. Data are represented as mean ± SEM. (D) Rap1 occupancy (qPCR-ChIP) on the indicated RPL30 promoter-YFP reporter constructs. Data are represented as mean ± SEM. See also Supplemental Figure S5.
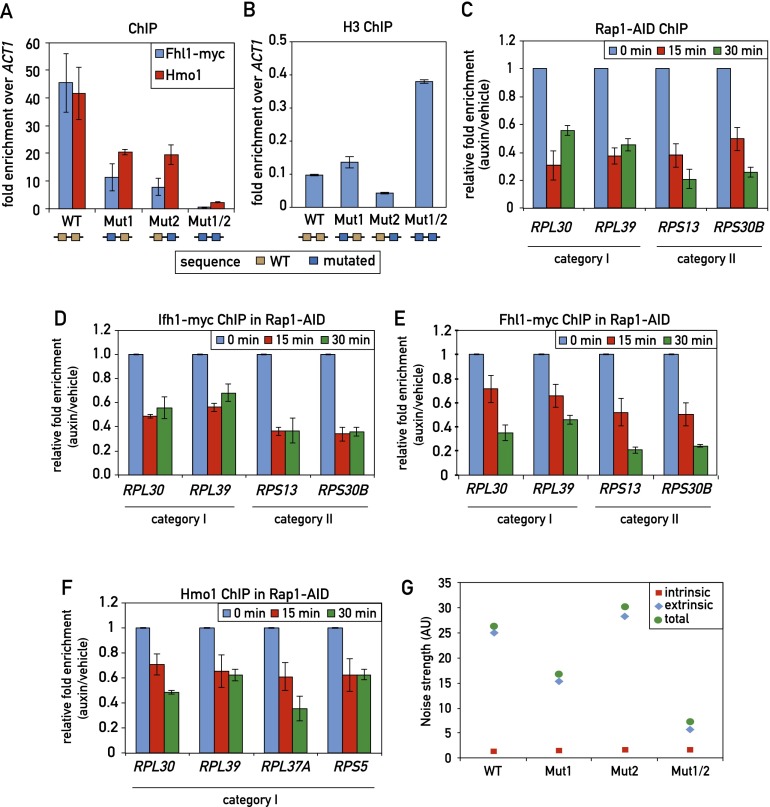
Rap1 is critical for Fhl1 and Hmo1 recruitment but not for noise suppression
Previous studies have implicated Rap1 binding in the recruitment of both Fhl1 (Zhao et al. 2006) and Hmo1 (Hall et al. 2006). However, these studies looked at large deletions in an RPG promoter or at the effect of mutating Rap1-binding sites outside of the context of an RPG promoter. To address this question more directly, we first examined the double Rap1 site mutation (Mut1/2) in the RPL30 promoter (Fig. 2) that abolishes detectable Rap1 binding in vivo. We found that this mutation also leads to a dramatic reduction (>10-fold) in both Fhl1 and Hmo1 association, whereas the single Rap1 mutations, which retain partial Rap1 binding, decrease Fhl1 binding by approximately fourfold and Hmo1 binding by approximately twofold (Fig. 3A). Rap1 is reported to evict nucleosomes (Ganapathi et al. 2011), and we also noted that the double-site mutant was unique in leading to a marked increase in histone H3 binding (Fig. 3B).
Figure 3.
Effect of Rap1 on TF recruitment and transcription noise strength. (A) Fhl1 and Hmo1 promoter occupancy (qPCR-ChIP) on the indicated RPL30 promoter alleles. Data are represented as mean ± SEM. (B) Histone H3 occupancy (qPCR-ChIP) on the indicated RPL30 promoter alleles. Data are represented as mean ± SEM. (C–F) RPG promoter occupancy of Rap1-AID (C), Ifh1-Myc (D), Fhl1-Myc (E), and Hmo1 (F) at the indicated times following auxin-induced depletion of AID-tagged Rap1. Data are plotted as auxin relative to vehicle treatment and normalized to t = 0. Data are represented as mean ± SEM. (G) The intrinsic and extrinsic noise strength of the wild-type and mutant versions of the RPL30 promoter were measured by microscopy using diploid yeast cells containing both RPL30 promoter-YFP reporter and RPL30 promoter-CFP reporter constructs.
In an effort to generalize the above observation, we used the auxin-induced degron (AID) system (Nishimura et al. 2009) to rapidly deplete the essential Rap1 protein. We found that introduction of the AID tag to a nonessential region in the N-terminal part of Rap1 had no obvious effect on cell growth but led to a rapid loss of ChIP signal (Fig. 3C) and total protein (data not shown) following auxin addition (see the Materials and Methods for details). Concomitant with the loss of Rap1, we detected a significant decrease in Ifh1, Fhl1, and Hmo1 binding (Fig. 3D-F) as well as that of RNAPII (Supplemental Fig. S5C), indicative of a decrease in transcription.
Yeast RPG expression displays exceptionally low levels of intrinsic noise (measured in our experiments by differences between YFP and CFP levels within individual cells) (see Raser and O’Shea 2004 for an introduction to intrinsic and extrinsic gene expression noise). This feature has been linked to promoter architecture (Newman et al. 2006) and, in one theoretical study, the binding of Rap1 (Muller and Stelling 2009). However, contrary to expectation, we found that reducing Rap1 binding at the RPL30 promoter did not increase intrinsic noise strength (Fig. 3G). Since reduced Rap1 binding also led to a strong drop in both Fhl1 and Hmo1 binding (Fig. 3A), these data suggest that the TFs themselves are not responsible for the observed noise characteristics of this and presumably other RPG promoters.
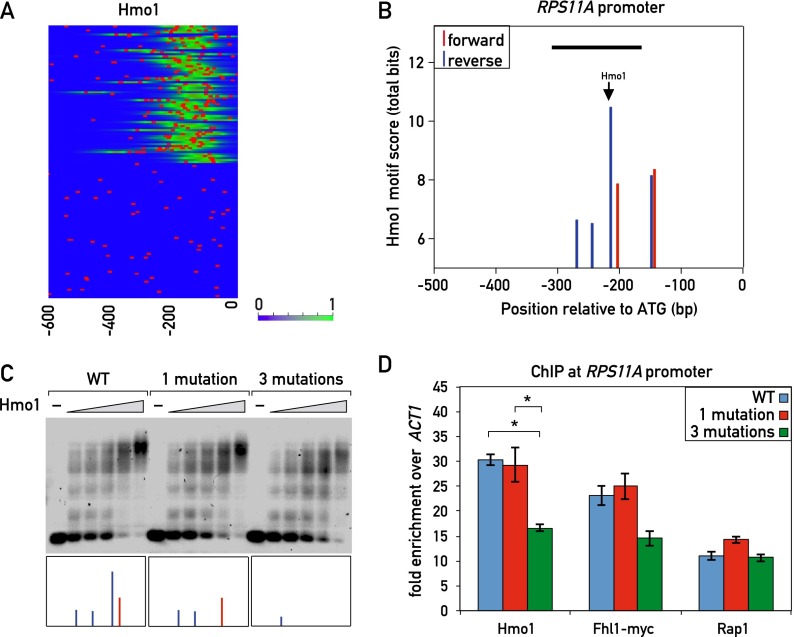
Hmo1 binds with sequence specificity and contributes to recruitment of Fhl1
Our MEME analysis of DNA sequences under Hmo1 ChIP-seq peaks identified a conserved motif similar to the IFHL motif (Hall et al. 2006; Lavoie et al. 2010), suggesting that the protein might bind at least in part through sequence-specific DNA contacts and/or structures. Consistent with this, as pointed out above, k-MITOMI analysis of Hmo1 binding using a De Bruijn library of all possible 8-mer sequences identified a similar G-rich motif (Supplemental Fig. S2A).
However, previous mutational analysis of a single IFHL motif in the context of an artificial promoter suggested that it plays little if any role in Hmo1 binding (Hall et al. 2006). Furthermore, we considered it unlikely that the prominent Hmo1 ChIP-seq peaks detected at category I RPG promoters arise from the contribution of a single Hmo1 motif. We thus searched for other sequence features at or near Hmo1 ChIP-seq peaks in category I genes. Strikingly, we found that searches for a degenerate version of the MEME- or MITOMI-derived Hmo1 motifs often revealed the presence of multiple copies of this motif at category I genes (and other Hmo1 target genes) (Supplemental Fig. S6A) but not at category II genes (Fig. 4A). For example, the category I RPS11A promoter displayed multiple motifs on both strands, with motif scores in the range of 6–11 bits (Fig. 4B). We mutated the highest scoring of these sites, which sits directly under the peak of Hmo1 binding (marked in Fig. 4B), but found no detectable difference in Hmo1 binding in vitro as measured by electrophoretic mobility shift assay (EMSA) (Fig. 4C) or in vivo binding as measured by ChIP (Fig. 4D). However, mutation of this site plus two weaker adjacent binding sites (see the Materials and Methods for details) resulted in a small but reproducible decrease in Hmo1 binding both in vitro (Fig. 4C; Supplemental Fig. S6B) and in vivo (Fig. 4D). We also observed multiple predicted Hmo1 sites within the rDNA locus, whose presence correlated with strong Hmo1 ChIP-seq peaks (Supplemental Fig. S6C).
Figure 4.
Sequence specificity of Hmo1 binding. (A) Hmo1 ChIP-seq signal (green) and Hmo1 motifs (red bars; derived with a low threshold of 5 bits from the ChIP-seq-derived position weight matrix [PWM] in Supplemental Fig. S2A). (B) Hmo1 motif scores (total bits based on ChIP-seq-derived PWM in Supplemental Fig. S2A) for both forward (red) and reverse (blue) sites at the RPS11A promoter. Peak Hmo1 binding by ChIP-seq is shown with an arrow. The black bar indicates the fragment of DNA that was used in the EMSA in C. (C) Cy5-labeled DNA templates of the wild-type RPS11A promoter fragment (−313 bp to −163 bp relative to the ATG; left) or the indicated mutant fragments (middle and right) were incubated with increasing amounts of 6xHis-Hmo1 protein and electrophoresed on a 0.7% agarose gel (see the Materials and Methods for details). The boxes at the bottom represent the forward (red) and reverse (blue) Hmo1 motifs that were present in the corresponding DNA fragment (see B for details). (D) Hmo1, Rap1, and Fhl1 occupancy, measured by qPCR-ChIP, on the wild-type and Hmo1 site mutant RPS11A promoters. Data are represented as mean ± SEM. A one-way ANOVA test was used to compare the means for each factor. (*) P < 0.05 using a Tukey post-hoc test. See also Supplemental Figure S6.
Mutation of the putative Hmo1-binding sites at RPS11A, in addition to causing a decrease in Hmo1 binding, also led to a small but reproducible decrease in Fhl1 ChIP compared with the wild type (P = 0.1, one-way ANOVA) but had no clear effect on Rap1 binding (Fig. 4D). This latter observation suggests that the correlation between Rap1–Hmo1 distance and Rap1 ChIP-seq signal strength noted above (Supplemental Fig. S3A) may not be due to Hmo1 binding per se. To further investigate the possible dependence of Fhl1 binding on Hmo1, we used the AID system to rapidly deplete Hmo1. We found that Fhl1 binding decreased significantly on several category I promoters following Hmo1 depletion but showed no change on the category II promoters tested (Supplemental Fig. S6D). We then performed the converse experiment in an Fhl1-AID strain. In contrast to the result with Hmo1-AID, depletion of Fhl1 had little or no effect on Hmo1 binding at any of the promoters tested (Supplemental Fig. S6E).
The FH domain of Fhl1 is required for promoter association at only a small subset of RPGs
Previous studies have suggested that Fhl1 is recruited to RPG promoters through physical interactions with Rap1 (Rudra et al. 2007; Gordan et al. 2009) or Hmo1 (Ito et al. 2001; Ho et al. 2002). Indeed, when the FH DNA-binding domain of Fhl1 is deleted, the mutant cells display only a mild growth phenotype compared with the extreme slow growth phenotype of the FHL1 deletion (Rudra et al. 2005). However, our own work (Supplemental Fig. S2A, “MITOMI”) and that of others (Badis et al. 2008; Zhu et al. 2009) have shown that the FH domain is able to recognize a specific DNA sequence in vitro.
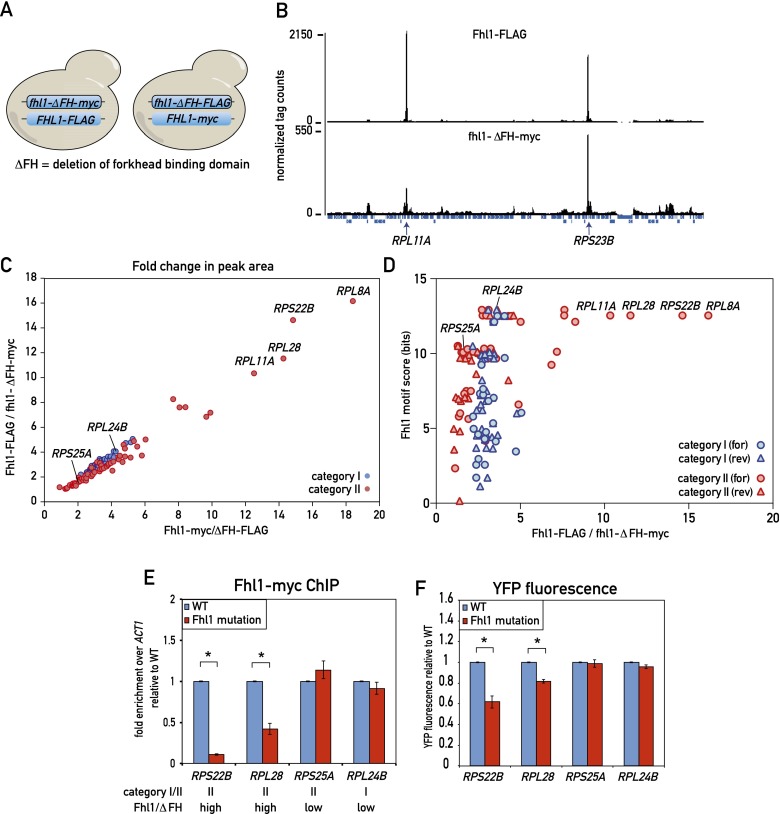
These apparently conflicting results prompted us to investigate the role of the Fhl1 FH domain in Fhl1 binding genome-wide. To this end, we first performed a ChIP-seq experiment using a diploid yeast strain containing one wild-type FHL1 allele marked with a C-terminal Flag epitope tag at the endogenous locus, with the homolog containing an FH domain deletion allele (ΔFH) carrying a C-terminal myc tag (Fig. 5A, left). Cross-linked chromatin from this heterozygous diploid strain was separately immunoprecipitated with anti-Flag and anti-myc antibody and analyzed by deep sequencing. A qualitative comparison of these parallel ChIP-seq experiments shows that most peaks display a remarkably similar ratio in height between the wild-type (Flag) and ΔFH (myc) samples, with the former consistently about twofold to fourfold higher than the latter. Although this rule holds true for all category I genes, a subset of category II genes displayed a significant deviation from this ratio, with the mutant protein peaks considerably lower (sixfold to nearly 20-fold) relative to the wild type (see Fig. 5B for one example, RPL11A). In order to account for possible tag effects, we also created a diploid strain with the tags swapped (Fhl1-myc and fhl1-ΔFH-Flag) (Fig. 5A, right) and carried out the same ChIP-seq analysis. The results were very similar, as shown by a plot of the wild-type versus ΔFH peak area ratios for the two strains (Fig. 5C). This analysis clearly shows that binding of wild-type protein at most RPGs (and all category I genes) is only twofold to fourfold higher than that of the ΔFH mutant, whereas a small subset of category II genes shows much stronger binding of the wild type relative to the mutant protein.
Figure 5.
Role of the Fhl1 FH domain in RPG promoter binding in vivo. (A) Schematic depicting the experimental setup. (Left) Diploid cells expressing both Fhl1-Flag and fhl1-ΔFH-myc were analyzed by parallel anti-Flag and anti-myc ChIP-seq from a single culture. (Right) The experiment was repeated with an isogenic tagged swapped strain (Fhl1-myc, fhl1-ΔFH-Flag). (B) ChIP-seq “tag count” plots from a region on chromosome VII for the indicated Fhl1 proteins. The positions of two RPGs in this region (RPL11A and RPS23B) are marked. (C) ChIP-seq peak area ratios (for the indicated tagged proteins) for all category I (blue) and category II (red) RPGs. Specific genes referred to below or in the text are marked. (D) The score of the strongest potential Fhl1-binding motif (either forward or reverse, as indicated) found in the 500-bp region upstream of the TSS for each RPG (bit scores calculated as in Fig. 4B) is plotted against the ChIP-seq peak area ratio for the wild type versus ΔFH (Fhl1-Flag-fhl1-ΔFH-myc) for the respective promoter. (E) Fhl1-myc ChIP signals for wild-type and Fhl1 site mutants of the indicated RPG promoters. The category and ChIP-seq ratio (wild type vs. fhl1-ΔFH) are indicated below the gene name for each promoter. Data are represented as mean ± SEM. A Student’s t-test was used to compare the means between wild type and the Fhl1 site mutants for each promoter. (*) P < 0.05. (F) The YFP fluorescence of the indicated promoter constructs (both wild type and Fhl1 site mutants) was measured by flow cytometry of exponentially growing cells and is reported relative to the wild-type value. Data are represented as mean ± SEM. A Student’s t-test was used to compare the means between wild type and the Fhl1 site mutants for each promoter. (*) P < 0.05. See also Supplemental Figure S7.
The simplest interpretation of these results is that the FH domain contributes only weakly to promoter binding at most genes, making sequence-specific contacts at a relatively small subset of category II genes. Consistent with this notion, all four promoters that show the highest ratio between the wild type and the ΔFH mutant are from category II and contain a strong Fhl1 motif (Fig. 5D). To directly test this hypothesis, we mutated a strong putative Fhl1-binding site found directly under the Fhl1 ChIP-seq peak in four RPG promoters: two category II promoters (RPS22B and RPL28) where the ChIP-seq data indicated that the FH domain plays a significant role in binding and two promoters where it does not (RPS25A, a category II promoter, and RPL24B, a category I promoter) (Supplemental Fig. S7A). Consistent with our prediction from the ChIP-seq data, site mutations at RPS22B and RPL28 caused a significant decrease in Fhl1 binding as measured by qPCR-ChIP (Fig. 5E), with no associated decrease in Rap1 binding at RPL28 (Supplemental Fig. S7B) and a milder decrease in promoter output (Fig. 5F). As expected, putative site mutations at RPS25A and RPL24B had no significant effect on Fhl1 binding or promoter output (Fig. 5E,F).
TF binding at both category I and category II promoters overlaps with an unusually MNase-labile nucleosome
We were intrigued by in vitro nucleosome assembly data (Kaplan et al. 2009) indicating the presence of a nucleosome positioned nearly coincident with the peak of Hmo1 binding at category I genes, particularly since this has been described as an unusually wide and highly conserved nucleosome-depleted region across yeast species (Tsankov et al. 2010). We thus decided to investigate nucleosome occupancy in vivo at an individual category I gene promoter. Preliminary analysis indicated the presence of a nucleosome in vivo at this position with higher sensitivity to MNase digestion than the immediately adjacent −1 and +1 nucleosomes (data not shown).
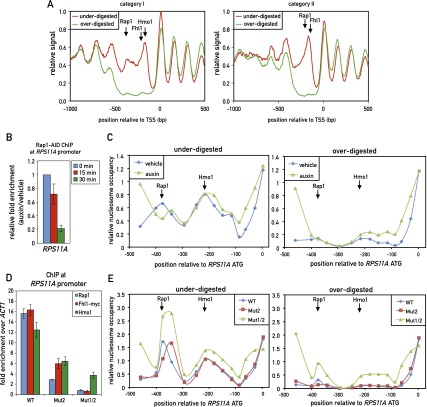
We next examined this phenomenon at the genome-wide level by carrying out deep sequencing of “incomplete” and “complete” MNase chromatin digests (Supplemental Fig. S8A), following a protocol that avoids DNA fragment size selection (Henikoff et al. 2011). Analysis of the resulting data to derive nucleosome occupancy maps and a comparison of the two different digests revealed a striking feature of all category I promoters; namely, the absence of signal from the overdigested sample in the region of Rap1 and Hmo1–Fhl1/Ifh1 binding but the clear presence of two nucleosome-like peaks at this location in the underdigested sample (Fig. 6A, left panel). It is important to note that the neighboring nucleosomes covering the gene bodies in the two samples (underdigested and overdigested) were strikingly similar in both position and peak area, further highlighting the unique property of the intervening promoter DNA. Remarkably, we observed a very similar effect at all of the category II promoters, where a single MNase-labile (“fragile”) nucleosome appears to be centered between the peaks of Rap1 and Fhl1/Ifh1 binding. Again, the neighboring nucleosomes at these promoters, in particular the adjacent +1 and −1 nucleosomes, were nearly identical in both occupancy and position between the two data sets (Fig. 6A, right panel; see Supplemental Fig. S8B,C for separate analyses of promoters with or without a linked divergent promoter; see Supplemental Fig. S9 for a plot showing Rap1 motifs in relation to the positions of fragile nucleosomes for individual promoters).
Figure 6.
TF binding at both category I and category II promoters overlaps with unusually MNase-labile chromatin. (A) Chromatin was underdigested or overdigested with MNase and sequenced (see the Materials and Methods). The average relative signal (a proxy for nucleosome occupancy) for category I (left) and category II (right) promoters aligned to their TSSs is plotted. Arrows mark the average positions of peak binding of Rap1, Fhl1, and Hmo1, as measured by ChIP-seq. (B) RPS11A promoter occupancy of Rap1 after auxin-induced depletion of AID-tagged Rap1. Data are plotted as auxin relative to vehicle treatment and normalized to t = 0. Data are represented as mean ± SEM. (C) Chromatin was underdigested (left panel) or overdigested (right panel) with MNase either before or 30 min after auxin-induced depletion of AID-tagged Rap1. Tiling qPCR reactions were used to measure DNA protection. (D) RPS11A promoter occupancy of Rap1, Fhl1-myc, and Hmo1 on the wild-type RPS11A promoter and promoters with one (Mut2) or two (Mut1/2) Rap1 sites mutated. The RPS11A promoter contains two forward Rap1-binding sites located at −404 bp and −384 bp upstream of the ATG. Mut2 corresponds to mutation of the −384-bp site, and Mut1/2 corresponds to mutation of both sites. Data are represented as mean ± SEM. (E) Chromatin from the indicated strains (RPS11A wild-type or mutant promoters, as described in C was underdigested (left panel) or overdigested (right panel) with MNase. DNA was measured as in C. See also Supplemental Figures S8 and S9.
To explore the factors responsible for the MNase sensitivity of these presumed nucleosomal particles at RPG promoters, we turned again to the AID system and examined the effect of Rap1-AID degradation on the RPS11A promoter. Genome-wide analysis had revealed two MNase-sensitive particles centered at −375 bp (coincident with Rap1 binding) and −200 bp (coincident with Hmo1 binding) from the ATG, whose properties were reproduced by qPCR analysis using a set of nested primer pairs covering this region. Strikingly, auxin-induced depletion of Rap1 had little or no effect on the MNase sensitivity of either of these particles (Fig. 6C) even at a time point (30 min following auxin addition) when Rap1 binding was reduced to less than a third of its normal level (Fig. 6B). In contrast, we observed increased protection upstream of the Rap1-binding site in both the underdigested and overdigested samples and a slight downstream shift (∼50 bp) of the protected region at the site of Rap1 binding in the underdigested sample. To more stringently test the effect of Rap1 on these two nucleosomes, we generated two mutations at the endogenous RPS11A promoter: one affecting the downstream Rap1-binding site (Mut2, position −384), and the other containing Mut2 and a second mutation of the upstream site at position –404 (Mut1/2). The Mut2 mutation showed significantly decreased levels of Rap1, Fhl1, and Hmo1, whereas the Mut1/2 mutation abolished binding of Rap1 and Fhl1 and drastically reduced Hmo1 (Fig. 6D). Interestingly, Mut1/2 caused partial stabilization (MNase resistance) at both the Hmo1- and Rap1-coincident nucleosomes (Fig. 6E, right panel). As noted in the case of Rap1-AID depletion, Mut1/2 also caused what appears to be the encroachment of the flanking stable nucleosomes into the fragile chromatin region, with the +1 nucleosome shifted upstream, and the −1 nucleosome shifted downstream, both by ∼50 bp (Fig. 6E).
Discussion
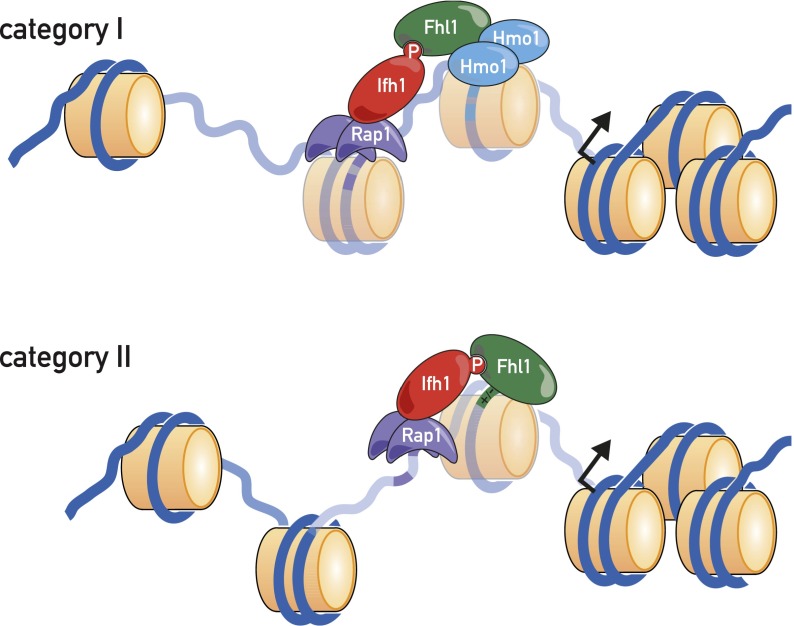
The major aim of the present study was to characterize as precisely as possible the architecture of the 138 RPG promoters with respect to TF location, explore the DNA sequence features of promoters that determine this architecture, establish a TF-binding hierarchy, and correlate the TF architecture with nucleosome occupancy patterns, transcriptional output, and regulation. Our principal findings regarding promoter architecture are summarized in schematic form in Figure 7.
Figure 7.
Schematic of TFs and nucleosomes present at category I and category II RPG promoters.
Two distinct RPG promoter architectures
The present study, which examines the four major RPG TFs by ChIP-seq, provides the first detailed picture of RPG promoter architecture. We show that Hmo1 is associated strongly with about half of all RPG promoters (category I) but displays little if any binding to the other half (category II) and that RPG promoters predominate in the list of strongest Hmo1 targets as measured by ChIP-seq, a feature not noted in previous hybridization-based binding studies (Hall et al. 2006; Kasahara et al. 2007). We also show that the RPG promoter “nucleosome-free regions,” previously grouped together (Tirosh and Barkai 2008; Zaugg and Luscombe 2012), are actually larger at category I compared with category II promoters. Our identification of unusually MNase-sensitive nucleosome-like particles in these regions is discussed below.
The ChIP-seq analysis described here arguably provides us with a more accurate measure of the spatial relationships between the TFs and their relative occupancy levels at different promoters. However, when combining this and other information (in vitro nucleosome binding data) with our steady-state transcriptional output measurements, we found that a simple linear regression model still explains only ∼40% of the measured expression variance for category II genes and even less at the more complex category I genes. This contrasts to a recent study in which the in vitro measured binding affinity of the Pho4 TF could account for 82% of the PHO5 promoter output variance that resulted from mutations in a single Pho4-binding site (Rajkumar et al. 2013). One possible reason for this difference could be that ChIP-seq signal strength correlates poorly with actual in vivo TF residence time (Lickwar et al. 2012). The present study thus points to areas where future work might lead to significant improvements toward the goal of predicting transcriptional output from promoter sequence (Segal and Widom 2009) and suggests that promoters that employ combinations of multiple TFs might be more challenging to model than those where a single TF plays a dominant role.
DNA sequence determinants of TF binding at RPG promoters
The relationship between RPG promoter DNA sequence TF landscape revealed here is surprisingly complex, even for the case of Rap1, whose in vitro binding affinity and specificity has been well documented (Konig et al. 1996; Taylor et al. 2000). We found that Rap1 binds RPG promoters at regions that typically contain a pair of tandem recognition sequences within a 20- to 50-bp window. Although this feature was predicted (Lascaris et al. 1999), a recent study using a nucleotide-resolution global mapping method (ChIP-exo) concluded that Rap1 uses a single strong binding site at most RPG promoters, including exclusive use of site 1 in the RPL30 promoter (Rhee and Pugh 2011). However, our mutational analysis of the RPL30 promoter clearly showed that both predicted sites at this promoter contribute to Rap1 binding and transcriptional output. Although further studies would be required to prove the generality of this observation, we note that multiple, closely spaced Rap1 sites have been shown to contribute to binding in vivo at a non-RPG promoter (De Sanctis et al. 2002). The mechanistic significance of tandem, closely spaced Rap1-binding sites is still unclear. We do not know, for example, whether the simultaneous occupation of both sites plays an important role in activation or whether the paired sites simply boost the probability that at least one Rap1 molecule will always be present at the promoter. In this regard, it is worth noting that a recent study found evidence that the “dwell time” of Rap1 at RPG promoters is high compared with other target genes (Lickwar et al. 2012). Finally, we note that tandem duplicated Rap1-binding sites (class 1) are more frequently associated with the moderately expressed category I genes, whereas promoters with a single Rap1 site (class 5) are overrepresented in category II promoters and frequently associated with high expression levels. These correlations warrant further experimental analysis.
Findings reported here provide new insight into the specificity of Hmo1 binding. As noted above, our ChIP-seq data indicate that Hmo1 binding at RPG promoters is unusually strong both in absolute terms and compared with non-RPG sites (45 of the top 50 Hmo1-binding sites by tag count number are at category I RPG promoters). This enhanced binding at RPG promoters is correlated with the presence of multiple near matches to a motif that emerges from both MEME analysis of Hmo1 ChIP-seq peaks and k-MITOMI in vitro analysis of Hmo1 binding. Significantly, a comparison of category I and category II promoters indicates a much higher occurrence of matches or near matches to this motif localized over an ∼150- to 200-bp region corresponding precisely to the peak of Hmo1 binding detected by ChIP-seq. We conclude that Hmo1 either makes sequence-specific contacts with this motif or recognizes a DNA duplex structural feature generated by motif repeats. In either case, the result would appear to be a much longer residency time for Hmo1 on RPG promoters relative to that of other HMG-B proteins, whose specific sites of DNA binding are often difficult to detect by ChIP (Dowell et al. 2010). Although our data are consistent with a role for motif repeats in Hmo1 binding, they also point to key roles for other factors. For example, a nearly equal number of non-RPG-binding sites for Hmo1 identified by ChIP-seq contain a similar number of motifs, yet their Hmo1 association, as measured by tag counts, is markedly lower (Supplemental Fig. S6A). One factor that might be responsible for this difference is Rap1, which binds to all category I RPGs and few, if any, of the non-RPG promoters bound by Hmo1. Consistent with this notion, rapid depletion of Rap1 from cells leads to a significant decrease in Hmo1 association. Whether this is due to a specific protein–protein interaction between Rap1 and Hmo1 or an effect of Rap1 on the chromatin environment at category I promoters is unknown.
Although Fhl1, a FH-like DNA-binding protein, is known to associate almost exclusively with RPG promoters in vivo (Lee et al. 2002; Harbison et al. 2004; Schawalder et al. 2004; Wade et al. 2004; Kasahara et al. 2007), the DNA sequence determinants of this specificity have been elusive. Our ChIP-seq experiments that directly measure competition between the wild type and fhl1-ΔFH help to clarify this issue, since they indicate that the Fhl1 FH domain plays a relatively minor role in promoter binding at most RPGs, consistent with a nonsequence-specific binding function at these genes. Indeed, our targeted mutagenesis experiments indicate that the predicted Fhl1-binding sites only play an important role in vivo at those few promoters where the FH domain is important for binding. This surprisingly limited role for the Fhl1 FH domain is perfectly consistent with the observation that deletion of the domain has only a minor effect on growth, particularly when compared with the severe growth defect caused by FHA domain deletion (Rudra et al. 2005).
Hierarchy of TF binding at RPG promoters and evolutionary implications
Both site mutation and depletion experiments reported here point to a key role of Rap1 in supporting both Fhl1/Ifh1 and Hmo1 promoter binding and the additional importance of Hmo1 in Fhl1/Ifh1 binding at category I promoters. Given the close overlap between Fhl1/Ifh1 and Hmo1 ChIP-seq peaks at category I genes, the limited role there of the Fhl1 FH domain, and an apparent physical interaction between the two proteins (Ho et al. 2002), we speculate that Hmo1 helps to recruit Fhl1/Ifh1 to these genes through a direct protein–protein interaction. Conversely, at category II promoters, where Rap1 binds much closer to Fhl1 and Hmo1 is not present, we propose that Rap1 interacts directly with Fhl1 and/or Ifh1, consistent with recently reported in vitro binding studies (Mallick and Whiteway 2013). Rap1 may also support Fhl1/Ifh1 and Hmo1 binding through a still poorly understood ability to exclude nucleosomes in its vicinity (Gandhi et al. 2011). Finally, we note that the assembly of Fhl1 together with Rap1 at nearly all RPG promoters, but essentially none of the roughly equal number of other promoters that also display robust Rap1 binding, is still enigmatic. Whereas a plausible argument can be made that interactions with both Hmo1 and Rap1 confer Fhl1-binding specificity at category I promoters, the situation at category II promoters is less clear, since our ChIP-seq data suggest that the FH domain of Fhl1 is unlikely to confer specificity at most of these genes.
The issue of Fhl1/Ifh1 specificity is particularly interesting in light of phylogenetic studies of the fungal RPG regulatory systems (Hogues et al. 2008; Lavoie et al. 2010), which show that in most ascomycete yeasts, the role of Rap1 is carried out by a different TF (Tbf1) and that only the Fhl1/Ifhl1 pair are common to all yeasts. In Saccharomyces cerevisiae, Tbf1 also plays a role in ribosome biogenesis, but only through activation of a large set of snoRNA genes (Preti et al. 2010). These findings suggest that the Fhl1/Ifh1 pair plays a primordial, highly conserved role in signaling pathways that relay growth and stress signals to RPG promoters. Evolutionary plasticity in this system has thus been most pronounced at the level of the highly sequence-specific TFs (Rap1 and Tbf1), with cells somehow managing to transition from exclusive use of Tbf1, thought to be the more ancient TF, to the widespread use of Rap1 in the clade containing S. cerevisiae. The requirement of Rap1 for Fhl1/Ifh1 binding suggests that Rap1 at the same time evolved a mechanism to directly recruit these proteins, and, indeed, one recent study indicates that this operates at least in part through an interaction between the N-terminal BRCT domain of Rap1 and the Ifh1 protein (Mallick and Whiteway 2013).
Interestingly, two category II RPG promoters, RPL41A and RPS22B, show binding of Tbf1 that is nearly coincident with that of Fhl1 (Preti et al. 2010). Rap1 also binds to the RPL41A promoter, whereas the RPS22B promoter shows no Rap1 signal. It is thus likely that the presence of Tbf1 at these two genes represents a snapshot in evolution—the last two S. cerevisiae RPG promoters that have yet to convert from Tbf1 to Rap1 dependency (Lavoie et al. 2010). Even more intriguing, we found that Fhl1 binds in a sequence-specific manner to the RPS22B promoter (Fig. 5E). We speculate that sequence-specific binding by Fhl1 played a more important role in its promoter localization in the ancestors of S. cerevisiae, perhaps facilitating the transition from a Tbf1-based promoter architecture to one using Rap1.
Dynamic FNs are a common feature of RPG promoter architecture
One of the more striking findings to emerge from this study is that Fhl1/Ifh1 and Hmo1 bind to regions that invariably contain one or more MNase-sensitive particles of near-nucleosome size that we speculate correspond to unstable (or “fragile”) nucleosomes (Weiner et al. 2010; Xi et al. 2011). At category I promoters, we observed two (or three) FNs, with Hmo1 binding near the center of the more TSS-proximal particle, and Fhl1/Ifh1 binding slightly upstream. At category II promoters, we observed a single FN whose center maps in between the ChIP-seq peaks of Rap1 and Fhl1/Ifh1. We still do not know how the FNs at RPGs are generated. Depletion and site mutagenesis experiments suggest that their instability is generated at least in part through the action of Rap1 but point to a role for other factors, perhaps in conjunction with Rap1. Candidates include the underlying DNA sequences at FNs [e.g., poly(dA:dT)] and trans-acting factors such as nucleosome remodelers and histone chaperones.
The colocalization of FNs with key TF-binding sites suggests that they play an important role in RPG activation and/or regulation through mechanisms yet to be uncovered. It remains to be determined, for example, whether the apparent dynamic property of these nucleosomes either promotes or inhibits TF binding and/or function. The histone modification state of these particles may also play an important regulatory function. Indeed, several studies have demonstrated recruitment or action of both NuA4 and SAGA histone acetyltransferase complexes at RPG promoters (Reid et al. 2000; Rohde and Cardenas 2003; Robert et al. 2004; Ghosh and Pugh 2011; Cai et al. 2013; Downey et al. 2013). In addition, binding of the RPD3L histone deacetylase complex coincides with binding of Fhl1 on RPG promoters and is markedly increased after specific inhibition of the TORC1 effector kinase Sch9 (Huber et al. 2011). A challenge for future studies will be to identify the molecular mechanism underlying the fragility of these RPG promoter nucleosomes and the specific role that they play in the transcription process and its regulation.
Materials and methods
Yeast strains
Yeast strains used in this study are listed in Supplemental Table S1.
ChIP and ChIP-seq
Sample preparation and DNA analysis for qPCR-ChIP (Ribaud et al. 2012) and ChIP-seq (Preti et al. 2010) were carried out as described.
ChIP-seq and motif-finding analysis
Promoter element identification was based on the list of S. cerevisiae RPGs reported in the Saccharomyces Genome Database (http://www.yeastgenome.org), with TSS annotations taken from Jiang and Pugh (2009). The promoter was defined as the 1-kb region upstream of the TSS. Methods for identifying TF ChIP-seq profiles and cross-correlations as well as motif-finding analyses are described in the Supplemental Material.
MITOMI measurements and motif-finding analyses
Rap1 and Fhl1 DNA-binding sequence motifs were derived by MITOMI analysis as previously described (Rockel et al. 2012). The dissociation constants (Kd) of all possible single-base substitutions starting from a high-affinity binding sequence were measured. The value of −log(Kd) corresponds to the binding energy. To keep the binding energy positive, its value is defined as −log(Kd) + log(maxKd). MatrixREDUCE software was used to calculate the position weight matrices (PWMs) for sequences with binding energies >3. The Hmo1 DNA-binding sequence motif was derived using a kinetic MITOMI approach (Geertz et al. 2012b). A De Bruijn sequence covering all 8-mer sequence variants was calculated (Philippakis et al. 2008) and then synthesized as overlapping oligonucleotide pairs that were converted to dsDNA using Cy3 and Cy5 extension primers (Geertz et al. 2012a). Kinetic MITOMI measurements were performed to derive dissociation rate constants for all 8-mer De Bruijn sequence oligonucleotides, which were then ranked by inferred koff values. The 40 highest-ranking sequences (with R2 > 0.6 and smallest koff) were used as “positive binders,” while 40 nonbinding oligonucleotides were used as negative controls. Taking a random motif PWM, the score of each sequence was calculated, and then the motif was iteratively improved by least-squares optimization between motif scores and 1/koff values. The procedure was repeated for 1000 different initial random matrices, and the best resulting PWM was kept (Foat et al. 2006).
AID protein depletion
Overnight cultures were diluted to OD600 0.1, grown at 30°C to exponential phase (OD600 ∼ 0.4), and then treated with auxin (3-indoloacetic acid) at 500 μM final concentration or vehicle alone (ethanol). Samples for ChIP were cross-linked with formaldehyde at 0, 15, and 30 min after auxin or vehicle treatment.
EMSA of Hmo1 binding
The DNA used in the EMSA was PCR-amplified from the genomic DNA. Reaction buffer contained 60 mM NaCl, 20 mM Tris-HCl (pH 7.5), 10% glycerol, 1 mM DTT, 0.1 mM EDTA, and 10 ng of poly d(I·C). The samples were incubated for 15 min on ice and loaded onto 0.7% agarose gel in 0.2× TB buffer. Gels were scanned using an Ettan DIGE imager. Preparation of Hmo1 protein and generation of Hmo1-binding site mutations in the RPS11A promoter are described in the Supplemental Material.
MNase digestion and nucleosome mapping
Chromatin for MNase digestion was prepared essentially as described (Kent and Mellor 1995). Spheroplasts derived from 100-mL cultures were treated with either 0.5 U (underdigested) or 2 U (overdigested) of MNase (Sigma) for 45 min at 37°C. Purified and precipitated DNA was sequenced using the paired-end TruSeq protocol (Illumina). Paired-end reads were aligned to sacCer2 (2008) genome assembly using HTSStation.
Acknowledgments
We thank members of the Shore laboratory for helpful discussions, and Nicolas Roggli for expert assistance with the figures. This study was supported by SystemsX.ch Grant DynamiX-RTD (2008/005), other grants from the Swiss National Science Foundation (to D.S.), and funds provided by the Republic and Canton of Geneva (to D.S.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.244434.114.
References
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–W373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DB, Gasch AP. 2008. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell 19: 4580–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. 2008. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell 19: 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, McCormick MA, Kennedy BK, Tu BP. 2013. Integration of multiple nutrient cues and regulation of lifespan by ribosomal transcription factor Ifh1. Cell Rep 4: 1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denervaud N, Becker J, Delgado-Gonzalo R, Damay P, Rajkumar AS, Unser M, Shore D, Naef F, Maerkl SJ. 2013. A chemostat array enables the spatio-temporal analysis of the yeast proteome. Proc Natl Acad Sci 110: 15842–15847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686 [DOI] [PubMed] [Google Scholar]
- De Sanctis V, La Terra S, Bianchi A, Shore D, Burderi L, Di Mauro E, Negri R. 2002. In vivo topography of Rap1p–DNA complex at Saccharomyces cerevisiae TEF2 UAS(RPG) during transcriptional regulation. J Mol Biol 318: 333–349 [DOI] [PubMed] [Google Scholar]
- De Virgilio C, Loewith R. 2006. The TOR signalling network from yeast to man. Int J Biochem Cell Biol 38: 1476–1481 [DOI] [PubMed] [Google Scholar]
- Dowell NL, Sperling AS, Mason MJ, Johnson RC. 2010. Chromatin-dependent binding of the S. cerevisiae HMGB protein Nhp6A affects nucleosome dynamics and transcription. Genes Dev 24: 2031–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Knight B, Vashisht AA, Seller CA, Wohlschlegel JA, Shore D, Toczyski DP. 2013. Gcn5 and sirtuins regulate acetylation of the ribosomal protein transcription factor ifh1. Curr Biol 23: 1638–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foat BC, Morozov AV, Bussemaker HJ. 2006. Statistical mechanical modeling of genome-wide transcription factor occupancy data by MatrixREDUCE. Bioinformatics 22: e141–e149 [DOI] [PubMed] [Google Scholar]
- Ganapathi M, Palumbo MJ, Ansari SA, He Q, Tsui K, Nislow C, Morse RH. 2011. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Res 39: 2032–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi SJ, Zenklusen D, Lionnet T, Singer RH. 2011. Transcription of functionally related constitutive genes is not coordinated. Nat Struct Mol Biol 18: 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geertz M, Rockel S, Maerkl SJ. 2012a. A high-throughput microfluidic method for generating and characterizing transcription factor mutant libraries. Methods Mol Biol 813: 107–123 [DOI] [PubMed] [Google Scholar]
- Geertz M, Shore D, Maerkl SJ. 2012b. Massively parallel measurements of molecular interaction kinetics on a microfluidic platform. Proc Natl Acad Sci 109: 16540–16545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Pugh BF. 2011. Sequential recruitment of SAGA and TFIID in a genomic response to DNA damage in Saccharomyces cerevisiae. Mol Cell Biol 31: 190–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves PM, Griffioen G, Minnee R, Bosma M, Kraakman LS, Mager WH, Planta RJ. 1995. Transcription activation of yeast ribosomal protein genes requires additional elements apart from binding sites for Abf1p or Rap1p. Nucleic Acids Res 23: 1475–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan R, Hartemink AJ, Bulyk ML. 2009. Distinguishing direct versus indirect transcription factor–DNA interactions. Genome Res 19: 2090–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DB, Wade JT, Struhl K. 2006. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol Cell Biol 26: 3672–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. 2011. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci 108: 18318–18323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Hogues H, Lavoie H, Sellam A, Mangos M, Roemer T, Purisima E, Nantel A, Whiteway M. 2008. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol Cell 29: 552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, French SL, Tekotte H, Yerlikaya S, Stahl M, Perepelkina MP, Tyers M, Rougemont J, Beyer AL, Loewith R. 2011. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J 30: 3052–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, Struhl K. 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J 14: 2570–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. 2009. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10: R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev 18: 2491–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamau E, Bauerle KT, Grove A. 2004. The Saccharomyces cerevisiae high mobility group box protein HMO1 contains two functional DNA binding domains. J Biol Chem 279: 55234–55240 [DOI] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara K, Ohtsuki K, Ki S, Aoyama K, Takahashi H, Kobayashi T, Shirahige K, Kokubo T. 2007. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol Cell Biol 27: 6686–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent NA, Mellor J. 1995. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res 23: 3786–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig P, Giraldo R, Chapman L, Rhodes D. 1996. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 85: 125–136 [DOI] [PubMed] [Google Scholar]
- Lascaris RF, Mager WH, Planta RJ. 1999. DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics 15: 267–277 [DOI] [PubMed] [Google Scholar]
- Lavoie H, Hogues H, Mallick J, Sellam A, Nantel A, Whiteway M. 2010. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol 8: e1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. 2012. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 484: 251–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat Genet 28: 327–334 [DOI] [PubMed] [Google Scholar]
- Macisaac KD, Gordon DB, Nekludova L, Odom DT, Schreiber J, Gifford DK, Young RA, Fraenkel E. 2006. A hypothesis-based approach for identifying the binding specificity of regulatory proteins from chromatin immunoprecipitation data. Bioinformatics 22: 423–429 [DOI] [PubMed] [Google Scholar]
- Mallick J, Whiteway M. 2013. The evolutionary rewiring of the ribosomal protein transcription pathway modifies the interaction of transcription factor heteromer Ifh1–Fhl1 (interacts with forkhead 1–forkhead-like 1) with the DNA-binding specificity element. J Biol Chem 288: 17508–17519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DE, Soulard A, Hall MN. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119: 969–979 [DOI] [PubMed] [Google Scholar]
- Muller D, Stelling J. 2009. Precise regulation of gene expression dynamics favors complex promoter architectures. PLoS Comput Biol 5: e1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922 [DOI] [PubMed] [Google Scholar]
- Philippakis AA, Qureshi AM, Berger MF, Bulyk ML. 2008. Design of compact, universal DNA microarrays for protein binding microarray experiments. J Comput Biol 15: 655–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti M, Ribeyre C, Pascali C, Bosio MC, Cortelazzi B, Rougemont J, Guarnera E, Naef F, Shore D, Dieci G. 2010. The telomere-binding protein Tbf1 demarcates snoRNA gene promoters in Saccharomyces cerevisiae. Mol Cell 38: 614–620 [DOI] [PubMed] [Google Scholar]
- Rajkumar AS, Denervaud N, Maerkl SJ. 2013. Mapping the fine structure of a eukaryotic promoter input-output function. Nat Genet 45: 1207–1215 [DOI] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK. 2004. Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JL, Iyer VR, Brown PO, Struhl K. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell 6: 1297–1307 [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. 2011. Comprehensive genome-wide protein–DNA interactions detected at single-nucleotide resolution. Cell 147: 1408–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribaud V, Ribeyre C, Damay P, Shore D. 2012. DNA-end capping by the budding yeast transcription factor and subtelomeric binding protein Tbf1. EMBO J 31: 138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell 16: 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockel S, Geertz M, Maerkl SJ. 2012. MITOMI: a microfluidic platform for in vitro characterization of transcription factor-DNA interaction. Methods Mol Biol 786: 97–114 [DOI] [PubMed] [Google Scholar]
- Rohde JR, Cardenas ME. 2003. The tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol Cell Biol 23: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Zhao Y, Warner JR. 2005. Central role of Ifh1p–Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J 24: 533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Mallick J, Zhao Y, Warner JR. 2007. Potential interface between ribosomal protein production and pre-rRNA processing. Mol Cell Biol 27: 4815–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schawalder SB, Kabani M, Howald I, Choudhury U, Werner M, Shore D. 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432: 1058–1061 [DOI] [PubMed] [Google Scholar]
- Segal E, Widom J. 2009. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat Rev Genet 10: 443–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Iyer VR. 2008. Stress-dependent dynamics of global chromatin remodeling in yeast: dual role for SWI/SNF in the heat shock stress response. Mol Cell Biol 28: 2221–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HO, O’Reilly M, Leslie AG, Rhodes D. 2000. How the multifunctional yeast Rap1p discriminates between DNA target sites: a crystallographic analysis. J Mol Biol 303: 693–707 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. 2008. Two strategies for gene regulation by promoter nucleosomes. Genome Res 18: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ. 2010. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol 8: e1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Hall DB, Struhl K. 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432: 1054–1058 [DOI] [PubMed] [Google Scholar]
- Warner JR 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. 2010. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20: 90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Yao J, Chen R, Li W, He X. 2011. Nucleosome fragility reveals novel functional states of chromatin and poises genes for activation. Genome Res 21: 718–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaugg JB, Luscombe NM. 2012. A genomic model of condition-specific nucleosome behavior explains transcriptional activity in yeast. Genome Res 22: 84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D, Sharon E, Lotan-Pompan M, Lubling Y, Shipony Z, Raveh-Sadka T, Keren L, Levo M, Weinberger A, Segal E. 2011. Compensation for differences in gene copy number among yeast ribosomal proteins is encoded within their promoters. Genome Res 21: 2114–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, McIntosh KB, Rudra D, Schawalder S, Shore D, Warner JR. 2006. Fine-structure analysis of ribosomal protein gene transcription. Mol Cell Biol 26: 4853–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. 2009. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]