Exopolysaccharide (EPS) is an extracellular matrix constituent of the B. subtilis biofilm. Here, Losick and colleagues report a previously unrecognized mechanism for the self-regulation of EPS production. EPS synthesis depends on a tyrosine kinase that consists of a membrane component (EpsA) and a kinase component (EpsB). EPS interacts with the extracellular domain of EpsA to control kinase activity. Further data show that EPS is a signaling molecule that controls its own synthesis. Importantly, tyrosine kinase-mediated self-regulation could be a widespread system of intercellular communication controlling exopolysaccharide production in bacteria.
Keywords: biofilm, exopolysaccharide, tyrosine kinase
Abstract
We report that the Bacillus subtilis exopolysaccharide (EPS) is a signaling molecule that controls its own production. EPS synthesis depends on a tyrosine kinase that consists of a membrane component (EpsA) and a kinase component (EpsB). EPS interacts with the extracellular domain of EpsA, which is a receptor, to control kinase activity. In the absence of EPS, the kinase is inactivated by autophosphorylation. The presence of EPS inhibits autophosphorylation and instead promotes the phosphorylation of a glycosyltransferase in the biosynthetic pathway, thereby stimulating the production of EPS. Thus, EPS production is subject to a positive feedback loop that ties its synthesis to its own concentration. Tyrosine kinase-mediated self-regulation could be a widespread feature of the control of exopolysaccharide production in bacteria.
Bacteria are frequently able to communicate with each other by mechanisms involving the production and secretion of signaling molecules. Among the best studied of these communication systems is quorum sensing, a system of intercellular signaling that is governed by cell population density (Ng and Bassler 2009). Signaling is mediated by small, secreted molecules (autoinducers), often modified peptides in Gram-positive bacteria and N-acyl homoserine lactones in Gram-negative bacteria. When the cell population density is low, the concentration of signaling molecules in the environment is too low to trigger a response. However, at high population densities (when the number of cells has attained a quorum), the concentration of signaling molecules reaches a threshold at which it activates the transcription of a suite of quorum-sensing genes, including genes involved in the production of the signaling molecule itself. This sets up a positive feedback loop that amplifies the response and triggers a coordinated response among cells in the population, such as the production of luciferase in bioluminescent bacteria and virulence factors in pathogens (Fuqua and Greenberg 2002; Novick and Geisinger 2008; Ng and Bassler 2009; Rutherford and Bassler 2012). Here we report the discovery of a novel system of intercellular communication that operates at a post-translational level and is mediated by direct sensing of a common good produced by cells in the community.
The Gram-positive bacterium Bacillus subtilis is capable of forming structurally complex communities known as biofilms on solid surfaces and at air/liquid interfaces (such floating biofilms are known as pellicles) (Branda et al. 2001). Cells in the biofilm are held together by an extracellular matrix that consists of amyloid-like fibers and an exopolysaccharide (henceforth EPS) (Branda et al. 2004; Chu et al. 2006; Romero et al. 2010). The genes for fiber production (a three-gene operon consisting of tapA, sipW, and tasA) and EPS production (the 15-gene epsA-O operon) are held silent in growing, motile cells by the SinR and AbrB repressors but are derepressed during biofilm formation, leading to high-level production of the protein and polysaccharide components of the extracellular matrix (Chu et al. 2006; Chai et al. 2008). An additional component of the matrix, BslA, confers hydrophobicity on the biofilm (Kobayashi and Iwano 2012; Hobley et al. 2013). Because mutants blocked in matrix production can be complemented extracellularly by matrix-producing cells (Branda et al. 2001), the matrix can be thought of as a common good that is shared among cells in the community. Here we focus on the production of EPS and the role of two genes in the epsA-O operon, epsA and epsB, that together encode a member of the family of bacterial tyrosine kinases.
Bacterial tyrosine kinases, which are widespread in Gram-positive and Gram-negative bacteria, are unrelated in mechanism and amino acid sequence to the tyrosine kinases found in eukaryotic cells (Grangeasse et al. 2012). Bacterial tyrosine kinases in Gram-positive bacteria consist of two components. One is a membrane-embedded protein with two transmembrane segments, an extracellular domain, and a cytoplasmic C-terminal domain. The other is a cytoplasmic protein that contains the tyrosine kinase in its N-terminal domain and a cluster of two to seven tyrosine residues in its C-terminal domain that are known to be sites of autophosphorylation (Grangeasse et al. 2012). Both components are needed for kinase activity. In Gram-negative bacteria, the kinase consists of a single polypeptide in which the membrane and cytoplasmic components are fused. The genes for bacterial tyrosine kinases are often associated with genes involved in the production of exopolysaccharides (Whitfield 2006). Moreover, in several cases, the kinases are known to play a regulatory role in exopolysaccharide production by phosphorylating and thereby activating a biosynthetic enzyme in the pathway for the exopolysaccharide (Grangeasse et al. 2007, 2010, 2012; Bechet et al. 2009; Jadeau et al. 2012). Thus, in Escherichia coli, the tyrosine kinases Wzc and Etk stimulate production of two kinds of capsular polysaccharides (group I and group IV, respectively) by phosphorylating a uridine diphosphate (UDP)–sugar dehydrogenase in their respective biosynthetic pathways (Wugeditsch et al. 2001; Grangeasse et al. 2003; Peleg et al. 2005; Lacour et al. 2008). Likewise, capsular polysaccharide production by Streptococcus thermophilus requires the tyrosine kinase EpsD, which phosphorylates a glycosyltransferase in the biosynthetic pathway (Minic et al. 2007). However, the physiological role of bacterial tyrosine kinases in exopolysaccharide production has been unclear, and the nature of the signal that controls their activity, if indeed there is one, has been mysterious.
We report here that the extracellular domain of the membrane component (EpsA) of the EpsA/EpsB tyrosine kinase (henceforth the EpsAB kinase) is a receptor that directly recognizes EPS. We show that the EpsAB kinase is inactivated by autophosphorylation and that EPS inhibits autophosphorylation, thereby enabling the kinase to phosphorylate other substrates in the biosynthetic pathway, such as a glycosyl transferase. Thus, EPS is both a common good in the biofilm community and a quorum-sensing molecule that promotes its own production. We therefore propose that bacterial tyrosine kinases mediate a previously unrecognized and perhaps widespread system of intercellular communication in bacteria for the control of exopolysaccharide production.
Results
EpsA and EpsB are required for robust matrix production
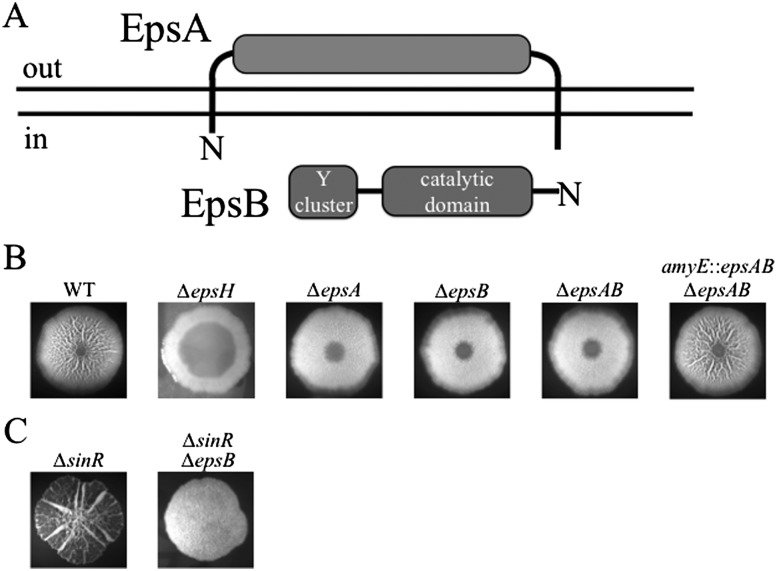
The first two genes (epsA and epsB) of the 15-gene eps operon encode homologs of the widely conserved family of two-component bacterial tyrosine kinases (Fig. 1A; Mijakovic et al. 2003). EpsA and its homologs are predicted to be integral membrane proteins with two transmembrane segments and an extracellular loop, whereas EpsB and its relatives are cytoplasmic proteins that contain the tyrosine kinase catalytic center. The presence of genes for homologs of both components of the bacterial tyrosine kinase family in the eps operon suggested that they function in the production of EPS.
Figure 1.
The EpsAB tyrosine kinase activity is required for biofilm formation. (A) Cartoon showing the disposition of EpsA in the membrane and the tyrosine (Y) cluster in EpsB. (B,C) Colonies of the wild type and the indicated mutants grown on biofilm-inducing medium. The strains were NCBI 3610 (WT), RL3852 (∆epsH), BAE250 (∆epsA), BAE251 (∆epsB), BAE252 (∆epsAB), BAE262 (∆epsAB amyE∷epsAB), RL3854 (∆sinR), and BAE388 (∆sinR∆epsB).
To investigate this hypothesis, we constructed single and double mutants of the two genes and tested their ability to form an extracellular matrix in biofilm-inducing medium. In B. subtilis, colony wrinkling is characteristic of EPS production (Vlamakis et al. 2013). In-frame deletion mutants of epsA and epsB as well as an epsAB double mutant displayed markedly decreased colony wrinkliness as compared with the wild type or an epsAB double mutant that harbored a wild-type copy of the two genes at an ectopic locus (amyE) (Fig. 1B). Nonetheless, colonies of the kinase mutants were not as flat as those of a biosynthetic mutant (epsH). We infer that epsA and epsB are each required for optimal EPS production but that production is not totally eliminated in the absence of the kinase. This was confirmed by direct measurements of EPS levels, which revealed that production was almost fourfold lower in an epsB mutant than in the wild type but not as low as in a biosynthetic gene mutant (Table 1). We conclude that production of EPS depends on the kinase but that a basal level of production takes place in its absence, as also recently noted by Gerwig et al. (2014).
Table 1.
Exopolysaccharide production depends on EpsAB

Interestingly, B. subtilis has a second bacterial tyrosine kinase, TkmA/PtkA (Mijakovic et al. 2003; Barbe et al. 2009), which has been reported to influence EPS production (Kiley and Stanley-Wall 2010). We therefore wondered whether PtkA activity accounts for the residual EPS production in an epsAB mutant. A ptkA mutant alone did not show a detectable effect on EPS production or biofilm formation under our experimental conditions (Supplemental Fig. S1). However, EPS production was abolished in a ptkA epsB double mutant (Table 1; Supplemental Fig. S1; Gerwig et al. 2014). We conclude that the contribution of PtkA to EPS production is largely masked by EpsB but that optimal production of the exopolysaccharide requires both tyrosine kinases. Henceforth, we focus on EpsA/EpsB.
Next, we asked whether EpsA and EpsB were acting upstream of the repressor SinR for eps gene expression. If so, epsA and epsB mutations should have no effect on colony wrinkling in cells mutant for the repressor. A sinR mutant normally forms hyperwrinkly colonies due to the constitutive expression of the matrix operons (Kearns et al. 2005). However, when an epsA (data not shown) or epsB (Fig. 1C) mutation was combined with the sinR mutant, the double-mutant cells formed colonies that lacked colony wrinkling, showing that the mutations were epistatic to the sinR mutation. We conclude that EpsA and EpsB function downstream from SinR, presumably acting at a post-transcriptional level in controlling EPS production.
Autophosphorylation of EpsB inactivates the kinase
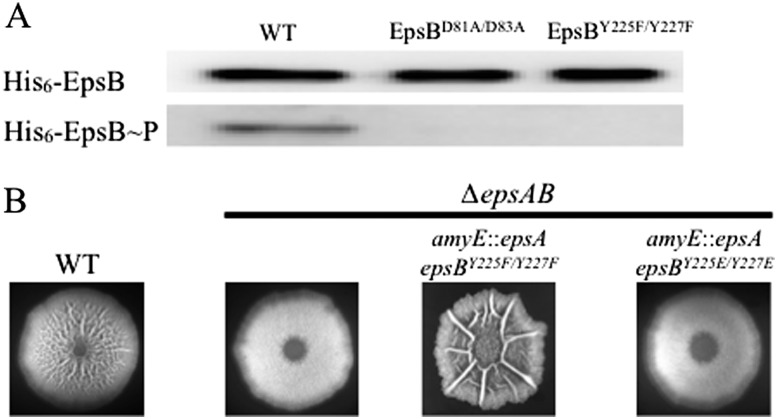
Bacterial tyrosine kinases are known to undergo autophosphorylation as well as phosphorylate other proteins (Grangeasse et al. 2012). To test whether EpsAB is able to phosphorylate itself, we introduced at amyE a copy of epsB that had been modified to contain six histidine codons at the 5′ end of its coding sequence. To determine the phosphorylation state of EpsB, we collected cells grown under biofilm-inducing conditions and affinity-purified the tagged EpsB variant (His6-EpsB) from a preparation of solubilized membrane proteins. Next, the affinity-purified protein was subjected to immunoblot analysis using anti-His6 antibodies (Fig. 2A, top panel) to detect the His6-EpsB protein and anti-phosphotyrosine antibodies (Fig. 2A, bottom panel) to detect the phosphorylation level of the tagged protein (Fig. 2A). In otherwise wild-type cells, the purified protein reacted with both antibodies, indicating that His6-EpsB had undergone phosphorylation at tyrosine residues. To determine whether this phosphorylation was indeed dependent on the kinase activity of EpsB, we analyzed a (His6-EpsB) variant that is expected to be catalytically inactive due to a substitution of two catalytic aspartate residues (81 and 83) with alanines. As expected, the mutant kinase reacted with the anti-His6 antibodies but not with the anti-phosphotyrosine antibodies.
Figure 2.
Autophosphorylation inactivates EpsAB. (A) Immunoblot of His6-tagged EpsB from the wild-type strain (BAE318) and strains producing EpsBD81A/D83A (BAE451) and EpsBY225F/Y227F (BAE450) grown in biofilm-inducing medium. The blot was probed with anti-His6 antibodies (top row) and anti-phosphotyrosine antibodies (bottom row). (B) Colony wrinkling on biofilm-inducing medium for strains NCBI 3610 (WT), BAE252 (∆epsAB), BAE320 (∆epsAB amyE∷epsABY225F/Y227F), and BAE406 (∆epsAB amyE∷epsABY225E/Y227E).
To identify the tyrosine residues that were phosphorylated, His6-EpsB was digested with trypsin, and the resulting peptides were enriched for phosphopeptides using TiO2 chromatography. The phosphopeptides were then analyzed by nano-liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS) on Orbitrap mass spectrometers (see the Supplemental Material). We detected two phosphotyrosine residues (Y225 and Y227) located in a C-terminal cluster of tyrosines (Supplemental Fig. S2), a region known to exhibit phosphorylation in other bacterial tyrosine kinases (Grangeasse et al. 2012). Phosphorylation of Y227 was also reported in a recent phosphoproteomic study (Ravikumar et al. 2014). As a further test of whether these tyrosines are the only or principal sites of phosphorylation in vivo, we purified a His6-EpsB variant in which the two tyrosines were substituted with phenylalanine. The purified protein was then subjected to immunoblot analysis, which revealed a signal with anti-His6 antibodies but not anti-phosphotyrosine antibodies (Fig. 2A).
What is the physiological function of autophosphorylation in EPS production? To address this question, we created strains harboring mutant copies of epsB in which the codons for the two phosphorylated tyrosine residues were replaced with either phenylalanine codons to create a mutant protein blocked from autophosphorylation or codons for glutamate, which, due to its negative charge, might mimic a phosphorylated residue. Cells with EpsB blocked from autophosphorylation formed hyperwrinkled colonies (Fig. 2B) and overproduced EPS (Table 1). In contrast, cells in which the phosphorylated tyrosines had been replaced with glutamates formed flat colonies that resembled the phenotype of an epsB-null mutant (Fig. 2B). These results are consistent with the idea that autophosphorylation inhibits both kinase activity and EPS production and, conversely, that the absence of autophosphorylation promotes kinase activity and EPS production.
The glycosyl transferase EpsE undergoes phosphorylation in an EpsAB-dependent manner
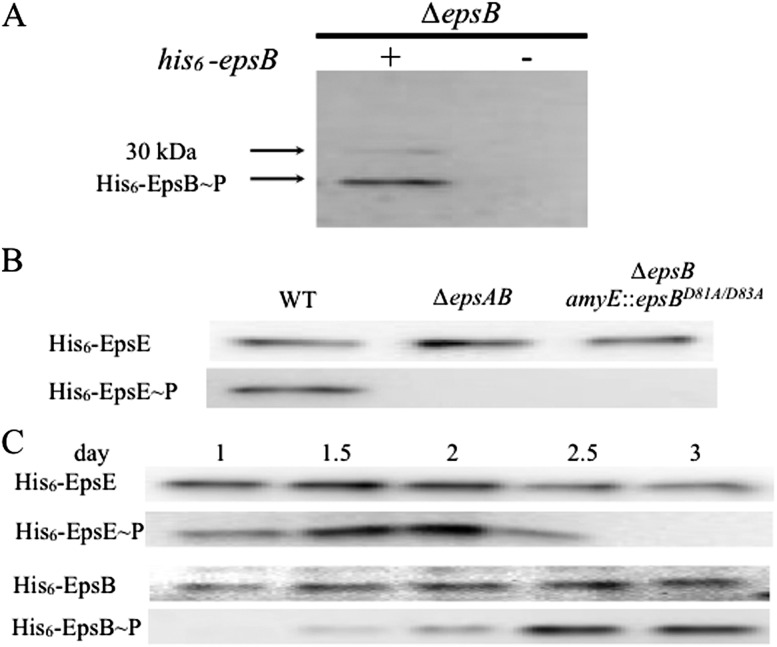
We hypothesized that the EpsAB kinase phosphorylates one of the biosynthetic enzymes in the EPS biosynthetic pathway to stimulate its activity and thereby promote EPS production. To search for substrates of the EpsAB kinase, we collected cells with His6-EpsB grown under biofilm-inducing conditions and affinity-purified the tagged kinase. His6-EpsB and associated proteins were visualized by immunoblot analysis using an antibody specific for phosphotyrosine residues, revealing immunoreactive proteins of ∼25 and 30 kDa (Fig. 3A). The 25-kDa species was approximately the size expected for His6-EpsB. The large species was approximately the size expected for the glycosyl transferase EpsE, the only protein in the EPS biosynthetic pathway with a predicted molecular weight (32 kDa) of ∼30 kDa.
Figure 3.
Glycosyl transferase EpsE is phosphorylated in an EpsAB-dependent manner. (A) Immunoblot probed with anti-phosphotyrosine antibodies of affinity-purified, His6-tagged EpsB and copurifying protein(s) from cells (BAE318) that had been grown on biofilm-inducing medium. Also shown is a control with wild-type (BAE262) cells that produced untagged EpsB. (B) Immunoblot of His6-tagged EpsE from wild-type cells (BAE560), ∆epsAB mutant cells (BAE561), and ∆epsB mutant cells (BAE563) harboring a mutant copy of the gene producing a catalytically inactive kinase, EpsBD81A/D83A. Cells were grown in biofilm-inducing medium, and the blot was probed with anti-His6 antibodies (top row) and anti-phosphotyrosine antibodies (bottom row). (C) Immunoblot showing the time course of phosphorylation of EpsE and EpsB. His-tagged protein was purified from cells producing His6-EpsE (BAE477) or cells producing His6-EpsB (BAE318). The cells had been harvested at the indicated times during growth on biofilm-inducing medium. The blots were probed with anti-His6 antibodies (top rows) and anti-phosphotyrosine antibodies (bottom rows).
To investigate whether EpsE is indeed phosphorylated and, if so, in an EpsAB-dependent manner, we introduced at sacA a xylose-inducible copy of epsE that had been modified to contain six histidine codons at the 5′ end. We then affinity-purified His6-EpsE from a solubilized preparation of membrane proteins from cells grown under biofilm-inducing conditions. The affinity-purified protein was then subjected to immunoblot analysis using anti-His6 antibodies (Fig. 3B, top panel) to detect the His6-EpsE protein and anti-phosphotyrosine antibodies (Fig. 3B, bottom panel). In agreement with our expectation, His6-EpsE that had been purified from otherwise wild-type cells reacted with both antibodies. However, His6-EpsE from cells that were deleted for epsA and epsB or cells that contained a mutant epsB gene that was expected to produce a catalytically inactive kinase (above) reacted only with the anti-His6 antibodies. We conclude that EpsE undergoes phosphorylation at one or more tyrosine residues and that this phosphorylation requires an active EpsAB kinase.
Phosphorylation of EpsE precedes EpsB autophosphorylation during biofilm formation
The results so far suggest that EpsB stimulates EPS production at least in part by phosphorylating EpsE and that EpsB is turned off by autophosphorylation. If so, then autophosphorylation of EpsB should be anti-correlated with phosphorylation of EpsE. To test this expectation, we collected cells producing His6-EpsB or His6-EpsE at various times during biofilm formation and monitored phosphorylation of the two proteins by immunoblot analysis. The time-course experiment of Figure 3C shows that EpsE was phosphorylated at early times during biofilm formation when EpsB was unphosphorylated. (Note that because cells were collected as floating biofilms [pellicles], biofilm formation and EPS production had already commenced at the earliest time point in the experiment.) Interestingly, at late times, EpsB became phosphorylated when the levels of phospho-EpsE markedly decreased. These results are consistent with a model in which EpsAB is unphosphorylated early in biofilm formation when it phosphorylates EpsE. Later, as the biofilm matures, EpsAB undergoes autophosphorylation when EpsE ceases to be phosphorylated and instead becomes dephosphorylated.
Purified EPS stimulates kinase activity
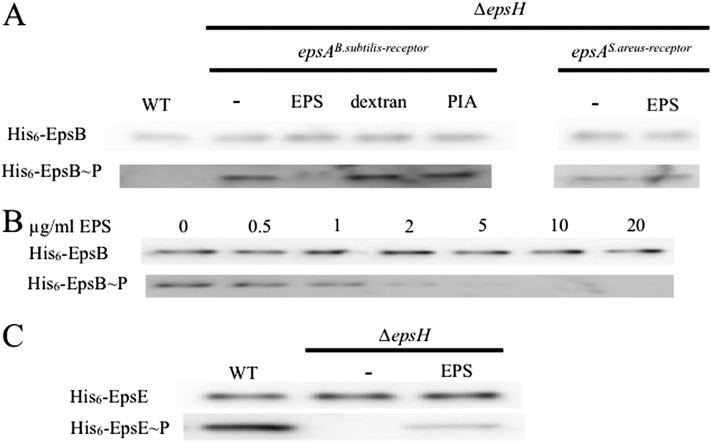
In toto, the above results indicate that EpsAB-mediated phosphorylation of EpsE stimulates EPS production at the start of biofilm formation. We speculated that EpsAB was sensing a signal that inhibits autophosphorylation of EpsB and allows phosphorylation of EpsE. An attractive candidate for such a signal was EPS itself, whose presence might indicate a high local density of EPS-producing cells. To investigate this possibility, we monitored EpsB autophosphorylation under biofilm-inducing conditions in EPS-producing cells and in cells blocked in EPS production due to a mutation in the biosynthetic gene epsH. Because epsAB is normally expressed at a low level, we enhanced expression using an IPTG-inducible promoter. The results show that His6-EpsB was phosphorylated in cells blocked in EPS production (due to a mutation in the biosynthetic gene epsH) but not in EPS-producing cells (Fig. 4A).
Figure 4.
EPS stimulates phosphorylation of EpsB and EpsB. (A) Immunoblot of purified His6-tagged EpsB from wild-type cells (BAE318) and cells mutant for epsH (BAE397) expressing the EpsAB (BAE397) or the chimeric CapA-EpsAB (BAE619) from an IPTG-inducible promoter grown in the presence or absence of polysaccharide. The wild-type cells and cells mutant for epsH were grown in biofilm-inducing medium to which IPTG was added. The epsH mutant cells were grown in LB medium to which IPTG was added at midexponential phase and that had been supplemented with no polysaccharide (−) or with purified B. subtilis EPS (20 μg/mL) or PIA (20 μg/mL) or 0.1% dextran as indicated. (B) Immunoblot showing the dose dependence of EpsB autophosphorylation on EPS. His6-tagged EpsB from epsH mutant cells that contained an IPTG-inducible copy of epsA and his6-epsB (BAE397) was grown in LB medium to which IPTG was added and treated with increasing concentrations of purified EPS. (C) Immunoblot of purified His6-tagged EpsE from wild-type cells (BAE560) or epsH mutant cells that carried a xylose-inducible copy of the gene for the tagged protein (BAE562). The cells were grown in biofilm-inducing medium in the presence of xylose for 24 h and either left untreated (−) or treated with 20 µg/mL EPS. For all three panels, the top row was probed with anti-His6 antibodies, and the bottom row was probed with anti-phosphotyrosine antibodies.
To ask whether EPS itself is the signal that inhibits EpsB autophosphorylation, we again used epsH mutant cells harboring the genes for EpsA and His6-EpsB fused to an IPTG-inducible promoter. The cells were grown under conditions (Luria-Bertani [LB] medium) that normally do not support biofilm formation. The results show that EpsB was highly phosphorylated and that the addition of EPS that had been extracted and purified from biofilms formed by wild-type cells (Kolodkin-Gal et al. 2012) prevented autophosphorylation (Fig. 4A) and did so in a dose-dependent manner that decreased with increasing concentrations of EPS (Fig. 4B). As controls, the addition of the polysaccharide dextran or the Staphylococcus aureus exopolysaccharide PIA, a homoglycan of β-(1,6)-linked N-acetylglucosamine residues (Rohde et al. 2010), did not prevent autophosphorylation (Fig. 4A). To further test the idea that EPS is part of a cell-to-cell signaling pathway, we cocultured a strain blocked in EPS production, which carries the gene for His6-EpsB, together with wild-type B. subtilis. The results show that cocultivation with the wild-type strain prevented autophosphorylation of His6-EpsB in the strain that was blocked in EPS production (Supplemental Fig. S3).
Finally, we investigated the effect of EPS on phosphorylation of EpsE under biofilm-inducing conditions using cells with a xylose-inducible copy of the gene for His6-EpsE. In mutant cells blocked in EPS production, EpsE was not phosphorylated, but phosphorylation was restored by the addition of purified EPS (Fig. 4C). We also tested the effect of purified EPS on EpsE phosphorylation under conditions that did not support biofilm formation using cells with an IPTG-inducible copy of epsAB and a xylose-inducible copy of the gene for His6-EpsE (Supplemental Fig. S4). Addition of purified EPS restored phosphorylation to a mutant blocked in EPS production and did so in a dose-dependent manner.
These results and those of Figure 3C support a model in which EPS stimulates EpsAB kinase activity by inhibiting autophosphorylation. At early times, this stimulation results in the phosphorylation and activation of the EpsE glycosyl transferase, promoting more EPS synthesis and setting up a positive feedback loop. This positive feedback loop does not persist indefinitely, however. As noted above, EpsB switches back to the inactive, phosphorylated state late in biofilm maturation. Evidently, when the biofilm matures, EPS ceases to inhibit autophosphorylation, preventing continued phosphorylation of EpsE. We do not know the mechanism by which EPS ceases to inhibit autophosphorylation late in development. (Conceivably, entrainment of EPS in the protein component of the matrix, the amyloid-like fiber TasA, late in development limits its ability to interact with EpsAB.)
Phosphorylated EpsB is proteolytically unstable
If EPS inhibits autophosphorylation, then how are pre-existing phosphorylated EpsB molecules replaced with unphosphorylated kinase molecules? One possibility is that the phosphate groups are removed by a tyrosine phosphatase. We consider this unlikely, however, because mutants of genes with homology with tyrosine phosphatases did not exhibit a measurable defect in biofilm formation (data not shown). Instead, our evidence indicates that the phosphorylation state of EpsB is controlled at the level of proteolysis. We found that phospho-EpsB in the absence of EPS was subject to rapid degradation, whereas unphosphorylated EpsB in the presence of EPS was relatively stable (Supplemental Fig. S5). Thus, when autophosphorylation is inhibited by EPS, phosphorylated and thus inactive EpsB molecules are rapidly replaced by newly synthesized, unphosphorylated, and active EpsB molecules.
The extracellular domain of EpsA determines the specificity of exopolysaccharide sensing
What feature of EpsAB is responsible for sensing and responding to EPS? Because EPS is present outside the cell, an attractive candidate for the receptor was the extracellular domain of the membrane-anchored component of the kinase. Cells entirely lacking EpsA were blocked in kinase activity. Also, cells producing a truncated EpsA that was missing the extracellular domain but retained the two transmembrane segments were similarly blocked in kinase activity (possibly simply because the truncated protein was unstable) (Supplemental Fig. S6). However, we were able to restore kinase activity by constructing a protein fusion in which the C-terminal, cytoplasmic domain of EpsA (lacking the transmembrane segments and the extracellular domain) was joined to the N terminus of EpsB. This fusion protein was functional as a kinase, but autophosphorylation was refractory to inhibition by EPS (Supplemental Fig. S7). Thus, fusion of EpsB to the cytoplasmic tail of EpsA is sufficient for kinase activity, but the extracellular domain is necessary for the response to EPS.
As a test of the idea that the extracellular domain is a receptor for EPS, we replaced the extracellular domain of EpsA with the corresponding domains of EpsA homologs from the Gram-positive bacteria Bacillus licheniformis, which shares an orthologous epsA-O operon with B. subtilis, and S. aureus, which produces the dissimilar exopolysaccharide PIA. The results show that the corresponding domain of the homologous receptor from B. licheniformis was able to restore the function of the EpsA receptor, both kinase activity and responsiveness to EPS (Supplemental Fig. S8). In contrast, when we tested a construct in which we replaced the extracellular domain with that of the corresponding homolog from S. aureus (CapA), we found that EpsB was phosphorylated and hence active as a kinase but that the addition of EPS did not inhibit autophosphorylation in cells grown under biofilm-forming conditions (Fig. 4A). Also, the chimeric protein was nonfunctional in that it did not support biofilm formation (Supplemental Fig. S8).
The extracellular domain of EpsA interacts directly and specifically with EPS
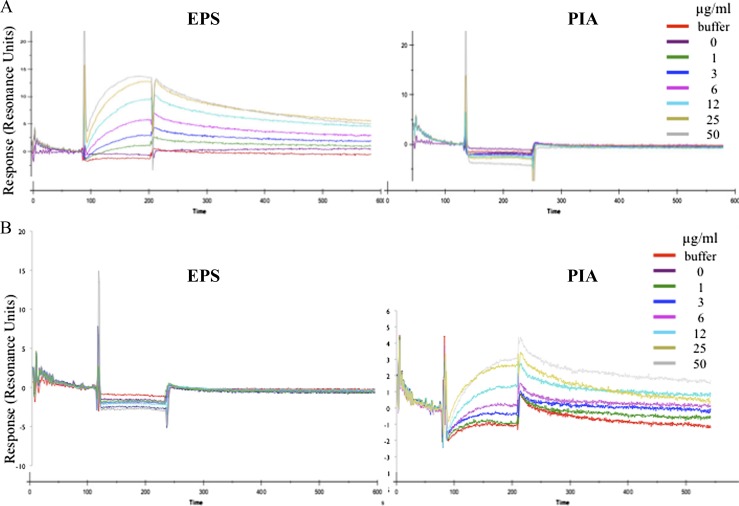
Finally, we used surface plasmon resonance to ask whether purified EPS directly binds to the extracellular domain of EpsA. Accordingly, we immobilized on the surface of a CM5 chip a segment of EpsA that corresponded to the extracellular domain. Various concentrations of purified EPS and PIA were passed over the chip, and binding to the extracellular domain was monitored. The results showed binding of EPS in a concentration-dependent manner but little or no binding of PIA (Fig. 5A). Also, dextran and the monosaccharides N-acetyl glucosamine and glucose did not show significant binding (data not shown).
Figure 5.
The extracellular domain of EpsA binds EPS. The His-tagged extracellular domain of EpsA and CapA was immobilized on a CM5 chip surface. Shown is the binding of EPS and PIA that had been passed over the chip surface at the indicated concentrations for the EpsA receptor (A) or the CapA receptor (B).
We then tested whether the extracellular domain of the CapA receptor was able to recognize its cognate exopolysaccharide, PIA. The results showed specific and dose-dependent binding of PIA to the CapA receptor (Fig. 5B).
These findings support the conclusion that the extracellular domain of EpsA is a receptor that interacts directly and specifically with EPS and that CapA is a receptor that specifically interacts directly and specifically with PIA. EPS is likely heterogeneous in length, and it will be of interest in future work to identify the size of polymers that bind with the highest affinity.
Discussion
Here we presented evidence for a novel system of intercellular signaling that controls the production of a common good in the tightly packed community of a biofilm by acting at the level of protein modification; namely, tyrosine phosphorylation. We showed that the levels of EPS produced by B. subtilis depend on not only the transcriptional regulatory circuitry that mediates derepression of the epsA-O operon under biofilm-inducing conditions but also the amount of EPS present at any given time. Our evidence indicates that the extracellular domain of the EpsA component of the EpsAB tyrosine kinase is a receptor that directly recognizes EPS and thereby regulates kinase activity by the cytoplasmic component EpsB. We therefore envision EPS as a short-range signaling molecule for cells in the developing biofilm that triggers a positive feedback loop that stimulates EPS production.
Because bacterial tyrosine kinases are frequently associated with exopolysaccharide biosynthetic production pathways in bacteria, we speculate that B. subtilis-like self-regulation may be a general feature of exopolysaccharide production at least in Gram-positive bacteria. Consistent with this idea, we show here that the extracellular domain of CapA specifically binds PIA, suggesting that it too is a receptor for controlling exopolysaccharide production in S. aureus. Bacterial tyrosine kinases, such as Wzc and Etk, are also found in Gram-negative bacteria, where they are associated with the pathways for the production of capsular polysaccharides I and IV, respectively (Wugeditsch et al. 2001; Peleg et al. 2005; Whitfield 2006). It is not obvious, however, how our generalization could apply in cases in which the receptor for the kinase is sequestered in the periplasm and hence unable to interact directly with the environment (Bechet et al. 2009). It seems unlikely that exopolysaccharide would be freely diffusible across the outer membrane and hence able to interact with the kinase. If bacterial tyrosine kinases also mediate community-based exopolysaccharide sensing in Gram-negative bacteria, then perhaps the enzyme is coupled to an additional component located in the outer membrane that serves as a receptor or to a channel for diffusion of exopolysaccharide into the periplasm.
The discovery of EPS self-regulation explains a paradox. Under non-biofilm-inducing conditions, such as during exponential phase growth in rich medium, B. subtilis exists in two alternative states and switches between them stochastically (Kearns and Losick 2005; Dubnau and Losick 2006; Norman et al. 2014). These are individual, motile cells, the predominant cell type, and long chains of sessile cells. The sessile chains are thought to be precursors of the biofilm, and, indeed, cells in the chains are derepressed for the expression of matrix genes (Kearns et al. 2005; Chai et al. 2010; Norman et al. 2014). Biofilms also consist of cell chains, but in this case, the chains are held together in parallel in dense assemblages by the extracellular matrix (Branda et al. 2001; Vlamakis et al. 2008). Why, then, would cells in the separate chains seen during growth express the epsA-O operon at high levels? Wouldn’t this lead to the wasteful production of EPS or in fact cause cells to clump together? Our present discovery of a community-based sensing system for controlling EPS production suggests a possible answer: Under conditions of exponential phase growth when the population is dominated by individual motile cells (which do not express the epsA-O operon), EPS production by a small number of separate chains is likely insufficient to enable the EPS to attain levels high enough to stimulate its own production via the tyrosine kinase-sensing system. If our reasoning is correct, then chains may indeed be precursors to biofilms that are primed to produce a matrix when appropriate environmental signals turn on the circuitry for biofilm formation in a proportion of the cell population high enough to activate the EpsAB kinase. Also, and unlike conventional quorum-sensing signaling molecules, EPS might be a relatively short-range signal that acts only when cells are in close proximity to each other. In this view, under biofilm-inducing conditions, a small number of chains that are near to each other might nucleate local production of EPS, stimulating other nearby chains to produce EPS and thereby stick together.
Interestingly, self-induced stimulation of exopolysaccharide production has previously been described for Psl, the polysaccharide component of the extracellular matrix of Pseudomonas aeruginosa biofilms (Irie et al. 2012). Importantly, Parsek and coworkers (Irie et al. 2012) have shown that purified Psl can stimulate its own production. In contrast to EPS, however, Psl works by stimulating synthesis of the regulatory molecule cyclic-di-GMP. Cyclic-di-GMP in turn acts at both the transcriptional and post-transcriptional levels to promote biofilm matrix production (Irie et al. 2012). Indeed, P. aeruginosa does not have a member of the bacterial tyrosine kinase family (Jadeau et al. 2012). Just how Psl stimulates cyclic-di-GMP synthesis is unknown, and the receptor for Psl remains elusive. Nonetheless, that both Psl and EPS regulate their own production, albeit by unrelated mechanisms, suggests that direct sensing of exopolysaccharide levels in biofilm communities confers a fitness benefit that has been selected independently more than once in bacteria.
An exopolysaccharide that plays both structural and signaling roles is also known in mammalian systems (Fraser et al. 1997; Toole 2000). Unlike the carbohydrate moiety of glycoproteins, the glycosaminoglycan hyaluronic acid is a freely occurring, high-molecular-weight polysaccharide found in the extracellular matrix (Comper 1996). Not only is hyaluronic acid a structural component of the matrix, but it is also a signaling molecule that is directly recognized by several receptors, including CD44, RHAMM, and Toll-like receptors (Aruffo et al. 1990; Schaefer 2010; Karbownik and Nowak 2013). For example, binding of hyaluronic acid to the CD44 receptor activates the NLRP3 inflammasome, leading to secretion of interleukin-1β (IL-1β) (Yamasaki et al. 2009).
Finally, we note that exopolysaccharides whose biosynthesis is associated with bacterial tyrosine kinases are sometimes important virulence factors. A well-known example is the capsular polysaccharide of Streptococcus pneumoniae (Morona et al. 2006). In such cases, the exopolysaccharide receptor may be an unusually attractive target for drug discovery (at least in Gram-positive bacteria) because it is displayed on the cell surfaces (hence circumventing issues of drug permeability) and readily lends itself to biochemical screens for receptor-binding molecules and whole-cell screens for kinase inhibitors.
Materials and methods
General methods
All B. subtilis strains used in the present study were derivatives of NCBI 3610 and are listed in Supplemental File 1A in the Supplemental Material. Oligonucleotides used in this study are listed in Supplemental File 1B. DNA manipulation and other molecular biological procedures were carried out according to standard protocols (Sambrook et al. 1989). Transformation of B. subtilis cells was performed by a two-step protocol (Wilson and Bott 1968). All plasmids were maintained in E. coli strains DH5α or New England Biolabs Turbo using 100 μg/mL ampicillin.
Bacterial strains and media
Cell cultivation was performed in LB medium (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl per liter of broth) or synthetic MSgg medium (5 mM potassium phosphate at pH 7, 100 mM MOPS at pH 7, 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 50 μM FeCl3, 1 μM ZnCl2, 2 μM thiamine, 0.5% glycerol, 0.5% glutamate, 50 μg/mL tryptophan, 50 μg/mL phenylalanine, 50 μg/mL threonine). The same medium with 1.5% Bacto-agar (Difco) was used for growth on plates. Antibiotics were added to the medium at the following concentrations: 17 μg/mL tetracycline, 100 μg/mL spectinomycin, 5 μg/mL kanamycin, MLS (1 μg/mL erythromycin, 25 μg/mL lincomycin), 100 μg/mL ampicillin, or 5 μg/mL chloramphenicol. Growth was monitored by measuring the optical density at 600 nm (OD600). Cultures were routinely grown in 500-mL Erlenmeyer flasks at 180 rpm and 37°C.
Strain construction
Plasmids were constructed using isothermal (Gibson) assembly method (Gibson et al. 2009). Each plasmid construct was sequenced to exclude point mutations and verify nucleotide substitutions. Markerless deletions of epsA, epsB, epsE, and epsAB were constructed according to Arnaud et al. (2004) using pMiniMAD (a gift of D. Kearns). For details of strain construction, see the Supplemental Material.
Biofilm assay
B. subtilis cells were cultured from a 1-d-old colony in LB broth at 37°C to midexponential phase. For colony formation, 2 μL of cells was spotted on MSgg agar plates (1.5%). Pellicle assays were performed in 6 mL of MSgg medium in a six-well plate to which 5 μL of cells was added. Both were incubated for 3 d at 30°C. All images were taken using either a Nikon Coolpix 950 digital camera or a SPOT camera (Diagnostic Instruments).
Protein expression and purification
For purification of His6-EpsB from colonies, the corresponding strains were grown on MSgg agar as described above. After 3 d, several colonies of the same strain were collected and gently sonicated to break up the biofilm. Cells were then collected by centrifugation. For purification of His6-EpsE from colonies (Fig. 3B), the corresponding strains were grown on MSgg agar as described above. After 2 d, several colonies of the same strain were collected, gently sonicated, and collected by centrifugation as above.
For purification of His6-EpsB and His6-EpsE from pellicles (Fig. 3C), the corresponding strains were grown in MSgg; pellicles were collected at the indicated times, washed, and gently sonicated; and cells were collected by centrifugation as above.
To purify His6-EpsB and His6-EpsE from MSgg medium (Fig. 4A,C), the corresponding strains were grown for 24 h (EpsE) or 48 h (EpsB) at 37°C. Next, cells were pelleted by centrifugation at 8000g for 10 min at 4°C.
To purify His6-EpsB, His6-EpsE, and His6-CapB, the corresponding strains were grown in LB until OD600 0.5. Next, 100 μM IPTG or 0.5% xylose was added, and cells were grown for another hour until the indicated polysaccharide was added. After another hour, cells were pelleted at 8000g for 10 min at 4°C.
Cell pellets were resuspended in 20 mL of lysis/binding buffer (20 mM Tris-HCl at pH 8.0, 150 mM NaCl, 20 mM imidazole, 1 mM PMSF, 1 mM β-glycerol phosphate, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, 10 mM sodium fluoride) and disrupted using a cell disruptor. The lysate was centrifuged at 1000g for 3 min to remove cell debris. Next, the supernatant was centrifuged for 30 min at 15,000g to separate the cell membrane from the soluble fraction. The membrane fraction was resuspended in lysis buffer containing 1% Triton X-100. The solution then was incubated overnight at 4°C. Next, it was centrifuged at 15,000g for 30 min to remove cell debris, and the supernatant was diluted with lysis buffer to <0.1% Triton X-100 and then incubated with Ni-NTA Dynabeads (Life Technologies) overnight at 4°C. Next, the beads were separated from the lysate using a magnet and washed three times with 1 mL of buffer W (50 mM Tris-HCl at pH 8, 150 mM NaCl, 20 mM imidazole, 1 mM PMSF, 1 mM β–glycerol phosphate, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, 10 mM sodium fluoride). The purified protein was eluted from the beads with 100 μL of elution buffer (50 mM Tris-HCl at pH 8, 150 mM NaCl, 250 mM imidazole).
Immunoblot analysis
To ensure loading of similar amounts of the purified proteins, we determined the concentration of the purified proteins using a Bradford assay and loaded the gels with 100 ng of protein. To confirm that we had loaded similar amounts of protein, we visualized the His6-tagged protein with an anti-hexahistidine (Abcam) antibody. Purified proteins were mixed with SDS-Laemmli buffer (Laemmli 1970) and separated by standard 12% SDS-PAGE with Mini-Protean cells (Bio-Rad). Proteins were transferred to Immobilon-P PVDF membrane (Millipore). The membrane was blocked with 5% BSA in TBST for 1 h and probed with anti-hexahistidine (Abcam) and anti-phosphotyrosine (Biolegend) antibodies as primary antibodies. A secondary antibody (goat anti-rabbit [His] or anti-mouse [phosphotyrosine] immunoglobulin G) conjugated to horseradish peroxidase was used at a 1:100,000 dilution (Bio-Rad). Immunoblots were developed with SuperSignal West Dura chemiluminescent substrate (Thermo) and imaged on a gel-doc (Bio-Rad).
EPS purification
Pellicles that had been formed in biofilm-inducing medium were collected at day 3, washed twice in phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), mildly sonicated, and then centrifuged to remove the cells. The supernatant was mixed with 5× ice-cold isopropanol and incubated overnight at 4°C. Samples were centrifuged at 8000 rpm for 10 min at 4°C. Pellets were resuspended in a digestion mix of 0.1 M MgCl2, 0.1 mg/mL DNase, and 0.1 mg/mL RNase; mildly sonicated; and incubated for 4 h at 37°C. Samples were extracted twice with phenol-chloroform. The aquatic fraction was dialyzed for 48 h with Slide-A-Lyzer dialysis cassettes by Thermo Fisher, 3500 MCWO, against dH2O. Samples were lyophilized. The remaining pellet was weighed and afterward dissolved in 500 μL of dH2O.
PIA purification
PIA purification was performed after a modified protocol (Mack et al. 1996). The SH1000 strain was grown in TBS medium with 0.5% glucose at 80 rpm for 24 h at 37°C. Cells were collected and gently sonicated to loosen the PIA exopolysaccharide. Next, cells were removed, and supernatant was precipitated with 5× ice-cold isopropanol overnight at 4°C. Samples were centrifuged at 8000 rpm for 10 min at 4°C. Pellets were resuspended in a digestion mix of 0.1 M MgCl2, 0.1 mg/mL DNase, and 0.1 mg/mL RNase; mildly sonicated; and incubated for 4 h at 37°C. Samples were extracted twice with phenol-chloroform. The aquatic fraction was dialyzed for 48 h with Slide-A-Lyzer dialysis cassettes by Thermo Fisher, 3500 MCWO, against dH2O. Samples were lyophilized. The remaining pellet was weighed and afterward dissolved in 500 μL of dH2O.
Protein production in and purification from E. coli
Genes were cloned into pRSETA (Life Technologies) using isothermal assembly (Gibson et al. 2009) with the primers Receptor_setA_for and Receptor_setA_rev for the EpsA receptor and the primers CapA_setA_ITA_for and CapA_setA_ITA_rev for the CapA receptor, which contained overlapping DNA sequences to the MCS of pRSETA. Plasmids were transformed into E. coli BL21. Cells were grown in LB medium at 37°C in the presence of 100 μg/mL ampicillin, and expression of the His6-tagged proteins was stimulated by the addition of 1 mM ITPG during the midexponential phase. Cells were grown for another 4 h at room temperature and then collected by centrifugation at 8000g at 4°C. Cell pellets were resuspended in 20 mL of lysis/binding buffer (50 mM NaH2PO4 at pH 8.0, 150 mM NaCl, 20 mM imidazole, 1 mM PMSF) and disrupted using a cell disruptor. Cell debris was removed with centrifugation at 15,000g for 20 min at 4°C. Supernatant was incubated with Ni-NTA Dynabeads (Life Technologies) overnight at 4°C. Next, the beads were separated from the lysate using a magnet and washed three times with 10 bed volumes of buffer W (50 mM NaH2PO4 at pH 8, 150 mM NaCl, 20 mM imidazole, 1 mM PMSF). The purified protein was eluted from the beads with 500 μL of elution buffer (50 mM NaH2PO4 at pH 8, 150 mM NaCl, 250 mM imidazole). Proteins were dialyzed against buffer containing PBS (pH 7.5).
Surface plasmon resonance
A Biacore X100 instrument was used for all experiments. The extracellular domain of EpsA (from residues 41–174) or CapA (from residues 43–171) that had been His-tagged was covalently coupled to the surface of a research-grade CM5 sensor chip using amine-coupling chemistry with coupling buffer (10 mM acetate buffer) adjusted to pH 4.5 for EpsA and pH 4 for CapA according to the manufacturer’s instructions. As a reference surface for the refractive index and correction of nonspecific binding, a second flow cell was treated in the same manner but without the addition of the EpsA extracellular domain. Solutions containing the indicated concentrations of the exopolysaccharide in HBS-EP buffer (10 mM 4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid [HEPES], 150 mM NaCl, 3.4 mM ethylenediaminetetraacetic acid [EDTA], 0.005% (v/v) surfactant polysorbate P20 at pH 7.4, from BIAcore) were passed over the sensor chip surface at a flow rate of 10 μL/min at 25°C. Association was measured for 150 sec, and dissociation was measured for 240 sec. After each injection, the surface was regenerated with a 15-sec injection of 10 mM glycine (pH 2.2) (purchased from Biacore) at a flow rate of 10 μL/min. The response of the same solutions on a mock-coupled surface was subtracted from all sensograms. Chips were reused as long as the RU response to test samples was reproducible.
Acknowledgments
We thank W. Lane for the mass spectrometry analysis, D. Andres for help with analysis of the Biacore experiments, Y. Chai for helpful discussions and strains, and R. Kolter, M. Parsek, J. Lee, and members of the Losick laboratory for advice on the manuscript. A.K.W.E. was an EMBO post-doctoral fellow. S.A.W. was a Jane Coffin Childs post-doctoral fellow. This work was supported by National Institutes of Health (NIH) grant GM18568 and NIH/National Institute of Allergy and Infectious Diseases grant 5P01AI083214.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.246397.114.
References
- Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl Environ Microbiol 70: 6887–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. 1990. CD44 is the principal cell surface receptor for hyaluronate. Cell 61: 1303–1313 [DOI] [PubMed] [Google Scholar]
- Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, Sekowska A, Vallenet D, Wang T, Moszer I, Médigue C, et al. . 2009. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155: 1758–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechet E, Guiral S, Torres S, Mijakovic I, Cozzone A-J, Grangeasse C. 2009. Tyrosine-kinases in bacteria: from a matter of controversy to the status of key regulatory enzymes. Amino Acids 37: 499–507 [DOI] [PubMed] [Google Scholar]
- Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci 98: 11621–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, González-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. 2004. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol 186: 3970–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Chu F, Kolter R, Losick R. 2008. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol 67: 254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Norman T, Kolter R, Losick R. 2010. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev 24: 754–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Kearns DB, Branda SS, Kolter R, Losick R. 2006. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol 59: 1216–1228 [DOI] [PubMed] [Google Scholar]
- Comper WD. 1996. Extracellular matrix. CRC Press, Boca Raton, FL. [Google Scholar]
- Dubnau D, Losick R. 2006. Bistability in bacteria. Mol Microbiol 61: 564–572 [DOI] [PubMed] [Google Scholar]
- Fraser JR, Laurent TC, Laurent UB. 1997. Hyaluronan: its nature, distribution, functions and turnover. J Intern Med 242: 27–33 [DOI] [PubMed] [Google Scholar]
- Fuqua C, Greenberg EP. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol 3: 685–695 [DOI] [PubMed] [Google Scholar]
- Gerwig J, Kiley TB, Gunka K, Stanley-Wall N, Stülke J. 2014. The protein tyrosine kinases EpsB and PtkA differentially affect biofilm formation in Bacillus subtilis. Microbiology 160: 682–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Grangeasse C, Obadia B, Mijakovic I, Deutscher J, Cozzone AJ, Doublet P. 2003. Autophosphorylation of the Escherichia coli protein kinase Wzc regulates tyrosine phosphorylation of Ugd, a UDP-glucose dehydrogenase. J Biol Chem 278: 39323–39329 [DOI] [PubMed] [Google Scholar]
- Grangeasse C, Cozzone AJ, Deutscher J, Mijakovic I. 2007. Tyrosine phosphorylation: an emerging regulatory device of bacterial physiology. Trends Biochem Sci 32: 86–94 [DOI] [PubMed] [Google Scholar]
- Grangeasse C, Terreux R, Nessler S. 2010. Bacterial tyrosine-kinases: structure–function analysis and therapeutic potential. Biochim Biophys Acta 1804: 628–634 [DOI] [PubMed] [Google Scholar]
- Grangeasse C, Nessler S, Mijakovic I. 2012. Bacterial tyrosine kinases: evolution, biological function and structural insights. Philos Trans R Soc Lond B Biol Sci 367: 2640–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobley L, Ostrowski A, Rao FV, Bromley KM, Porter M, Prescott AR, MacPhee CE, van Aalten DMF, Stanley-Wall NR. 2013. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc Natl Acad Sci 110: 13600–13605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie Y, Borlee BR, O’Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. 2012. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci 109: 20632–20636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadeau F, Grangeasse C, Shi L, Mijakovic I, Deléage G, Combet C. 2012. BYKdb: the bacterial protein tyrosine kinase database. Nucleic Acids Res 40: D321–D324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbownik MS, Nowak JZ. 2013. Hyaluronan: towards novel anti-cancer therapeutics. Pharmacol Rep 65: 1056–1074 [DOI] [PubMed] [Google Scholar]
- Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev 19: 3083–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55: 739–749 [DOI] [PubMed] [Google Scholar]
- Kiley TB, Stanley-Wall NR. 2010. Post-translational control of Bacillus subtilis biofilm formation mediated by tyrosine phosphorylation. Mol Microbiol 78: 947–963 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Iwano M. 2012. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol 85: 51–66 [DOI] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Cao S, Chai L, Böttcher T, Kolter R, Clardy J, Losick R. 2012. A self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell 149: 684–692 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lacour S, Bechet E, Cozzone AJ, Mijakovic I, Grangeasse C. 2008. Tyrosine phosphorylation of the UDP–glucose dehydrogenase of Escherichia coli is at the crossroads of colanic acid synthesis and polymyxin resistance. PLoS ONE 3: e3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol 178: 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I, Poncet S, Boël G, Mazé A, Gillet S, Jamet E, Decottignies P, Grangeasse C, Doublet P, Le Maréchal P, et al. . 2003. Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J 22: 4709–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minic Z, Marie C, Delorme C, Faurie J-M, Mercier G, Ehrlich D, Renault P. 2007. Control of EpsE, the phosphoglycosyltransferase initiating exopolysaccharide synthesis in Streptococcus thermophilus, by EpsD tyrosine kinase. J Bacteriol 189: 1351–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona JK, Morona R, Paton JC. 2006. Attachment of capsular polysaccharide to the cell wall of Streptococcus pneumoniae type 2 is required for invasive disease. Proc Natl Acad Sci 103: 8505–8510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W-L, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet 43: 197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman TM, Lord ND, Paulsson J, Losick R. 2014. Memory and modularity in cell-fate decision making. Nature 503: 481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42: 541–564 [DOI] [PubMed] [Google Scholar]
- Peleg A, Shifrin Y, Ilan O, Nadler-Yona C, Nov S, Koby S, Baruch K, Altuvia S, Elgrably-Weiss M, Abe CM, et al. . 2005. Identification of an Escherichia coli operon required for formation of the O-antigen capsule. J Bacteriol 187: 5259–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar V, Shi L, Krug K, Derouiche A, Jers C, Cousin C, Kobir A, Mijakovic I, Macek B. 2014. Quantitative phosphoproteome analysis of Bacillus subtilis reveals novel substrates of the kinase PrkC and phosphatase PrpC. Mol Cell Proteomics doi: 10.1074/mcp.M113.035949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde H, Frankenberger S, Zähringer U, Mack D. 2010. Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur J Cell Biol 89: 103–111 [DOI] [PubMed] [Google Scholar]
- Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci 107: 2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Bassler BL. 2012. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2: a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schaefer L 2010. Extracellular matrix molecules: endogenous danger signals as new drug targets in kidney diseases. Curr Opin Pharmacol 10: 185–190 [DOI] [PubMed] [Google Scholar]
- Toole BP 2000. Hyaluronan is not just a goo! J Clin Invest 106: 335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R, Kolter R. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22: 945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 75: 39–68 [DOI] [PubMed] [Google Scholar]
- Wilson GA, Bott KF. 1968. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J Bacteriol 95: 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wugeditsch T, Paiment A, Hocking J, Drummelsmith J, Forrester C, Whitfield C. 2001. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J Biol Chem 276: 2361–2371 [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Muto J, Taylor KR, Cogen AL, Audish D, Bertin J, Grant EP, Coyle AJ, Misaghi A, Hoffman HM, et al. . 2009. NLRP3/cryopyrin is necessary for interleukin-1β (IL-1β) release in response to hyaluronan, an endogenous trigger of inflammation in response to injury. J Biol Chem 284: 12762–12771 [DOI] [PMC free article] [PubMed] [Google Scholar]