Background: How oncogenic signals attenuate mitochondrial function and promote the switch to glycolysis remains unclear.
Results: Tyr-94 phosphorylation inhibits mitochondrial PDP1 and is important for the glycolytic switch and tumor growth.
Conclusion: Phosphorylation at different tyrosine residues inhibits PDP1 through independent mechanisms to promote the Warburg effect.
Significance: These data provide novel insights into the molecular mechanisms underlying the Warburg effect.
Keywords: Cell Proliferation, Phosphoprotein Phosphatase, Phosphotyrosine Signaling, Tumor Metabolism, Warburg Effect
Abstract
Many cancer cells rely more on aerobic glycolysis (the Warburg effect) than mitochondrial oxidative phosphorylation and catabolize glucose at a high rate. Such a metabolic switch is suggested to be due in part to functional attenuation of mitochondria in cancer cells. However, how oncogenic signals attenuate mitochondrial function and promote the switch to glycolysis remains unclear. We previously reported that tyrosine phosphorylation activates and inhibits mitochondrial pyruvate dehydrogenase kinase (PDK) and phosphatase (PDP), respectively, leading to enhanced inhibitory serine phosphorylation of pyruvate dehydrogenase (PDH) and consequently inhibition of pyruvate dehydrogenase complex (PDC) in cancer cells. In particular, Tyr-381 phosphorylation of PDP1 dissociates deacetylase SIRT3 and recruits acetyltransferase ACAT1 to PDC, resulting in increased inhibitory lysine acetylation of PDHA1 and PDP1. Here we report that phosphorylation at another tyrosine residue, Tyr-94, inhibits PDP1 by reducing the binding ability of PDP1 to lipoic acid, which is covalently attached to the L2 domain of dihydrolipoyl acetyltransferase (E2) to recruit PDP1 to PDC. We found that multiple oncogenic tyrosine kinases directly phosphorylated PDP1 at Tyr-94, and Tyr-94 phosphorylation of PDP1 was common in diverse human cancer cells and primary leukemia cells from patients. Moreover, expression of a phosphorylation-deficient PDP1 Y94F mutant in cancer cells resulted in increased oxidative phosphorylation, decreased cell proliferation under hypoxia, and reduced tumor growth in mice. Together, our findings suggest that phosphorylation at different tyrosine residues inhibits PDP1 through independent mechanisms, which act in concert to regulate PDC activity and promote the Warburg effect.
Introduction
Although aerobic glycolysis appears to be a key metabolic factor in human cancers (1) and leukemia (2, 3), the detailed mechanisms by which cancer/leukemia cells switch to this metabolic fate and how crucial this is for tumorigenesis/leukemogenesis and disease development in vivo remain unknown. It is believed that oncogenes including Myc and HIF1 up-regulate gene expression levels of glycolytic enzymes to promote glycolysis in cancer/leukemia cells. On the other hand, the metabolic switch for cancer cells to rely less on oxidative phosphorylation and more on glycolysis is also suggested to be, in part, due to functional attenuation of mitochondria in cancer cells (4). However, how oncogenic signals attenuate mitochondrial function and promote the switch to glycolysis to provide a metabolic advantage to cancer development remains unclear.
In mammalian cells, pyruvate dehydrogenase complex (PDC)4 is responsible for conversion of pyruvate to acetyl-CoA (pyruvate decarboxylation). PDC is a complex of predominantly three enzymes including pyruvate dehydrogenase (PDH), the most important enzyme component of PDC that transforms pyruvate into acetyl-CoA, and its upstream pyruvate dehydrogenase kinase (PDK) and phosphatase (PDP). PDC is organized around a 60-meric dodecahedral core formed by acetyltransferase (E2p) and E3-binding protein (E3BP) (5), which binds PDH (aka E1p), PDK, PDP, and dihydrolipoamide dehydrogenase (E3) (6). PDK1 inhibits PDH and consequently PDC by phosphorylating PDH at several serine residues including Ser-293, Ser-300, and Ser-232, whereas dephosphorylation of PDH by PDP restores its enzyme activity as well as PDC activity (7).
The Warburg effect describes a unique metabolic phenomenon of cancer cells where cancer cells uptake glucose at a high rate but prefer glycolysis by converting pyruvate to lactate regardless of the presence of oxygen. This may be in part due to up-regulation of PDK activity and inhibition of PDH/PDC in cancer cells. PDK1 is believed to be up-regulated by Myc and HIF1 to achieve functional inhibition of mitochondria by phosphorylating and inactivating PDH in cancer cells (8–10). However, how oncogenic signals inhibit PDC to regulate cancer cell metabolism is not quite clear. We recently reported that oncogenic tyrosine kinases promote the Warburg effects in cancer and leukemia cells by attenuating mitochondria function via phosphorylation and activation of PDK1 (11). In addition, we found that acetylation at Lys-321 and Lys-202 inhibits PDHA1 and PDP1, respectively (12). Moreover, lysine acetylation of PDHA1 and PDP1 is common in EGF-stimulated cells and diverse human cancer cells, which is regulated by Tyr-381 phosphorylation of PDP1 that simultaneously dissociates deacetylase SIRT3 and recruits acetyltransferase ACAT1 to PDC (12). Here we report that phosphorylation of PDP1 at an additional tyrosine residue Tyr-94 is also common in human cancer cells, which promotes the Warburg effect by inhibiting PDP1 through a distinct and independent molecular mechanism.
EXPERIMENTAL PROCEDURES
Reagents
PDP1 cDNA image clone (Open Biosystems) was used to engineer several PDP1 variants with a FLAG epitope tag and were subsequently subcloned into pDEST27 and pET60 vectors (Invitrogen) for GST-tagged PDP1 expression and purification in mammalian cells and bacteria, respectively. Point mutations were introduced using QuikChange-XL site-directed mutagenesis kit (Stratagene). [5-3H]glucose and [1-14C]pyruvate were purchased from PerkinElmer, and R-[3H]lipoic acid was purchased from American Radiolabeled Chemicals. Stable knockdown of endogenous PDP1 was achieved using a lentiviral vector harboring an shRNA construct (Open Biosystems; 5′-GTTCAGTTCAATTCTCATGTT-3′). PDP1 rescue H1299 cell lines were generated as described previously (12). Briefly, retroviral vector pLHCX (Clontech) containing shRNA-resistant, FLAG-tagged human PDP1 wild type (WT) or mutant Y94F forms harboring silent mutations in the shRNA targeted region were transfected into H1299 cells containing shRNA directed against endogenous PDP1. Antibody against PDP1 was purchased from Abgent. Phospho-Tyr antibody phospho-Tyr-99 was purchased from Santa Cruz Biotechnology. PDC E2 antibody was purchased from Invitrogen. Anti-FLAG, actin, and GST antibodies were purchased from Sigma. Specific antibody against phospho-PDP1 (phospho-Tyr-94) was generated by Cell Signaling Technology specifically for the current project and is not currently commercially available.
Cell Culture
H1299, A549, HEL, KG-1a, MO91, EOL1, Molm14, and K562 cells were cultured in RPMI 1640 medium with 10% bovine serum (FBS). 293T, MDA-MB231, MCF7, human foreskin fibroblast, and HaCaT cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS. TKI258 (generously provided by Novartis Pharma) treatment was performed by incubating cells with 1 μm TKI258 for 4 h. For the cell proliferation assay, 125 nm oligomycin was added in the cell culture medium as described previously (13). Imatinib was purchased from Santa Cruz Biotechnology.
Purification of PDP1 Proteins
GST-FLAG-PDP1 protein was purified by sonication of high expressing BL21(DE3)pLysS cells obtained from a 250-ml culture subjected to isopropyl-1-thio-β-d-galactopyranoside induction for 16 h at 30 °C. Cell lysates were harvested after centrifugation and then incubated with glutathione-Sepharose beads (GE Healthcare) for 2 h at 4 °C. After washing twice, the final purity of the PDP1 protein was examined by Coomassie Brilliant Blue staining and Western blot using anti-PDP1 (Abgent) antibodies.
PDC Activity Assay
The PDC activity was measured by conversion rate of [1-14C]pyruvate (PerkinElmer) to 14CO2 as described (12, 14). In brief, 14CO2 production through PDC was measured using isolated mitochondria (1 mg) in radioactive mitochondria resuspension buffer (1 ml) containing [1-14C]pyruvate (0.1 μCi/ml), 200 mm sucrose, 10 mm HEPES-HCl (pH 7.4), 1 mm pyruvate, 1 mm malate, 2 mm sodium monophosphate, 1 mm EGTA. The incubation mixture was placed at the bottom of a vial with a rubber stopper and maintained in agitation. The 14CO2 produced during incubation was trapped by hyamine hydroxide placed in an Eppendorf tube in the vial. The reaction was terminated by adding 0.5 ml of 50% TCA to the reaction after 1 h. Twenty minutes after the TCA injection, all the samples in hyamine hydroxide were transferred to mini-vials together with 5 ml of scintillation fluid, and radioactivity was assayed on a scintillation counter. The results were normalized based on mitochondrial protein levels assayed by Bradford assay using BSA as a standard.
In Vitro Kinase Assay
For FGFR1 kinase assay, 200 ng of purified recombinant PDP1 WT, Y381F, or Y94F proteins were incubated with 250 ng of recombinant active FGFR1 in FGFR1 kinase buffer (10 mm Hepes (pH 7.5), 150 mm NaCl, 10 mm MnCl2, 0.01% Triton X-100, 5 mm DTT, 200 μm ATP) for 1 h at 30 °C. For ABL kinase assay, purified recombinant PDP1 proteins were incubated with 175 ng of recombinant active ABL in ABL kinase buffer (50 mm Tris (pH 7.5), 10 mm MgCl2, 0.01% Nonidet P-40, 1 mm DTT, 200 μm ATP) for 1 h at 30 °C. For JAK2 kinase assay, purified recombinant PDP1 proteins were incubated with 200 ng of recombinant active JAK2 in JAK2 kinase buffer (25 mm Hepes (pH 7.5), 10 mm MgCl2, 0.5 mm EGTA, 0.01% Triton X-100, 2.5 mm DTT, 0.5 mm sodium orthovanadate, 5 mm glycerophosphate, 200 μm ATP) for 1 h at 30 °C. The samples were electrophoresed on 10% SDS-polyacrylamide gel, transferred on a nitrocellulose membrane, and then detected with the phospho-PDP1 (phospho-Tyr-94) and anti-phospho-tyrosine (phospho-Tyr-99) antibodies.
Lipoic Acid Binding Assay
Purified recombinant GST-FLAG-PDP1 proteins including WT, Y94F, and control Y381F that were immobilized on anti-FLAG beads were treated with or without recombinant FGFR1 (rFGFR1) in an in vitro kinase assay as described above. The beads were incubated in the presence or absence of 100 μm CaCl2 (15) at room temperature for 5 min followed by incubation with 10 μm R-[3H]lipoic acid (American Radiolabeled Chemicals) for 30 min at room temperature. The beads were then washed twice with TBS to remove the unbound R-[3H]lipoic acid. The PDP1 proteins were eluted, and the retained R-[3H]lipoic acid on PDP1 was measured using a scintillation counter.
PDC E2 Binding Assay
GST-tagged PDP1 WT, Y94F, and Y381F proteins were incubated with active rFGFR1 in the in vitro FGFR1 kinase assay (described above) followed by incubation with whole cell lysates from 293T cells. GST-FLAG-PDP1 proteins were pulled down with beads and resolved by Western blot. Binding of PDP1 with PDC E2 protein was assessed by comparison with the amount of E2 bound to GST-FLAG-PDP1 in GST pulldown samples.
PDP1 Assay
The PDP1 activity was determined by the rate of the dephosphorylation reaction catalyzed by PDP1 using purified PDC complex (Sigma) as a substrate. In brief, recombinant PDP1 was incubated with active rFGFR1 in the in vitro FGFR1 kinase assay (described above) followed by a 1-h incubation with purified PDC complex in PDP1 buffer containing 10 mm Hepes, pH 7.5, 100 μm EDTA, 100 μm EGTA, 0.5% BSA, and 1 mm DTT (Sigma) at room temperature. The dephosphorylation of Ser-293 of PDHA in the PDC complex was assessed by Western blot using specific antibody recognizing phospho-PDHA (phospho-Ser-293).
Lactate Production, Oxygen Consumption, and Intracellular ATP Assays
Cellular lactate production under normoxia was measured using a fluorescence-based lactate assay kit (MBL International). Phenol red-free RPMI medium without FBS was added to a 6-well plate of subconfluent cells and was incubated for 1 h at 37 °C. After incubation, 1 μl of medium from each well was assessed using the lactate assay kit. Cell numbers were determined by cell counting using a microscope (×40). Oxygen consumption rates were measured with a Clark type electrode equipped with a 782 oxygen meter (Strathkelvin Instruments). 1 × 107 cells were resuspended in RPMI 1640 medium with 10% FBS and placed into a water-jacketed chamber RC300 (Strathkelvin Instruments), and recording was started immediately. Intracellular ATP concentration was measured by an ATP bioluminescent somatic cell assay kit (Sigma). Briefly, 1 × 106 cells were trypsinized and resuspended in ultrapure water. Luminescence was measured with a spectrofluorometer (SpectraMax Gemini; Molecular Devices) immediately after the addition of ATP enzyme mix to cell suspension.
Cell Proliferation Assays
Cell proliferation assays were performed by seeding 5 × 104 cells in a 6-well plate and culturing the cells at 37 °C in normoxia (5% CO2 and 95% air). Twenty-four hours after seeding, cells that were used for further culture under hypoxia were cultured at 37 °C in a sealed hypoxia chamber filled with 1% O2, 5% CO2, and 94% N2. Cell proliferation was determined by cell numbers recorded by a TC10 automated cell counter (Bio-Rad) at the indicated days.
Xenograft Studies
Approval of use of mice and designed experiments was given by the Institutional Animal Care and Use Committee of Emory University. Nude mice (nu/nu, male 4–6 weeks old, Harlan Laboratories) were subcutaneously injected with 20 × 106 rescue H1299 cells stably expressing human PDP1 (hPDP1) WT and hPDP1 Y94F with stable knockdown of endogenous hPDP1 on the left and right flanks, respectively. Tumor growth was recorded by measurement of two perpendicular diameters using the formula 4π/3 × (width/2)2 × (length/2). The tumors were harvested and weighed at the experimental end point, and the tumor masses were compared between tumors (g) derived from rescue cells expressing hPDP1 WT or hPDP1 Y94F with stable knockdown of endogenous hPDP1. Statistical analyses were performed using a two-tailed paired Student's t test.
RESULTS
FGFR1 Inhibits PDP1 via Phosphorylation at Tyr-94
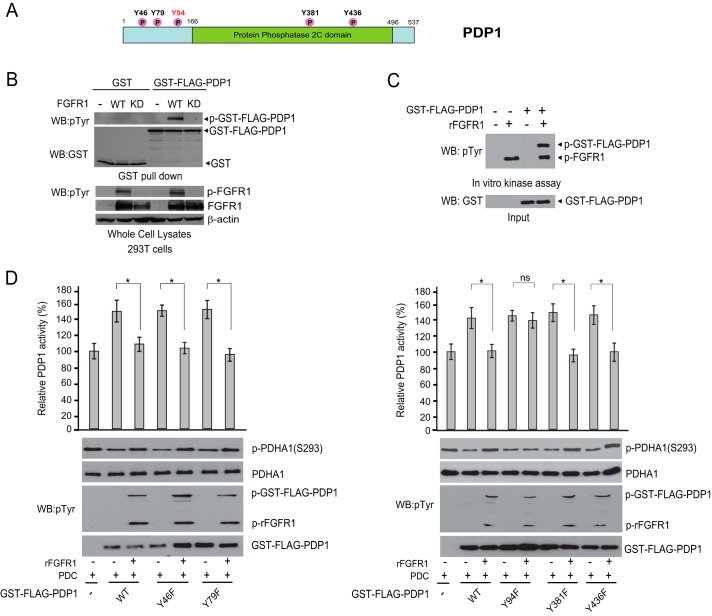
We recently reported that tyrosine phosphorylation activates the upstream kinase of PDH, PDK1, to promote the Warburg effect in cancer cells and tumor growth (11). Our phospho-proteomics studies (11, 12) and multiple proteomics-based studies performed by our collaborators at Cell Signaling Technology revealed that the upstream phosphatase of PDH, PDP1, is phosphorylated at a group of tyrosine residues in human cancer cells (Fig. 1A). Consistently, we found that coexpression of FGFR1 wild type (WT) but not a kinase dead form resulted in tyrosine phosphorylation of GST-tagged PDP1 (Fig. 1B). Moreover, treatment with active rFGFR1 resulted in tyrosine phosphorylation of purified GST-tagged PDP1 in an in vitro kinase assay (Fig. 1C). To examine the effect of tyrosine phosphorylation on PDP1, we next performed mutational analysis and generated diverse phospho-deficient Tyr → Phe mutants of PDP1 to replace each of the tyrosine residues that were identified as phosphorylated. We found that treatment with FGFR1 significantly reduced PDP1 phosphatase activity, whereas only substitution of PDP1 Tyr-94 but not Tyr-46, Tyr-79, Tyr-381, or Tyr-436 abolished FGFR1-dependent inhibition of PDP1 (Fig. 1D). These results together suggest that FGFR1 inhibits PDP1 by phosphorylating Tyr-94.
FIGURE 1.
FGFR1 inhibits PDP1 via phosphorylation at Tyr-94. A, schematic representation of PDP1. Five identified FGFR1-direct tyrosine phosphorylation sites are shown. B, Western blot (WB) detecting tyrosine phosphorylation (p) of purified GST-FLAG-PDP1 from cells co-expressing FGFR1 wild type (WT) or an FGFR1 kinase dead form (KD) using a pan phospho-Tyr (pTyr) antibody. C, active rFGFR1 (p-FGRF1) directly phosphorylates purified GST-FLAG-PDP1 at tyrosine residues in an in vitro kinase assay. D, purified FLAG-tagged PDP1 variants were incubated with rFGFR1 followed by PDP1 phosphatase assay in the presence of purified PDC. PDP1 phosphatase activity was assessed by the altered phospho-Ser-293 levels of PDHA1 (p-PDHA1(S293)). ns, not significant. Error bars indicate mean ± S.D. *, p < 0.05.
Tyr-94 Phosphorylation Inhibits PDP1 by Blocking Lipoic Acid Binding
Tyr-94 is evolutionarily conserved (Table 1). Structural analysis revealed that Tyr-94 is ∼20 Å distal from the catalytic cleft, suggesting that Tyr-94 phosphorylation may not directly affect the catalytic activity of PDP1 (Fig. 2A). However, Tyr-94 is more proximal to the structural features that enable lipoic acid binding to PDP1 (Fig. 2A). Phosphatase activity of PDP1 is dependent on binding to the L2 domain of dihydrolipoyl acetyltransferase (E2) of PDC. Such binding is mediated by the lipoic acid covalently attached to the L2 domain of E2, which is calcium-dependent. In the absence of calcium, PDP1 cannot form a stable complex with E2 and has a significant reduction in catalytic activity (16). Thus, we hypothesized that phosphorylation of Tyr-94 may potentially impact PDP1 activity by reducing the ability of PDP1 to bind lipoic acid and consequently E2. Indeed, we found that incubation of purified recombinant rPDP1 with purified rFGFR1 resulted in decreased 3H-labeled lipoic acid binding to rPDP1 WT and control Y381F mutant, but not Y94F mutant (Fig. 2B). Consistent with this finding, incubation with rFGFR1 resulted in decreased PDC E2 binding to GST-FLAG-PDP1 and control Y381F mutant, whereas substitution of Tyr-94 abolished FGFR1-dependent attenuation of PDP1-E2 association (Fig. 2C).
TABLE 1.
Phylogenetic analysis of PDP1 in different species
| Species | PDP1 sequence |
|---|---|
| Homo sapiens (human) | SILKANEYSFKVPEF |
| Pan troglodytes (chimpanzee) | SILKANEYSFKVPEF |
| Macaca mulatta (Rhesus monkey) | SILKANEYSFKVPEF |
| Canis lupus familiaris (dog) | SILKANEYSFKVPEF |
| Bos taurus (cattle) | SILKANEYSFKVPEF |
| Mus musculus (house mouse) | SILKANEYSFKVPEF |
| Rattus norvegicus (Norway rat) | SILKANEYSFKVPEF |
| Gallus gallus (chicken) | SILKANEYSFKVPEF |
| Danio rerio (zebrafish) | SILKANEYNFKVPEF |
| Xenopus (Silurana) tropicalis (western clawed frog) | SILKANEYSFKVPEF |
a Bold sequence indicates Tyr-94.
FIGURE 2.
Tyr-94 phosphorylation inhibits PDP1 by blocking lipoic acid binding. A, graphic representation of PDP1 structure (PDB ID: 2PNQ (16)). Tyr-94 is proximal to the structural features that enable lipoic acid binding to PDP1 as indicated. B, purified GST-FLAG-PDP1 variants were incubated with rFGFR1 followed by incubation with 3H-labeled lipoic acid. PDP1-bound [3H]lipoic acid was assessed by scintillation counting. C, GST-FLAG-PDP1 variants were incubated with rFGFR1 followed by incubation with H1299 whole cell lysates and GST pulldown assay. Right panel shows quantitation results of relative E2 binding ability based on the densities of signals in Western blots (WB) of bound E2 to GST pulldown, normalized to the density of signals of input E2 (left). ns, not significant. p, phosphorylation. Error bars indicate mean ± S.D. *, p < 0.05.
Tyr-94 Phosphorylation of PDP1 Is Common in Human Cancer Cells
We next generated a specific phospho-PDP1 antibody that recognizes Tyr-94-phosphorylated PDP1. Using this antibody, we found that EGF treatment resulted in increased phosphorylation levels of PDP1 at Tyr-94 as well as previously reported Tyr-381 (12) in 3T3 cells (Fig. 3A). In addition, we found that purified rFGFR1, rABL, and rJAK2 (Fig. 3, B–D, left panels, respectively) directly phosphorylated purified rPDP1 WT and control Y381F mutant at Tyr-94 in vitro, whereas substitution at Tyr-94 of PDP1 abolished such phosphorylation. Consistently, inhibition of FGFR1 by TKI258, BCR-ABL by imatinib, and JAK2 by AG490 resulted in decreased Tyr-94 phosphorylation of PDP1 in the pertinent human leukemia cell lines (Fig. 3, B–D, right panels, respectively).
FIGURE 3.
Oncogenic tyrosine kinase including FGFR1, ABL, and JAK2 directly phosphorylate PDP1 at Tyr-94. A, 3T3 cells treated with EGF for increasing time were examined for phospho-PDP1 (p-PDP1) leve1 using immunoblotting. Y94, Tyr-94; Y381, Tyr-381. B–D, left panels: purified GST-FLAG-PDP1 WT, Y94F, or Y381F was incubated with rFGFR1 (B), rABL (C), or rJAK2 (D) followed by immunoblotting (WB) to detect Tyr-94 phosphorylation (p) of PDP1. Right panels: immunoblotting shows phosphorylation levels of PDP1 Tyr-94 in FGFR1-expressing human lung cancer H1299 cells treated with FGFR1 inhibitor TKI258 (B), BCR-ABL-expressing leukemia K562 cells treated with ABL inhibitor imatinib (C), or JAK2 V617F-expressing leukemia HEL cells treated with JAK2 inhibitor AG490 (D).
In addition, we found that Tyr-94 phosphorylation of PDP1 is common in diverse human tumor cells including MDA-MB-231 breast cancer cells, H1299 and A549 lung cancer cells, as well as a group of leukemia cells associated with distinct leukemogenic tyrosine kinases including EOL1 (HIP1L1-PDGFRA), HEL (JAK2 V617F), K562 (BCR-ABL), KG-1a (FOP2-FGFR1), Molm14 (FLT3-ITD), and Mo91 (TEL-TrkC) cells (Fig. 4A), but not in normal proliferating human foreskin fibroblast and HaCaT keratinocyte cells and MCF7 breast cancer cells (Fig. 4A). Furthermore, we observed that Tyr-94 phosphorylation levels of PDP1 were increased in primary leukemia cells from three acute myeloid leukemia and one B-cell acute lymphoid leukemia patients as compared with peripheral blood cells from two healthy donors (Fig. 4B).
FIGURE 4.

Tyr-94 phosphorylation of PDP1 is common in human cancer cells. A and B, immunoblotting to detect phosphorylation levels of PDP1 Tyr-94 (p-PDP1 (Y94)) in diverse human tumor and leukemia cells (A) as well as human primary leukemia cells isolated from peripheral blood (PB) or bone marrow (BM) samples from representative patients (B). Normal proliferating human foreskin fibroblasts (HFF), HaCaT keratinocyte cells, and peripheral blood cells from healthy human donors were included as controls. AML, acute myeloid leukemia; ALL, acute lymphoid leukemia.
Expression of PDP1 Y94F Mutant in H1299 Cells Leads to Decreased Proliferation under Hypoxia and Increased Oxidative Phosphorylation
We next tested whether Tyr-94 phosphorylation-dependent inhibition of PDP1 is important for glycolysis and cancer cell proliferation. We generated rescue H1299 cells with stable knockdown of endogenous PDP1 followed by rescue expression of shRNA-resistant FLAG-PDP1 WT and Y94F that harbor silent mutations in the target regions of shRNA (Fig. 5A). As shown in Fig. 5B, knockdown or rescue expression of PDP1 WT did not affect H1299 cell proliferation under normoxic or hypoxic conditions. In contrast, rescue expression of phospho-deficient PDP1 Y94F significantly attenuated cell proliferation under hypoxia but not normoxia, whereas neither normoxia nor hypoxia affected Tyr-94 phosphorylation of PDP1 in H1299 cells (Fig. 5B, right). Consistent with these findings, PDP1 Y94F-expressing cells demonstrated increased PDC flux rate (Fig. 5C, left) that is consistent with decreased Ser-293 phosphorylation of PDHA1 as compared with the control PDP1 WT rescue cells (Fig. 5C; right). Consistently, PDP1 Y94F-expressing cells also showed decreased lactate production under normoxia as compared with WT cells (Fig. 5D). In addition, PDP1 Y94F cells were more sensitive to treatment with ATP synthase inhibitor oligomycin in terms of inhibition of ATP production (Fig. 5E, left) and oxygen consumption rate (Fig. 5F) as compared with control PDP1 WT rescue cells, whereas oligomycin treatment did not affect Tyr-94 phosphorylation of PDP1 in H1299 cells (Fig. 5E, right). These data together suggest that abolishment of Tyr-94 phosphorylation PDP1 resulted in a metabolic change to allow cells rely more on oxidative phosphorylation with decreased glycolysis, providing a metabolic disadvantage to cell proliferation under hypoxia.
FIGURE 5.
Expression of PDP1 Y94F mutant in H1299 cells leads to decreased proliferation under hypoxia and increased oxidative phosphorylation. A, generation of H1299 cells with stable knockdown of endogenous hPDP1 (endo-PDP1) followed by stable rescue expression of FLAG-tagged hPDP1 variants, which harbor silent mutations that confer PDP1 shRNA resistance. WB, Western blot. B, distinct PDP1 rescue H1299 cells as well as parental H1299 and control knockdown (KD; empty vector rescue) cells were tested for cell proliferation rate under normoxia (left; 17% oxygen) or hypoxia (middle; 1% oxygen). Cell proliferation was determined based on cell numbers counted daily. Right: phosphorylation level of PDP1 Tyr-94 (PDP1 p-Y94) was examined in H1299 cells under normoxic or hypoxic conditions using immunoblotting. C, PDP1 WT or Y94F rescue H1299 cells were tested for PDC flux rate (left) and Ser-293 phosphorylation levels of PDHA1 (p-PDHA1 (S293)) in immunoblotting (middle) under normoxia. D, PDP1 rescue cells were tested for lactate production under normoxia. E and F, PDP1 rescue cells were tested for intracellular ATP level (E) and oxygen consumption (F) in the presence and absence of ATP synthase inhibitor oligomycin under normoxia. E, right, phosphorylation level of PDP1 Tyr-94 was examined in H1299 cells in the presence or absence of oligomycin. G, PDP1 rescue cells were tested for PDC flux rate (left), lactate production (middle), and intracellular ATP level (right) under normoxic or hypoxic conditions. Error bars indicate mean ± S.D. *, p < 0.05, **, p < 0.01.
Moreover, we found that, although hypoxic condition did not affect the increased PDC activity (Fig. 5G, left) nor the decreased lactate production (Fig. 5G, middle) in PDP1 Y94F-expressing cells, such a low oxygen condition resulted in decreased ATP levels in these cells (Fig. 5G, right) as compared with control PDP1 WT rescue cells. This is consistent with our hypothesis that cells expressing PDP1 Y94F mutant rely more on oxidative phosphorylation for ATP production as compared with control WT cells, so under hypoxic condition where oxygen is insufficient to sustain oxidative phosphorylation level, PDP1Y94F cells showed decreased ATP levels and subsequently reduced cell proliferation.
Tyr-94 Phosphorylation of PDP1 Is Important for Tumor Growth
We next performed xenograft experiments and found that the growth rate (Fig. 6A) and masses of tumors (Fig. 6B) derived from PDP1 Y94F rescue H1299 cells were significantly reduced with decreased Tyr-94 phosphorylation of PDP1 (Fig. 6C) in tumor cells as compared with those of tumors formed by the control PDP1 WT rescue cells. Together, these data demonstrate an important role for Tyr-94 phosphorylation of PDP1 in tumor growth.
FIGURE 6.
Tyr-94 phosphorylation of PDP1 is important for tumor growth. A and B, tumor growth (A) and (B) masses in xenograft nude mice injected with PDP1 Y94F rescue cells as compared with mice injected with control PDP1 WT rescue cells are shown. p values were determined by a two-tailed paired Student's t test. C, top panels show dissected tumors in two representative nude mice injected with PDP1 WT and Y94F rescue cells on the left and right flanks, respectively. Bottom panels show detection of Tyr-94 phosphorylation levels of PDP1 in tumor lysates using specific phospho-PDP1 (Tyr-94) (p-PDP1 (Y94)) antibody. Error bars indicate mean ± S.D.
DISCUSSION
Our findings provide new insight into the molecular mechanisms by which oncogenic signals attenuate mitochondrial function by inhibiting PDP1 through tyrosine phosphorylation and consequently PDC activity. Reduced PDC activity thus in part promotes cancer cells to rely more on glycolysis instead of mitochondrial oxidative phosphorylation, providing a metabolic advantage to cancer cell proliferation and tumor growth. We previously reported that, in cancer cells where tyrosine kinase signaling is commonly up-regulated, Tyr-381 phosphorylation of PDP1 results in simultaneous dissociation of deacetylase SIRT3 and recruitment of acetyltransferase ACAT1 to PDC (12). ACAT1 acetylates PDHA1 at Lys-321 and PDP1 at Lys-202, which results in recruitment of active PDK1 to PDHA1 and dissociation of PDP1 from PDHA1, respectively, leading to increased inhibitory Ser-293 phosphorylation of PDHA1 and consequently inhibition of PDC. Our current results suggest that oncogenic signals also inhibit PDP1 through phosphorylation at an additional tyrosine site, Tyr-94, which results in decreased binding ability of PDP1 to lipoid acid that is required for PDP1 phosphatase activity via recruitment of PDP1 to PDC through E2 association. Thus, this finding represents a distinct and independent molecular mechanism underlying tyrosine phosphorylation-dependent inhibition of PDP1. Consistently, abolishment of Tyr-94 phosphorylation did not alter Tyr-381 phosphorylation or Lys-202 acetylation levels of PDP1, whereas Tyr-94 phosphorylation levels of PDP1 Y381F and K202R mutants were not altered as compared with PDP1 WT (Fig. 7A). These findings together showcase the beauty of precise and organized signal transduction-based regulation of cellular processes, in which tyrosine phosphorylation at different residues including Tyr-94 and previously reported Tyr-381 (12) acts in concert to provide independent and parallel regulation of PDP1 that ensures appropriate control of PDC that is, unfortunately, “hijacked” by the Warburg effect in cancer cells (Fig. 7B).
FIGURE 7.

Tyr-94 phosphorylation represents a distinct mechanism underlying regulation of PDP1. A, distinct PDP1 rescue cells were tested in immunoblotting to detect phosphorylation levels of Tyr-94 (p-PDP1(Y94)) or Tyr-381 (p-PDP1(Y381)) or acetylation levels of Lys-202 (Acetyl- PDP1 (K202)). B, proposed model shows that phosphorylation (P) at Tyr-94 (Y94)and Tyr-381 (Y381) inhibits PDP1 through distinct and independent molecular mechanisms in EGF-stimulated cells and cancer cells where tyrosine kinase signaling is commonly up-regulated.
Our findings also suggest that tyrosine phosphorylation of PDP1 is common in human tumor and leukemia cells, which represents an acute mechanism to mediate upstream oncogenic tyrosine kinase signaling-dependent regulation to mitochondrial PDC. Tyrosine phosphorylation of PDP1 may occur in mitochondria as we and others previously reported mitochondrial localization of oncogenic tyrosine kinases including EGF receptor (EGFR), FGFR1, BCR-ABL, and JAK2 (11, 12, 17). These findings add to emerging evidence which has shown that tyrosine phosphorylation of metabolic enzymes is common and important to link cell signaling pathways to metabolic pathways in cancer cells. These metabolic enzymes include PGAM1 (18), PKM2 (19), lactate dehydrogenase A (LDH-A) (13), PDP1 (12), and PDK1 (11), which are able to be phosphorylated by a similar set of oncogenic tyrosine kinases in cancer cells. It will be interesting to explore the molecular mechanisms by which tyrosine phosphorylation of these enzymes could coordinate and/or crosstalk to each other to provide an ultimately optimized metabolic advantage to cancer cell proliferation and tumor growth.
Lastly, our findings suggest that functional activation of PDP1 could lead to attenuated cancer cell proliferation and tumor growth due to metabolic defects. Future studies are warranted to explore PDP1 activators as novel anticancer agents.
Acknowledgment
We thank Dr. Seth Brodie for critical review and edit of the manuscript.
This work was supported in part by National Institutes of Health grants including the Career Development Fellow Award (to J. F.) from National Institutes of Health SPORE in Head and Neck Cancer P50CA128613 (to D. M. S.), the Pharmacological Sciences Training Grant T32 GM008602 (to S. E.), and the Hematology Tissue Bank of the Emory University School of Medicine and the Georgia Cancer Coalition (to H. J. K.).
- PDC
- pyruvate dehydrogenase complex
- PDK
- pyruvate dehydrogenase kinase
- PDP
- pyruvate dehydrogenase phosphatase
- PDH
- pyruvate dehydrogenase
- hPDP1
- human PDP1
- r
- recombinant.
REFERENCES
- 1. Kroemer G., Pouyssegur J. (2008) Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell 13, 472–482 [DOI] [PubMed] [Google Scholar]
- 2. Elstrom R. L., Bauer D. E., Buzzai M., Karnauskas R., Harris M. H., Plas D. R., Zhuang H., Cinalli R. M., Alavi A., Rudin C. M., Thompson C. B. (2004) Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 64, 3892–3899 [DOI] [PubMed] [Google Scholar]
- 3. Gottschalk S., Anderson N., Hainz C., Eckhardt S. G., Serkova N. J. (2004) Imatinib (STI571)-mediated changes in glucose metabolism in human leukemia BCR-ABL-positive cells. Clin. Cancer Res. 10, 6661–6668 [DOI] [PubMed] [Google Scholar]
- 4. Kim J. W., Dang C. V. (2006) Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 66, 8927–8930 [DOI] [PubMed] [Google Scholar]
- 5. Hiromasa Y., Fujisawa T., Aso Y., Roche T. E. (2004) Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus the E3-binding protein and their capacities to bind the E1 and E3 components. J. Biol. Chem. 279, 6921–6933 [DOI] [PubMed] [Google Scholar]
- 6. Read R. J. (2001) Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D 57, 1373–1382 [DOI] [PubMed] [Google Scholar]
- 7. Roche T. E., Baker J. C., Yan X., Hiromasa Y., Gong X., Peng T., Dong J., Turkan A., Kasten S. A. (2001) Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog. Nucleic Acid Res. Mol. Biol. 70, 33–75 [DOI] [PubMed] [Google Scholar]
- 8. Papandreou I., Cairns R. A., Fontana L., Lim A. L., Denko N. C. (2006) HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 3, 187–197 [DOI] [PubMed] [Google Scholar]
- 9. Kim J. W., Gao P., Liu Y. C., Semenza G. L., Dang C. V. (2007) Hypoxia-inducible factor I and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol. Cell. Biol. 27, 7381–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim J. W., Tchernyshyov I., Semenza G. L., Dang C. V. (2006) HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 [DOI] [PubMed] [Google Scholar]
- 11. Hitosugi T., Fan J., Chung T. W., Lythgoe K., Wang X., Xie J., Ge Q., Gu T. L., Polakiewicz R. D., Roesel J. L., Chen G. Z., Boggon T. J., Lonial S., Fu H., Khuri F. R., Kang S., Chen J. (2011) Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol. Cell 44, 864–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fan J., Shan C., Kang H. B., Elf S., Xie J., Tucker M., Gu T. L., Aguiar M., Lonning S., Chen H., Mohammadi M., Britton L. M., Garcia B. A., Alečković M., Kang Y., Kaluz S., Devi N., Van Meir E. G., Hitosugi T., Seo J. H., Lonial S., Gaddh M., Arellano M., Khoury H. J., Khuri F. R., Boggon T. J., Kang S., Chen J. (2014) Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol. Cell 53, 534–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fan J., Hitosugi T., Chung T. W., Xie J., Ge Q., Gu T. L., Polakiewicz R. D., Chen G. Z., Boggon T. J., Lonial S., Khuri F. R., Kang S., Chen J. (2011) Tyrosine phosphorylation of lactate dehydrogenase A is important for NADH/NAD+ redox homeostasis in cancer cells. Mol. Cell. Biol. 31, 4938–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pezzato E., Battaglia V., Brunati A. M., Agostinelli E., Toninello A. (2009) Ca2+-independent effects of spermine on pyruvate dehydrogenase complex activity in energized rat liver mitochondria incubated in the absence of exogenous Ca2+ and Mg2+. Amino Acids 36, 449–456 [DOI] [PubMed] [Google Scholar]
- 15. Morrison R. S., Yamaguchi F., Bruner J. M., Tang M., McKeehan W., Berger M. S. (1994) Fibroblast growth factor receptor gene expression and immunoreactivity are elevated in human glioblastoma multiforme. Cancer Res. 54, 2794–2799 [PubMed] [Google Scholar]
- 16. Vassylyev D. G., Symersky J. (2007) Crystal structure of pyruvate dehydrogenase phosphatase 1 and its functional implications. J. Mol. Biol. 370, 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boerner J. L., Demory M. L., Silva C., Parsons S. J. (2004) Phosphorylation of Y845 on the epidermal growth factor receptor mediates binding to the mitochondrial protein cytochrome c oxidase subunit II. Mol. Cell. Biol. 24, 7059–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hitosugi T., Zhou L., Fan J., Elf S., Zhang L., Xie J., Wang Y., Gu T. L., Alečković M., LeRoy G., Kang Y., Kang H. B., Seo J. H., Shan C., Jin P., Gong W., Lonial S., Arellano M. L., Khoury H. J., Chen G. Z., Shin D. M., Khuri F. R., Boggon T. J., Kang S., He C., Chen J. (2013) Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation. Nat. Commun. 4, 1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hitosugi T., Kang S., Vander Heiden M. G., Chung T. W., Elf S., Lythgoe K., Dong S., Lonial S., Wang X., Chen G. Z., Xie J., Gu T. L., Polakiewicz R. D., Roesel J. L., Boggon T. J., Khuri F. R., Gilliland D. G., Cantley L. C., Kaufman J., Chen J. (2009) Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci. Signal. 2, ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]