Background: Isatin hydrolase is involved in bacterial indole-3-acetic acid degradation and belongs to a structurally uncharacterized family.
Results: We determined the first crystal structure of a functionally characterized metal dependent hydrolase of this overall fold.
Conclusion: The new hydrolytic fold reveals a transient water wire that allow proton release.
Significance: Proton transfer is a fundamental process important in the understanding of enzymes and membrane transporters.
Keywords: Bacterial Metabolism, Enzyme Structure, Hydrolase, Metalloenzyme, Proton Transport, Grotthuss, Indole-3-acetic Acid Degradation, Kynurenine Formamidase, Gut Flora, Proton Wire
Abstract
The high resolution crystal structures of isatin hydrolase from Labrenzia aggregata in the apo and the product state are described. These are the first structures of a functionally characterized metal-dependent hydrolase of this fold. Isatin hydrolase converts isatin to isatinate and belongs to a novel family of metalloenzymes that include the bacterial kynurenine formamidase. The product state, mimicked by bound thioisatinate, reveals a water molecule that bridges the thioisatinate to a proton wire in an adjacent water channel and thus allows the proton released by the reaction to escape only when the product is formed. The functional proton wire present in isatin hydrolase isoform b represents a unique catalytic feature common to all hydrolases is here trapped and visualized for the first time. The local molecular environment required to coordinate thioisatinate allows stronger and more confident identification of orthologous genes encoding isatin hydrolases within the prokaryotic kingdom. The isatin hydrolase orthologues found in human gut bacteria raise the question as to whether the indole-3-acetic acid degradation pathway is present in human gut flora.
Introduction
Isatin hydrolase is a bacterial manganese-dependent enzyme that catalyzes the hydrolysis of the highly conjugated and heterocyclic compound isatin (1H-indole-2,3-dione) to isatinate (2-(2-aminophenyl)-2-oxoacetate) (Fig. 1). Isatin has been identified as an intermediate in the degradation of indole-3-acetic acid in Bradyrhizobium japonicum (now Bradyrhizobium diazoefficiens), the microsymbiont of soybean (1). Isatin hydrolase activity was demonstrated in different rhizobial species (2). Isatin, the substrate of isatin hydrolase, is not confined to symbiotic bacteria but is found in both eukaryotic and prokaryotic organisms. In humans, isatin is the major component of the endogenous non-peptide drug group tribulin (3). Isatin is found in the bloodstream at a concentration of 0.4 μm (4). A study performed on rats showed that isatin is also present in the brain with the highest levels, of 0.9 μm, found in the hippocampus and the cerebellum (5). Isatin has been shown to inhibit the breakdown of neurotransmitter by inhibiting monoamine oxidase B with a Ki of 3–20 μm (6, 7). The neurologic role of isatin is still unclear. However, isatin levels in blood have been reported to be elevated in persons with Parkinson disease (8), although it is not believed to be part of the etiology of the disorder. Increased levels of isatin have also been detected under certain kinds of stress (9). Rodents or primates administered isatin exhibit anxiety at low doses but experience a sedative effect at higher doses (10). In prokaryotes, isatin has been identified as a signaling molecule in biofilm formation and during chemotactic response. Biofilm formation by enterohemorrhagic Escherichia coli increased 4-fold when exposed to isatin (11). Cellular migration toward higher isatin concentration was observed when E. coli were subjected to isatin concentration gradients (12). Isatin is an unwanted byproduct in the microbial biosynthesis of indigo. The use of isatin hydrolase to remove isatin has been patented (13, 14).
FIGURE 1.

Isatin hydrolase catalyzes the hydrolysis of isatin to isatinate.
Here, we present the first crystal structure of isatin hydrolase isoform b (IH-b)2 from Labrenzia aggregata (formerly Stappia aggregata). The structure revealed a novel catalytic fold that may represent a general hydrolytic fold common to this new class of hydrolases. Elements identified from structural analysis important for the overall fold were found to be conserved in e.g. bacterial kynurenine formamidase (KynB) that clearly show structural homology. In the process of structure analysis, a water substrate chain and proton wire was discovered. The role of a key serine located in the path of the proton wire was studied through mutagenesis. Metalloenzymes require efficient removal of a proton to form a reactive hydroxyl group (15). A number of proton wires have been identified and studied in other systems, e.g. gramicidin A (16), carbonic anhydrase II, and green fluorescent protein (17). To facilitate the proton transfer in biological systems, proton wires were formed. They included a chain of water molecules coordinated to allow a hydronium ion to form at each water position along the chain. The mechanism was originally proposed in 1806 by Theodor von Grotthuss (18) and since been reviewed in 2006 (19). In recent years, research has focused on the structure and regulation of these wires in macromolecules. Proton wires used by biochemical catalysts remain a highly active field of research and add another layer of complexity to the understanding of the catalytic machinery of enzymes.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
The open reading frame encoding IH-b (UniProtKB code A0NLY7) was amplified by PCR from a boiled colony of L. aggregata IAM12614 using the primers 5′-GCTGGATCCGCACAGAGCGCGCTC-3′ and 5′- GCTGAATTCTTATTAGGCTTTGGGGACCAG-3′. DNA fragments were isolated, cut with BamHI and EcoRI, and cloned into the T7-RNA polymerase-dependent E. coli expression plasmid pT7H6 (20) yielding pT7His6-IH-b. This represents the wild-type (wt) enzyme IH-b-wt. The IH-b-S225A and IH-b-S225C mutants were created using the QuikChange Lightning mutagenesis kit (Agilent Technologies), with pT7H6 IH-b plasmid DNA as template and the following oligonucleotides as primers (for S225A, 5′- CTGGCCGCTCTTGCCAATCTCG-3′ and 5′-CGAGATTGGCAAGAGCGGCCAG-3′; for S225C, 5′-CTGGCCTGTCTTGCCAATCTCG-3′ and 5′-GGCAAGACAGGCCAGACCGAAG-3′). The His6-containing IH-b was expressed in E. coli BL21 AI cells (Invitrogen) in 2× tryptone yeast extract medium for 4 h in accordance with the protocol provided by the manufacturer. After expression, cells were harvested by centrifugation and resuspended and lysed by sonication in a buffer containing 50 mm Tris-HCl, pH 8.0, and 0.5 m NaCl supplemented with a protease inhibitor mixture (Sigma-Aldrich). Insoluble material was removed by centrifugation, and the soluble protein extract was batch-adsorbed onto 25 ml of nickel-nitrilotriacetic acid agarose resin (Qiagen) per liter of original culture and loaded into empty liquid chromatography glass columns. The protein-loaded nickel-nitrilotriacetic acid columns were washed with more than 20 column volumes of washing buffer (50 mm Tris-HCl, pH 8.0, 0.5 m NaCl, and 50 mm imidazole). Bound IH was eluted in elution buffer (50 mm Tris-HCl pH 8.0, 0.5 m NaCl, and 10 mm Na2EDTA). The purified recombinant proteins eluted after the blue nickel EDTA fraction. Fractions containing the recombinant proteins were pooled, and the buffer was changed in to 10 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 1 mm Na2EDTA on a Sephadex G-25 column (GE Healthcare).
Prior to crystallization, IH-b was treated with 10 mm EDTA and dialyzed into 5 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 1 mm DTT. The protein solutions were centrifuged at 180,000 × g for 10 min at 4 °C, and the supernatant could hereafter be stored in a stable condition at 4 °C for at least 12 months.
Protein sample homogeneity was assessed by size-exclusion chromatography using a Superdex 200 10/300 GL column connected to an Äkta Purifier system (GE Healthcare). The sample buffer was 25 mm HEPES, pH 7.0, 100 mm NaCl, and 2 mm DTT.
Crystallization and Structure Determination
Apo-IH-b was crystallized by vapor diffusion using sitting drop trays with a protein concentration of 20 mg/ml. Crystals appeared in 0.2 m calcium acetate, 16% (w/v) PEG3350, and 1 mm MnCl2 and grew within 48 h. Glycerol was added to a final concentration of 12% in the drop, before the crystals were mounted and flash-cooled in liquid nitrogen. Data collection was performed at 100 K. A data set extending to 2.25 Å resolution was collected at 9.54 keV (1.30 Å). Data collection strategy was performed with iMOSFLM (21), and the data were processed with XDS (22). A molecular replacement solution was found using a putative metal-dependent hydrolase (Protein Data Bank (PDB) code 1R61) as search model and combined with experimental phases extracted from the two manganese sites. Phased refinement was performed in PHENIX (23). Thioisatinate IH-b crystals were produced by co-crystallization of IH-b with 1 mm thioisatin using crystal condition established for apo-IH-b. The crystals were cryo-protected as for apo-IH-b. A data set extending to 1.90 Å was collected at 9.54 keV (1.30 Å) and processed as apo-IH-b. A molecular replacement solution was found using apo-IH-b as a search model. Initial model building was done using Phenix.Autobuild, and the partially built structure was refined and manually built using Phenix.Refine and Coot (24). The Ramachandran plot was produced and inspected using RAMPAGE (25). Pictures were produced using PyMOL Molecular Graphics System (version 1.5.0.3, Schrödinger, LLC) and Caver PyMOL (plugin 2.1.2) (26). All models and structure factors were deposited to the PDB, given the PDB codes 4J0N (apo-IH-b) and 4M8D (thioisatinate IH-b).
Enzyme Assays
The enzymatic activity of IH-b was assayed as a function of the variation in absorbance spectrum between isatin and isatinate. The development of isatinate was monitored as an increase in light absorption at 368 nm. Measurements were done in a JASCO V-630 spectrophotometer with a cuvette light path of 1 cm. Enzyme reactions to determine kinetic parameters of the IH-b-wt, IH-b-S225A, and IH-b-S225C were performed in reaction buffer A (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 0.2 mm MnCl2). The IH-b concentrations were determined using a NanoDrop instrument at 280 nm (Thermo Scientific). A dilution series of isatin in reaction buffer A of the following concentrations was prepared (100, 50, 25, 12.5, 6.3, 3.3, and 1.6 μm). The reaction was started by mixing 900 μl of the isatin-containing reaction buffer A, with 100 μl of IH-b stock (100 nm) in a 1-ml cuvette. To calculate the absorbance units to concentration, an extinction coefficient of 4.5 × 103 cm−1 mol−1 liter was used for isatinate (2). All measurements were carried out in triplicate. The reaction rate was determined from linear regression of the measured curves at T0. A fit of Michaelis-Menten functions to the three data sets (wild-type and mutant) were produced using non-linear regression to determine Vmax, Km, and kcat. All data treatment and statistics were performed with MS Excel and GraphPad Prism (version 6).
IC50 Measurement of Thioisatinate
At a concentration of 4 nm, IH-b was added to 80 mm isatin in 25 mm Tris-HCl, pH 8.0, 100 mm NaCl, 0.2 mm MnCl2 supplemented by increasing concentrations of thioisatin. A stock of 25 mm thioisatin was prepared in 96% EtOH. It was observed that thioisatin would auto-hydrolyze over time. In this article, we assumed that thioisatin spontaneously hydrolyzes to thioisatinate when exposed to alkaline pH.
Circular Dichroism
All CD experiments were performed on a Jasco J-810 circular dichroism spectropolarimeter, equipped with a Peltier-controlled cuvette holder. IH-b concentrations of 6 μm in 5 mm Tris-HCl, pH 8.0, and 50 mm NaCl, was placed in 2-mm sample length Quartz cuvette during measurement.
Thermal denaturation was monitored by the change in CD signal at 220 nm while heating the sample from 25 to 95 °C at a ramp speed of 1 °C/min. The melting point, Tm, was determined from the inflection point of a nonlinear sigmoidal curve fit between the low (native) and high (unfolded) temperature plateaus.
RESULTS
The Structure of Isatin Hydrolase
The crystal structures of apo-IH-b and thioisatinate-bound IH-b were determined at 2.25 and 1.90 Å resolution, respectively (Table 1). IH-b is a homodimer composed of a core fold region and a region involved in domain swapping with the opposite monomer (Fig. 2, a and b). In terms of Structural Classification of Proteins database classification, IH-b belongs to the class of α/β hydrolases of which the central fold resembles that known as the swiveling β/α/β domain (Fig. 2c) (27). There are two domain swaps that amalgamate the dimer. Domain swap 1 (residues 2–16) is formed by a complete α-helix that exchanges between the monomers. Residues 17–30 act as a linker region between domain swaps 1 and 2 while delivering β1 to the central β-barrel-like structure. The active site pocket is formed by residues 31–48 from the core monomer, whereas residues 49–71 are contributed by the opposite monomer via domain swap 2. Domain swap 2 is stabilized through the formation of an anti-parallel β-sheet by β2 and β3 from each monomer. The conserved metal binding motif is initiated by an inverse γ-turn consisting of residues Glu-72, His-73, and Ser-74, important for positioning the metal coordinating His-73. Two related but functionally uncharacterized structures of proposed metal-dependent hydrolases from Geobacillus stearothermophilus (formerly Bacillus stearothermophilus) (PDB code 1R61) and Methanocaldococcus jannaschii (formerly Methanococcus jannasschii) (PDB code 2B0A) had been deposited to the PDB, without accompanying publication. The central β-barrel in IH-b seems slightly distorted toward the dimer interface and is not closed completely. Three general structural features are conserved among the above-mentioned hydrolases. 1) The central β-barrel structure contains both parallel (β5–β8)) and anti-parallel (β4, β10 and β1, β11, β9) parts. 2) Two α-helices, α1 and α2, are found on the outside of the β barrel of which α2 is always found as a β-α-β motif between strands β7 and β8. 3) Two antiparallel β-strands form a β-hairpin structure that constitutes domain swap 2 (residues 49–71). Conserved structural elements are highlighted in Fig. 2c.
TABLE 1.
X-ray crystallographic data collection and refinement statistics
The necessary diffraction data were obtained from one crystal. Values in parentheses are for the highest resolution shell.
| Apo-IH-b | Thioisatinate-IH-b | |
|---|---|---|
| Data collection | ||
| PDB code | 4J0N | 4M8D |
| Wavelength (Å) | 1.30 | 1.30 |
| Space group | P32 | P21 |
| Unit cell parameters | a = 51.94, b = 51.94, and c = 176.69 Å; α = 90, β = 90, and γ = 120° | a = 84.98, b = 51.82, and c = 357.09 Å; α = 90, β = 91.92, and γ = 90° |
| Resolution (Å) | 43.59–2.25 (2.33-2.25) | 49.51-1.90 (2.00-1.90) |
| Rmeasa | 0.096 (0.765) | 0.108 (0.530) |
| Average I/σI | 11.12 (2.12) | 8.53 (2.83) |
| Anomalous completeness (%) | 98.6 (95.3) | 98.0 (96.5) |
| Redundancy | 4.8 (4.6) | 3.9 (3.8) |
| Wilson B-factor | 29.56 | 27.59 |
| Refinement | ||
| Resolution (Å) | 2.25 | 1.90 |
| No. of reflections | 121,242 (8403) | 471,486 (67179) |
| Rworkb/Rfreec | 0.168/0.218 | 0.152/0.192 |
| No. atoms | 4254 | 27,261 |
| Protein | 3941 | 23,674 |
| Ligand/ion | 10 | 203 |
| Water | 204 | 3383 |
| B-factors (Å2) | ||
| Protein | 32.7 | 25.4 |
| Water | 34.7 | 35.2 |
| r.m.s.d. | ||
| Bond lengths (Å) | 0.006 | 0.010 |
| Bond angles | 0.90° | 1.06° |
| Ramachandran plot (%) | ||
| Favored region | 98.1 | 98.0 |
| Allowed region | 1.5 | 1.6 |
| Outlier region | 0.4 | 0.4 |
a Rmeas is the redundancy independent R-factor calculated as Rmeas = (Σhkl√((n)/(n−1))Σj|Ihkl,j − 〈Ihkl〉|)/(ΣhklΣjhkl,j) (36).
b Rwork was calculated as Rwork = (Σhkl|Fhklobs−Fhklcalc|)/(ΣhklFhklobs).
c Rfree was calculated similarly to Rwork using 5% randomly selected reflections omitted from refinement.
FIGURE 2.
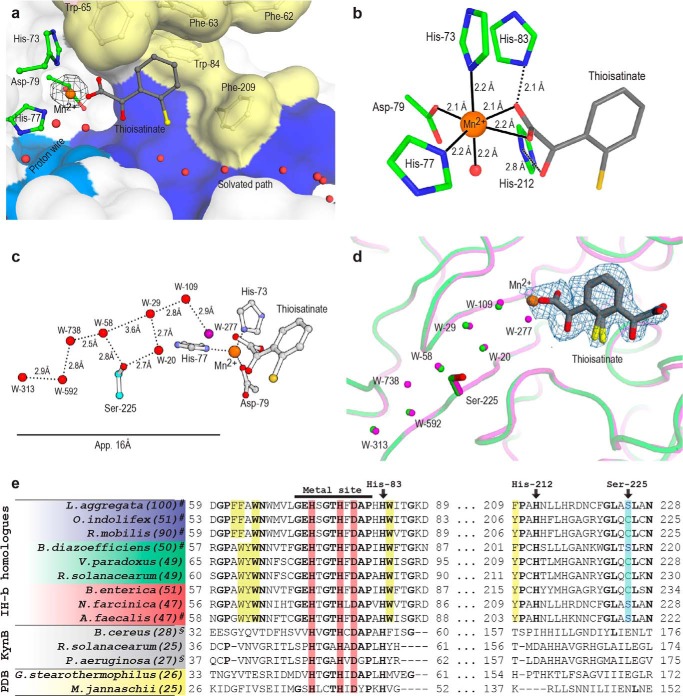
Crystal structure overview and domain swaps of isatin hydrolase. a, isatin hydrolase dimer. Each monomer is represented as surface in shades of gray. The exchanging domain swaps are represented as schematic (blue and red). The manganese atoms and coordinating residues are represented as ball-and-stick diagrams. b, the isatin entry cavity in green and the separate water channel in light blue, orthogonal to the isatin entry channel. c, conserved secondary structure elements between the IH-b and the related putative metal-dependent hydrolases 1R61 and 2B0A. The central β-barrel is conserved along with the β-sheet structure of the exchanging loops and helices α1 and α2. d, the active site and binding pocket in the thioisatinate-bound form. The product analogue is coordinating directly to the manganese ion. The aromatic ring is held in place through π-π interaction with the aromatic residues Trp-65 (red), Phe-63 (red), Trp-84 (white), and Phe-209 (white). The residues Trp-65 and Phe-63 are donated by the domain swap of the opposite monomer.
Thioisatinate Is a Product Analogue
The binding pocket with the product analogue thioisatinate reveals the position and orientation of isatinate post-hydrolysis. Thioisatinate was trapped in a primary and secondary conformation in the active site pocket. In the primary conformation, thioisatinate is held in place through nonspecific dispersion forces of the π-conjugated aromatic moiety of thioisatinate and the aromatic side chains of Trp-84 and Phe-209 of the core monomer, and Trp-65 and Phe-63 of the domain swap 2 (Fig. 2d). The low occupancy secondary conformation of thioisatinate was only visible in the difference density map (Fo − Fc) in three of the 12 monomers present in the asymmetric unit. The primary conformation shows that the thioisatinate carboxyl group coordinates directly to the Mn2+ ion (Figs. 2d and 3b), whereas in the secondary conformation, the carboxyl group is oriented toward the exit of the binding pocket. This indicates that isatinate may flip 180° and make contact with the solvated pathway in the substrate pocket before exiting.
FIGURE 3.
A transient proton wire in the product state of IH-b. a, primary conformation of thioisatinate, found in all 12 active sites in the asymmetric unit. Hydrophobic (yellow) and solvated (blue) regions are colored. Electron density found in the anomalous map verifying the presence of manganese over zinc is contoured at 8.0σ (black). b, manganese coordination in IH-b. The manganese is found in an octahedral arrangement (coordination number 6) with three bonds to the protein, two occupied bidentate by the thioisatinate carboxylate, and the last position occupied by water. The relative locations and proximities of His-83 and His-212 are shown. c, a ball-and stick model of the manganese binding site and the water channel with distances shown. d, alignment of waters in the proton wire channel. Water molecules in the two structures are found at almost exact same positions. W-277 and W-738 are only found in thioisatinate-IH-b. Fo − Fc electron density of thioisatinate in both primary and secondary conformations is contoured at 2.5σ (blue). e, alignment of IH-b putative orthologues found in marine bacteria (blue), plant associated symbionts and pathogens (green), human pathogens (red), and structurally related proteins (gray). Conserved (identity) residues among the IH sequence homologues are marked in boldface type. The functional conserved Ser-225 and the two non-metal coordinating histidine residues of the active site, His-83 and His-212, are marked along with the conserved hydrolase motif found in all sequences. Manganese coordinating histidine residues are highlighted in red. Residues forming a part of the hydrophobic binding pocket are highlighted in yellow. UniProt accession code/ORF names are as follows: Labrenzia aggregata (A0NLY7/SIAM614_09648), O. indolifex (A9EG38/OIHEL45_18466), Ruegeria mobilis (R0DM09/K529_21167), B. diazoefficiens (G7DNG8/BJ6T_62090), Variovorax paradoxus (C5CSQ1/Vapar_1498), R. solanacearum (Q8XYC3/RSc1835), B. enterica (GenBankTM accession no. ERF84741.1), Norcardia farcinica (Q5Z0F8/NFA_12380), A. faecalis (J0UVS6/QWA_04169), Bacillus cereus (Q81CK1/BC_2758), R. solanacearum (Q8Y1D0/RSc760), Pseudomonas aeruginosa (Q9I234/PA2081) Geobacillus stearothermophilus (P84132/-), Methanocaldococcus jannaschii (Q58193/MJ0783). #, indicates that isatin hydrolase activity has been verified. §, indicates verified kynurenine formamidase activity. The values in parentheses indicate sequence similarity to IH-b.
Thioisatinate Inhibits the Isatin Hydrolase
Thioisatinate was identified as an inhibitor with an IC50 of ∼10 μm (data not shown). The identification of a product analogue inhibitor enabled us to determine the crystal structure of the enzyme-thioisatinate complex. The crystals structure of thioisatinate-bound IH-b determined at 1.9 Å resolution allows detailed description of the product state. Thioisatinate shares strong structural similarities to isatinate with the amino group substituted by a thiol.
The IH-b Active Site
The crystallographic data were collected at 9.54 keV, above the x-ray absorption (k) edge for zinc, to allow clear distinction between Mn2+, Zn2+, and Ca2+. The manganese-binding site was formed by His-73, His-77, and Asp-79. A strong peak (17σ) in the anomalous difference density map at this position (Fig. 3a) confirmed the presence of manganese over calcium in the binding site, although calcium is present in large excess in the crystallization conditions. The calcium sites in the crystal contact regions have a reduced anomalous signal peak of 7σ. The anomalous signal also ruled out the presence of zinc with an expected anomalous signal comparable with that of sulfur. In comparison, the sulfur of Cys-122 has a weak anomalous signal of 4σ. The manganese is found in an octahedral arrangement with three bonds occupied by the above-mentioned residues. Thioisatinate is coordinated bidentate to the manganese in the crystal structure. In the absence of thioisatinate, two water molecules occupy these positions. In addition, a water molecule chelates manganese to complete the octahedron (Fig. 3b). His-83 and His-212 are found in close proximity to the manganese binding site, but they do not coordinate. The conformation of the manganese binding residues and the proximal histidine residues resembles that found in subtype III amidohydrolases (28). However, a structural comparison of the manganese-binding site of subtype III amidohydrolases, e.g. cytosine deaminase, to that of IH-b shows clearly different geometries. The position of the metal binding site in relation to the β-barrel also differs from the amidohydrolases.
Isatin Hydrolase Orthologues
The IH-b carries the amino acid sequence motif HXGTHXDXPXH characteristic of hydrolases of the Protein families database superfamily PF04199 and is a member of the Class Orthologues Group 1878. In addition to the direct metal-binding residues (shown in boldface type), His-83 is also conserved in the motif and may serve a role in proton donor/acceptor along with the conserved His-212 (Fig. 3e). A protein BLAST search with IH-b as query revealed >100 bacterial species that might encode one or two isatin hydrolases. Selected parts of the aligned sequences are shown. Three bacterial species L. aggregata (100), Ruegeria mobilis (90), and Oceanibulbus indolifex (51) represent a marine habitat. Some species are plant-associated, such as Bradyrhizobium diazoefficiens (formerly japonicum) (50), Ralstonia solanacearum (49), and Variovorax paradoxus (49), whereas others are found in humans, such as pathogenic bacteria isolated from the human gut microbiota Bradyrhizobium enterica (51), Nocardia farcinica (47), and Alcaligenes faecalis (47). The numbers in parentheses indicate % identity with the L. aggregata amino acid sequence.
The Substrate Channel
The substrate entry and product exit cavity is divided into a solvated/polar surface and a hydrophobic surface (Fig. 3a). Water molecules are shown lining the solvated surface. The hydrophobic surface is formed by Trp-84 and Phe-209 from the core of the monomer, whereas Phe-63, Phe-62, and Trp-65 are contributed through domain swap 2. The exact mechanism and molecular mode of isatin recognition are not revealed by the crystal structures. However, the polarized nature of the entry region could reflect the polarized structure of isatin with the carbonyls recognized by the solvated surface, whereas the aromatic ring is recognized by the hydrophobic surface.
Structural Evidence for a Proton Wire
A remarkable feature of IH-b is that two separate entry channels lead to the active site, the main substrate entry, and a narrow water-filled channel that forms a proton wire (Fig. 2b). The water molecules in the proton wire are found at regular intervals separated by a distance compatible with hydrogen bonding (Fig. 3, b and c). The dimensions of the water channel are too narrow to accommodate an isatin or isatinate molecule. The water coordinates in the channel are conserved between apo and thioisatinate-bound IH-b with two exceptions: W-277 is found only in the product state bridging the water chain directly to the thioisatinate carboxyl group by hydrogen bonding. W-738 is likewise only found in the product state but carries relatively low scattering power in the electron density map. An alignment of the water positions with manganese and thioisatinate placed for reference is shown in Fig. 3d. The water molecules found in the channel are coordinated by the backbone carbonyls of residues Pro-33, His-73, and Ile-195 and the amide group from Leu-35 and Gly-75. The only side chain to participate in direct water coordination is Ser-225. Ser-225 may act as a gating residue involved in coordinating the first water molecule with access to the channel. This residue is functionally conserved as either a serine or a cysteine residue in all isatin hydrolase orthologues (Fig. 3e). The proton wire is disrupted between W-58 and W-29 with a 3.6 Å gap, but it is bridged by the hydroxyl group of Ser-225.
Enzyme Kinetic Support for a Proton Wire
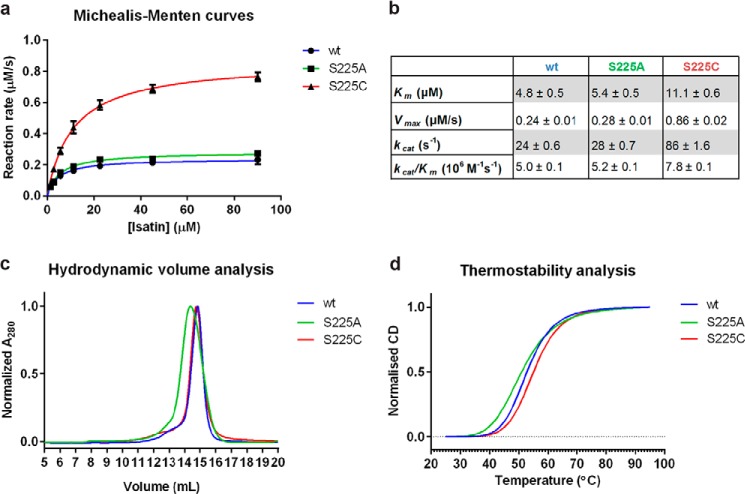
To investigate the role of Ser-225, IH-b mutants carrying the S225A or S225C mutation were generated. The S225C mutation was based on the observation that cysteine commonly appears at this position in the isatin hydrolase orthologues. A S225A mutant was produced to remove the water coordinating hydroxyl of the serine. Determination of the kinetic parameters for the wt and mutant enzymes shows that the S225C mutant is catalytically activated by the mutation, whereas no significant change is observed for the S225A mutation. The turnover number kcat was found to be 86 s−1 for S225C in comparison with 24 s−1 and 28 s−1 for the wt and S225A, respectively. The S225C mutation increased Km only 2-fold compared with the wt Km. No significant difference in Km or kcat/Km was observed between wt and S225A (Fig. 4, a and b). Based on this, S225C seemed activated by >50% as compared with wt. To assess the structural impact of the mutations, a comparative study of hydrodynamic volumes by size exclusion chromatography and thermostabilities by CD was performed. Special attention was given to determine the protein concentration in the experiments. The size exclusion chromatography measurements revealed no significant change in hydrodynamic volume between wt and S225C. A small decrease in retention volume of S225A could indicate a slightly increased hydrodynamic volume (Fig. 4c). A small decrease in melting temperature (Tm) was observed for the S225A compared with the wt by CD. No significant change in Tm was observed between wt and S225C (Fig. 4d). This indicated that the S225C mutation had minimal structural impact on the protein, whereas S225A was slightly destabilized. As the biophysical properties of the of the wt and S225C enzyme appeared unchanged, we assumed that the activating effect observed is due to an enhanced proton transfer rate or water shuttling, in the proton wire, gated by Ser-225.
FIGURE 4.
The S225C mutant is kinetically activated. a, Michealis-Menten curves for verification of kinetic model and determination of kinetic parameters Km and Vmax. IH-b-wt (R2 = 0.96), IH-b-S225A (R2 = 0.97), and IH-b-S225C (R2 = 0.99) display Michaelis-Menten kinetics. b, kinetic parameters for comparison of activity, including kcat and kcat/Km for wt and mutants. c, size-exclusion chromatography of wt and the S225C mutant revealed peaks of similar profile and retention volumes, indicating identical hydrodynamic volumes. The S225A mutation resulted in a slight decrease in retention volume corresponding to a small increase in hydrodynamic volume. d, averaged (n = 3) and normalized CD melting curves of IH-b-wt, IH-b-S225A, and IH-b-S225C. No significant change in melting temperature (Tm) was found between wt and S225C (p = 0.067), and the Tm was determined to 53 °C. A significant decrease in Tm of 2 °C for the S225A mutant compared with wt was found (p = 0.016).
DISCUSSION
The crystal structure of IH-b represents the first enzymatically characterized hydrolase with the swiveling β/α/β domain fold. The IH-b structure represents a new general hydrolytic fold for a dimerized metal-dependent amidohydrolases. The amidohydrolase superfamily comprises a large number of structurally related enzymes with common motifs in catalytic architecture and a large diversity in substrate specificities. The superfamily was first identified in 1997 through structural comparison of urease (EC 3.5.1.5), phosphotriesterase (EC 3.1.8.1), and adenosine deaminase (EC 3.5.4.4) (29). Amidohydrolase superfamily enzymes contain a mono- or binuclear metal-binding motif and a central (α/β)8 triose-phosphate isomerase barrel for stability of the fold (28). Apart from a central barrel-like structure found in each monomer of IH-b, there is no structural homology to the amidohydrolase superfamily. IH-b catalyzes the hydrolysis of a cyclic amide bond (lactam) and is thus classified as an enzyme of EC 3.5.2. The side chains of the conserved metal-binding motif locate to the surface of the β-barrel, unlike the common fold of amidohydrolase superfamily enzymes that often cluster the metal binding site at the end of the β-barrel structure (29).
A product of bacterial tryptophan degradation, kynurenine, has recently received attention due to its role in human physiology, as elevated levels of kynurenine have been found in human tumor cells (30). In bacteria, a three-step aerobic degradation pathway of tryptophan to anthranilate via kynurenine has been identified (31). The complete reaction is carried out by the bacterial enzymes tryptophan-2,3-dioxygenase, KynB, and kynureninase. The bacterial enzyme KynB was characterized as a metal-dependent amidohydrolase in contrast to its eukaryotic counterpart, which has an Asp-His-Ser catalytic triad (32), KynB catalyzes the hydrolysis of a linear amide and is therefore found in EC 3.5.1. By combining sequence alignment data with structural information, we are able expand the structural analysis to include KynB homologues and two close structural homologues deposited as PDB codes 1R61 and 2B0A. Both are uncharacterized in terms of enzymatic activity. Both structures share an overall similar fold to that of IH-b.
During revision of this paper, crystal structures of KynB from three bacterial species were deposited to the PDB: Bacillus anthracis (codes 4COA and 4CO9), Pseudomonas aeruginosa (code 4COB), and Burkholderia cenocepacia (code 4COG). These confirm the similarity of the overall fold.
The determinants for substrate specificity are largely found in the domain swap 2 of IH-b. This region contains aromatic residues important for the coordination of isatin in the binding pocket, and they are conserved as either FFAWN or WYWN in IH-b homologues. These motifs are not found in either KynB or in PDB codes 1R61 or 2B0A. No conserved motif is found between PDB codes 1R61 or 2B0A and KynB, leaving the substrate specificity of PDB codes 1R61 and 2B0A unpredictable. However, the analysis allows us to separate regions important for structural integrity and fold from regions of importance to substrate specificity. As an example, a Pro-Leu-Lys (PLK) motif found in the C-terminal region of IH-b is conserved among all proteins of this fold and locates to the central dimer interface, possibly for stabilization.
The metal binding motif is highly conserved among all aligned sequences, which may be a result of intense selective pressure on metal-binding capability. The metal-binding motif, including His-83, is conserved, whereas His-212 seems to be isatin hydrolase-specific (Fig. 3e). No metal ion is modeled in the corresponding site in PDB code 2B0A, whereas the metal in the PDB code 1R61 structure is modeled as zinc. Interestingly, KynB is modeled with a binuclear metal binding site and with no manganese present in any structure, but an in-depth structural comparison with IH-b is premature as the KynB structures remain unpublished. His-83 and His-212 are found in close proximity to the active site with His-212 being found at a distance compatible with hydrogen bonding to the thioisatinate carbonyl oxygen O-11. This orientates His-83 and His-212 in favorable positions for the acceptance or donation of a proton during the catalysis. The coordination of thioisatinate with manganese is likely equivalent to that expected of isatinate. We therefore hypothesize that the initial coordination of isatin to manganese would position the carbonyl (C-8=O-10) to make it susceptible to a nucleophilic attack from the activated water. The above observations indicate that manganese likely fulfills a dual role in the catalysis event with importance both in the activation of water prior to the nucleophilic attack and in the direct coordination of isatin upon substrate binding.
The functionally conserved cysteine/serine is found only in IH-b. Structural alignment of IH-b to PDB codes 2B0A (overall r.m.s.d. = 2.6 Å) and 1R61 (overall r.m.s.d. = 2.1 Å) identify a water channel-like structure as a conserved feature. This path of water molecules is modeled at the approximate position of the water channel in IH-b (Fig. 5b). There is no apparent sequence conservation of the proton wire water channel within the identified isatin hydrolase homologues; however, as the channel pathway is mainly lined by backbone carbonyls and amides, this may not be a surprise.
FIGURE 5.
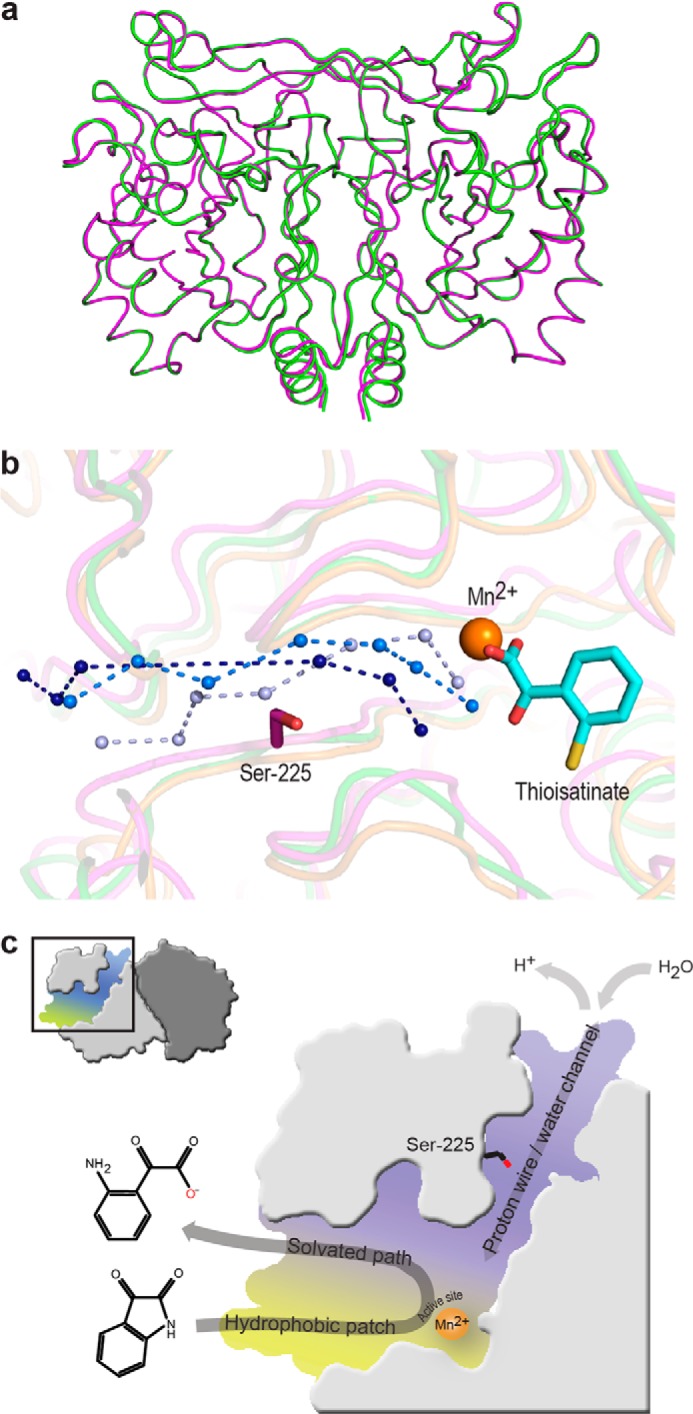
Alignment and overview of catalytically involved structural features of IH-b. a, structural superposition reveals no significant differences between the apo (green) and thioisatinate (magenta) bound IH-b (overall r.m.s.d., 0.17 Å). b, structural alignment of IH-b against 1R61 and 2B0A reveals conservation of the proton wire water channel. Ser-225, Mn2+, and thioisatinate are shown for structural orientation. The proton wire water channels are for IH-b (light blue) and PDB codes 1R61 (marine) and 2B0A (deep blue). c, schematic representation of structural features important for substrate recognition and catalytic function of IH-b.
The identification of thioisatinate as an inhibitor allowed us to capture the enzyme in the product-bound state and trap the product state of the catalytic cycle. The crystal structures of the apo-IH-b and thioisatinate bound IH-b are structurally equivalent, with an r.m.s.d. of 0.17 Å on all protein atoms (Fig. 5a). A significant difference is the additional water molecule (W-277) only present in IH-b with thioisatinate bound. This water molecule connects the active site with a proton wire through a novel water channel separate from the main entry site for isatin. The W-277 is missing in the apo structure and thus allows us to conclude that the complete proton wire must be a transient structure that is definitely present in the product state and possibly also in the substrate-binding state. The water channel is interrupted only by Ser-225, the only side chain residue that participates in hydrogen bonding to the solvent molecules.
Within the orthologous isatin hydrolase sequences, Ser-225 is only substituted with the structurally equivalent residue cysteine (Fig. 3e). To test the hypothesis that protons may be transferred through this wire, a mutant S225C was purified and tested for activity. The S225C mutant showed an increase in kcat/Km of >50%. Most of this increase was due to a 3-fold increase in kcat. This significant activation of the mutant IH-b-S225C emphasizes the possible importance of this residue to this subclass of hydrolases. In addition, it establishes the biochemical evidence that the water channel may indeed be a proton wire with functional relevance to substrate turnover and not just a structural feature important for stability. Proton-conducting water wires have been shown to be involved in protein stability as they form a structural element (33). The pKa value of the thiol group of the cysteine side chain has been determined to 8.6 (34), which is presumably highly dependent on the local environment. This is lower than would be expected for the hydroxyl group of serine, very likely with a pKa value above 12. The influence of the lowered pKa value of the thiol group could be what is reflected in the increased turnover, suggesting that Ser-225 has an active role in proton transport. The S225A mutant did not show a significant change in kinetic parameters as compared with wt. However, the biophysical studies of hydrodynamic volume suggest that the structure is destabilized by this mutation and may have an increased flexibility that results in a small increase of hydrodynamic volume; and thus, a water molecule could have replaced the missing hydroxyl group in S225A.
Isatin hydrolase activity was first found in the endophytic symbiont B. diazoefficiens. Based on structural insights and alignment data, we have identified putative isatin hydrolase orthologues in numerous bacteria; among those, the plant pathogen R. solanacearum, the marine O. indolifex, the human pathogens A. faecalis, and the recently discovered B. enterica. B. enterica has been isolated from the gastrointestinal tract and is associated with cord colitis syndrome (35). This leads to the question whether the indole-3-acetic acid degradation pathway is present in microbiota found in the gastrointestinal tract and what roles intermediates such as isatin formed in this pathway may serve. Isatin has been shown to induce a chemotaxic response. Another intriguing question is whether microbiota of the gastrointestinal tract, encoding isatin hydrolase activity, may benefit from endogenous isatin present in blood and tissue.
Acknowledgments
We thank Anette Kjems for help in the cloning and purification of wt and mutant forms of IH-b. Part of this work was carried out at beam line I911, MAX-lab synchrotron radiation source (Lund University, Lund, Sweden) with help from Thomas Ursby and Derek Logan. At the MX1 beam line at PETRA III storage ring located at EMBL (Hamburg, Germany), we are grateful for the assistance of Johanna Kallio. We are also grateful for fruitful discussion and critical proofreading by Drs. Christine Schar and Paul Tucker.
The atomic coordinates and structure factors (codes 4J0N and 4M8D) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- IH-b
- isatin hydrolase isoform b
- KynB
- bacterial kynurenine formamidase
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Jensen J. B., Egsgaard H., Van Onckelen H., Jochimsen B. U. (1995) Catabolism of indole-3-acetic acid and 4- and 5-chloroindole-3-acetic acid in Bradyrhizobium japonicum. J. Bacteriol. 177, 5762–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Olesen M. R., Jochimsen B. U. (1996) Identification of enzymes involved in indole-3-acetic acid degradation. Plant and Soil 186, 143–149 [Google Scholar]
- 3. Medvedev A. E., Glover V. (2004) Tribulin and endogenous MAO-inhibitory regulation in vivo. Neurotoxicology 25, 185–192 [DOI] [PubMed] [Google Scholar]
- 4. Mawatari K., Segawa M., Masatsuka R., Hanawa Y., Iinuma F., Watanabe M. (2001) Fluorimetric determination of isatin in human urine and serum by liquid chromatography postcolumn photoirradiation. Analyst. 126, 33–36 [DOI] [PubMed] [Google Scholar]
- 5. Watkins P., Clow A., Glover V., Halket J., Przyborowska A., Sandler M. (1990) Isatin, regional distribution in rat brain and tissues. Neurochem. Int. 17, 321–323 [DOI] [PubMed] [Google Scholar]
- 6. Binda C., Li M., Hubalek F., Restelli N., Edmondson D. E., Mattevi A. (2003) Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high-resolution crystal structures. Proc. Natl. Acad. Sci. U.S.A. 100, 9750–9755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Medvedev A., Igosheva N., Crumeyrolle-Arias M., Glover V. (2005) Isatin: role in stress and anxiety. Stress. 8, 175–183 [DOI] [PubMed] [Google Scholar]
- 8. Hamaue N., Yamazaki N., Terado M., Minami M., Ohno K., Ide H., Ogata A., Honma S., Tashiro K. (2000) Urinary isatin concentrations in patients with Parkinson's disease determined by a newly developed HPLC-UV method. Res. Commun. Mol. Pathol. Pharmacol. 108, 63–73 [PubMed] [Google Scholar]
- 9. Norman T. R., Burrows G. D., Mcintyre I. M. (1992) Stress and Isatin-Effects on the Serotonergic System. Stress Med. 8, 141–145 [Google Scholar]
- 10. Glover V., Bhattacharya S. K., Chakrabarti A., Sandler M. (1998) The psychopharmacology of isatin: A brief review. Stress Med. 14, 225–229 [Google Scholar]
- 11. Lee J., Bansal T., Jayaraman A., Bentley W. E., Wood T. K. (2007) Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl. Environ. Microbiol. 73, 4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Englert D. L., Manson M. D., Jayaraman A. (2009) Flow-based microfluidic device for quantifying bacterial chemotaxis in stable, competing gradients. Appl. Environ. Microbiol. 75, 4557–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berry A., Dodge T. C., Pepsin M., Weyler W. (2002) Application of metabolic engineering to improve both the production and use of biotech indigo. J. Ind. Microbiol. Biotechnol. 28, 127–133 [DOI] [PubMed] [Google Scholar]
- 14. Weyler W., Dodge T. C., Lauff J. L., Wendt D. J. (February 2, 1999) Microbial production of indigo, U. S. Patent 5,866,396
- 15. Christianson D. W., Cox J. D. (1999) Catalysis by metal-activated hydroxide in zinc and manganese metalloenzymes. Annu. Rev. Biochem. 68, 33–57 [DOI] [PubMed] [Google Scholar]
- 16. Chernyshev A., Cukierman S. (2006) Proton transfer in gramicidin water wires in phospholipid bilayers: attenuation by phosphoethanolamine. Biophys. J. 91, 580–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shinobu A., Agmon N. (2009) Mapping proton wires in proteins: carbonic anhydrase and GFP chromophore biosynthesis. J. Phys. Chem. A. 113, 7253–7266 [DOI] [PubMed] [Google Scholar]
- 18. Grotthuss C. J. (1806) Sur la décomposition de l'eau et des corps q'uelle tient en dissolution à l'aide de l'électricité galvanique. Ann. Chim. 58, 54–74 [Google Scholar]
- 19. Cukierman S. (2006) Et tu, Grotthuss! and other unfinished stories. Biochim. Biophys. Acta 1757, 876–885 [DOI] [PubMed] [Google Scholar]
- 20. Christensen J. H., Hansen P. K., Lillelund O., Thøgersen H. C. (1991) Sequence-specific binding of the N-terminal three-finger fragment of Xenopus transcription factor IIIA to the internal control region of a 5S RNA gene. FEBS Lett. 281, 181–184 [DOI] [PubMed] [Google Scholar]
- 21. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kabsch W. (2010) XDS. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 26. Chovancova E., Pavelka A., Benes P., Strnad O., Brezovsky J., Kozlikova B., Gora A., Sustr V., Klvana M., Medek P., Biedermannova L., Sochor J., Damborsky J. (2012) CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 8, e1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Herzberg O., Chen C. C., Kapadia G., McGuire M., Carroll L. J., Noh S. J., Dunaway-Mariano D. (1996) Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites. Proc. Natl. Acad. Sci. U.S.A. 93, 2652–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seibert C. M., Raushel F. M. (2005) Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 44, 6383–6391 [DOI] [PubMed] [Google Scholar]
- 29. Holm L., Sander C. (1997) An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins 28, 72–82 [PubMed] [Google Scholar]
- 30. Opitz C. A., Litzenburger U. M., Sahm F., Ott M., Tritschler I., Trump S., Schumacher T., Jestaedt L., Schrenk D., Weller M., Jugold M., Guillemin G. J., Miller C. L., Lutz C., Radlwimmer B., Lehmann I., von Deimling A., Wick W., Platten M. (2011) An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478, 197–203 [DOI] [PubMed] [Google Scholar]
- 31. Kurnasov O., Jablonski L., Polanuyer B., Dorrestein P., Begley T., Osterman A. (2003) Aerobic tryptophan degradation pathway in bacteria: novel kynurenine formamidase. FEMS Microbiol. Lett. 227, 219–227 [DOI] [PubMed] [Google Scholar]
- 32. Han Q., Robinson H., Li J. (2012) Biochemical identification and crystal structure of kynurenine formamidase from Drosophila melanogaster. Biochem. J. 446, 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thomaeus A., Naworyta A., Mowbray S. L., Widersten M. (2008) Removal of distal protein-water hydrogen bonds in a plant epoxide hydrolase increases catalytic turnover but decreases thermostability. Protein Sci. 17, 1275–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grimsley G. R., Scholtz J. M., Pace C. N. (2009) A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci. 18, 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bhatt A. S., Freeman S. S., Herrera A. F., Pedamallu C. S., Gevers D., Duke F., Jung J., Michaud M., Walker B. J., Young S., Earl A. M., Kostic A. D., Ojesina A. I., Hasserjian R., Ballen K. K., Chen Y. B., Hobbs G., Antin J. H., Soiffer R. J., Baden L. R., Garrett W. S., Hornick J. L., Marty F. M., Meyerson M. (2013) Sequence-based discovery of Bradyrhizobium enterica in cord colitis syndrome. N. Engl. J. Med. 369, 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Diederichs K., Karplus P. A. (1997) Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4, 269–275 [DOI] [PubMed] [Google Scholar]